Abstract
Interactions between glioma cells and their local environment are critical determinants of brain tumor growth, infiltration and neovascularisation. Communication with host cells and stroma via microvesicles represents one pathway by which tumors can modify their surroundings to achieve a tumor-permissive environment. Here we have taken an unbiased approach to identifying RNAs in glioma-derived microvesicles, and explored their potential to regulate gene expression in recipient cells. We find that glioma microvesicles are predominantly of exosomal origin and contain complex populations of coding and noncoding RNAs in proportions that are distinct from those in the cells from which they are derived. Microvesicles show a relative depletion in microRNA compared with their cells of origin, and are enriched in unusual or novel noncoding RNAs, most of which have no known function. Short-term exposure of brain microvascular endothelial cells to glioma microvesicles results in many gene expression changes in the endothelial cells, most of which cannot be explained by direct delivery of transcripts. Our data suggest that the scope of potential actions of tumor-derived microvesicles is much broader and more complex than previously supposed, and highlight a number of new classes of small RNA that remain to be characterized.
Keywords: exosome, microparticle, glioblastoma, small noncoding RNA, vault RNA, gene expression
Introduction
Gliomas are the most common and deadly of adult primary brain tumors. They typically grow as widely infiltrative lesions, evade local innate immune surveillance, and induce proliferation of local vasculature.1 These features indicate that gliomas extensively modulate their local environment to achieve successful invasive growth. While tumor-stroma interactions are in part mediated by the release of soluble factors such as growth factors, there is growing interest in the role of small membrane-bound vesicles in mediating such interactions.
Membrane vesicles are generally classified into three groups based on their size, surface membrane composition, and mechanism of release from the cell: exosomes, microparticles and apoptotic bodies.2 Microparticles (100–1,000 nm) and apoptotic bodies (1–5 µm) are formed from cytoplasmic blebs that bud from the cell membrane and carry surface markers of their cell of origin. Exosomes (50–100 nm) carry distinct membrane proteins and are formed internally in multivesicular bodies that fuse with the plasma membrane to exit the cell. Microvesicles (MV), defined here as a mixed population of microparticles and exosomes, are capable of interacting with a variety of other cell types, and their target-cell specificity may be imparted by a distinct repertoire of membrane surface proteins.3-5 Given their near ubiquity in multicellular organisms, MV appear to represent a phylogenetically ancient form of intercellular communication, relevant to normal biology as well as pathology (reviewed in refs. 6 and 7).
Shedding of MV occurs in many (perhaps most) somatic cells in humans, and the process is increased in cancer cell lines and primary tumors.8 Tumour-derived MV have been shown to suppress the immune response, increase tumor progression, promote tumor invasiveness, and metastasis and confer multidrug resistance.9-15 Gliomas produce MV that contain angiogenic proteins capable of stimulating endothelial cell growth and proliferation in vitro.16 Glioma MV also carry mRNA, including that derived from mutant oncogenes, as well as microRNA and even DNA,16 suggesting the scope and mechanisms of their actions on recipient cells may extend beyond the promotion of local angiogenesis. Furthermore, detection of glioma-derived MV in peripheral blood and cerebrospinal fluid suggest that they may be useful tumor biomarkers.16,17
Small noncoding RNAs (sncRNAs) play a major role in the regulation of gene expression. The most abundant and best characterized group are microRNAs (miRNAs), which typically regulate the stability of mRNA transcripts and their translation into protein.18 Other sncRNAs can regulate DNA transcription, chromatin structure and genome integrity.19 SncRNAs have been identified in MV from various cell types, including gliomas;8,16,20,21 however, most studies have focused on miRNA characterization, hence the full range and complexity of small RNA species in MV derived from gliomas is yet-to-be determined. In this study we used microarray and deep sequencing to profile mRNAs and ncRNAs in glioma-derived MV. We find that specific RNA species are selectively packaged in glioma MV, and that many of these are previously undescribed and of unknown function. Microvascular endothelial cells exposed to glioma MV exhibit significant gene expression changes, indicative of the capacity of these particles to modulate gene expression in recipient cells.
Results
Isolation and characterization of glioma microvesicles
Microvesicles (MV) purified from the media of the U251 glioblastoma cell line are shown in Figure 1A, as visualized by transmission electron microscopy (TEM). TEM revealed a relatively uniform population of membrane-bound vesicles ranging between 85–154 nm in diameter, with an average diameter of 103 nm (Fig. 1B). Most MV exhibited a “cup” shaped morphology typical of exosomes under TEM.22 MALDI-TOF mass spectrometry of the proteins extracted from MV preparations identified a total of 112 proteins, of which 111 are known to be of exosomal origin (ExoCarta database,23 exocarta.org; Table S1). Of the top 25 exosomal proteins in ExoCarta, 17 were present in our MV preparation, including the tetraspanin CD9, an “exosomal marker” protein.24 Functional analysis of MV proteins using Ingenuity Pathway Analysis (IPA) identified a large number of significantly enriched pathways, among which cancer- and apoptosis-related ontologies were prominent (Fig. 1C).
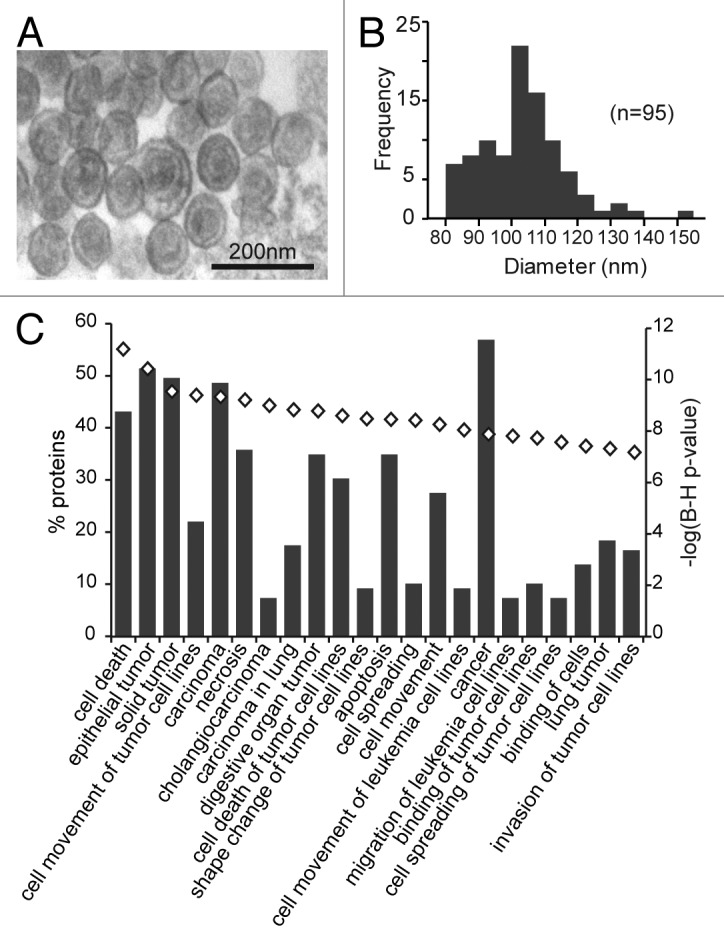
Figure 1. Characterization of microvesicles from U251 glioma cells. (A) Representative transmission electron micrograph of U251 microvesicle preparation. (B) Size distribution of a typical U251 microvesicle population as determined from electron micrographs. (C) Functional ontology analysis of microvesicle proteins identified by mass spectrometry. The proportion of microvesicle proteins in each ontology is shown on the left y-axis and the bar graph. Functional categories over-represented in the microvesicle proteins are presented in decreasing order significance with the lowest being p < 1 × 10−7. The right y-axis shows the statistical significance of each functional category (represented by the open diamonds).
Overall, the size distribution, ultrastructural morphology and protein composition strongly suggest that the glioma-derived MV in our preparations are largely exosomes. However, as we cannot discount the possibility that there may also be a contribution by other membrane-bound vesicles (such as small microparticles), we will continue to refer to the vesicles in this study using the collective term, microvesicles (MV).
Glioma MV contain a complex population of small RNAs and an under-representation of miRNA
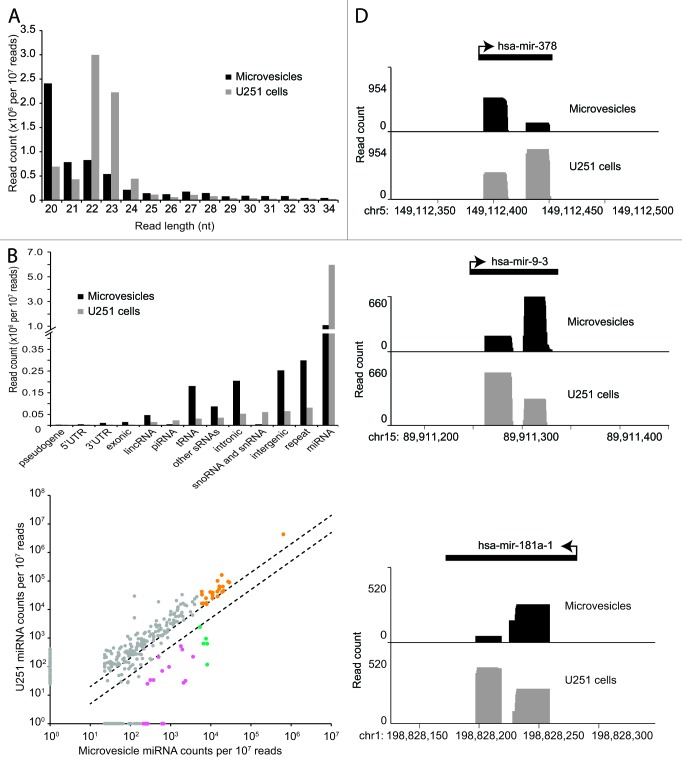
To characterize the small RNA content of glioma MV, we made small RNA libraries from RNA extracted from MV, as well as from the parent U251 glioma cells, and subjected them to SOLiD sequencing. Reads were mapped to the hg19 version of the human genome using mapreads, annotated using custom scripts, and normalized for comparison (see Materials and Methods). The length distribution of small RNAs from U251 glioma cells (modal length 22 nt; Fig. 2A) was characteristic of the small RNA content of somatic cells and cancer cells, whose dominant small RNA species is miRNA,25 and the vast majority of these small RNA sequences were confirmed to be miRNAs (90.4%), with only very few reads mapping to other annotation categories (Fig. 2B). In contrast, the size distribution of reads in MV was skewed toward smaller sequence length (modal length 20; Fig. 2A), suggesting an enrichment of other non-miRNA small RNAs. Annotation confirmed that a surprisingly modest proportion of MV small RNAs (38.7%) map to miRNAs, and there was a striking increase in the proportion of reads mapping to intergenic, repetitive and intronic regions of the genome, as well as to tRNA and other miscellaneous RNA species (Fig. 2B). These findings indicate that glioma MV are depleted in miRNAs relative to the cell from which they are derived, and that other, shorter small RNA species are selectively targeted to MV for export.
Figure 2. Specific miRNAs and miRNA variants are selectively packaged in glioma microvesicles. (A) Distribution of small RNA lengths in microvesicles and U251 cells. (B) Distribution of small RNA annotations in microvesicles and U251 cells. (C) Scatter plot of relative miRNA abundance in U251 parent cells and their microvesicles. Dashed lines indicate 2-fold enrichment/depletion; pink data points indicate enriched miRNAs with greater than 200 counts in the microvesicles as well as greater than 2-fold enrichment in the microvesicles compared with the U251 cells; orange points indicate abundant miRNAs with greater than 5,000 counts in the microvesicles; green indicates the overlap between these two groups. miRNAs that were present but did not meet the threshold of ≥ 10 reads are represented with an abundance of 1. (D) Plots of three representative miRNAs that exhibit 5p/3p arm-switching in microvesicles relative to the U251 parent cells.
Specific miRNAs are enriched in MV relative to the parent glioma cells
Although miRNAs were relatively depleted in MV, a number of microRNA species were still abundant in both U251 glioma cells and their MV (Fig. 2C, Table 1), the most abundant being miR-21. This is unsurprising as miR-21 is one of the most consistently overexpressed microRNAs in cancer, including gliomas, and has been characterized as an “oncomir.”26 In addition, a small number of miRNAs were significantly enriched in the MV compared with parent U251 cells, indicative of selective packaging. The most abundant of these are shown in Table 2A. Among those selectively packaged miRNAs, several have been reported in MV before: miR-451 was previously identified as a highly enriched miRNA in MV from HEK293 cells, endothelial cells and in breast cancer cell lines;27,28 mir-1246 has also previously been reported to be selectively released in MV from breast cancer cell lines.28 Gene ontology analysis of the predicted targets of the top 10 selectively packaged miRNAs revealed significant enrichment of several functional pathways (Table 2B); most highly enriched was the KEGG pathway of glycosaminoglycan (GAG) biosynthesis, a pathway that plays a critical role in tumor growth and angiogenesis.29
Table 1. Most abundant miRNAs in glioma microvesicles.
| microRNA | Normalized MV reads |
Normalized U251 reads |
Fold enrichment in MV |
|---|---|---|---|
| hsa-mir-21 |
641159 |
4352797 |
0.15 |
| hsa-mir-99a |
28983 |
88832 |
0.33 |
| hsa-mir-23a |
27191 |
95794 |
0.28 |
| hsa-mir-30a |
20775 |
45509 |
0.46 |
| hsa-mir-30d |
20084 |
63697 |
0.32 |
| hsa-mir-30b |
19312 |
41509 |
0.47 |
| hsa-mir-22 |
18736 |
162910 |
0.12 |
| hsa-mir-125a |
16350 |
65083 |
0.25 |
| hsa-let-7b |
16188 |
53137 |
0.30 |
| hsa-mir-25 |
14731 |
28718 |
0.51 |
| hsa-mir-221 |
14722 |
101775 |
0.14 |
| hsa-mir-92b |
14560 |
42586 |
0.34 |
| hsa-mir-135b |
11213 |
39642 |
0.28 |
| hsa-mir-29a |
11084 |
32056 |
0.35 |
| hsa-mir-222 |
11003 |
25199 |
0.44 |
| hsa-mir-100 |
9704 |
41055 |
0.24 |
| hsa-mir-451a |
8131 |
118 |
68.91 |
| hsa-mir-4301 |
8111 |
636 |
12.75 |
| hsa-mir-27b |
7868 |
15314 |
0.51 |
| hsa-mir-15b |
7836 |
17002 |
0.46 |
| hsa-mir-23b |
7620 |
25071 |
0.30 |
| hsa-mir-5096 |
7613 |
952 |
8.00 |
| hsa-mir-3676 |
6575 |
643 |
10.23 |
| hsa-mir-30e |
6481 |
16546 |
0.39 |
| hsa-mir-374b |
6026 |
33347 |
0.18 |
| hsa-mir-339 |
6004 |
16376 |
0.37 |
| hsa-mir-191 |
5979 |
42344 |
0.14 |
| hsa-mir-4454 | 5358 | 2437 | 2.20 |
Table 2A. Selectively packaged miRNAs in glioma microvesiclesa .
| microRNA | Fold enrichment in MV | Normalized MV reads |
Normalized U251 reads |
|---|---|---|---|
| hsa-mir-451a |
69 |
8131 |
118 |
| hsa-mir-4301 |
13 |
8111 |
636 |
| hsa-mir-5096 |
8 |
7613 |
952 |
| hsa-mir-3676–5p |
10 |
6575 |
643 |
| hsa-mir-4454 |
2 |
5358 |
2437 |
| hsa-mir-1303 |
73 |
2350 |
32 |
| hsa-mir-1273a |
78 |
2100 |
27 |
| hsa-mir-619 |
5 |
1954 |
398 |
| hsa-mir-448 |
3 |
1799 |
520 |
| hsa-mir-1246 |
9 |
914 |
98 |
| hsa-mir-4792 |
666 |
666 |
< 10 |
| hsa-mir-5095 |
9 |
615 |
71 |
| hsa-mir-1273 g |
610 |
610 |
< 10 |
| hsa-mir-4256 |
2 |
502 |
218 |
| hsa-mir-4255 |
11 |
376 |
34 |
| hsa-mir-5100 |
9 |
313 |
34 |
|
AC068946.1 |
270 |
270 |
< 10 |
| hsa-mir-1285–1 |
11 |
266 |
25 |
| hsa-mir-1269b |
248 |
248 |
< 10 |
| hsa-mir-4500 |
227 |
227 |
< 10 |
| hsa-mir-1273d |
207 |
207 |
< 10 |
| hsa-mir-4443 | 171 | 171 | < 10 |
a With > 2-fold enrichment in MV, as shown in Figure 2C
Table 2B. KEGG pathways targeted by the top 10 selectively packaged miRNAs.
| KEGG pathway | P valueb |
|---|---|
| Glycosaminoglycan biosynthesis - heparan sulfate |
5.077E-26 |
| Endocrine and other factor-regulated calcium reabsorption |
2.920E-03 |
| ECM-receptor interaction | 9.994E-03 |
b DIANA-Mirpath, with Benjamini-Hochberg correction
In addition to the selective packaging of specific miRNAs, we also observed a number of miRNA processing variants in glioma MV compared with U251 cells (Fig. 2D; Table S2). These cases involved either a switch in arm bias when both 5p and 3p microRNAs were present, or a dominance of the “star” form of the miRNA (relative to the mature) in the MV. Considering that the seed sequences of 5p and 3p (and of mature and star) species will almost certainly be distinct, it is likely that the miRNA variants in the MV target a different repertoire of mRNAs to the miRNA in the glioma cells themselves. Differential packaging of miRNA variants derived from the same precursor also suggests that exosome biogenesis intersects at some point with miRNA biogenesis pathways.
Transposon-derived small RNAs are prominent in glioma MV
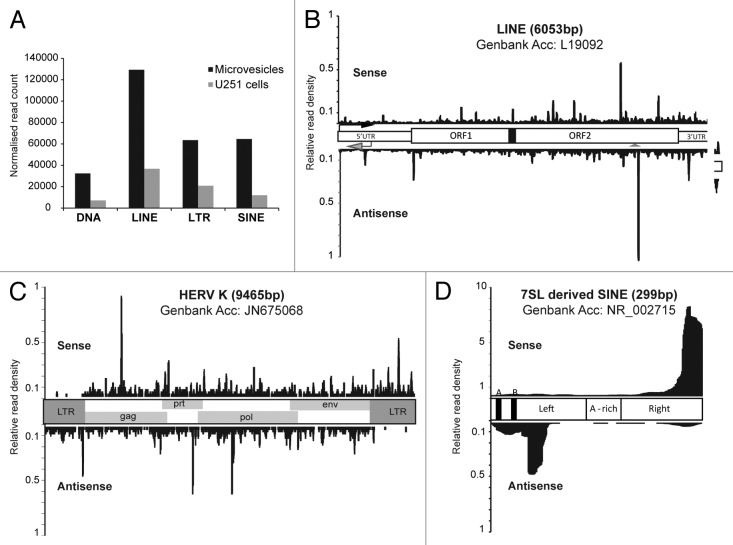
In contrast to the U251 data set where a minority (5%) of small RNA reads mapped to repetitive elements, more than 20% of reads in the MV data set mapped to repetitive elements (Fig. 2B), the majority of which were DNA transposons, or retrotransposons: LINEs, LTR-type and SINEs (Fig. 3A). Increased transcriptional activity of transposable elements in cancer is well documented (reviewed in ref. 30) and retrotransposon transcripts have previously been detected in microvesicles by microarray.31 We sought to determine whether these transposon-derived small (20 nt) RNAs exhibited signatures of processing (as opposed to being fragments of longer transcripts) by mapping to canonical sequences representative of each retrotransposon class. With LINEs we observed a fairly even distribution of reads mapping both sense and antisense with respect to the element, with the exception of a higher density around an antisense polyadenylation site known to be involved in gene breakage32 (Fig. 3B, arrowhead). The abundance of antisense LINE-derived small RNAs argues against these RNAs being a product of degradation. A canonical HERV-K sequence was used to determine the pattern of small RNA distribution for the LTR class of retrotransposons. Here we observed sense and antisense reads mapping along the open reading frames of the element, but the 5′ and 3′ LTR regions that act as promoters for these elements exhibited predominantly only antisense, or sense reads, respectively (Fig. 3C). Mapping to SINE elements (derived from 7SL sequence) produced the most striking pattern: the majority of reads mapped in a sense direction to the distal 3′end of the element, and a smaller proportion concentrated in an antisense direction just downstream of the internal RNA Pol III promoter (Fig. 3D). Taken together, these data indicate that retrotransposon transcripts are processed into small RNA fragments for export out of the tumor cell.
Figure 3. Repeat element-derived small RNAs are abundant in microvesicles. (A) Abundance of repeat-derived small RNAs in microvesicles and U251 cells. The distribution of small RNA reads across a canonical element (sense, above; antisense, below) is shown for (B) LINEs, (C) LTRs and (D) SINEs.
Novel small RNAs are selectively packaged in glioma MV
A large proportion of reads in the MV small RNA libraries were novel, and mapped to otherwise unannotated intergenic, as well as intronic locations (Fig. 2B). Loci with abundant reads showed “stacking” of sequences with clearly defined start and end positions (representative examples are shown as insets in Fig. 4A‒C). Their abundance and precisely defined distribution suggests that these novel intergenic and intronic reads are processed small RNA species, as opposed to randomly distributed sequencing errors or degradation products. Furthermore, 50% (1,056) of intron-derived small RNAs mapped exclusively in an antisense direction relative to the parent gene (e.g., Fig. 4C), militating against these RNAs being a product of mRNA degradation or splicing. The majority of intergenic and intronic small RNAs were enriched in MV relative to the parent U251 cells (1,976/2,284 (87%) and 1,723/2,092 (82%) with > 2-fold enrichment in MV relative to the parent U251 cells, respectively), indicating that most of these novel small RNAs are selectively packaged into MV. Interestingly, 1,904 intergenic and 1,668 intronic small RNA species were not detected in the parent glioma cells at all, occurring exclusively in the MV, some at very high abundance. This suggests that these specific small RNAs have functions only through MV trafficking.
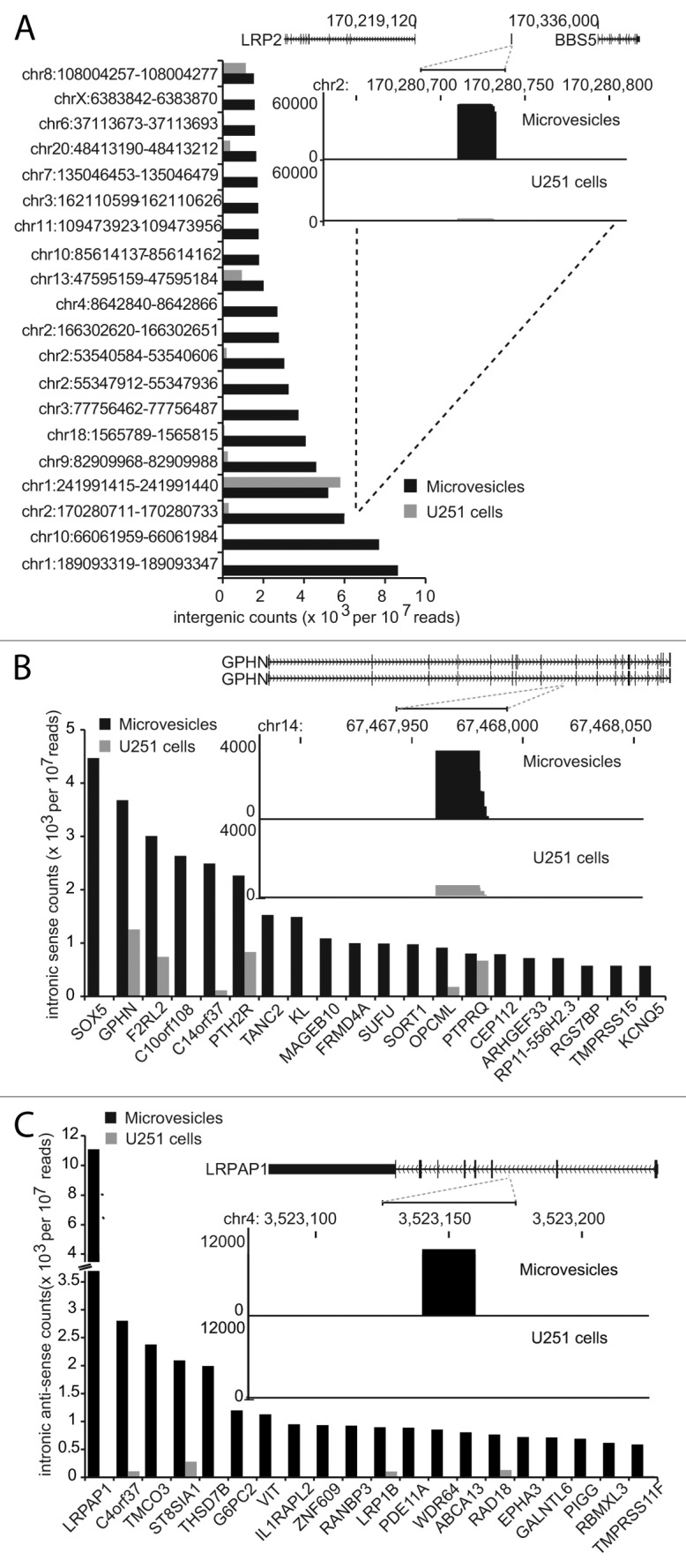
Figure 4. Novel small RNAs derived from intergenic regions and introns are selectively packaged in glioma microvesicles. (A) Bar graph showing the abundance of the top 20 intergenic loci producing small RNAs found almost exclusively in microvesicles. Inset histogram shows the characteristic stacked distribution of reads at a representative locus. (B) Bar graph showing the abundance of small RNAs mapping to introns in a sense direction to the gene shown on the y-axis. Inset histogram shows the characteristic stacked distribution of reads at representative locus. (C) Same as in (B) except the small RNAs map in an antisense direction to the genes indicated on the x-axis. Note that these antisense intronic small RNAs are found almost exclusively in the microvesicles and not the parent U251 cells.
The functions of these intergenic and intron-derived small RNAs are at present unknown, but they are unlikely to be novel microRNAs: they have an average length of 20 nt, and we did not detect any significant hairpin structures typical of miRNA precursors in the surrounding sequence.
Microvesicles are highly enriched in vault RNAs
Some of the most abundant and enriched small RNA species in our MV data set were 20mers that mapped to loci encoding an RNA family known as “vault” RNA; these small RNAs were virtually absent from the parent U251 cells (Fig. 5A). Vault RNAs complex with three proteins to form the vault organelle, and although the function of these RNAs is not understood, the vault organelle has been implicated in multidrug resistance in cancer.33 Vault RNA has recently been shown capable of being processed by Dicer into six distinct small vault RNAs (svRNAs),34 and at least some of these have been shown to exhibit miRNA-like Ago2-dependent repression of semi-complementary targets. In our data set we observed svRNAs mapping predominantly to the 3′end of the vault transcripts VTRNA1-1 and VTRNA2-1, although for VTRNA1-1 (the most abundant) we also observed reads mapping from the 5′end (Fig. 5B). This contrasts with the original description of svRNAs, in which the majority of svRNAs were derived from the 5′ end.34 We did not detect the protein components of the vault complex in our mass spectrometry analysis of MV proteins, but the coverage of small RNA reads at VTRNA1-1 prompted us to perform northern blots to assess whether full-length vault RNAs were also present in the MV. We found that full-length VTRNA1-1 was very abundant in total RNA from the MV, much more so than in the total RNA from the parent U251 glioma cells (Fig. 5C). We did not observe the small vault RNA species on the northern blot, indicating that full-length vault RNA is present in MV in vast excess to its processed forms.
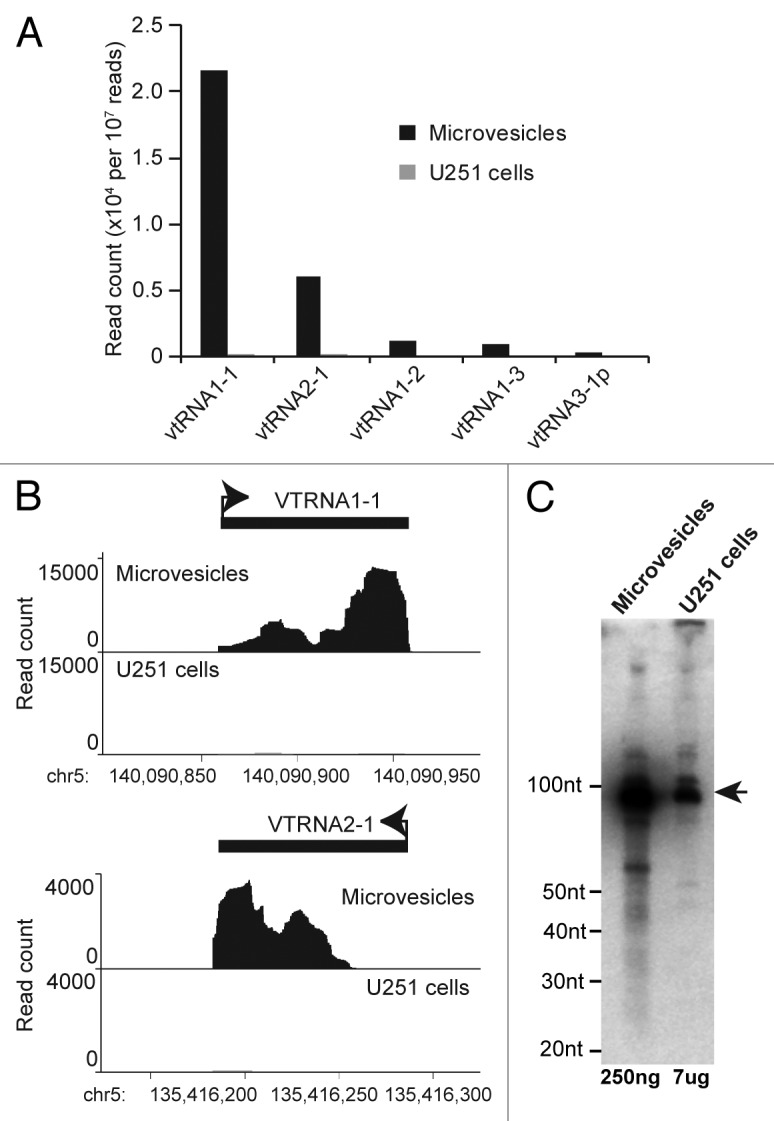
Figure 5. Glioma microvesicles are highly enriched in vault RNAs. (A) Bar graph showing abundance of small RNAs derived from vault RNA family members in microvesicles; these processed vault RNAs are absent from the parent U251 cells. (B) Histograms showing the distribution of microvesicle small RNAs across VTRNA1-1 and VTRNA2-1. (C) Northern blot of total RNA from microvesicles and U251 cells using a probe against the 3′ end of VTRNA1-1. Mature vault transcript is indicated by the arrow.
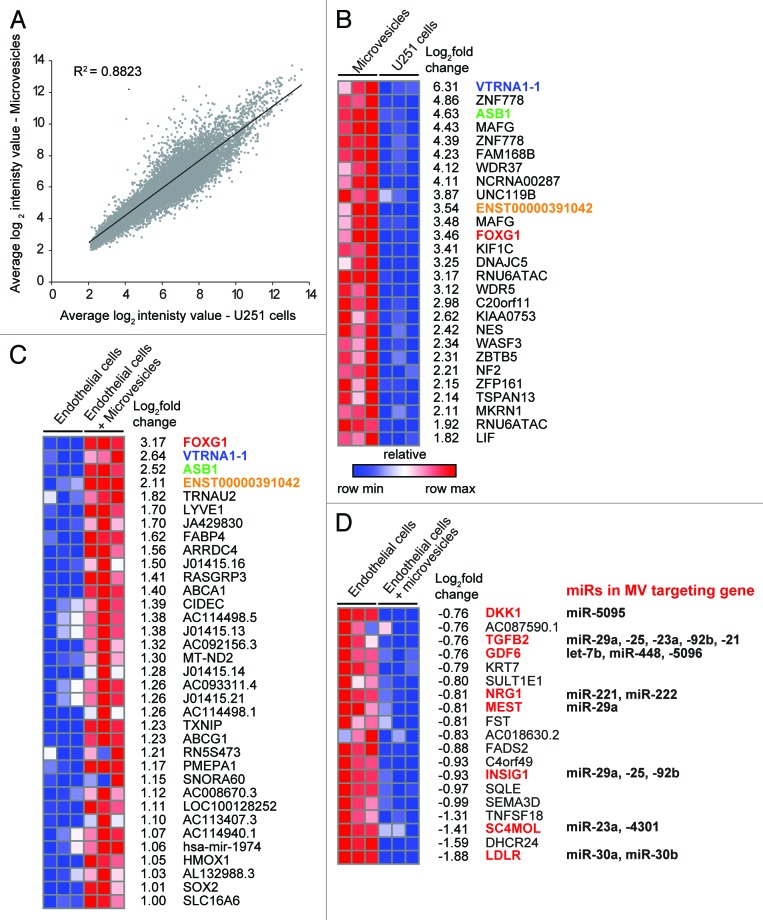
The presence of full-length vault RNA in MV prompted us to then ask what other longer RNAs are present in glioma MV. We compared the expression profiles of MV and their parent U251 cell cultures by hybridizing total RNA from triplicate MV/U251 preparations to Affymetrix 1.0 ST arrays. Overall, there was a good correlation between the RNA profiles of MV and their parent U251 cells (r2 = 0.88; Fig. 6A). However, a subset of transcripts was selectively enriched in MV; those most enriched (> 3-fold) are shown in Figure 6B. Consistent with our observations with northern blotting (Fig. 5C), we again find that VTRNA1-1 is the most enriched transcript in MV. Gene ontology analysis of these MV transcripts obtained a number of enriched functions related to cell morphology, growth, signaling and development (Fig. S1).
Figure 6. Glioma microvesicles alter the transcriptional profile of exposed endothelial cells. (A) Scatterplot comparing transcriptional profiles of U251 cells and their microvesicles. (B) Heat map showing transcripts that are enriched in microvesicles relative to U251 cells (> 1.5 log2 enrichment, FDR < 0.15). (C) Heat map showing the relative expression levels of transcripts that are significantly upregulated in endothelial cells in response to exposure to glioma microvesicles. Transcripts which are in common between B and C are colored. (D) Heat map showing transcripts that are significantly downregulated in endothelial cells in response to microvesicle exposure. Transcripts that are predicted targets of miRNAs in the microvesicles are shown in red.
Glioma MV induce gene expression changes in cultured vascular endothelial cells
We exposed cultured human brain microvascular endothelial cells (EC) to glioma MV to determine whether the molecular cargo of MV could alter the transcriptome of recipient cells. After 24 h of exposure the EC were washed and RNA harvested for expression profiling using Affymetrix 1.0 ST arrays; RNA from untreated EC of the same confluence were used for comparison and the experiment was performed in triplicate. We found 54 transcripts that had a greater than 2-fold expression change in EC upon exposure to MV: 35 were upregulated, while 19 were downregulated. Of the 35 upregulated transcripts only four corresponded to transcripts that were abundant in MV (Fig. 6C; Fig. S2), indicating that the increases in EC gene expression are unlikely to be due to direct delivery of mRNA from the MV in most cases. Of the genes that were downregulated in the EC in response to MV exposure, many are predicted targets of microRNAs that are abundant or selectively packaged in the MV (Fig. 6D).
Interestingly, we also observed that the majority (17/20) of genes that harbored antisense intron-derived sRNAs in the MV (Fig. 4C) were also downregulated in EC exposed to MV (Table 3), suggesting that these intronic antisense small RNAs may be able to modulate their parent gene’s expression when delivered to recipient cells.
Table 3. Response of EC genes to MV exposure where the gene has antisense intron-derived sRNAs in MV.
| Gene with MV-associated antisense sRNA | Log2fold change in EC+MV |
|
|---|---|---|
| LRPAP1 |
-0.012 |
|
| C4orf37 |
-0.054 |
|
| TMCO3 |
-0.190 |
|
| ST8SIA1 |
-0.100 |
|
| THSD7B |
-0.146 |
|
| G6PC2 |
-0.272 |
|
| VIT |
-0.252 |
|
| IL1RAPL2 |
0.008 |
|
| ZNF609 |
-0.056 |
|
| RANBP3 |
0.016 |
|
| LRP1B |
-0.004 |
|
| PDE11A |
-0.088 |
|
| WDR64 |
-0.159 |
|
| ABCA13 |
-0.165 |
|
| RAD18 |
-0.094 |
|
| EPHA3 |
-0.086 |
|
| GALNTL6 |
-0.102 |
|
| PIGG |
-0.139 |
|
| RBMXL3 |
0.162 |
|
| TMPRSS11F | -0.154 | |
Discussion
In this study we have found that microvesicles (MV) released from U251 glioma cells are predominantly of exosomal origin and carry a complex cargo of RNA species, many of which are novel and of unknown function. Taking an unbiased approach with small RNA deep sequencing, we have found that the repertoire of small noncoding RNAs in MV is strikingly distinct from the repertoire in their cell of origin, indicating that many RNA species are produced within tumor cells specifically for export out of the cell. Exposure of brain microvascular endothelial cells to glioma MV resulted in significant changes in endothelial cell gene expression, which reflects the capacity of these particles to mediate intercellular communication. Our data suggest that gliomas can modify the transcriptional landscape of their local environment through a variety of microvesicle-delivered RNA-based pathways. Many of these RNA-based mechanisms remain to be characterized.
Most previous studies on the RNA content of MV have focused on miRNAs;16,21,35-39 these small (~22 nt) noncoding RNAs are post-transcriptional regulators formed by the processing of a larger precursor transcript with a hairpin intermediate.18 With our unbiased approach, we have found that miRNAs are relatively depleted in MV, although a small subset is specifically enriched (Table 2B). The most enriched miRNA, miR-451, plays an important role in glioma cell proliferation and migration,40 but it has been reported to be selectively enriched in MV from non-neoplastic cell lines as well,27,41 indicating that selective export of this particular miRNA is not tumor-specific. However, available evidence suggests that selective export of most other miRNA species in glioma MV is unique; this points to potential tumor-specific modulatory roles of these miRNAs.
Most miRNAs show predominant accumulation of one “arm” of the precursor transcript after processing, and this is generally thought to be the mature functional effector miRNA, stabilized by incorporation into the RISC complex.18 In a number of cases we found that the dominant/mature miRNA in MV was derived from the opposite arm of the miRNA precursor to that which was dominant in the parent U251 cells. This surprising finding suggests that miRNA biogenesis and endosomal/exosomal processing may be interlinked, and that both arms of miRNA precursors are utilized more commonly than is currently appreciated, albeit in different contexts.
A previous study on glioma MV used antibody arrays to identify angiogenic proteins (such as angiogenin, IL-6 and IL-8) within MV that were capable of stimulating endothelial cell growth and tubule formation in vitro.16 We did not find these angiogenic proteins in our mass spectrometry analysis of glioma MV proteins, but we did find that many transcripts downregulated in endothelial cells exposed to glioma MV are predicted targets of miRNAs that are either abundant or specifically enriched in the MV. KEGG pathway analysis of these predicted miRNA targets identified GAG synthesis, Ca2+ reabsorption and ECM-receptor interactions as highly significant, over-represented functions. All of these pathways are relevant to glioma biology: GAG biosynthesis is a powerful modulator of tumor growth and angiogenesis,29 Ca2+ signaling modulates cell death and proliferation42 and ECM-receptor interactions are critical for tumor infiltration.43 Our findings suggest that modulation of vascular endothelial cell behavior by MV can also occur through small RNA pathways, in addition to protein-based mechanisms. In addition to any sequence-specific effects of MV miRNA, recent reports suggests that these (and perhaps other) small RNA species may activate Toll-like receptors in a sequence-independent manner, leading to multiple downstream immunomodulatory events in the recipient cells.44,45
Perhaps the most striking finding is the abundance of novel small RNAs in glioma MV. Nearly half of the small RNA population identified are novel small RNAs that map to otherwise unannotated intronic and intergenic regions. The intergenic RNAs are particularly intriguing: many are found in gene-poor regions whose sequence conservation extends only to primates. The function of these RNAs is completely unknown. Intronic-derived sRNAs are found both sense and antisense to the parent gene, and exhibit processing features that indicate they are not simply degradation products of their parent transcripts. Furthermore, the overwhelming majority are undetectable in the cells from which the MV are derived, suggesting that the primary function of these small RNAs is achieved via MV-trafficking. In the case of antisense intron-derived reads, some function can be inferred by the endothelial cell co-culture experiments, where parent transcripts in the recipient cells are downregulated (Table 3); this suggests a role for these small RNAs in gene repression that is not intended in the cell in which the small RNA is produced.
Repeat-derived small RNAs are the second most abundant class of small RNA in glioma MV and at least in the case of SINEs and LTR elements, the processing features of these small RNAs suggests that they are not the product of degradation. Retrotransposon RNAs have previously been described in glioma MV, although they were detected by microarray and presumed to be full-length transcripts.31 While it is possible that the small RNA fragments may have contributed to the microarray signals in that study, it is also possible that, like the vault RNAs, both full-length and processed RNAs are packaged into glioma MV. The function of these small RNAs is again completely unknown, however similar retrotransposon-derived small RNAs have been observed in murine oocytes and pre-implantation embryos,46,51 where there is some evidence that they are involved in gene silencing during early development.47 It is tempting to speculate that these RNAs may have silencing roles in host cells exposed to MV that could promote a tumor-permissive environment; however, given the vast numbers of retrotransposons interspersed among all the genes in the genome, it may be impossible to identify specific targets.
Vault complexes are very large (13 MDa) ribonucleoprotein organelles, three times the size of ribosomes, whose function is not well characterized.48 They are composed of three proteins and several non-coding RNAs. The major vault protein accounts for 70% of the mass of vaults, and has been implicated in multidrug resistance in a variety of tumors, including gliomas.49 Although it is abundant in glioma cell lines, we did not identify the major vault protein (or other vault proteins) in glioma MV by mass spectrometry. We did however identify an abundance of full-length vault RNA, VTRNA1-1, in MV by microarray and northern blotting, and deep sequencing revealed a large proportion of small RNA reads that were consistent with processed vtRNA. Both full-length and small vtRNAs were highly enriched in MV relative to the U251 parent cells; similar processed vtRNAs have also been identified in vesicles obtained from the media of murine T-cell and dendritic cell co-cultures.50 The processing of vtRNA into functional fragments has been demonstrated in breast cancer cells,34 but in contrast to this report we did not see repression of predicted targets of small vtRNAs in the EC exposed to the MV. Thus, it is likely that small vtRNAs have additional targets or different functions yet to be characterized. Taken together with the abundance of full-length vtRNA in the absence of major vault protein, vault RNAs and their derivatives likely have functions in MV separate to the vault ribonucleoprotein complex.
The study of tumor MV is relatively young, but those studies that have focused on their RNA content have already shown that MV-derived miRNAs are likely to play a significant role in regulation of gene expression in host cells (reviewed in refs. 39 and 51). The most striking finding of this study is that glioma MV contents are relatively depleted in miRNAs and instead are highly enriched for unusual or completely novel noncoding RNAs, including small RNAs from primate-specific regions of the genome. The functions of most of these are yet to be characterized, but point to unexpected complexities in intercellular communication mediated by membrane bound vesicles. Microvesicle RNAs represent a new and intriguing mechanism of tumor-host interactions that may provide novel therapeutic opportunities in the treatment of glioma and other tumors.
Materials and Methods
Cell culture and MV preparation
Human Glioblastoma (Astrocytoma) cells (U251-MG; U251) were obtained from the ATCC. They were cultured in RPMI-Glutamax (Invitrogen) with 10% fetal calf serum, 1% glucose and 1% penicillin/streptomycin filtered through a 0.1 µM filter. U251 cells were seeded at 2 × 106 cells in a T75 culture flask and media collected from cultures after the cells reached 80–90% confluence. Media was centrifuged at 1,800 g for 10 min to remove cellular debris. The supernatant was then collected and centrifuged at 18,000 g for 45 min at 15°C. Pelleted microvesicles were resuspended in 500 µL of phosphate buffered saline, centrifuged again at 18,000 g for 45 min and the pellet snap-frozen in liquid nitrogen. Microvesicle samples were verified by flow cytometry analysis, as described previously,52 and electron microscopy as described below. Multiple microvesicle preparations were then pooled for RNA and protein extraction. From a total of 225 mL of U251 conditioned media we obtained approximately 1.3 × 106 MV; this number of MV yielded a total of 2.36 µg RNA. The contribution of any bovine MV from the fetal calf serum remaining after filtration was determined to be minimal: identical preparations from equivalent amounts of media alone yielded no obvious MV pellet, and less than 50 ng of RNA.
Transmission electron microscopy
Microvesicles were harvested as above, and the final pellet was resuspended in 2.5% glutaraldehyde in 0.1M cacodylate buffer pH7.4 and fixed for 24 h at 4°C. The sample was centrifuged at 3,000 rpm for 20 min and the pellet was processed routinely for electron microscopy. Ultrathin sections were stained with uranyl acetate and lead citrate and examined on a Philips CM10 electron microscope.
Protein analysis
Microvesicle pellets were denatured in LDS Sample Buffer (Life Technologies) diluted in PBS and 100 mM DTT. Samples were heated to 95°C for 5 min and centrifuged at 17,000 g to precipitate any insoluble material and separated using a NuPAGE® Novex 4–12% Bis-Tris Gel (Life Technologies). The entire lane was then excised from the gel and sent to the Bioanalytical Mass Spectrometry Facility at the University of New South Wales for protein identification. Scaffold software (Proteome Software53)was used to probabilistically validate protein identifications from Mascot scores using the X!Tandem and ProteinProphet computer algorithms. Validated proteins were defined as those with at least five peptides detected with a minimum identification probability of 95%, and a minimum protein identification probability of 99.9%. Functional ontology analysis of validated microvesicle proteins was performed using Ingenuity Pathways Analysis software (Ingenuity Systems; www.ingenuity.com). Right-tailed Fisher’s exact test was used to calculate a p value determining the probability that each biological function had occurred due to chance alone, followed by Benjamini-Hochberg correction for multiple testing.
Small RNA sequencing
Total RNA was extracted from frozen microvesicle and U251 cell pellets with TRIzol® (Life Technologies) according to the manufacturer’s instructions. Small RNA libraries were generated using the NEBNext® Small RNA Sample Prep 3 kit (New England Biolabs) according to kit protocols. Libraries were sequenced on an Applied Biosystems SOLiD 5,500 × l using 35 bp chemistry.
Small RNA mapping
SOLiD color space reads (35mers) were mapped to the human genome using LifeScope (Life Technologies). First reads were filtered through a default filter file using a 25 base seed allowing up to three errors (25.3.0) identifying tRNA, rRNA and repetitive elements including LINE, SINE and LTR elements. Tags were then mapped to mirBase v18 (18.2.0). Remaining reads were mapped to human genome build 19 (20.1.0).
Annotation of mapped reads
Mapped reads were annotated using custom Perl scripts and the following databases: mirBase v18, UCSC hg19 Repeat Masker track, UCSC piRNA annotations and the Ensembl database version 67. Each read was assigned a gene accession, a biotype/RNA class and orientation if its coordinates were found to overlap with those recorded in one of the databases. This was performed in a hierarchical manner where the mirBase data set was queried first, followed by the Repeat Masker track, the piRNA track and finally the Ensembl database through a local installation of the ensemble API. Reads with no known annotations in these data sets were assigned as “intergenic.” Reads mapping to or near protein coding genes were further grouped into the following subclasses upstream/downstream, exonic, intronic, antisense, 5′UTR or 3′ UTR.
Counts and enrichment analysis
All counts were expressed as normalized values, per 10 million mapped reads, prior to filtering. A two-pass filter was applied to each of the reads before being included in further analysis. First, reads were required to have a Map Quality (MAPQ) of 10 or more before progressing to the second filter step where the number of reads starting at a given map position should be greater than or equal to 10. After filtering the number of mapped reads was 2,856,057 for U251 cells and 1,725,859 for MV.
Repeat element analysis
Reads mapping to three of the major classes of repeats LINE, SINE and LTR were remapped against a single representative full-length sequence of that repeat element with SHRiMP54 using a 14 base seed and allowing up to five mismatches to account for their characteristic degeneracy. The elements chosen for remapping were LINE L19092, SINE7SL NR002715 and LTR HERV K JN675068. The resulting alignments were plotted against a schematic of the representative sequence, separating tags mapping to the positive or negative strand.
Co-cultures
Primary human brain microvascular endothelial cells (EC) were obtained from Cell Systems, Inc. EC were cultured on plasticware coat with 3% collagen and were maintained in EBM-2 complete media (Lonza) with 5% fetal calf serum, ascorbic acid (5 μg/ml), hydrocortisone (Sigma, 1.4 μmol/L), chemically defined lipid concentrate (Invitrogen, 1:100 dilution), HEPES (Invitrogen, 10 mmol/L) and β-Fibroblast Growth Factor (Sigma, 1 ng/ml). EC were used from four to passage 11. Purified U251 MV were added to EC cultures at ~50% confluence at a ratio of 10MV/EC. EC were co-cultured with MV for 24 h and then washed extensively with PBS prior to harvesting for RNA extraction.
Microarray
Total RNA was extracted from frozen microvesicles, U251 cell or endothelial cell pellets with TRIzol® (Life Technologies) according to the manufacturer’s instructions and analyzed with Human Gene 1.0 ST arrays (Affymetrix). Labeling, hybridization and scanning was performed by the Ramaciotti Centre for Gene Function Analysis (University of New South Wales), with each sample hybridized to an individual array. Microarray results were analyzed using GenePattern software (Broad Institute, MIT55). Data was first preprocessed and normalized by robust multichip average (RMA) using the NormalizeAffymetrixST module (version 2.0, available at pwbc.garvan.unsw.edu.au/gp), followed by differential gene expression analysis using the LimmaGP module (version 19.3, available at pwbc.garvan.unsw.edu.au/gp). This module makes use of the Limma algorithm,56 which combines linear models with an empirical Bayes, moderated t-statistic which has more power than the Student’s t-test or ANOVA for analyzing microarray data.
Supplementary Material
Acknowledgments
The authors thank David IK Martin for helpful comments on the manuscript.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by a Cancer Council NSW Innovator Grant (IG 11-15); Cancer Institute NSW, and the Victor Chang Cardiac Research Institute (VCCRI). CMS is an Australian Research Council Future Fellow (FT120100097).
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/25281
References
- 1.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 2.Simpson RJ, Mathivanan S. Extracellular Microvesicles: The Need for Internationally Recognised Nomenclature and Stringent Purification Criteria. Journal of Proteomics and Bioinformatics. 2012;5:ii. [Google Scholar]
- 3.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–5. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 4.Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–93. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 5.Combes V, El-Assaad F, Faille D, Jambou R, Hunt NH, Grau GE. Microvesiculation and cell interactions at the brain-endothelial interface in cerebral malaria pathogenesis. Prog Neurobiol. 2010;91:140–51. doi: 10.1016/j.pneurobio.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 6.D’Souza-Schorey C, Clancy JW. Tumor-derived microvesicles: shedding light on novel microenvironment modulators and prospective cancer biomarkers. Genes Dev. 2012;26:1287–99. doi: 10.1101/gad.192351.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.György B, Szabó TG, Pásztói M, Pál Z, Misják P, Aradi B, et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci. 2011;68:2667–88. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 9.Millimaggi D, Mari M, D’Ascenzo S, Carosa E, Jannini EA, Zucker S, et al. Tumor vesicle-associated CD147 modulates the angiogenic capability of endothelial cells. Neoplasia. 2007;9:349–57. doi: 10.1593/neo.07133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janowska-Wieczorek A, Wysoczynski M, Kijowski J, Marquez-Curtis L, Machalinski B, Ratajczak J, et al. Microvesicles derived from activated platelets induce metastasis and angiogenesis in lung cancer. Int J Cancer. 2005;113:752–60. doi: 10.1002/ijc.20657. [DOI] [PubMed] [Google Scholar]
- 11.Liu C, Yu S, Zinn K, Wang J, Zhang L, Jia Y, et al. Murine mammary carcinoma exosomes promote tumor growth by suppression of NK cell function. J Immunol. 2006;176:1375–85. doi: 10.4049/jimmunol.176.3.1375. [DOI] [PubMed] [Google Scholar]
- 12.Ginestra A, La Placa MD, Saladino F, Cassarà D, Nagase H, Vittorelli ML. The amount and proteolytic content of vesicles shed by human cancer cell lines correlates with their in vitro invasiveness. Anticancer Res. 1998;18(5A):3433–7. [PubMed] [Google Scholar]
- 13.Clayton A, Mitchell JP, Court J, Mason MD, Tabi Z. Human tumor-derived exosomes selectively impair lymphocyte responses to interleukin-2. Cancer Res. 2007;67:7458–66. doi: 10.1158/0008-5472.CAN-06-3456. [DOI] [PubMed] [Google Scholar]
- 14.Wieckowski E, Whiteside TL. Human tumor-derived vs dendritic cell-derived exosomes have distinct biologic roles and molecular profiles. Immunol Res. 2006;36:247–54. doi: 10.1385/IR:36:1:247. [DOI] [PubMed] [Google Scholar]
- 15.Bebawy M, Combes V, Lee E, Jaiswal R, Gong J, Bonhoure A, et al. Membrane microparticles mediate transfer of P-glycoprotein to drug sensitive cancer cells. Leukemia. 2009;23:1643–9. doi: 10.1038/leu.2009.76. [DOI] [PubMed] [Google Scholar]
- 16.Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–6. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teplyuk NM, Mollenhauer B, Gabriely G, Giese A, Kim E, Smolsky M, et al. MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro Oncol. 2012;14:689–700. doi: 10.1093/neuonc/nos074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(Spec No 1):R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 20.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 22.Théry C, Boussac M, Véron P, Ricciardi-Castagnoli P, Raposo G, Garin J, et al. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166:7309–18. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- 23.Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012;40(Database issue):D1241–4. doi: 10.1093/nar/gkr828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–20. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 25.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 27.Guduric-Fuchs J, O’Connor A, Camp B, O’Neill CL, Medina RJ, Simpson DA. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics. 2012;13:357. doi: 10.1186/1471-2164-13-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pigati L, Yaddanapudi SC, Iyengar R, Kim DJ, Hearn SA, Danforth D, et al. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS One. 2010;5:e13515. doi: 10.1371/journal.pone.0013515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sasisekharan R, Shriver Z, Venkataraman G, Narayanasami U. Roles of heparan-sulphate glycosaminoglycans in cancer. Nat Rev Cancer. 2002;2:521–8. doi: 10.1038/nrc842. [DOI] [PubMed] [Google Scholar]
- 30.Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1:239–59. doi: 10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nature communications 2011; 2:180. [DOI] [PMC free article] [PubMed]
- 32.Wheelan SJ, Aizawa Y, Han JS, Boeke JD. Gene-breaking: a new paradigm for human retrotransposon-mediated gene evolution. Genome Res. 2005;15:1073–8. doi: 10.1101/gr.3688905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheffer GL, Schroeijers AB, Izquierdo MA, Wiemer EA, Scheper RJ. Lung resistance-related protein/major vault protein and vaults in multidrug-resistant cancer. Curr Opin Oncol. 2000;12:550–6. doi: 10.1097/00001622-200011000-00007. [DOI] [PubMed] [Google Scholar]
- 34.Persson H, Kvist A, Vallon-Christersson J, Medstrand P, Borg A, Rovira C. The non-coding RNA of the multidrug resistance-linked vault particle encodes multiple regulatory small RNAs. Nat Cell Biol. 2009;11:1268–71. doi: 10.1038/ncb1972. [DOI] [PubMed] [Google Scholar]
- 35.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA. 2010;107:6328–33. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101:2087–92. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montecalvo A, Larregina AT, Shufesky WJ, Stolz DB, Sullivan ML, Karlsson JM, et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119:756–66. doi: 10.1182/blood-2011-02-338004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diehl P, Fricke A, Sander L, Stamm J, Bassler N, Htun N, et al. Microparticles: major transport vehicles for distinct microRNAs in circulation. Cardiovasc Res. 2012;93:633–44. doi: 10.1093/cvr/cvs007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaiswal R, Luk F, Gong J, Mathys JM, Grau GE, Bebawy M. Microparticle conferred microRNA profiles--implications in the transfer and dominance of cancer traits. Mol Cancer. 2012;11:37. doi: 10.1186/1476-4598-11-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Godlewski J, Nowicki MO, Bronisz A, Nuovo G, Palatini J, De Lay M, et al. MicroRNA-451 regulates LKB1/AMPK signaling and allows adaptation to metabolic stress in glioma cells. Mol Cell. 2010;37:620–32. doi: 10.1016/j.molcel.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jaiswal R, Gong J, Sambasivam S, Combes V, Mathys JM, Davey R, et al. Microparticle-associated nucleic acids mediate trait dominance in cancer. FASEB J. 2012;26:420–9. doi: 10.1096/fj.11-186817. [DOI] [PubMed] [Google Scholar]
- 42.Monteith GR, McAndrew D, Faddy HM, Roberts-Thomson SJ. Calcium and cancer: targeting Ca2+ transport. Nat Rev Cancer. 2007;7:519–30. doi: 10.1038/nrc2171. [DOI] [PubMed] [Google Scholar]
- 43.Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol. 2012;196:395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. 2012;109:E2110–6. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lehmann SM, Krüger C, Park B, Derkow K, Rosenberger K, Baumgart J, et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat Neurosci. 2012;15:827–35. doi: 10.1038/nn.3113. [DOI] [PubMed] [Google Scholar]
- 46.Ohnishi Y, Totoki Y, Toyoda A, Watanabe T, Yamamoto Y, Tokunaga K, et al. Small RNA class transition from siRNA/piRNA to miRNA during pre-implantation mouse development. Nucleic Acids Res. 2010;38:5141–51. doi: 10.1093/nar/gkq229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohnishi Y, Totoki Y, Toyoda A, Watanabe T, Yamamoto Y, Tokunaga K, et al. Active role of small non-coding RNAs derived from SINE/B1 retrotransposon during early mouse development. Mol Biol Rep. 2012;39:903–9. doi: 10.1007/s11033-011-0815-1. [DOI] [PubMed] [Google Scholar]
- 48.van Zon A, Mossink MH, Scheper RJ, Sonneveld P, Wiemer EA. The vault complex. Cell Mol Life Sci. 2003;60:1828–37. doi: 10.1007/s00018-003-3030-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berger W, Spiegl-Kreinecker S, Buchroithner J, Elbling L, Pirker C, Fischer J, et al. Overexpression of the human major vault protein in astrocytic brain tumor cells. Int J Cancer. 2001;94:377–82. doi: 10.1002/ijc.1486. [DOI] [PubMed] [Google Scholar]
- 50.Nolte-’t Hoen EN, Buermans HP, Waasdorp M, Stoorvogel W, Wauben MH, ’t Hoen PA. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012;40:9272–85. doi: 10.1093/nar/gks658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee Y, El Andaloussi S, Wood MJ. Exosomes and microvesicles: extracellular vesicles for genetic information transfer and gene therapy. Hum Mol Genet. 2012;21(R1):R125–34. doi: 10.1093/hmg/dds317. [DOI] [PubMed] [Google Scholar]
- 52.Pankoui Mfonkeu JB, Gouado I, Fotso Kuaté H, Zambou O, Amvam Zollo PH, Grau GE, et al. Elevated cell-specific microparticles are a biological marker for cerebral dysfunctions in human severe malaria. PLoS One. 2010;5:e13415. doi: 10.1371/journal.pone.0013415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Searle BC. Scaffold: a bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics. 2010;10:1265–9. doi: 10.1002/pmic.200900437. [DOI] [PubMed] [Google Scholar]
- 54.Rumble SM, Lacroute P, Dalca AV, Fiume M, Sidow A, Brudno M. SHRiMP: accurate mapping of short color-space reads. PLoS Comput Biol. 2009;5:e1000386. doi: 10.1371/journal.pcbi.1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–1. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 56.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:e3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.