Abstract
Background
Crystal structures of the tobacco mosaic virus (TMV) coat protein (CP) in its helical and disk conformations have previously been determined at the atomic level. For the helical structure, interactions of proteins and nucleic acids in the main chains were clearly observed; however, the conformation of residues at the C-terminus was flexible and disordered. For the four-layer aggregate disk structure, interactions of the main chain residues could only be observed through water–mediated hydrogen bonding with protein residues. In this study, the effects of the C-terminal peptides on the interactions of TMV CP were investigated by crystal structure determination.
Methodology/Principal Findings
The crystal structure of a genetically engineered TMV CP was resolved at 3.06 Å. For the genetically engineered TMV CP, a six-histidine (His) tag was introduced at the N-terminus, and the C-terminal residues 155 to 158 were truncated (N-His-TMV CP19). Overall, N-His-TMV CP19 protein self-assembled into the four-layer aggregate form. The conformations of residues Gln36, Thr59, Asp115 and Arg134 were carefully analyzed in the high radius and low radius regions of N-His-TMV CP19, which were found to be significantly different from those observed previously for the helical and four-layer aggregate forms. In addition, the aggregation of the N-His-TMV CP19 layers was found to primarily be mediated through direct hydrogen-bonding. Notably, this engineered protein also can package RNA effectively and assemble into an infectious virus particle.
Conclusion
The terminal sequence of amino acids influences the conformation and interactions of the four-layer aggregate. Direct protein–protein interactions are observed in the major overlap region when residues Gly155 to Thr158 at the C-terminus are truncated. This engineered TMV CP is reassembled by direct protein–protein interaction and maintains the normal function of the four-layer aggregate of TMV CP in the presence of RNA.
Introduction
The tobacco mosaic virus (TMV) coat protein (CP) assembly system is composed of inlaid hydrophobic and hydrophilic patches with TMV RNA associated via multiple aromatic interactions between neighboring subunits to stabilize a helical virus structure [1]–[5]. Several experiments to crystallize TMV CP have demonstrated a dimer of bilayer disks with 17 subunits per layer, and the structures have been resolved to show the total form of 34 subunits [6]–[10]. Each subunit can be split into three regions: the high radius (HR), the middle radius (MR), and the low radius (LR) regions [11]. The HR region is far from the central axis of TMV CP, whereas the MR and LR regions are close to the central axis. The first structural determination of TMV CP, solved by X-ray fiber diffraction at a resolution of 2.8 Å in 1978 [11], reveals a similar conformation of protein subunits in the two layers. In 1998, a four-layer aggregate of TMV CP without RNA was reported [12], revealing an important role of water in its biological macromolecular assembly. The TMV viral structure was determined in 1989 through X-ray fiber diffraction [13], suggesting that viral disassembly is driven by electrostatic repulsions between nucleic acids and proteins. However, the electron densities of the C-terminal residues 155 to 158 are poor and display a high B factor. In 2007, the molecular assembly of TMV CP and RNA was described using cryo-electron microscopy (cryo-EM) at 4.6 Å. Notably, this assembly was more ordered: the residues clearly showed secondary structure near the RNA molecule in the LR region [14]. In this complex, the electrostatic interactions between the phosphate backbone of the RNA and the arginine residues (Arg112 and Arg113) significantly contributed to the stability of the complex. Taken together, the main differences between the previously determined structures of the virus and TMV CP protein lie in the LR region. In the virus particle, the interactions between the RNA and Arg90 and Arg92 could be observed by the cryo-EM method, but not by the X-ray crystallographic method. The interactions were more ordered in the LR region of the virus. In addition, the contact pair of Glu50 and Asp77 in the MR region was found to play an important role in the assembly and disassembly of the virus [15]–[19].
Here, we identify the effect of the C-terminal peptides on the molecular packaging and interactions of subunits within TMV CP aggregates. In order to purify and crystallize TMV CP, we constructed a TMV CP variant with a His-tag at the N-terminus and a truncation of residues Gly155 to Thr158 at the C-terminus. The engineered N-His-TMV CP19 protein was crystallized using hanging-drop vapor diffusion and the micro-seeding method. The crystal structure of the four-layer aggregate of N-His-TMV CP19 was determined at 3.06 Å. The major differences between N-His-TMV CP19 and the previously reported TMV CP structures are the manner of subunit assembly and the protein-protein interactions in the major overlap region. It should be noted that this engineered truncation mutant still maintains the normal function of the four-layer aggregate of TMV CP in the presence of RNA.
Results and Discussion
The Oligomeric State of N-His-TMV CP19 in Solution
To prepare N-His-TMV CP19 for crystallization, four amino acids (Gly155, Pro156, Ala157 and Thr158) were truncated at the C-terminus, and a 6-His-tag was incorporated at the N-terminus. Freshly purified protein was examined in solution using size exclusion chromatography (SEC). In 20 mM sodium phosphate and 100 mM NaCl (pH 8.0) (phosphate buffer; PB), the retention time of the N-His-TMV CP19 protein revealed its oligomeric state as a dimer and monomer. After equilibration in a buffer containing 300 mM ammonium sulfate and 100 mM Tris-HCl (pH 8.0), the oligomeric state of N-His-TMV CP19 was converted to a 20S structure. These results show that the assembly of N-His-TMV CP19 is influenced by the environmental conditions.
Crystallization of N-His-TMV CP19 Using a Micro-Seeding Method
Several TMV CPs have previously been crystallized by the micro-dialysis method against a buffer containing 300 mM ammonium sulfate and 100 mM Tris-HCl (pH 8.0) [20]–[22]. However, crystallization using this method is proven to be erratic in terms of reproducibility and crystal morphology. Instead, we crystallized N-His-TMV CP19 using hanging-drop vapor diffusion and a micro-seeding method. The micro-seeding method facilitated efficient and reproducible crystal growth of N-His-TMV CP19 [23]. This crystallization procedure was successfully used to produce large crystals of N-His-TMV CP19 with a typical maximal dimension of 0.2 mm ×0.3 mm within several days [24].
Overall Structure of N-His-TMV CP19
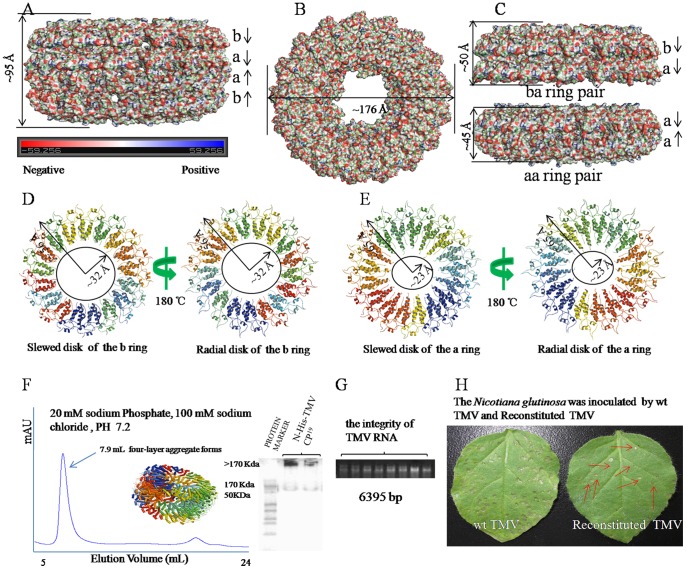
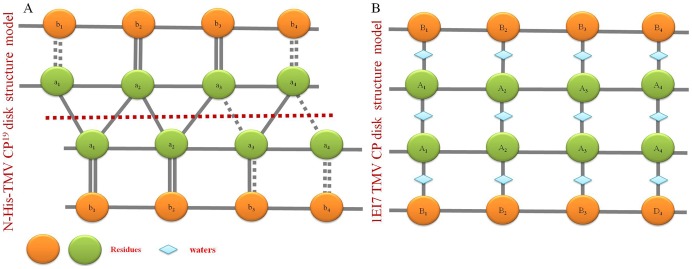
The N-His-TMV CP19 crystals belong to an orthorhombic space group P21212, with unit cell dimensions a = 173 Å, b = 222 Å, and c = 226 Å (Table S1 and Figure S1). The structure was solved by molecular replacement and refined to a resolution of 3.06 Å (PDB code 4GQH). The local noncrystallographic symmetry (NCS) axis was refined with a grid search wherein we varied the orientation or translation of the asymmetrical unit using XPLOR rigid body dynamics [12]. By using the NCS restraints option within the XPLOR refinement program, the 34 subunits of the asymmetrical unit were individually refined by simulated annealing. There were no departures from the NCS found during this procedure of refinement, as indicated by the Root Mean Square Deviation (RMSD) between the individual subunits (0.01–0.02 Å), The secondary structures of the a-chain, the b-chain, and the b- and a-chain in our refined atomic model are very similar to ones described by Bhyravbhatla et al. [12], which have RMSDs of ∼0.6 Å, ∼1.6 Å and ∼1.6 Å for the carbon-alpha backbone (not including the loop region and carboxyl-terminal four residues) (Figure S2A, Figure S2B and Figure S2C). The well-ordered portions of the virus subunit [19] have similar secondary structure when compared with either of the a-chains in the disk aggregate; superposition of the core of the protein backbone carbon-alpha atoms (RMSD of ∼1.1 Å) establishes that the overall folding of the protein is similar in the TMV CP helical structure and the TMV CP disk structure (Figure S2D). The ordered segments of the a- and b-chains can be superimposed on each other by a rigid body translation with a RMSD of 0.2 Å for the carbon-alpha atoms (Figure S3). The present atomic model also details the locations and orientations of the side chains involved in the inter- and intra-subunit interactions based on the error estimated by Luzzati statistics [25]. The refinement statistics after adding NCS restraints or constraints are listed in Table S1. The HR and LR residue densities corresponding to fitted coordinates are supplied in Figure S4 and Figure S5. It is clear that the density for the large hydrophobic residues, some of the small hydrophobic residues and the basic side chains are in agreement with the fitted coordinates. The N-His-TMV CP19 can form disks and infectious viruses (Figure S6 and Figure S7). The density corresponding to the side chains of N-His-TMV CP19 is mapped in Figure S8 and Figure S9. The 2Fo-Fc electron density maps have been contoured at 1.0 σ level, and the residues have been shown according to structure sequence. Similar to previous studies, one asymmetric unit was observed containing four aggregate disks, with every disk containing 17-fold monomers [11], [12]. The structure of the N-His-TMV CP19, which contains a hexahistidine tag at the N-terminus to facilitate purification [26], is similar to the four-layer aggregate [12]. The N-His-TMV CP19 four-layer aggregate was about ∼176 Å in diameter ( Figure 1A ) and ∼95 Å in height ( Figure 1B ). The four-layer aggregate disk was packed into a ba-ring pair outside (∼50 Å in height) and an aa-ring pair in the centre (∼45 Å in height) ( Figure 1C ). In the HR region, a well-ordered electron density map corresponding to the protein was observed (Figure S8A, Figure S4 and Figure S5). In the LR region, a poor-quality electron density corresponding to the protein was mapped (Figure S8B, Figure S4 and Figure S5). Therefore, residues 91–113 are absent in the b-ring atomic model, and residues 92–106 are absent in the a-ring atomic model ( Figure 1D and Figure 1E ). The slewed (radial) disk of the b-ring defines an axial pore of 64 Å in diameter based on residue Val114 (bottom panel); and the slewed (radial) disk of the a-ring defines an axial pore of 46 Å in diameter based on residue Thr107 (bottom panel). Residues Gly155, Pro156, Ala157 and Thr158 at the C-terminus of the N-His-TMV CP19 chains are truncated, which may facilitate the formation of the four-layer aggregate detected by SEC (20 mM PB, 100 mM NaCl, PH 7.2), and native-PAGE and TEM, after equilibration with an ammonium sulfate solution as above ( Figure 1F and Figure S7). N-His-TMV CP19 refolding and further self-assembly was carried out at a protein concentration of 1.8 mg/mL. When the N-His-TMV CP19 was incubated at a concentration of 1.8 mg/mL at 22°C for 5 hr, the N-His-TMV CP19 disk or helical structures were formed as observed using TEM in solution ( Figure 2 ). When TMV RNA ( Figure 1G ) isolated from wild type (WT) TMV was added to the N-His-TMV CP19 disk or helical structure, a reconstituted TMV particle was formed ( Figure 3 ). This virus particle can infect Nicotiana glutinosa ( Figure 1H and Figure S6). Thus, the N-His-TMV CP19 four-layer aggregate can effectively package and protect the nucleic acid under conditions of 20 mM PB, 100 mM NaCl and pH 7.2. For infection of tobacco leaves, the TMV CP disks undergo disassembly and reassembly mediated by RNA, and the reconstituted virus-like particles are made up of the helical form as determined by TEM ( Figure 3 and Figure S6). In other words, the RNA can stabilize the disordered residues in the LR region.
Figure 1. Crystal structure and function of the four-layer aggregate disk of N-His-TMV CP19.
(A) Electrostatic surface presentation of the N-His-TMV CP19 four-layer aggregate of ∼95 Å in height. The electrostatic surface was calculated in pyMOL. The complete four-layer aggregate is shown in four rings in the following order: b-ring, a-ring, a-ring, b-ring. (B) Electrostatic surface presentation of the N-His-TMV CP19 four-layer aggregate disk of ∼176 Å in diameter. (C) Electrostatic surface presentation of the ba-ring and aa-ring pairs. The aa-ring pair is sandwiched between the two b-rings. The a-ring and a-ring chains are anti-parallel and pack more tightly to ∼45 Å in height; the b-ring and a-ring chains are parallel and pack to ∼50 Å in height. (D) The slewed (radial) disk of the b-ring defines a 64 Å diameter axial pore based on residue Val 114 (bottom panel); (E) the slewed (radial) disk of the a-ring defines a 46 Å diameter axial pore based on residue Thr107 (bottom panel). (F) Assembly of the N-His-TMV CP19 four-layer aggregate, as measured by SEC and native-PAGE electrophoresis. (G) TMV RNA integrity was examined by 1% agarose gel electrophoresis. (H) The N. glutinosa was inoculated with WT TMV and reconstituted TMV. The local lesions that were inoculated with reconstituted TMV are labeled with red arrows.
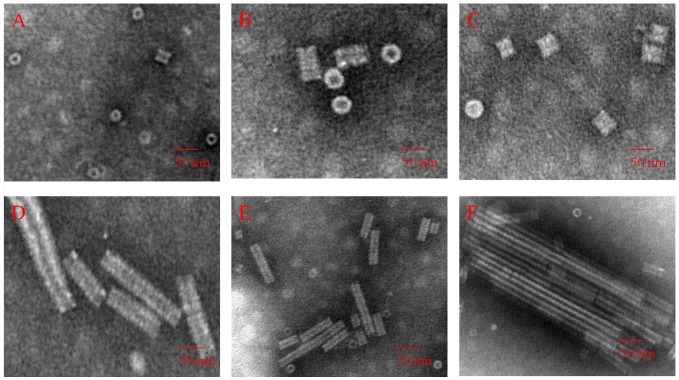
Figure 2. Characterization of the disk and helical stack height using TEM in 10 mM PB, 100 mM NaCl, pH 7.2 with increasing concentrations of protein, temperature and time.
As concentration, temperature and time increased, the dominant structure changed from disks to rods. (A) 1.8 mg/mL N-His-TMV CP19, 4°C, 10 h: monomers, dimers and disks. (B) 1.8 mg/mL N-His-TMV CP19, 20°C, 15 h: disks. (C) 1.8 mg/mL N-His-TMV CP19, 20°C, 20 h: disks. (D) 14 mg/mL N-His-TMV CP19, 20°C, 10 h: disks and rods. (E) 14 mg/mL N-His-TMV CP19, 20°C, 15 h: disks and rods. (F) 14 mg/mL N-His-TMV CP19, 20°C, 20 h: rods.
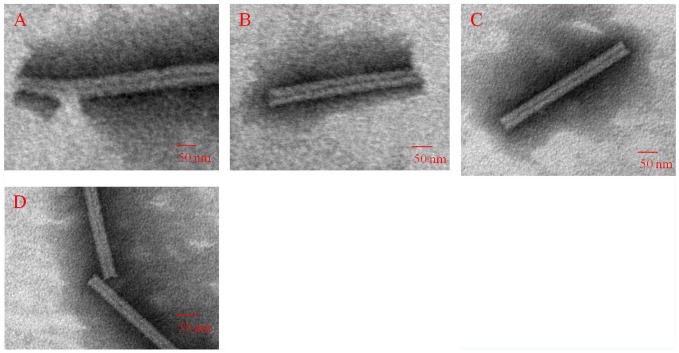
Figure 3. Characterization of reconstituted N-His-TMV CP19 virus using TEM in 10 mM PB, 100 mM NaCl, pH 7.2 after 20 h at 22°C.
1 mL purified self-assembled N-His-TMV CP19 disks (1.8 mg/mL) in 10 mM PB, 100 mM NaCl, pH 7.2 was mixed with 0.2 mL purified TMV RNA for 20 h at 22°C. Four TEM images of the reconstituted virus are shown in A–D.
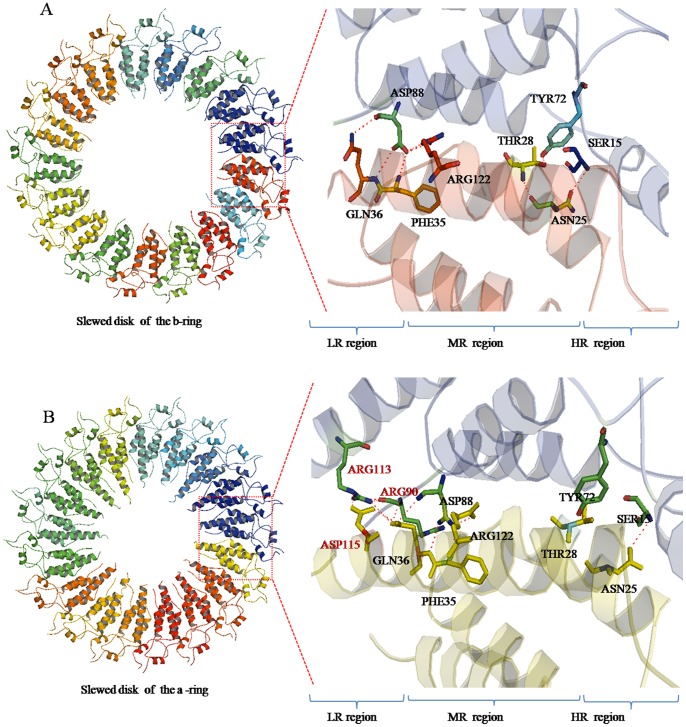
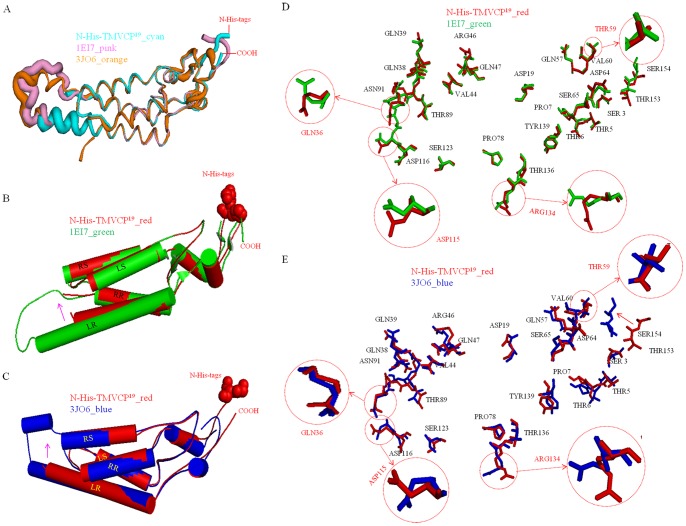
In the atomic model, the b-ring and a-ring in the HR and MR region are packed in order with observable inter-chain interactions between Asn25-Ser15, Tyr72-Thr28, Phe35-Asp88, Gln36-Asp88 and Arg122-Asp88 ( Figure 4A ), and the residue densities corresponding to fitted coordinates are supplied in Figure S10, The inter-molecular hydrogen bonds between residues are indicated by magenta dashed lines, The residues are shown according to N-His-TMV CP19 sequence. The a-ring subunits in the LR region loop residues are of higher order than the b-ring subunits with observable inter-chain interactions between Asn25-Ser15, Tyr72-Thr28, Phe35-Asp88, Gln36-Asp88 and Arg122-Asp88 ( Figure 4B ). In the previous study by Bhyravbhatla et al [12], no observable inter-chain interactions were observed in the LR region. In the helical structure, the Arg113-Arg90 interaction was observed and these residue loops were stabilized by RNA base-binding pockets [13], [19]. There are obvious differences in the four-layer assembly in the current structure and the previously reported one. For N-His-TMV CP19, a six-histidine (His) tag was introduced at the N-terminus, and the C-terminal residues 155 to 158 were truncated; direct inter-chain interactions between Asn25-Ser15, Tyr72-Thr28, Phe35-Asp88, Gln36-Asp88 and Arg122-Asp88 were observed in N-His-TMV CP19 (Figure S10). Because the oligomer terminal residues were changed, the conformations of residues Gln36, Thr59, Asp115 and Arg134 in the HR and LR regions of N-His-TMV CP19 were found to be significantly different from those observed in the helical structure (PDB code: 1EI7) (Figure S11) and in the four-layer aggregate disk structure of TMV CP (PDB code: 3JO6) (Figure S12). Protein–protein interactions on the salt bridges between Asp19 and Arg134, and between Asp66 and Arg134 were observed in the dihedral-related ba-chain pair. In the dihedral related aa-chain pair, the protein–protein interactions on the salt bridges between Lys53 and Glu22 and the hydrogen-bonding interactions between Thr59 and Thr59 were observed in the middle between trans-layers. All interactions between the four-layer aggregate disk had similar bond lengths (∼3.5 Å) ( Figure 5A ). A particular slide range was observed because of the interaction force between the a1-chain subunit Thr59 and the a2-chain subunit Thr59 in the HR region ( Figure 5B ). In contrast to the earlier model (PDB code: 1EI7), the protein–protein interactions within the salt bridges were mediated in most part by solvent molecules [12]. The electron density maps and fitted coordinates of direct protein–protein interactions involving salt-bridges between Asp19 and Arg134 are depicted in Figure S13A. The electron density map and fitted coordinates corresponding to the direct protein–protein interactions between Asp19-Arg134 and Asp66-Arg134 for the b-pairs and a-pairs of N-His-TMV CP19 and the protein–protein interactions between Asp19-Arg134 and Asp66-Arg134 for the previously reported TMV CP disk structures (PDB code: 1EI7) are depicted in Figure S13B. For the previously reported TMV CP disk structure, protein–protein interactions between Asp19-Arg134 and Asp66-Arg134 for the b-pairs and a-pairs were mediated by water. The electron density maps of protein–protein interactions involving salt-bridges between Lys53 and Glu22 are depicted in Figure S13C. The electron density maps of hydrogen-bonding interactions between Thr59 and Thr59 were observed in the middle trans-layers (Figure S13D). As shown in Figure S11 and Figure S14, a close-up electron density map and fitted coordinates show the inter-chain residues of N-His-TMV CP19 and the previously reported TMV CP (PDB code: 1EI7) subunits. Compared to the previously reported structures, these direct hydrogen-bonding interactions between the protein–protein trans-layers were not observed. In the previous structure (PDB code: 1EI7), the protein–protein interactions involved salt-bridges mediated by water. For the N-His-TMV CP19 structure, the conformations of Asp19, Arg134, Asp66, Lys53, Glu22 and Thr59 were different from the previously reported structure (Figure S11 and S14).
Figure 4. N-His-TMV CP19 interactions involved in b-ring and a-ring inter-subunits.
(A) Inter-chain interactions between Asn25-Ser15, Tyr72-Thr28, Phe35-Asp88, Gln36-Asp88 and Arg122-Asp88 in slewed disks of the b-ring were observed. These interactions are different from the previously reported disk inter-chain interactions (mediated by water). (B) Because the aa-ring is sandwiched between the b-rings, the a-ring inter-subunits in the LR region loop residues have stronger interactions than the b-ring inter-subunits, except for the observed inter-chain interactions between Asn25-Ser15, Tyr72-Thr28, Phe35-Asp88, Gln36-Asp88 and Arg122-Asp88.
Figure 5. Side view of the major overlap region between layers of atomic chains.
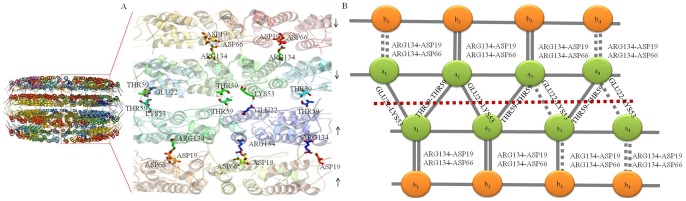
(A) The Asp19-Arg134 and Asp66-Arg134 protein–protein interactions between the b-pair and a-pair of atomic chains; the Lys53-Glu22 and Thr59-Thr59 protein–protein interactions between the a-pair and a-pair of atomic chains. (B) The N-His-TMV CP19 four-layer aggregate disk interaction model involves inter-subunits between layers. The horizontal dotted line is the central axis of the aa-pair. A particular slide range is observed because of the Thr59-Thr59 interaction.
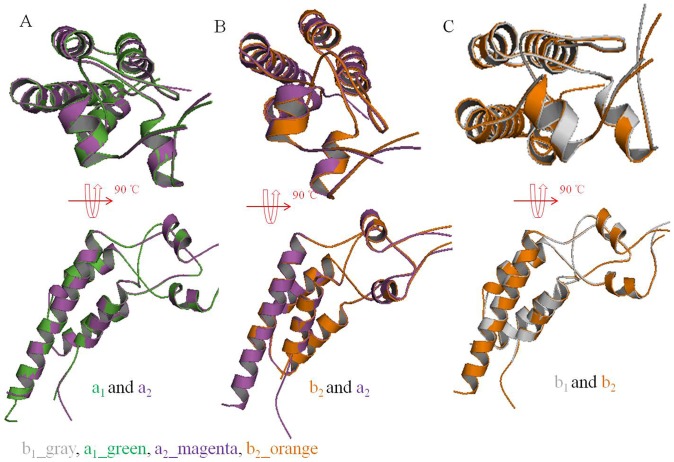
However, there were no differences in the N-His-TMV CP19 protein monomers. The N-His-TMV CP19 protein monomers could assemble into dimers, oligomers, a or b disks and four-layer aggregates. The LR region loops of the b disk were lost more than the a disk. Every two monomers were aligned between the aa disks, ab disks and bb disks. We found that there were no differences for the CP protein monomers ( Figure 6 ).
Figure 6. Superposition of two monomers of N-His-TMV CP19.
(A) Superposition of monomers between the a1-chain and a2-chain in N-His-TMV CP19. (B) Superposition of monomers between the b2-chain and a2-chain in N-His-TMV CP19. (C) Superposition of monomers between the b1-chain and b2-chain in N-His-TMV CP19.
Comparison with the Reported TMV CP Structures
The amino acid sequence of N-His-TMV CP19 is consistent with the previously reported sequence of the TMV CP disks and TMV CP helices except that a six-His tag was included at the N-terminus and four residues were truncated at the C-terminus, the atomic model of N-His-TMV CP19 presented here includes a newly formed section between the LR loop region and water molecules as observed by electron density mapping (Figure S4). Compared with the previously reported disk structure (PDB code: 1EI7) and the helical structure (PDB code: 3JO6), the major differences are the manner of subunit assembly and protein-protein interactions in the major overlap region. In the N-His-TMV CP19 structure, most protein–protein interactions were observed in the N-His-TMV CP19 inter-chains within 3.5 Å, including salt bridges between Asp19-Arg134 (∼3.12 Å), Asp66-Arg134 (∼3.29 Å), Lys53-Glu22 (∼3.38 Å), Arg122-Asp88 (∼2.43 Å), Arg113-Asp115 (∼3.24 Å) and hydrogen bonding of Asn25-Ser15 (∼2.73 Å), Tyr72-Thr28 (∼2.76 ), Phe35-Asp88 (∼3.03 Å), Gln36-Asp88 (∼3.50 Å) and Thr59-Thr59 (∼3.28 Å). In the previously reported disk structure (PDB code: 1EI7), the charged groups of the interactions were more than 4.0 Å (for example, the Asp66-Arg134 pair was ∼4.2 Å, and the Glu22-Lys53 pair was ∼5.4 Å), and most side-chains of the residues interacted via salt bridges and hydrogen-bonding through a network mediated by water [12], A model of these CP disks is provided in Figure 7 . Furthermore, for our structure, the N-His-TMV CP19 b-ring residues 91–113 (a-ring residues 92–106) in the low-radius inner loop region were missing, which is dissimilar to the previously reported TMV CP disk and helical structures. In the TMV CP disk structure (PDB code: 1EI7), the a-ring and b-ring residues 90–110 in the low-radius inner loop region were uniformly disrupted [12]. In the helical structure (PDB code: 3JO6), the residues 85–103 in the low-radius inner loop region were stabilized by RNA binding, and the residues Asp116-Arg92 conformation was affected by calcium-driven switches [19].
Figure 7. The inter-chain and intra-chain interaction model of N-His-TMV CP19 and the previously reported TMV CP disk (PDB code: 1EI7).
(A) The inter-chain and intra-chain interaction model of N-His-TMV CP19. (B) The inter-chain and intra-chain interaction model of the previously reported TMV CP disk structure (PDB code: 1EI7).
The structures of the side chains differed: a figure of the electron density map is depicted with an average resolution of 3 Å (Figure S4). In the atomic model, the region between 0 and 30 Å in radius, corresponding to the central 90-Å-diameter hole in the macromolecular assembly, was completely filled with solvent. In the region between 30-Å and 40-Å in radius, the atomic chain was poorly ordered with no observable inter-chain or intra-chain interactions. In the initial study done by Bhyravbhatla et al. [12], no electron density corresponding to the protein was observed in this region. Therefore, the N-His-TMV CP19 b-ring residues 91–113 (a-ring residues 92–106) are absent in the atomic model. Collecting good low-resolution data and using 17-fold redundancy allowed us to map out the backbone of the polypeptide chain in this region. The side chains are well defined in the electron density map in Figure S5.
Atoms of the inner loop region (residues 90–110) and the last four residues (154–158) had high B factors both in the previously reported disk structure (PDB code: 1EI7) and helical structure (PDB code: 3JO6), implying that these regions were disordered compared with the average structure [12], [19]. After truncating the last four residues (154–158) and introducing a six-His tag at the TMV CP N-terminus, the modified TMV CP, N-His-TMV CP19, had a higher atomic B factor near the missing region (LR region residues 104–111 of the a-ring; residues 107–111 of the b-ring), indicating a poorly ordered structure in the LR region; the N-His-TMV CP19 had a lower atomic B factor in the central helix (right slewed helical residues 38–51 and right radial helical residues 76–87) and the C-terminal loop region (residues 148–154), indicating a more tightly packed, well-ordered structure in these regions ( Figure 8A , Figure 8B and Figure 8C ). For the N-His-TMV CP19, the poorly ordered structure was stabilized by protein–protein hydrogen-bonding interactions in the adjacent subunit, such as Asn25-Ser15, Tyr72-Thr28, Phe35-Asp88, Gln36-Asp88 and Arg122-Asp88. These interactions resulted in conformational changes of residues, such as Gln36 and Asp115 in the LR region ( Figure 8D and Figure 8E ), which differ from the previously reported disk structure (PDB code: 1EI7) and helical structure (PDB code: 3JO6). The close-up electron density map and fitted coordinates between N-His-TMV CP19 and the previously reported disk structures are described in Figure S11 and Figure S12. In addition, the direct protein–protein interactions by the salt-bridges and hydrogen-bonding networks between layers of the N-His-TMV CP19 structure were different from the previously reported disk structure (PDB code: 1EI7) and helical structure (PDB code: 3JO6), and these interactions could be interpreted as conformational changes affected by Thr59 and Arg134, as shown in Figure 8D and Figure 8E . A cross-section of the X-ray map and fitted coordinates of the HR residues and LR loop residues of N-His-TMV CP19 and the previously reported TMV CP disk (PDB code 1EI7) are provided in Figure S15, the N-His-TMV CP19 residues (Ser1, Tyr2, Val58, Pro63, Asp64, Thr89, Arg90, Asn91, Arg141, Trp152, Thr153, Ser154) of the HR and the LR loop regions are different from those residues in the previous structure. To summarize, the main differences between N-His-TMV CP19 and TMV CP disks involve trans-layers proteins.
Figure 8. Structure comparisons between the previously reported TMV CP structure (PDB code 1EI7) and N-His-TMV CP19; and virus structure (PDB code 3JO6) and N-His-TMV CP19.
(A) The B factors of the atomic model are based on the atomic structure. Compared with TMV CP (PDB code 1EI7) and TMV CP (PDB code 3JO6), the N-His-TMV CP19 residues at the C-terminus loop (residues 148–154) have a low atomic B factor; the N-His-TMV CP19 in the low-radius inner loop [residues 104–111 of the a-ring (residues 107–111 of the b-ring)] have a high atomic B factor, indicating that the residues in these loops are disordered. In this structure, the disordered residues are stabilized by protein–protein hydrogen bonding interactions in the adjacent subunit. (B) Structure comparison between the TMV CP monomer (PDB code 1EI7) and the N-His-TMV CP19 monomer. The shifting N-His-TMV CP19 LR cylindrical helix is highlighted by an arrow. (C) Structure comparison between the TMV CP monomer (PDB code 3JO6) and the N-His-TMV CP19 monomer. The shifting N-His-TMV CP19 LR cylindrical helix is highlighted by an arrow. (D) Alignment of the side view of the TMV CP monomer (PDB code 1EI7) and the N-His-TMV CP19 monomer to show the different conformations in the main- and side-chain residues. The key residues in oligomer assembly are highlighted and labeled. (E) Alignment of the side view of the TMV CP monomer (PDB code 3JO6) and the N-His-TMV CP19 monomer to show the different conformations in the main- and side-chain residues. The key residues in oligomer assembly are highlighted and labeled.
Conclusions
Full-length N-His-TMV CP19 is easily overexpressed and purified to large scale; however, it is difficult to crystallize. After truncating four residues at the N-His-TMV CP19 C-terminus, high-quality crystals of N-His-TMV CP19 were easily obtained by hanging-drop vapor diffusion and micro-seeding methods. This phenomenon shows the conformational flexibility of the four residues at the C-terminus, which affects the crystal packing. The structure of N-His-TMV CP19 showed that the HR loop region (residues148–154) at the C-terminus had a low atomic B factor, which indicates a well-ordered structure in the HR loop region. Compared with the helical and disk structures of the previously reported TMV CP, the N-His-TMV CP19 subunits displayed different conformations in the LR and HR regions, and its four-layer aggregate disk was mainly retained by protein–protein salt-bridges and hydrogen-bonding networks between the trans-layers. In addition, the N-His-TMV CP19 could package RNA effectively and could assemble into the infectious virus particle with RNA. Thus, the N-His-TMV CP19 protein can form a reassembled disk or helical structure, and this structure exists in an RNA base-binding pocket. This pocket can be stabilized by TMV RNA and reassembled into a complete virus, and thus can serve as a potential molecular target of anti-plant virus drugs. These principles can also be applied in biology and structure-based drug design.
Materials and Methods
Isolation of TMV RNA and Construction of TMV CPs
TMV (common strain) was isolated from infected Nicotiana tabacum K326 leaves cultivated in a greenhouse at the Center for Research and Development of Fine Chemicals of Guizhou University. The virus was purified using the method described by Gooding [27], [28] and modified by Shire [29]. TMV RNA was extracted from purified virus by treatment with phenol and sodium dodecyl sulfate [30], [31]. The full-length viral cDNA sequence was generated by reverse transcription of TMV RNA using primer 1 (5′-GGAATTCCATATGTCTTACAGTATCACTACTCC-3′, the restriction site is underlined) in 50 mM Tris-HCl (pH 8.0), 8.0 mM magnesium chloride, 75 mM potassium chloride, 10 mM DL-dithiothreitol, 1.0 mM dNTPs, 0.5 unit/μL, AMV reverse transcriptase (TaKaRa), and 1.0 unit/μL RNase inhibitor (TaKaRa) for 1.5 hr at 85°C. The N-His-TMV CP19 gene was amplified by PCR using viral cDNA as template, with primer 1 and primer 9 (5′-CCGCTCGAGTCAAGAGGTCCAAACCAAACC-3′, the restriction site is underlined).
N-His-TMV CP19 Protein Expression and Purification
A six-His tag was introduced at the N-terminus to enable protein purification [25], and four amino acids (Gly155, Pro156, Ala157 and Thr158) were deleted at the C-terminus of WT TMV CP for crystallization [24]. The N-His-TMV CP19 protein was constructed by cloning common strains of the TMV CP gene into the Nde I and Xho I sites of pET28a vector (Novagen) and was expressed in E. coli BL21 (DE3) RIL cells (Novagen). The cells were grown in Luria-Bertani medium supplemented with 50 μg/mL kanamycin at 37°C to an OD600 of 0.65, shaking at 220 rpm. Then, the expression of protein was induced with 0.1 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG) at 16°C overnight. The cells were harvested by centrifugation and then stored at −80°C. Cell pellets were resuspended in lysis buffer (30 mM PB, 300 mM NaCl, 1 mM β-mercaptoethanol, pH 8.0) and then lysed at 4°C by sonication. After centrifugation at 15,000 g for 30 min to remove any insoluble material, the supernatant was loaded onto a 5 mL HisTrap High-Performance column (GE Healthcare), and the protein was eluted with a single gradient of 50 mM to 400 mM imidazole (pH 8.0). The C-terminally truncated protein was further purified by gel filtration (SEC) using a superdex 200 column (GE Healthcare, 120 mL) in a buffer containing 20 mM PB (pH 8.0) and 100 mM NaCl. Gel filtration was performed at 4°C using a calibrated superdex 200 10/300 GL column (GE Healthcare) attached to an AKTA purifier fast protein liquid chromatography system (GE Healthcare). The column was equilibrated with a buffer containing 10 mM PB (pH 7.2) or 100 mM NaCl depending on the TMV CP disk buffer [32]. Molecular mass standards (Bio-Rad) included thyroglobulin (669 kDa), ferritin (440 kDa), bovine serum albumin (67 kDa), β-lactoglobulin (35 kDa), ribonuclease A (13.7 kDa), cytochrome (13.6 kDa), aprotinin (6.51 kDa), and vitamin B12 (1.36 kDa). The proteins were monitored by absorbance at a wavelength of 280 nm. Native Polyacrylamide Gel Electrophoresis (PAGE) was performed on ice with the N-His-TMV CP19 samples, which were equilibrated overnight in a buffer containing 10 mM PB (pH 7.2) and 100 mM NaCl. A 20 µL sample was treated with 20 µL 2× loading buffer [12.5% 0.5 M Tris-HCl (V/V), pH 6.8, 0.5% bromophenol blue (W/V) and 30% glycerin (V/V)], and then, 8 μL of sample and 4 μL of protein marker were loaded onto a native-PAGE gel (4% stacking and 17% separating gel). Electrophoresis was run in 1× native page buffer (Tris-Gly, PH 8.8) at 0°C for about 1 hr [33]. After electrophoresis, the lanes were stained with coomassie blue [34], [35] to locate the protein and then destained with methanol and glacial acetic acid.
Reconstituted N-His-TMV CP19 Virus
One milliliter of purified self-assembled N-His-TMV CP19 disk (1.8 mg/mL), incubated in 10 mM PB, 100 mM NaCl, pH 7.2 solution, was mixed with 0.2 mL purified TMV RNA (0.5 mg/mL), and then the mixture was incubated at 22°C for 20 h. Suspensions were centrifuged at 5,000 rpm for 1 min, and then the reconstituted virus was obtained [4], [8], [29]. A 20 μL portion of protein solution was deposited onto a 300-mesh formvar-carbon-coated copper grid for 2 min, followed by rinsing with ddH2O. The grid was then stained with 20 μL of 2% aqueous solution of tungstophosphoric acid for 90 s as a negative stain [35]–[40]. Images were obtained at the Zunyi Medical University Electron Microscope Lab using a Hitachi H-7650 transmission electron microscope (TEM) with 80 kV accelerating voltage. The pattern of RNA packaging is very important for the reconstitution of infectious virus, which has a similar mechanism to other species [41]–[43]. Growing leaves of N. glutinosa were mechanically inoculated with reconstructed virus or the WT virus as a control. The local lesion numbers were recorded 3−4 days after inoculation.
Crystallization of N-His-TMV CP19
Crystallization was performed at 22°C by using the hanging-drop vapor diffusion method. N-His-TMV CP19 protein was concentrated to 14.0 mg/mL in a buffer containing 20 mM PB, pH 8.0, 100 mM NaCl, and 1 mM DTT, and then crystallization conditions were carefully screened [24]. Briefly, 1 µL protein solution was mixed with 1 µL reservoir solution (300 mM ammonium sulfate, 100 mM Tris-HCl, pH 7.7) and equilibrated against 0.6 mL reservoir solution. The micro-crystals were grown for 3 days and collected and deposited with a seeding tool (Hampton Research) [23]. Optimization of crystallization was performed in reservoir solution at different protein concentrations, ion concentrations (100 mM to 300 mM ammonium sulfate, 100 mM Tris-HCl) and pH (7.5 to 8.5). High-quality crystals of N-His-TMV CP19 were mounted and flash-frozen in liquid nitrogen following cryo-protection with reservoir solution containing an additional 30% (v/v) glycerol [44].
Data Collection and Structure Determination
Diffraction data were collected at the Shanghai Synchrotron Radiation Facility (SSRF) beamline 17U. All X-ray data were processed using the HKL2000 program [45] and converted to structure factors within the CCP4 program [46]. A typical octahedral-shaped crystal belongs to the space group P21212. The structure was solved by molecular replacement in Phaser [47] using the published WT TMV CP monomer as a search model (PDB code 1EI7). The N-His-TMV CP19 protein model was manually built in COOT [48], and computational refinement was conducted with the program REFMAC5 [49] in the CCP4 suite. Molecular graphic figures were prepared in PyMOL [50]. The local noncrystallographic symmetry (NCS) axis was refined by a grid search and varied orientation or translation of the asymmetrical unit was done using XPLOR rigid body dynamics. After using the NCS restraints option of the XPLOR refinement program [51], the 34 subunits of the asymmetrical unit were refined individually using simulated annealing. There were no departures from the noncrystallographic symmetry found during this procedure of refinement, as indicated by the RMSD between the individual subunits (0.01–0.02 Å) [12].
Supporting Information
Ramachandran Plot of the N-His-TMV CP19 monomer.
(TIF)
Comparison of the structure of the truncated TMV CP and previous structures by pyMOL. The following structural comparisons are shown: (A) The N-His-TMV CP19 a-chain and the TMV CP (PDB code: 1EI7) a-chain; (B) The N-His-TMV CP19 a-chain and the TMV CP (PDB code: 1EI7) b-chain; (C) The N-His-TMV CP19 a-chain and the TMV CP (PDB code: 3JO6) a-chain; (D) The N-His-TMV CP19 a- and b-chains and TMV CP (PDB code: 1EI7) a- and b-chains.
(TIF)
Superimposition of the N-His-TMV CP19 a- and b-chains by pyMOL. (A) Carbon-alpha trace showing a superimposition of the N-His-TMV CP19 a- and b-chains; (B) Bonds showing the superimposition of the N-His-TMV CP19 a- and b-chains.
(TIF)
Electron density map of the N-His-TMV CP19 by Coot. (A) The electron density map of N-His-TMV CP19viewed looking perpendicular to the crystallographic twofold axis, (B) An overview of the electron density map of N-His-TMV CP19 is provided at different angles.
(TIF)
A well-defined electron density map of the N-His-TMV CP19 monomer by Coot. (A) Carbon-alpha trace showing a well-defined electron density map of the N-His-TMV CP19 monomer; (B) Bonds showing a well-defined electron density map of the N-His-TMV CP19 monomer.
(TIF)
Electron microscopic analysis of tobacco leaves infected with reconstituted particles. (A) Original image of the tobacco leaves infected with reconstituted particles. (B) Magnified view of the tobacco leaves infected with reconstituted particles. N-His-TMV CP19 discs are enclosed in red boxes.
(TIF)
Assembly of the four-layer aggregate disks of N-His-TMV CP19 and wild type TMV CP as assessed by native-PAGE and SEC. (A) Disks of N-His-TMV CP19 eluted at 7.9 mL on SEC. (B) Disks of N-His-TMV CP19 were analyzed by native-PAGE. (C) Disks of wild TMV CP eluted at 7.9 mL on SEC. (D) Disks of wild TMV CP were analyzed by native-PAGE.
(TIF)
X-ray density map and fitted coordinates of a cross-section corresponding to the HR residues and LR loop residues of N-His-TMV CP19. (A) Cross-section of an X-ray map and fitted coordinates at the HR residues of N-His-TMV CP19. (B) Cross-section of an X-ray map and fitted coordinates at the LR loop residues of N-His-TMV CP19.
(TIF)
Side chain density map and fitted coordinates corresponding to the side chain residues of N-His-TMV CP19.
(TIF)
Electron density map and fitted coordinates corresponding to the inter-chain interactions within the N-His-TMV CP19 b-ring subunit. The electron density map and fitted coordinates corresponding to the inter-chain interactions between the following residues in the N-His-TMV CP19 disk of the b-ring are shown: (A) Phe35-Asp88 and Gln36-Asp88. (B) Tyr72-Thr28. (C) Arg113-Asp115. (D) Asn25-Ser15.
(TIF)
Close-up electron density map and fitted coordinates showing the side chain residues of N-His-TMV CP19 and the reported TMV CP (PDB code 1EI7) subunits from Figure 8D .
(TIF)
Close-up electron density map and fitted coordinates showing the side chain residues of N-His-TMV CP19 and the reported TMV CP (PDB code 3JO6) subunits from Figure 8E .
(TIF)
Electron density map and fitted coordinates corresponding to the protein–protein interactions of the N-His-TMV CP19 trans-layer. The electron density map and fitted coordinates between the following N-His-TMV CP19 protein–protein interactions are shown: (A) The Asp19-Arg134 and Asp66-Arg134 between the b-pair and a-pair. (B) The Asp19-Arg134 and Asp66-Arg134 between the b-pair and a-pair and the Asp19-Arg134 and Asp66-Arg134 between the previously reported TMV CP disk structures (PDB code 1EI7). In the previously reported TMV CP disk structure, the protein–protein interactions involved in Asp19-Arg134 and Asp66-Arg134 between the b-pairs and a-pairs were mediated by water. (C) The Thr59-Thr59 between the a-pair and a-pair. (D) The Lys53-Glu22 between the a-pair and a-pair.
(TIF)
Close-up electron density map and fitted coordinates showing the inter-chain residues of N-His-TMV CP19 and the previously reported TMV CP (PDB code 1EI7) subunits.
(TIF)
Cross-section of the X-ray map and fitted coordinates in the HR residues and LR loop residues of N-His-TMV CP19 and the previously reported TMV CP disk (PDB code 1EI7). Cross-section of the X-ray map and fitted coordinates of N-His-TMV CP19 and the TMV CP disk structure (PDB code 1EI7) are shown for the region of: (A) the HR residues; and (B) the LR loop residues.
(TIF)
Data collection and refinement statistics.
(DOCX)
Acknowledgments
Data collection was performed at beamline BL17U at the Shanghai Synchrotron Radiation Facility, China. We thank the staff for the data collection support. We thank Professor Chuan He from the University of Chicago for his guidance in experiments, and we also thank Dr. Baoen Chen from the Shanghai Institute of Materia Medica for his assistance in crystallography.
Funding Statement
This work was supported by the National Basic Research Program of China (Grant No. 2010CB126105), the Key Technologies R& D Program (Grant No. 2011BAE06B05-6), and the National Natural Science Foundation of China (Grant No. 21132003). The funders had no role in the study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
References
- 1. Stubbs G, Kendall A (2012) Helical viruses. Adv Exp Med Biol 726: 631–58. [DOI] [PubMed] [Google Scholar]
- 2.Bloomer AC, Butler PJG (1986) Tobacco mosaic virus structure and self-assembly: The plant viruses. In: Regenmortel MHV, Fraenkel-Conrat H. editors. New York: Plenum Press. 19–57.
- 3. Mandelkow E, Holmes KC, Gallawitz U (1976) A new helical aggregate of tobacco mosaic virus protein. J Mol Biol 102: 265–285. [DOI] [PubMed] [Google Scholar]
- 4. Jonathan P, Butler G, Durham AC (1977) Tobacco mosaic virus protein aggregation and the virus assembly. Adv Protein Chem 31: 188–251. [DOI] [PubMed] [Google Scholar]
- 5. Mandelkow E, Stubbs G, Warren S (1981) Structures of the helical aggregates of tobacco mosaic virus protein. J Mol Biol 152: 375–386. [DOI] [PubMed] [Google Scholar]
- 6. Butler PJG (1984) The current picture of the structure and assembly of tobacco mosaic virus. J Gen Virol 65: 253–279. [DOI] [PubMed] [Google Scholar]
- 7. Caspar DLD, Namba K (1990) Switching in the self-assembly of tobacco mosaic virus. Adv Biophys 26: 157–185. [DOI] [PubMed] [Google Scholar]
- 8. Okada Y (1986) Molecular assembly of tobacco mosaic virus in vitro. Advances in Biophysics 22: 95–145. [DOI] [PubMed] [Google Scholar]
- 9. Schuster TM, Scheele RB, Adams ML, Shire SJ, Steckert JJ, et al. (1980) Studies on the mechanism of assembly of tobacco mosaic virus. Biophys J 10: 313–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kegel WK, Schoot P (2006) Physical regulation of the self-assembly of tobacco mosaic virus coat protein. Biophys J 91: 1501–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bloomer AC, Champness JN, Bricogne G, Stoden R, Klug A (1978) Protein disk of tobacco mosaic virus at 2.8 Å resolution showing the interaction within and between subunits. Nature 276: 362–368. [DOI] [PubMed] [Google Scholar]
- 12. Bhyravbhatla B, Stanley JW, Caspar DLD (1998) Refined atomic model of the four-layer aggregate of the tobacco mosaic virus coat protein at 2.4 Å resolution. Biophys J 74: 604–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Namba K, Pattanegek R, Stubbs G (1989) Visualization of protein RNA interaction in a virus: Refined structure of TMV at 2.9 Å resolution by X-ray fiber diffraction. J Mol Biol 208: 307–325. [DOI] [PubMed] [Google Scholar]
- 14. Sachse C, Chen JZ, Coureux PD, Stroupe ME, Fändrich M, et al. (2007) High-resolution electron microscopy of helical specimens: A fresh look at tobacco mosaic virus. J Mol Biol 371: 812–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pattanayek R, Stubbs G (1992) Structure of the U2 strain of tobacco mosaic virus refined at 3.5 Å resolution using x-ray fiber diffraction. J Mol Biol 228: 516–528. [DOI] [PubMed] [Google Scholar]
- 16. Stubbs G, Warren S, Holmes K (1997) Structure of RNA and RNA binding site in tobacco mosaic virus from a 4 Å map calculated from X-ray fiber diagrams. Nature 267: 216–221. [DOI] [PubMed] [Google Scholar]
- 17. Namba K, Stubbs G (1986) Structure of tobacco mosaic virus at 3.6 Å resolution: implications for assembly. Science 231: 1401–1406. [DOI] [PubMed] [Google Scholar]
- 18. Clare DK, Orlova EV (2010) 4.6 Å cryo-EM reconstruction of tobacco mosaic virus from images recorded at 300 keV on a 4k×4k CCD camera. J Struct Biol 171: 303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ge P, Zhou ZH (2011) Hydrogen-bonding networks and RNA bases revealed by cryo electron microscopy suggest a triggering mechanism for calcium switches. Proc Natl Acad Sci USA 1008: 9637–9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sperling R, Klug A (1975) States of aggregation of the Dahlemense strain of tobacco mosaic virus protein and their relation to crystal formation. J Mol Biol 96: 425–430. [DOI] [PubMed] [Google Scholar]
- 21. Raghavendra K, Adams ML, Schuster TM (1985) Tobacco mosaic virus protein aggregates in solution: Structural comparison of 20S aggregates with those near conditions for disk crystallization. Biochemistry 24: 3298–3304. [DOI] [PubMed] [Google Scholar]
- 22. Raghavendra K, Kelly JA, Khairallah L, Schuster TM (1988) Structure and function of disk aggregates of the coat protein of tobacco mosaic virus. Biochemistry 27: 7583–7588. [DOI] [PubMed] [Google Scholar]
- 23.Bergfors TE (2009) Protein crystallization. La Jolla: International University Line. 95–108.
- 24.Li XY, Song BA, Hu DY, Chen X, Wang ZC, et al.. (2012) The development and application of new crystallization method for tobacco mosaic virus coat protein. Virology J In press. [DOI] [PMC free article] [PubMed]
- 25. Luzzati V (1952) Traitement statistique des erreurs dans la determination des structures cristallines. Acta Crystallogr 5: 802–810. [Google Scholar]
- 26. Bruckman MA, Soto CM, McDowell H, Liu JL, Ratna BR, et al. (2011) Role of hexahistidine in directed nanoassemblies of tobacco mosaic virus coat protein. Nano Lett 5(3): 1606–1616. [DOI] [PubMed] [Google Scholar]
- 27. Gooding GV, Hebert TT (1967) A simple technique for purification of tobacco mosaic virus in large quantities. Phytopathology 57(11): 1285. [PubMed] [Google Scholar]
- 28. Hebert TT (1963) Precipitation of plant viruses by polyethylene glycol. Phytopathology 53: 36. [Google Scholar]
- 29. Shire SJ, Steckert JJ, Schuster TM (1979) Mechanism of self assembly of tobacco mosaic virus protein. II. Characterization of the metastable polymerization nucleus and the initial stages of helix formation. J Mol Biol 127: 487–506. [DOI] [PubMed] [Google Scholar]
- 30. Fraenkel-Conrat H, Singer B, Tsugita A (1961) Purification of viral RNA by means of bentonite. Virology 14: 54–58. [DOI] [PubMed] [Google Scholar]
- 31. Wilcockson JRH (1974) The rapid isolation of plant virus RNAs using sodium perchlorate. J Gen Virol 23: 107–111. [DOI] [PubMed] [Google Scholar]
- 32. Shire SJ, Steckert JJ, Adams ML, Schuster TM (1979) Kinetics and mechanism of tobacco mosaic virus assembly: Direct measurement of relative rates of incorporation of 4S and 20S protein. Proc Natl Acad Sci USA 76(6): 2745–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem 72: 248–254. [DOI] [PubMed] [Google Scholar]
- 34. Neuhoff V, Arold N, Taube D, Ehrhardt W (1988) Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9: 255–262. [DOI] [PubMed] [Google Scholar]
- 35. Castellanos-Serra LR, Fernandez-Patron C, Hardy E, Huerta V (1996) A procedure for protein elution from reverse-stained polyarcylamide gels applicable at the low picomole level: An alternative route to the preparation of low abundance proteins for microanalysis. Electrophoresis 17: 1564–1572. [DOI] [PubMed] [Google Scholar]
- 36. Bruckman MA, Soto CM, McDowell H, Liu JL, Ratna BR, et al. (2011) Role of hexahistidine in directed nanoassemblies of tobacco mosaic virus coat protein. ACS Nano 5: 1606–1616. [DOI] [PubMed] [Google Scholar]
- 37. Geiger FC, Eber FJ, Eiben S, Mueller A, Jeske H, et al. (2013) TMV nanorods with programmed longitudinal domains of differently addressable coat proteins. Nanoscale 5: 3808–3816. [DOI] [PubMed] [Google Scholar]
- 38. Kadri A, Maiss E, Amsharov N, Bittner AM, Balci S, et al. (2011) Engineered Tobacco mosaic virus mutants with distinct physical characteristics in planta and enhanced metallization properties. Virus Res 157: 35–46. [DOI] [PubMed] [Google Scholar]
- 39. Kadri A, Wege C, Jeske H (2013) In vivo self-assembly of TMV-like particles in yeast and bacteria for nanotechnological applications. J Virol Methods 189: 328–340. [DOI] [PubMed] [Google Scholar]
- 40. Mueller A, Kadri A, Jeske H, Wege C (2010) In vitro assembly of Tobacco mosaic virus coat protein variants derived from fission yeast expression clones or plants. J Virol Methods 166: 77–85. [DOI] [PubMed] [Google Scholar]
- 41. Mancini EJ, Tuma R (2012) Mechanism of RNA packaging motor. Adv Exp Med Biol 726: 609–629. [DOI] [PubMed] [Google Scholar]
- 42. Kainov DE, Pirttimaa M, Tuma R, Butcher SJ, Thomas GJ Jr, et al. (2003) RNA packaging device of double-stranded RNA bacteriophages, possibly as simple as hexamer of P4 protein. J Biol Chem 278: 48084–48091. [DOI] [PubMed] [Google Scholar]
- 43. Kainov DE, Lisal J, Bamford DH, Tuma R (2004) Packaging motor from double-stranded RNA bacteriophage phi12 acts as an obligatory passive conduit during transcription. Nucleic Acids Res 32: 3515–3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dedeo MT, Duderstadt KE, Berger JM, Francis MB (2010) Nanoscale protein assemblies from a circular permutant of the tobacco mosaic virus. Nano Lett 10: 181–186. [DOI] [PubMed] [Google Scholar]
- 45. Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology 276: 307–326. [DOI] [PubMed] [Google Scholar]
- 46. Collaborative Computational Project N (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763. [DOI] [PubMed] [Google Scholar]
- 47. Read RJ (2001) Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D Biol Crystallogr 57: 1373–1382. [DOI] [PubMed] [Google Scholar]
- 48. Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- 49. Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255. [DOI] [PubMed] [Google Scholar]
- 50.DeLano WL (2002) The PyMOL Molecular Graphics System.
- 51. Kleywegt GJ, Brunger AT (1996) Checking your imagination: applications of the free R value. Structure 4: 897–904. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ramachandran Plot of the N-His-TMV CP19 monomer.
(TIF)
Comparison of the structure of the truncated TMV CP and previous structures by pyMOL. The following structural comparisons are shown: (A) The N-His-TMV CP19 a-chain and the TMV CP (PDB code: 1EI7) a-chain; (B) The N-His-TMV CP19 a-chain and the TMV CP (PDB code: 1EI7) b-chain; (C) The N-His-TMV CP19 a-chain and the TMV CP (PDB code: 3JO6) a-chain; (D) The N-His-TMV CP19 a- and b-chains and TMV CP (PDB code: 1EI7) a- and b-chains.
(TIF)
Superimposition of the N-His-TMV CP19 a- and b-chains by pyMOL. (A) Carbon-alpha trace showing a superimposition of the N-His-TMV CP19 a- and b-chains; (B) Bonds showing the superimposition of the N-His-TMV CP19 a- and b-chains.
(TIF)
Electron density map of the N-His-TMV CP19 by Coot. (A) The electron density map of N-His-TMV CP19viewed looking perpendicular to the crystallographic twofold axis, (B) An overview of the electron density map of N-His-TMV CP19 is provided at different angles.
(TIF)
A well-defined electron density map of the N-His-TMV CP19 monomer by Coot. (A) Carbon-alpha trace showing a well-defined electron density map of the N-His-TMV CP19 monomer; (B) Bonds showing a well-defined electron density map of the N-His-TMV CP19 monomer.
(TIF)
Electron microscopic analysis of tobacco leaves infected with reconstituted particles. (A) Original image of the tobacco leaves infected with reconstituted particles. (B) Magnified view of the tobacco leaves infected with reconstituted particles. N-His-TMV CP19 discs are enclosed in red boxes.
(TIF)
Assembly of the four-layer aggregate disks of N-His-TMV CP19 and wild type TMV CP as assessed by native-PAGE and SEC. (A) Disks of N-His-TMV CP19 eluted at 7.9 mL on SEC. (B) Disks of N-His-TMV CP19 were analyzed by native-PAGE. (C) Disks of wild TMV CP eluted at 7.9 mL on SEC. (D) Disks of wild TMV CP were analyzed by native-PAGE.
(TIF)
X-ray density map and fitted coordinates of a cross-section corresponding to the HR residues and LR loop residues of N-His-TMV CP19. (A) Cross-section of an X-ray map and fitted coordinates at the HR residues of N-His-TMV CP19. (B) Cross-section of an X-ray map and fitted coordinates at the LR loop residues of N-His-TMV CP19.
(TIF)
Side chain density map and fitted coordinates corresponding to the side chain residues of N-His-TMV CP19.
(TIF)
Electron density map and fitted coordinates corresponding to the inter-chain interactions within the N-His-TMV CP19 b-ring subunit. The electron density map and fitted coordinates corresponding to the inter-chain interactions between the following residues in the N-His-TMV CP19 disk of the b-ring are shown: (A) Phe35-Asp88 and Gln36-Asp88. (B) Tyr72-Thr28. (C) Arg113-Asp115. (D) Asn25-Ser15.
(TIF)
Close-up electron density map and fitted coordinates showing the side chain residues of N-His-TMV CP19 and the reported TMV CP (PDB code 1EI7) subunits from Figure 8D .
(TIF)
Close-up electron density map and fitted coordinates showing the side chain residues of N-His-TMV CP19 and the reported TMV CP (PDB code 3JO6) subunits from Figure 8E .
(TIF)
Electron density map and fitted coordinates corresponding to the protein–protein interactions of the N-His-TMV CP19 trans-layer. The electron density map and fitted coordinates between the following N-His-TMV CP19 protein–protein interactions are shown: (A) The Asp19-Arg134 and Asp66-Arg134 between the b-pair and a-pair. (B) The Asp19-Arg134 and Asp66-Arg134 between the b-pair and a-pair and the Asp19-Arg134 and Asp66-Arg134 between the previously reported TMV CP disk structures (PDB code 1EI7). In the previously reported TMV CP disk structure, the protein–protein interactions involved in Asp19-Arg134 and Asp66-Arg134 between the b-pairs and a-pairs were mediated by water. (C) The Thr59-Thr59 between the a-pair and a-pair. (D) The Lys53-Glu22 between the a-pair and a-pair.
(TIF)
Close-up electron density map and fitted coordinates showing the inter-chain residues of N-His-TMV CP19 and the previously reported TMV CP (PDB code 1EI7) subunits.
(TIF)
Cross-section of the X-ray map and fitted coordinates in the HR residues and LR loop residues of N-His-TMV CP19 and the previously reported TMV CP disk (PDB code 1EI7). Cross-section of the X-ray map and fitted coordinates of N-His-TMV CP19 and the TMV CP disk structure (PDB code 1EI7) are shown for the region of: (A) the HR residues; and (B) the LR loop residues.
(TIF)
Data collection and refinement statistics.
(DOCX)