Abstract
Rigid spinning: Site-directed spin-labeling studies using a rigid nitroxide spin label (Ç) reveal that both length and sequence of a single-stranded junction (J1/2) modulate nanosecond motional ordering of the substrate-recognition duplex (P1) of the 120 kD group I ribozyme. The studies demonstrate an approach for experimental measurements of nanosecond dynamics in high-molecular-weight RNA complexes.
Keywords: dynamics, EPR spectroscopy, motional ordering, RNA, spin labeling
With the rapid advances in our knowledge of the cellular functions of RNA, it is increasingly important to elucidate how an RNA’s primary sequence dictates the chemical and physical attributes that exact its function. One ubiquitous physical attribute is dynamics, which spans vast timescales, encompasses a large number of modes, and is dictated by an intimate interplay between intrinsic RNA features and external factors.[1] Information on pathways and kinetics of conformational transformations has been obtained for a number of RNA molecules, yet less is known about the intrinsic dynamics of individual equilibrium states. Intrinsic dynamics dictates the ability of RNA to sample different conformations of an equilibrium ensemble, thereby shaping pathways and kinetics of transformations.[1] As such, intrinsic dynamics is crucial to elucidating the relationship between structure and function.
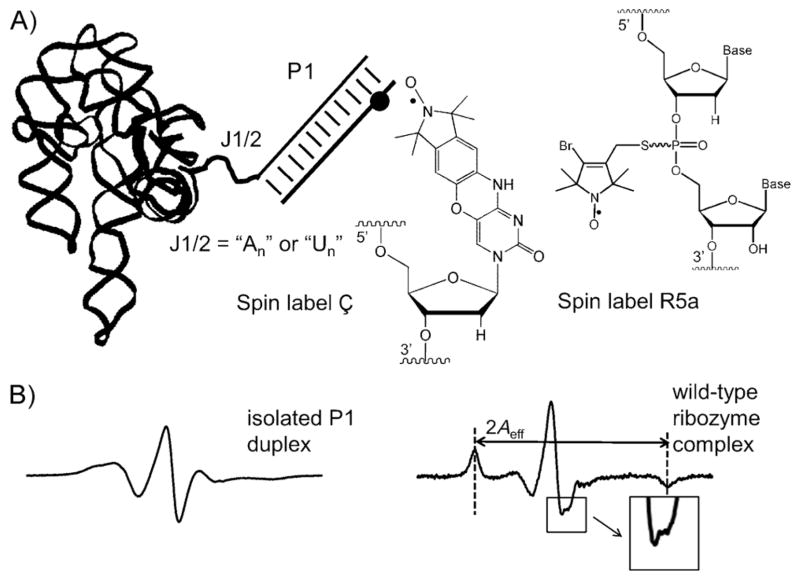
Here we used site-directed spin labeling (SDSL)[2] to examine how an RNA junction modulates equilibrium dynamics of a duplex attached to a large folded RNA. Studies were carried out on a 120 kD group I ribozyme derived from Tetrahymena thermophila, which has been used extensively to investigate RNA structure, folding, and catalysis.[3] By using a nitroxide probe (Ç, Figure 1A) rigidly fused to a modified cytosine,[4] dynamics of the ribozyme substrate recognition duplex (P1) was assessed in the “open complex”, in which the P1 duplex forms no tertiary contacts with the folded ribozyme core (Figure 1A). Data obtained by X-band continuous-wave (cw) electron paramagnetic resonance (EPR) spectroscopy (which reports nitroxide rotational motions in the 0.5–50 nanosecond range) revealed that motional ordering of P1 is modulated by both the length and sequence of the single-stranded junction (J1/2) that connects P1 to the ribozyme core. This work established an incisive method for examining the local nanosecond dynamics of specific secondary structure elements within a large folded RNA.
Figure 1.
A) Ribozyme open complex. The black dot marks the location of the spin label; chemical structures of Ç and R5a are shown to the right. B) X-band cw-EPR spectra of Ç measured in aqueous buffer at 25°C.
SDSL provides structural and dynamic information of biomolecules by using a site-specifically attached stable radical probe (usually a nitroxide).[2] Previously, we used a flexible R5a nitroxide (Figure 1A) attached to a phosphorothioate at a specific site on the RNA to study nanosecond equilibrium dynamics of P1 in the folded ribozyme.[5] However, in this previous study, the nitroxide pyrroline ring (at which the unpaired electron is localized) was connected to the RNA backbone by single bonds. Rotations of the pyrroline ring with respect to the P1 duplex are convoluted with motions of P1 with respect to the ribozyme core. This convolution not only limits the probe’s sensitivity to P1 motion, but also complicates the analysis of P1 motion by using the observed EPR spectra.
To overcome the problem associated with a flexible probe, we incorporated the rigid spin label Ç[4] into a deoxyribose oligonucleotide analogue of the ribozyme substrate (dSÇ; see Section S1 in the Supporting Information). Pairing of dSÇ with the ribozyme’s internal-guide-sequence resulted in the formation of the open complex (Figure 1A, Section S1). The EPR spectrum of dSÇ obtained in the presence of the wild-type ribozyme was dramatically different from that of dSÇ in an isolated P1 duplex (Figure 1B, Section S2), thus demonstrating successful assembly of a ribozyme with a Ç-labeled P1 analogue. The ribozyme spectrum shows an apparent splitting at the center line, and well-resolved hyperfine extrema in the low- and high-field regions (Figure 1B, right). These features are characteristics of a probe with limited mobility,[2a] which would be expected for a nitroxide rigidly fused to the P1 duplex. Furthermore, moving Ç to a different location of the P1 duplex yielded identical spectra (Section S3). This provides strong support that Ç and the P1 duplex behave as a single rigid body, such that the observed Ç spectrum reports motions of the entire P1 duplex and is not influenced by local motions at or near the nitroxide labeling site.
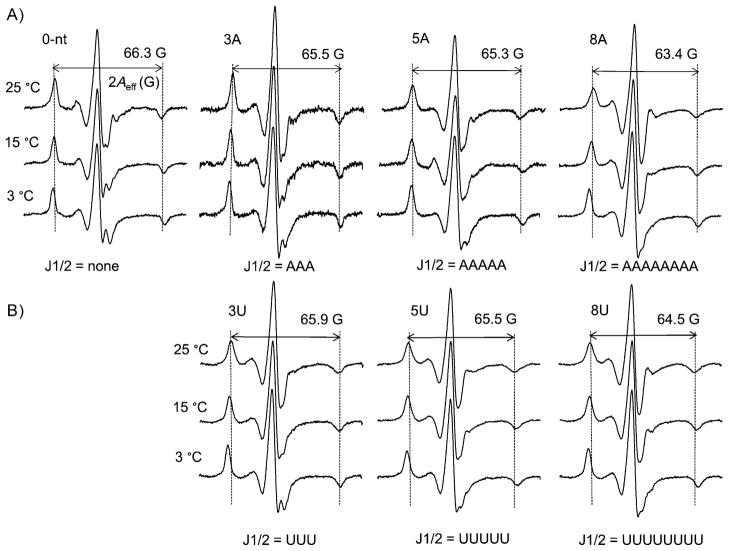
Previous studies using (flexible) R5a revealed increases in P1 nanosecond mobility upon lengthening of J1/2.[5] These experiments were repeated with the rigid Ç. Figure 2A shows ribozyme-bound dSÇ spectra in which the J1/2 sequence (i.e., base composition) was kept as polyadenosine (same as the wild-type), while its length is either shortened (0 nt, i.e., no J1/2), remains the same as the wild-type (the “3A” ribozyme, J1/2 = “AAA”) or is lengthened (5A and 8A). For each variant, lowering the temperature led to spectral broadening, which reflected the expected reduction in Ç mobility, as decreasing temperature tempers all modes of motion (e.g., ribozyme tumbling, P1 motion). More importantly, at each temperature, a similar trend of spectral change is observed with increasing J1/2 length: the central line shows less prominent splitting and becomes narrower; the amplitudes of the low- and high-field peaks reduce, and the splitting between the outer extrema (2Aeff) decreases slightly (Figure 2A). These changes indicate increased averaging of the nitroxide magnetic tensors resulting from increased Ç motion. As the ribozyme variants differ only in J1/2 length, their overall tumbling motions and the environment in the immediate vicinity of Ç were expected to be nearly identical, and were not expected to cause spectral variation. Therefore, the observed Ç mobility changes report increasing P1 motion as J1/2 lengthens. This conclusion is in complete agreement with that reached when using the flexible R5a.[5]
Figure 2.
Ç spectra of ribozyme variants with J1/2 being varying length of A) polyadenosine and B) polyuridine. For each variant, the 2Aeff of the 25°C spectrum is shown; and pairs of vertical dotted lines are shown to aid comparison of 2Aeff.
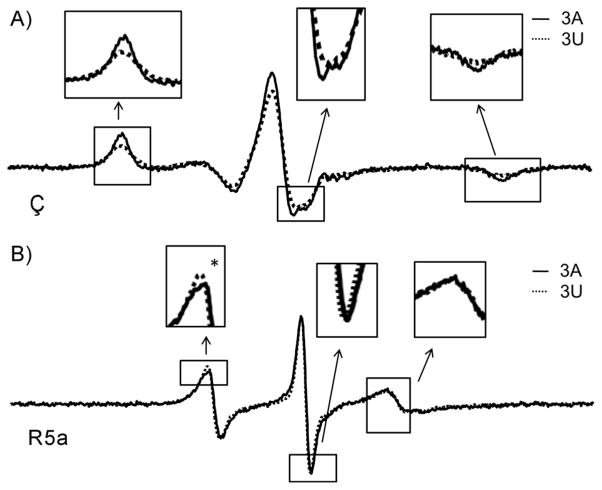
Expanding on previous studies that focused on only J1/2 with poly-A sequences, we examined how the J1/2 sequence influences P1 motion, by obtaining EPR spectra of a series of ribozymes in which J1/2 was mutated to poly-U of varying length (Figure 2B). Significant differences in Ç spectra were observed between the 3A and the 3U ribozymes (Figures 2 and 3A, Section S4), thus indicating that the J1/2 sequence also modulates P1 nanosecond motions, and that the rigid Ç is able to report such differences. For comparison, spectra were also obtained for 3A and 3U ribozymes with the flexible R5a (Figure 3B). The Ç spectra are substantially broader than those for the corresponding R5a (Figure 3), as expected from probe mobility reduction by removal of the rotatable bonds (Figure 1A). More importantly, differences between the Ç spectra are much larger than those for R5a (Figure 3). Thus, Ç indeed provides higher sensitivity of P1 motion.
Figure 3.
Comparisons of spectra obtained at 25°C between 3A (black line) and 3U (dotted line) ribozymes, with A) Ç, and B) R5a.
To obtain insight into how mutating J1/2 sequence alters the characteristics of P1 motion, we analyzed lineshape differences in the Ç spectra. Most noticeably, at 25°C, splitting at the central line is present in the 3A ribozyme, but is barely detectable for 3U (Figure 3A, middle inset). Splitting of the central line arises from incomplete g-tensor averaging.[2a, 6] Its presence indicates less motional averaging in the 3A ribozyme. In addition, at either the low- or the high-field manifold, the 3A ribozyme has narrower features with larger amplitude relative to 3U (Figure 3A, left and right insets), and the corresponding integrated spectrum shows more prominent outer shoulders and larger spectral breath (Section S5). These spectral features reveal less averaging of the hyperfine tensor, another indication of reduced motion in the 3A ribozyme. Interestingly, with the difference in hyperfine tensor averaging, the slopes at the outer edges of the integrated spectra differ between the 3A and 3U ribozymes (Section S5), thereby giving rise to a slightly smaller 2Aeff value in the 3A ribozyme (Figure 2).
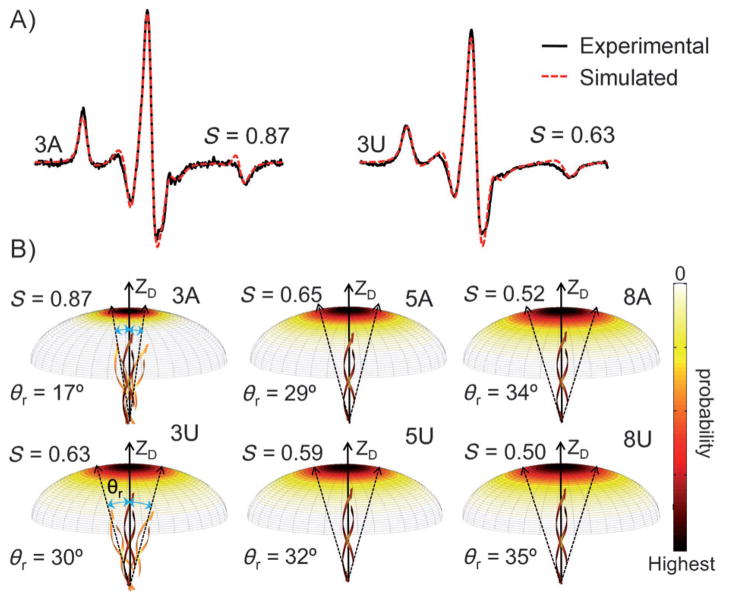
To further assess differences in P1 motions between the 3A and 3U ribozymes, we simulated the Ç spectra. The simulations used a model that describes the nitroxide motion within the macromolecular environment as a Brownian rotational diffusion constricted by an ordering potential.[6, 7] Figure 4A shows the resulting best-fit spectra from simulations of the 3A and 3U ribozymes. These simulations recapitulate the key spectral differences between the samples, i.e., the 3A ribozyme has narrower lines with larger amplitudes at both the low- and the high-field manifolds, and its 2Aeff is smaller than that of the 3U ribozyme (vide supra). The simulations revealed that in the 3A ribozyme, Ç (and consequently the P1 duplex) experiences a higher ordering potential with a faster rate than for 3U (Section S6). We note that “increasing ordering with increasing rate” is considered one form of “reducing mobility”, as has been observed in protein studies.[8] The simulations therefore indicate lower Ç mobility in the 3A ribozyme, which is consistent with the lineshape analysis (Figure 3A).
Figure 4.
A) Simulated spectra of 3A and 3U at 25°C. The order parameter S obtained from simulations is shown. Additional details are reported in Section S6. B) Plots of the probability for the P1 diffusion axis to rotate to a given angle with respect to the overall averaged axis (ZD). Dotted arrows are drawn based on θr to depict the effective amplitude of motion.
Although the motional model used in the simulations was a first-order approximation, it allowed us to visualize qualitatively differences in P1 motion within the 0.5–50 ns timeframe. By using the potentials obtained from simulations, we plotted the probability for the instantaneous P1 diffusion axis to rotate to a given angle relative to the overall averaged axis (Figure 4B). In addition, from the potentials we computed an order parameter S and derived a value (θr) that represents the effective amplitude of the P1 motion[6, 9] (Section S1). The best-fit spectra yielded S values of 0.87 for the 3A ribozyme and 0.67 for the 3U ribozymes (θr = 17° and 30°, respectively; Figure 4). These values reflect a significant increase in the motional range of P1 upon mutating J1/2 from “AAA” to “UUU”. Furthermore, control studies showed that uncertainty in the simulation parameters[10] does not alter the conclusion that Ç motion in the 3A ribozyme is more ordered (Section S6).
In addition to the differences between the 3A and 3U ribozymes, EPR spectra also report increasing Ç mobility (and consequently P1 motion) as J1/2 is changed from 3U to 5U and 8U, as indicated by narrowing of the central line, amplitude reduction of low- and high-field manifolds, and decreasing 2Aeff (Figure 2B). These data follow the same trend as that observed for the ribozymes with poly-A J1/2 (Figure 2A). However, sequence-dependent effects diminish with increasing J1/2 length. The 5A/5U set shows smaller spectral differences than the 3A/3U set (Figures 2 and 4B, Section S4). For the 8A/8U set, the spectra are nearly indistinguishable (Figure 2, Section S4), and differences in P1 motion are barely detectable (Figure 4B).
The observed dependence of P1 mobility on J1/2 sequence and length can be accounted for by the intrinsic properties of the single-stranded J1/2. Single-stranded poly-A is known to exhibit base stacking[11] and adopt dynamic helical conformations,[12] whereas single-stranded poly-U exhibits negligible base stacking[11] and behaves as an unstructured random coil.[13] Thus the poly-A ribozymes have a “stiffer” J1/2, thereby resulting in higher motional ordering of P1 (Figures 3 and 4). Furthermore, with the additional freedom of a longer J1/2, P1 mobility increases as J1/2 lengthens (Figure 2). Previous studies have reported that for single-stranded RNAs, their persistent lengths range from 1 to 2 nm,[12a, 13, 14] and the corresponding Kuhn segment (which can be considered “freely joined” in a polymer chain) would consist of 4 to 7 nucleotides.[12a] As J1/2 changes from 3 to 5 and 8 nucleotides, it exceeds the Kuhn length, and differences between poly-A and poly-U are predicted to diminish, as is indeed borne out in the data in Figures 2 and 4B.
Previous studies using a fluorophore rigidly fused to P1 have revealed that fluorescence anisotropy measured in the open complex decreases as J1/2 was extended or mutated from poly-A to poly-U, thus suggesting alteration of P1 motion.[15] The EPR data obtained for Ç are in agreement with the fluorescence results. Analyses presented here further suggest that J1/2 modulates motional ordering of P1 in the 0.5–50 ns range. Motional ordering dictates the probability of attaining a particular configuration (Figure 4B), and is one of the key factors that impact on sampling of the conformational space. Such information is important in studies of RNA dynamics, but is currently lacking, especially for large folded RNAs.
Nevertheless, a number of obstacles remain to be overcome before one can achieve quantitative descriptions of RNA motion by SDSL. For example, even with the rigid Ç and the use of a highly simplified ordering potential, a large number of parameters are required to fully describe the motion, and uniquely determining the motional parameters is challenging and requires further investigation (e.g., multiple-frequency EPR). Furthermore, the work presented here focused on nanosecond dynamics, and much remains to be learned regarding the connection with motions at slower timescales.
In summary, the rigid Ç enhances our ability to distinguish variations in RNA motions, and allowed us to reveal that in the ribozyme open complex both the sequence and length of the single-stranded J1/2 junction modulate nanosecond motional ordering of the P1 duplex. The SDSL method provides unique measurement of motions in the 0.5–50 ns range in large RNAs, and can be further combined with other techniques in our quest to understand RNA dynamics and its functional implication. Particularly, synergetic integrations of SDSL and molecular dynamics simulation have shown promise in studies of model protein and DNA,[16] and might be fruitful in investigations of RNA dynamics.
Experimental Section
Experimental details on sample preparation, EPR spectral acquisition and analyses are reported in the Supporting Information.
Supplementary Material
Acknowledgments
We thank Dr. P. Cekan for help with sample preparation, and NIH (R01GM069557, P.Z.Q.; P01GM66275, D.H.) and NSF (CHE1213673, P.Z.Q.) for financial support.
Footnotes
We would like to dedicate this article to the late Professor Ivano Bertini for his seminal contributions on developing magnetic resonance methods to study paramagnetic species in biological systems.
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/cbic.201300376.
References
- 1.Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM. Nature. 2012;482:322– 330. doi: 10.1038/nature10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a) Sowa GZ, Qin PZ. Prog Nucleic Acid Res Mol Biol. 2008;82:147–197. doi: 10.1016/S0079-6603(08)00005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Nguyen P, Qin PZ. Wiley Interdiscip Rev: RNA. 2012;3:62–72. doi: 10.1002/wrna.104. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Shelke SA, Sigurdsson S Th. Eur J Org Chem. 2012;2012:2291–2301. [Google Scholar]; d) Zhang X, Qin P. In: Biophysics of RNA Folding. Russell R, editor. Vol. 3. Springer; New York: 2013. pp. 69–87. [Google Scholar]
- 3.Hougland JL, Piccirilli JA, Forconi M, Lee J, Herschlag D. In: The RNA World. 3. Gesteland RF, Cech TR, Atkins JF, editors. CSHL Press; Cold Spring Harbor: 2006. pp. 133–205. [Google Scholar]
- 4.Barhate N, Cekan P, Massey AP, Sigurdsson S Th. Angew Chem. 2007;119:2709– 2712. doi: 10.1002/anie.200603993. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2007;46:2655– 2658. doi: 10.1002/anie.200603993. [DOI] [PubMed] [Google Scholar]
- 5.Grant GPG, Boyd N, Herschlag D, Qin PZ. J Am Chem Soc. 2009;131:3136– 3137. doi: 10.1021/ja808217s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Earle KA, Budil DE. In: Advanced ESR Methods in Polymer Research. Schlick S, editor. Wiley; New York: 2006. pp. 53–83. [Google Scholar]
- 7.Meirovitch E, Nayeem A, Freed JH. J Phys Chem. 1984;88:3454–3465. [Google Scholar]
- 8.Meirovitch E, Zerbetto M, Polimeno A, Freed JH. J Phys Chem B. 2011;115:143– 157. doi: 10.1021/jp107553j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LaConte LEW, Voelz V, Nelson W, Enz M, Thomas DD. Biophys J. 2002;83:1854– 1866. doi: 10.1016/S0006-3495(02)73950-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Columbus L, Kalai T, Jeko J, Hideg K, Hubbell WL. Biochemistry. 2001;40:3828–3846. doi: 10.1021/bi002645h. [DOI] [PubMed] [Google Scholar]; b) Zhang Z, Fleissner MR, Tipikin DS, Liang Z, Moscicki JK, Earle KA, Hubbell WL, Freed JH. J Phys Chem B. 2010;114:5503– 5521. doi: 10.1021/jp910606h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turner DH. In: Nucleic Acids: Structures, Properties, and Functions. Bloomfield VA, Crothers DM, Tinoco I Jr, editors. University Science Books; Sausalito: 2000. pp. 259–334. [Google Scholar]
- 12.a) Seol Y, Skinner GM, Visscher K, Buhot A, Halperin A. Phys Rev Lett. 2007;98:158103. doi: 10.1103/PhysRevLett.98.158103. [DOI] [PubMed] [Google Scholar]; b) Eichhorn CD, Feng J, Suddala KC, Walter NG, Brooks CL, Al-Hashimi HM. Nucleic Acids Res. 2012;40:1345– 1355. doi: 10.1093/nar/gkr833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seol Y, Skinner GM, Visscher K. Phys Rev Lett. 2004;93:118102. doi: 10.1103/PhysRevLett.93.118102. [DOI] [PubMed] [Google Scholar]
- 14.Chen H, Meisburger SP, Pabit SA, Sutton JL, Webb WW, Pollack L. Proc Natl Acad Sci USA. 2012;109:799– 804. doi: 10.1073/pnas.1119057109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a) Shi X, Solomatin SV, Herschlag D. J Am Chem Soc. 2012;134:1910–1913. doi: 10.1021/ja2083575. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Shi X, Mollova ET, Pljevaljčić G, Millar DP, Herschlag D. J Am Chem Soc. 2009;131:9571– 9578. doi: 10.1021/ja902797j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.a) Budil DE, Sale KL, Khairy KA, Fajer PG. J Phys Chem A. 2006;110:3703–3713. doi: 10.1021/jp054738k. [DOI] [PubMed] [Google Scholar]; b) Sezer D, Freed JH, Roux B. J Am Chem Soc. 2009;131:2597–2605. doi: 10.1021/ja8073819. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Popova AM, Hatmal MM, Frushicheva MP, Price EA, Qin PZ, Haworth IS. J Phys Chem B. 2012;116:6387–6396. doi: 10.1021/jp303303v. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.