Abstract
Background:
Many micro-RNAs (miRNAs) are differentially expressed in Helicobacter pylori-infected gastric mucosa and in gastric cancer tissue and previous reports have suggested the possibility of serum miRNAs as complementary tumour markers. The aim of the study was to investigate serum miRNAs and pepsinogen levels in individuals at high risk for gastric cancer both before and after H. pylori eradication.
Methods:
Patients with recent history of endoscopic resection for early gastric cancer and the sex- and age-matched controls were enrolled. Serum was collected from subjects before or after eradication and total RNA was extracted to analyse serum levels of 24 miRNAs. Serum pepsinogen (PG) I and II levels were measured using enzyme-linked immunosorbent assay kits.
Results:
Using miR-16 as an endogenous control, the relative levels of miR-106 and let-7d before and after H. pylori eradication and miR-21 after eradication were significantly higher in the high-risk group than in the controls. H. pylori eradication significantly decreased miR-106b levels and increased let-7d only in the control group. After eradication, the combination MiR-106b with miR-21 was superior to serum pepsinogen and the most valuable biomarker for the differentiating high-risk group from controls.
Conclusion:
Serum miR-106b and miR-21 may provide a novel and stable marker of increased risk for early gastric cancer after H. pylori eradication.
Keywords: H. pylori eradication, miR-106b, miR-21, pepsinogen
Gastric cancer is the fourth most common cancer and second leading cause of cancer deaths (∼700 000 deaths annually) worldwide (Melton et al, 2010). Helicobacter pylori is now well known as an important pathogen related to the development of gastric cancer, and the effect of H. pylori eradication on reducing gastric cancer incidence is related to the risk existing at the time of eradication therapy (Graham and Shiotani, 2005; Shiotani et al, 2008). In a Japanese multicenter clinical study, the incidence of new cancers after endoscopic mucosal resection of an early gastric cancer was reduced by one-third among those with H. pylori eradication compared with no eradication therapy (Fukase et al, 2008). However, the study also confirmed that H. pylori eradication did not completely prevent development of gastric cancer, and the prevalence of metachronous gastric cancer within 3 years after successful eradication was 3.3%. The previous studies reported the prevalence of metachronous gastric cancer within 3 years about 9% (Shiotani et al, 2008; Maehata et al, 2012), and the patients with recent history of endoscopic treatment for gastric cancer are known to have evidently greater risk compared with the patients with mild atrophy related to persistent H. pylori infection in Japan. The high prevalence of gastric cancer in Japan has led to a gastric cancer screening program that has shown benefits in terms of a decreased risk of mortality from gastric cancer through the detection of malignant lesions in the early stage in the screened population (Sano et al, 1993). Cancer screening has proven to be a high-cost endeavour and thus there has been continuing interest in developing non-invasive screening tests to identify the populations most likely to benefit. Measurement of serum pepsinogen (PG) or serum antibody against H. pylori is now considered as valid methods for identification of patients for screening for gastric cancer in terms of cost benefit and accuracy (Kitahara et al, 1999; Kudo et al, 2011; Miki, 2011). Pepsinogen I is produced by the chief and mucous neck cells in the fundic glands and PG II is produced by these cells and also by the cells in the pyloric glands and Brunner's glands (Samloff, 1971a, 1971b). Progressive mucosal damage results in a progressive decline in PG I levels and in the ratio of PG I to PG II (PG I/II) such that measuring PG levels can non-invasively identify the presence of atrophic gastritis and thus enhance the detection of positive cases in gastric screening programs (Miki et al, 1987, 2003). However, cumulative data have shown some limitations, one of which is that the predictive value of PG as gastric cancer risk marker is reduced following H. pylori eradication as H. pylori eradication can significantly change PG levels with a decrease of PG I and PG II and an increase of PG I/II even in those at high risk for gastric cancer (Kawai et al, 2007; Kaise et al, 2013). As such no validated predictive non-invasive biomarkers for gastric cancer after H. pylori eradication are yet available. In a longitudinal cohort study conducted in H. pylori-infected middle-aged Japanese men to evaluate the preventive effects of H. pylori eradication on the development of gastric cancer using PG test-positive criteria PG I ⩽70 ng ml−1 and PG I/II ⩽3.0. Significant reduction (P<0.05; log-rank test) in cancer incidence after H. pylori eradication was observed only in PG test-negative subjects (PG I >70 ng ml−1 or PG I/II >3.0) (Yanaoka et al, 2009). Cancer development both before and after H. pylori eradication seems to depend on the presence of extensive atrophy such that patients with a medical history of endoscopic submucosal dissection (ESD) are thought to be high-risk group for gastric cancer even after H. pylori eradication. Non-invasive biomarkers to identify those at highest risk among this already high risk group are needed.
Micro-RNAs (miRNAs) are 18–25 nucleotide noncoding RNA sequences that are transcribed but not translated into proteins. Some miRNAs have been shown to possess oncogenic or tumour suppressor activity and relate to apoptosis, proliferation, differentiation, metastasis, angiogenesis and immune response, which are all potentially involved in cancer initiation, progression and treatment response (Lu et al, 2005; Wang et al, 2010). There have been an increasing number of studies showing overexpression or downregulation of specific miRNAs in various malignant tumours including gastric cancer and H. pylori-infected gastric mucosa (Okubo et al, 2010; Hishida et al, 2011; Kamada et al, 2012; Shiotani et al, 2012a).
MiRNAs can be detected circulating in a cell-free form in blood, most probably in exosomes which protect them against degradation by ribonuclease (Rnase), and their signatures in blood are similar in men and women, as well as individuals of different age (Chim et al, 2008; Hunter et al, 2008; Mitchell et al, 2008). Furthermore, miRNA levels are similar in plasma and serum, and freeze/thaw as well as prolonged storage at room temperature does not affect their levels (Mitchell et al, 2008). Thus, serum miRNAs have the potential of a novel biomarker for many cancers, including gastric cancer (Mitchell et al, 2008; Ng et al, 2009; Liu et al, 2011; Chen et al, 2012). Accumulating evidence also suggest the potential of miRNAs in the early detection of patients with malignancies such as lymphoma, colorectal cancer, tongue cancer and ovarian cancer (Lawrie et al, 2008; Wong et al, 2008; Ng et al, 2009; Resnick et al, 2009). However, there are few papers reporting the value of serum miRNAs in early detection of gastric cancer (Liu et al, 2012; Song et al, 2012b). Our goal was to examine serum miRNAs as markers to individuals at high risk for gastric cancer not only before H. pylori eradication but also after eradication.
Materials and Methods
This was a case–control study of patients with a recent history of ESD for early gastric cancer and non-cancer controls. Patients were enrolled for the study between October 2006 and September 2012.
Patients
Patients with recent medical history of ESD for early stage, non-cardia intestinal-type gastric cancer without lymph node metastasis were enrolled as the high-risk group for gastric cancer. The control group consisted of subjects who had been previously diagnosed as H. pylori-positive gastric ulcer or atrophic gastritis. Exclusion criteria included H. pylori eradication within 1 year, gastrectomy, renal failure, cirrhosis or other malignant diseases. Demographic data collected at study entry included age, gender, smoking habit, alcohol consumption, drug treatments and eradication history. Drinking and smoking were defined as ‘regular' when consumption was more than 35 g for ethanol or 5 cigarettes per day, respectively. The study was approved by the Osaka Medical Center for Cancer and Cardiovascular Diseases Ethical Committee and Kawasaki Medical School Ethical Committee and written informed consent was obtained from each patient.
RNA extraction and quantitative polymerase chain reaction
Five ml of peripheral blood was collected just before endoscopy, and 1–4 h after collection the serum was separated by centrifugation and were stored at −80 °C until use. Total naked circulating RNA was extracted from 800 μl of serum using QIAamp Circulating Nucleic Acid kits (QIAGEN K. K., Tokyo, Japan), and finally eluted into 25 μl of elution solution according to the manufacturer's protocol. We examined 20 miRNAs (Let-7, miR-17-92 cluster, miR-21, miR-106b-25 cluster, miR-141, miR-143, miR-194, miR-203, miR-204, miR-205, miR-214, miR-223, miR-375) that were evaluated in the previous study (Shiotani et al, 2012b) and additionally miR-221 and miR-744 that had been recently reported as serum biomarkers for early detection of gastric cancer in China (Song et al, 2012b). Expression levels of miRNAs were analysed by quantitative reverse-transcriptase PCR (qRT–PCR). cDNA was prepared from total RNA using a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). Predesigned TaqMan MicroRNA Assays including primer set and TaqMan probe were purchased from Applied Biosystems. PCR was performed using TaqMan Universal PCR Master Mix II, and all reactions were run in triplicate on the 7900HT Fast Real-Time PCR system (Applied Biosystems). The cycle passing threshold (Ct) was recorded for each candidate miRNAs, and U6snRNA and miR-16 were used as the endogenous control for data normalisation. Relative expression of the target miRNAs was calculated using the comparative threshold cycle method.
Assay for serum levels of PG I and II
Serum levels of PG I and II were measured using enzyme-linked immunosorbent assay (ELISA) kits (LS test Eiken Kagaku, Inc., Tokyo, Japan).
Diagnosis of H. pylori
Venous blood samples were analysed for specific IgG H. pylori antibodies with ELISA kits using the E plate test (Eiken Kagaku, Inc.). Patients were considered to be infected with H. pylori, if the serum test was positive combined with evidence of chronic gastritis or atrophy with H. pylori on histopathological examination. Post eradication was confirmed by a negative 13C-urea breath test (13C-UBT) or negative serum H. pylori antibodies combined with history of the completion of H. pylori eradication therapy.
Statistical analyses
Values are expressed as the mean±s.d. or the median with a 25–75% range, whichever was appropriate depending on whether the data were normally distributed. Mantel–Haenszel χ2-analysis and the unpaired t-test were performed to measure differences in demographic and clinical characteristics. Statistical analyses for significant differences of miRNA levels were performed using the nonparametric Mann–Whitney U-test between the two groups and the Wilcoxon's signed rank test for paired data. Receiver–operator characteristic curves (ROC) were obtained to establish the most suitable cutoff point. Mantel–Haenszel statistics were used to assess the relationship between the high-risk group and the serum levels of miRNAs and PG showing the odds ratio (OR) and 95% confidence interval (CI). A two-sided P-value of less than 0.05 was considered statistically significant. All statistical computations were performed using SPSS (SPSS Inc., Chicago, IL, USA).
Results
Eighty-seven patients with a prior ESD history for non-cardiac early gastric cancer as a high risk group and 114 controls who were matched to the high-risk group by gender and age were enrolled. Serum samples were taken from those patients before and 1 year after eradication. In the high-risk group, serum samples were taken before eradication in 25 patients, after eradication in 23 and both in 39. In the controls, samples were taken before eradication in 44 patients, after eradication in 50 and both in 29. Demographic and clinical characteristics of the study groups are shown in Table 1.
Table 1. Demographic and clinical characteristics of the patients.
| Controls | Cancer | P-values | |
|---|---|---|---|
| Before eradication |
n=64 |
n=64 |
|
| Age, mean (s.d.) |
68.4 (8.3) |
67.9 (8.2) |
0.72a |
| Gender, male (%) |
41 (64.1) |
45 (70.3) |
0.57b |
| Regular alcohol intake (%) |
19 (28.1) |
31 (48.4) |
0.06b |
| Current smokers (%) |
11 (17.1) |
16 (25) |
0.46b |
| After eradication |
n=70 |
n=62 |
|
| Age, mean (s.d.) |
66.7 (7.5) |
67.8 (8.7) |
0.44a |
| Gender, male (%) |
47 (67.1) |
44 (71.0) |
0.71b |
| Regular alcohol intake (%) |
27 (38.6) |
25 (40.3) |
0.85b |
| Current smokers (%) | 11 (15.7) | 19 (30.6) | 0.08b |
P-values calculated using the unpaired t-test and Mantel–Haenszel χ2-analysis.
Unpaired t-test.
Mantel–Haenszel χ2-analysis.
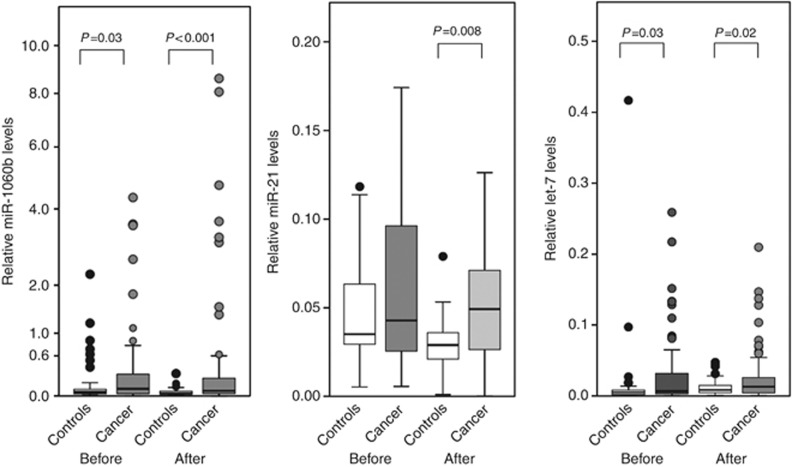
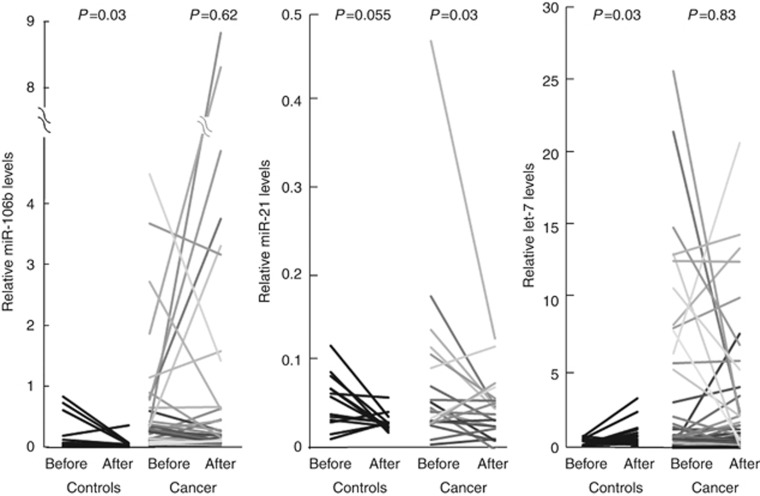
Using U6snRNA as an endogenous control for data normalisation, none of the miRNAs levels were significantly different between the two groups. In contrast, using miR-16 as an endogenous control, the relative levels of miR-106b before (8.46 × 10−2 vs 4.70 × 10−2, P=0.03) and after (7.19 × 10−2 vs 3.11 × 10−2, P<0.001) H. pylori eradication, miR-21 after eradication (4.79 × 10−2 vs 2.90 × 10−2, P=0.003), and let-7d before (0.63 × 10−2 vs 0.46 × 10−2, P=0.03) and after (1.33 × 10−2 vs 0.79 × 10−2, P=0.02) were significantly higher in the high-risk group than those in the controls (Table 2, Figure 1). The other examined miRNAs levels were not significantly different between the two groups (Table 2). In the controls, miR-106b significantly decreased (P=0.03) and let-7d increased (P=0.03) after H. pylori eradication (Figure 2), although other miRNAs did not significantly change after eradication. In the high-risk group, miR-21 significantly decreased (P=0.03) after H. pylori eradication (Figure 2), although other miRNAs did not significantly change.
Table 2. Comparisons of serum miRNAs levels between the controls and the high-risk group.
| Controls | Cancers | P-values | |
|---|---|---|---|
|
miR-106 (1 × 10−2) | |||
| Before | 4.70 (2.14–9.02) | 8.46 (2.73–29.7) | 0.03 |
| After |
3.11 (2.15–6.33) |
7.19 (3.08–29.1) |
<0.001 |
|
miR-21 (1 × 10−2) | |||
| Before | 3.51 (2.81–6.36) | 4.28 (2.41–10.0) | 0.91 |
| After |
2.90 (2.03–3.61) |
4.79 (2.51–7.21) |
0.003 |
|
Let-7 (1 × 10−2) | |||
| Before | 0.46 (0.18–0.82) | 0.63 (0.27–2.41) | 0.03 |
| After |
0.79 (0.36–1.49) |
1.33 (0.44–3.73) |
0.02 |
|
miR-214 (1 × 10−2) | |||
| Before | 7.93 (0.83–43.1) | 3.87 (1.11–25.9) | 0.34 |
| After |
1.83 (0.57–17.6) |
4.18(1.14–16.6) |
0.14 |
|
miR-221 (1 × 10−2) | |||
| Before | 1.24 (0.84–2.81) | 0.94 (0.37–1.70) | 0.11 |
| After |
3.11 (2.15–5.00) |
2.66 (1.19–6.13) |
0.26 |
|
miR-744 (1 × 10−2) | |||
| Before | 1.20 (0.50–1.68) | 0.85 (0.30–1.88) | 0.30 |
| After |
1.44 (0.80–3.01) |
1.20 (0.48–2.10) |
0.22 |
|
miR-143 (1 × 10−2) | |||
| Before | 0.33 (0.14–0.71) | 0.45 (0.20–1.28) | 0.29 |
| After |
0.28 (0.13–0.57) |
0.41 (0.14–0.78) |
0.40 |
|
miR-223 | |||
| Before | 0.74 (0.22–3.94) | 0.96 (0.18–2.11) | 0.73 |
| After |
1.28 (0.75–1.94) |
0.85 (0.28–1.86) |
0.26 |
|
miR-375 (1 × 10−2) | |||
| Before | 0.04 (0.02–0.23) | 0.20 (0.02–0.45) | 0.24 |
| After |
0.13 (0.08–0.25) |
0.19 (0.03–0.37) |
0.86 |
|
miR-17 (1 × 10−2) | |||
| Before | 5.39 (3.89–6.86) | 5.83 (4.03–8.18) | 0.67 |
| After |
4.35 (3.57–5.96) |
5.62 (3.60–6.78) |
0.35 |
|
miR-194 (1 × 10−2) | |||
| Before | 0.08 (0.05–0.22) | 0.16 (0.11–0.29) | 0.18 |
| After |
0.10 (0.05–0.19) |
0.12 (0.08–0.32) |
0.35 |
|
miR-25 (1 × 10−2) | |||
| Before | 7.48 (1.87–13.3) | 8.54 (5.90–16.0) | 0.44 |
| After |
11.8 (8.43–16.8) |
8.99 (5.47–11.5) |
0.10 |
|
miR-92 | |||
| Before | 2.04 (0.41–4.36) | 2.37 (1.52–3.07) | 0.80 |
| After |
3.16 (2.60–5.45) |
2.33 (1.30–3.80) |
0.09 |
|
miR-93 (1 × 10−2) | |||
| Before | 3.84 (2.74–6.95) | 5.18 (4.12–5.90) | 0.32 |
| After | 5.22 (4.10–6.60) | 6.15 (4.85–7.33) | 0.52 |
Abbreviation: miRNA=micro-RNA.
P-values were calculated using the nonparametric Mann–Whitney U-test between the two groups.
The values indicated relative expression of miRNAs using miR-16 as an endogenous control.
Figure 1.
Comparisons of serum miRNAs between the control group (Controls) and the high-risk group (Cancer) before and after eradication. Horizontal bar=median; Box=25th–75th interquartile range; vertical lines=range of values. The cycle passing threshold (Ct) was recorded for each miRNA, and miR-16 was used as the endogenous control for data normalisation. P-values were calculated using the nonparametric Mann–Whitney U-test.
Figure 2.
Changes of serum miRNAs by H. pylori eradication in the control group (Controls) and the high-risk group (Cancer). The cycle passing threshold (Ct) was recorded for each miRNA, and miR-16 was used as the endogenous control for data normalisation. P-values were calculated using the Wilcoxon's signed rank test.
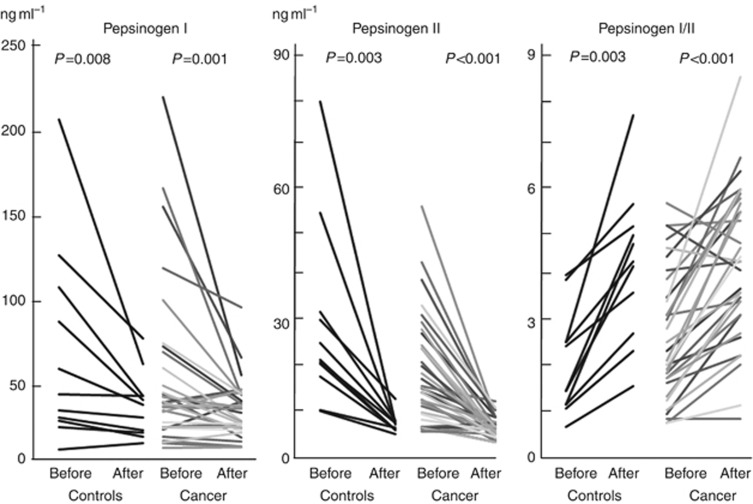
Serum PG I levels (before eradication 35.1 vs 55.3, P=0.01, after 31.7 vs 41.9, P=0.01) and PG II were significantly lower in the high-risk group than those in the controls both before and after eradication. Pepsinogen I/II were significantly lower (4.70 vs 5.25, P=0.01) in the high-risk group than those in the controls after eradication, but nor before eradication (Table 3). Serum PG I (P=0.008 in the controls and P=0.001 in the high-risk group) and PG II levels significantly decreased and PG I/II significantly increased (P=0.003 in the controls and P<0.001 in the high-risk group) in the both groups after eradication (Figure 3).
Table 3. Comparisons of serum pepsinogen levels between the controls and the high-risk group.
| Controls | Cancers | P-values | |
|---|---|---|---|
|
Pepsinogen I | |||
| Before | 55.3 (37.3–86.0) | 35.1 (19.1–79.3) | 0.01 |
| After |
41.9 (27.3–58.7) |
31.7 (19.3–43.0) |
0.01 |
|
Pepsinogen II | |||
| Before | 22.0 (15.7–33.5) | 16.7 (8.40–26.4) | 0.02 |
| After |
8.15 (6.13–10.8) |
6.70 (5.10–8.80) |
0.04 |
|
Pepsinogen I/II | |||
| Before | 2.50 (1.50–3.45) | 2.20 (1.70–3.70) | 0.97 |
| After | 5.25 (4.30–6.60) | 4.70 (3.50–5.70) | 0.01 |
P-values were calculated using the nonparametric Mann–Whitney U-test between the two groups.
Figure 3.
Changes of serum pepsinogen levels by H. pylori eradication in the control group (Controls) and the high-risk group (Cancer). P-values were calculated using the Wilcoxon's signed rank test.
Determination of cutoff points for screening for gastric cancer
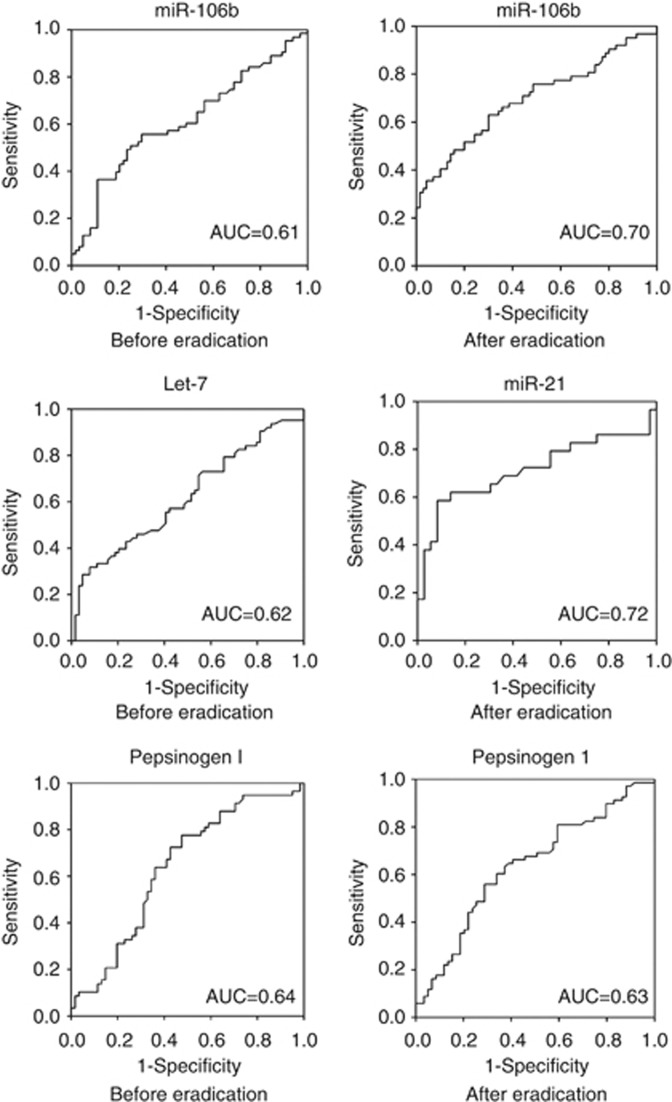
Receiver–operator characteristic curve analyses revealed that miR-106b levels before (the area under the ROC curve (AUC) 0.61; 95% CI: 0.51–0.71, P=0.03) and after eradication (AUC 0.70, 95% CI: 0.60–0.79, P<0.001), let-7d before (AUC 0.62, 95% CI: 0.52–0.72, P=0.02) and miR-21 after (AUC 0.72, 95% CI: 0.57–0.85, P=0.004) were the valuable biomarkers for differentiating high-risk individuals from controls (Figure 4). The suitable cutoff points of miR-106b for differentiating high-risk individuals obtained by an ROC were 0.075 and 0.032 before and after H. pylori eradication, respectively. Using the suitable cutoff point, the sensitivity and specificity of screening for high-risk group were 55.6% and 70.3% before H. pylori eradication (OR=2.96, 95% CI: 1.42–6.15, P=0.004) and 75.8% and 51.4% after eradication, respectively (OR=3.32, 95% CI: 1.57–7.00, P=0.002). The most suitable cutoff points of miRNAs for differentiating high-risk individuals obtained by an ROC after eradication was a miRNA-21more than 0.04, and the sensitivity and specificity were 58.6% and 86.1%, respectively (OR=8.78, 95% CI: 2.65–29.1, P<0.001). The most suitable cutoff points of miR-106b combined with miR-21 after eradication were a miR-106b more than 0.032 and miR-21 more than 0.02, and the sensitivity and specificity were 69.0% and 69.4%, respectively (OR=5.05, 95% CI: 1.75–14.6, P=0.003) (Table 4).
Figure 4.
Receiver–operator characteristic (ROC) curve analysis using serum miRNAs and pepsinogen for discriminating the high-risk group from the controls before or after H. pylori eradication. Abbreviation: AUC=area under the ROC curve.
Table 4. Cutoff points of serum miRNAs and pepsinogen for screening for high risk of gastric cancer.
| Cutoff points | Sensitivity (%) | Specificity (%) | Odds ratio (95% CI) |
|---|---|---|---|
|
Before eradication | |||
| miR-106b >0.075 | 55.6 | 70.3 | 2.96 (1.42–6.15) |
| miR-106b >0.035 and Let-7 >0.0035 | 60.3 | 59.4 | 2.22 (1.09–4.52) |
| Pepsinogen I<45 |
62.3 |
63.8 |
2.91 (1.38–6.13) |
|
After eradication | |||
| miR-106b >0.032 | 75.8 | 51.4 | 3.32 (1.57–7.00) |
| miR-21>0.04 | 58.6 | 86.1 | 8.78 (2.65–29.1) |
| miR-106b >0.032 and miR-21>0.02 | 69.0 | 69.4 | 5.05 (1.75–14.6) |
| Pepsinogen I<40 | 69.5 | 55.9 | 2.89 (1.39–6.00) |
| Pepsinogen I <40 and I/II <6 | 67.8 | 61.8 | 3.40 (1.63–7.08) |
Abbreviations: CI=confidence interval; miRNA=micro-RNA.
The most suitable cutoff points of PG were a PG I concentration less than 45 ng ml−1 before eradication and PG I less than 40 ng ml−1 combined with PG I/II less than 6.0 after eradication. Using the most suitable cutoff point, the sensitivity and specificity of screening for high-risk group were 62.3% and 63.8%, before eradication (OR=2.91, 95% CI: 1.38–6.13, P=0.005) and 67.8% and 61.8% after eradication, respectively (OR=3.40, 95% CI: 1.63–7.08, P=0.001) (Table 4).
Discussion
Lawrie et al (2008) first discovered tumour-specific deregulation of circulating miRNAs and subsequently, circulating miRNAs have been suggested great potential as biomarkers for many cancers including gastric cancer (Mitchell et al, 2008; Ng et al, 2009; Liu et al, 2011; Chen et al, 2012). Moreover, accumulating reports suggest the potential of miRNAs in the early detection of patients with several malignancies including gastric cancer (Ng et al, 2009; Resnick et al, 2009; Liu et al, 2012); however, the value of circulating miRNAs as biomarker for detection of high risk of gastric cancer has not been reported yet. In the present study, we first indicated the association between circulating miRNAs and high risk for gastric cancer demonstrating that the miR-106b and let-7d before and after H. pylori eradication and miR-21 after eradication were significantly higher in the high-risk group than those in the controls.
The miR-106b-25 cluster (miR-106b, miR-93 and miR-25) and miR-21 are oncogenic and are abnormally upregulated in gastric cancer (Belair et al, 2009; Wang et al, 2010; Wu et al, 2010). Targets of miR-106b-25 include cyclin-dependent kinase inhibitor (CDKN1A; p21CIP1/WAF1), E2F1, and proapoptoic factor Bim leading to a decreased response of gastric cells to TGF-β, (Petrocca et al, 2008; Kim et al, 2009) and miR-21 targets and inhibits tropomyosin 1 (alpha) (TPM1), programmed cell death 4 (neoplastic transformation inhibitor) (PDCD4), and phosphatase and tensin homologue (PTEN) and other tumour-related genes. (Zhu et al, 2007; Frankel et al, 2008; Wittmann and Jack, 2010) and (Tsujiura et al, 2010) reported that the plasma concentrations of miRNAs, miR-17-5p, miR-21, miR-106a and miR-106b were significantly higher in 69 gastric cancer patients than in 30 healthy controls indicating the greatest AUC value for miR-106b by ROC analysis. In contrast, the let-7 family acts as tumour suppressors (Bueno et al, 2008; Furuta et al, 2010), and the target genes are oncogenes such as high mobility group A2 (HMGA2), Ras, C-Myc and so on, and reduction of let-7 leads to overexpression of the oncogenes contributing to carcinogenesis (Johnson et al, 2005; Mayr et al, 2007; Sampson et al, 2007).
The results of serum miR-106b in the present study were similar to the results in our previous study investigating miRNAs expression in the gastric mucosa. The oncogenic miRNAs (miR-17/92 and miR-106b-93-25 cluster, miR-21, miR-194 and miR-196) levels were significantly higher in the gastric mucosa of the high-risk group compared with controls irrespective of H. pylori eradication. H. pylori eradication resulted in a significant fall in the oncogenic miRNAs only in the gastric mucosa of the controls, whereas let-7d expression increased in both groups (Shiotani et al, 2012b). In the present study, serum miR-21 expression levels even in the high-risk group decreased after eradication and associated with cancer risk, and the other oncogenic serum miRNAs were not significantly associated with risk for gastric cancer. Interestingly, a recent study detected increased levels of serum miR-21 in patients with chronic hepatitis C virus infection (CHC) and no difference compared with CHC-associated hepatocellular carcinoma. H. pylori by itself or related inflammation may strongly associate with serum miR-21 levels, and serum miR-21 levels could be an available biomarker to detect high-risk group only after successful eradication.
In our study, serum let-7d levels were significantly higher in the high-risk group than in controls irrespective of H. pylori eradication and increased after eradication in the controls. Let-7 family expression levels have been shown to be negatively associated with histological scores for activity, chronic inflammation and H. pylori density (Matsushima et al, 2011). H. pylori containing the cagPAI have been speculated to decrease let-7 expression in the gastric epithelium via the pivotal mitogen-activated protein kinase and nuclear factor-κB (Roush and Slack, 2008; Peter, 2009; Matsushima et al, 2011). One study suggested that serum let-7a concentrations (not normalised by endogenous controls) were significantly lower in those with cancer compared with controls (Tsujiura et al, 2010). However, another study reported that let-7e levels (normalised to the spiked-in cel-miR-39) were significantly higher in cancer thus confirming our results (Song et al, 2012b). We suspect that the different results in let-7 levels in relation of cancer are likely related to normalisation with endogenous controls.
More recently, Song et al (2012b) using miRNA profiling in a population-based study in China identified a panel of three serum miRNAs (miR-221, miR-744 and miR-376c) as potential biomarkers for early detection of gastric cancer indicating that only miR-221 was significantly elevated in those with dysplasia. However, there is no association between the miR-221 levels and high risk for gastric cancer both before and after eradication. The suitable biomarker to detect high risk of gastric cancer may differ depending on populations. However, the results of another profiling study identifying specific miRNAs in China were not consistent and miR-378 was distinguished as a novel biomarker in early detection of gastric cancer (Liu et al, 2012).
In the present study, H. pylori eradication significantly changed serum PG levels even in the high-risk group, whereas eradication did not significantly alter miR-106b and let-7 levels in the high-risk group. The serum miRNAs seem to be equivalent or even superior to serum PG as the biomarkers to detect a group at high risk for gastric cancer, especially after eradication. In a recent Japanese multicenter clinical study to investigate the incidence of new gastric cancers occurring after endoscopic treatment of an early gastric cancer, the incidence of new gastric cancers was reduced by one-third among those with H. pylori eradication compared with no eradication therapy. The study also confirmed that the incidence of new gastric cancer was 3.3% and quite high even after successful eradication (Fukase et al, 2008). Our previous study, which reported the prevalence of metachronous gastric cancer within 3 years was about 9%, indicated that severe atrophy assessed by histology and lower levels of serum PG I before eradication were predictive factors for metachronous gastric cancer in high-risk patients after successful eradication. Non-invasive biomarkers to identify those at highest risk among this already high-risk group are needed, and continued surveillance (secondary prevention) should be considered (Shiotani et al, 2013). The serum miR-106b and miR-21 seem to be suitable non-invasive markers before and after eradication, respectively, and the combination of these miRANs after eradication seems to be superior to serum pepsinogen and the most useful to detect those at high risk for gastric cancer among the examined miRNAs.
MiR-16 and U6snRNA are often used for normalisation to evaluate serum miRNAs as diagnostic markers for different tumours (Brase et al, 2010). We used miRNA-16 as the endogenous control for data normalisation based on the fact that none of the miRNAs levels were significantly different between the two groups when using U6snRNA. Previous studies have also recommended miR-16 and miR-93 as suitable reference genes for serum miRNA analysis for gastric cancer and healthy controls (Song et al, 2012a).
The major limitation of the present study is that it was a case–control study and a prospective cohort study should be done to confirm the association between the serum miRNAs levels and risk of gastric cancer development. Another limitation is that our investigation was not based on microarray profiling and there will be significant abnormalities of expression of other miRNAs related to development of gastric cancer. Further investigations in vivo and in vitro are required to obtain a comprehensive understanding role of serum miRNAs and the associated abnormalities in cellular signalling in carcinogenesis.
In conclusion, serum miRNAs may provide a new and stable biomarker to detect those at high risk for gastric cancer, and serum miR-106b and miR-21 may provide a novel and stable marker of increased risk for early gastric cancer after H. pylori eradication.
Acknowledgments
We thank Professor David Y Graham for helpful comments and Ms Maki Nomura for assistance of laboratory work.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
References
- Belair C, Darfeuille F, Staedel C. Helicobacter pylori and gastric cancer: possible role of microRNAs in this intimate relationship. Clin Microbiol Infect. 2009;15:806–812. doi: 10.1111/j.1469-0691.2009.02960.x. [DOI] [PubMed] [Google Scholar]
- Brase JC, Wuttig D, Kuner R, Sultmann H. Serum microRNAs as non-invasive biomarkers for cancer. Mol Cancer. 2010;9:306. doi: 10.1186/1476-4598-9-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno MJ, Perez de Castro I, Gomez de Cedron M, Gomez de Cedron M, Santos J, Calin GA, Cigudosa JC, Croce CM, Fernandez-Piqueras J, Malumbres M. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell. 2008;13:496–506. doi: 10.1016/j.ccr.2008.04.018. [DOI] [PubMed] [Google Scholar]
- Chen X, Hu Z, Wang W, Ba Y, Ma L, Zhang C, Wang C, Ren Z, Zhao Y, Wu S, Zhuang R, Zhang Y, Hu H, Liu C, Xu L, Wang J, Shen H, Zhang J, Zen K, Zhang CY. Identification of ten serum microRNAs from a genome-wide serum microRNA expression profile as novel noninvasive biomarkers for non-small cell lung cancer diagnosis. Int J Cancer. 2012;130:1620–1628. doi: 10.1002/ijc.26177. [DOI] [PubMed] [Google Scholar]
- Chim SS, Shing TK, Hung EC, Leung TY, Lau TK, Chiu RW, Lo YM. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem. 2008;54:482–490. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- Fukase K, Kato M, Kikuchi S, Inoue K, Uemura N, Okamoto S, Terao S, Amagai K, Hayashi S, Asaka M, Japan Gast Study G Effect of eradication of Helicobacter pylori on incidence of metachronous gastric carcinoma after endoscopic resection of early gastric cancer: an open-label, randomised controlled trial. Lancet. 2008;372:392–397. doi: 10.1016/S0140-6736(08)61159-9. [DOI] [PubMed] [Google Scholar]
- Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766–776. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- Graham DY, Shiotani A. The time to eradicate gastric cancer is now. Gut. 2005;54:735–758. doi: 10.1136/gut.2004.056549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishida A, Matsuo K, Goto Y, Naito M, Wakai K, Tajima K, Hamajima N. Combined effect of miR-146a rs2910164 G/C polymorphism and Toll-like receptor 4 +3725G/C polymorphism on the risk of severe gastric atrophy in Japanese. Dig Dis Sci. 2011;56:1131–1137. doi: 10.1007/s10620-010-1376-1. [DOI] [PubMed] [Google Scholar]
- Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, Xiao T, Schafer J, Lee ML, Schmittgen TD, Nana-Sinkam SP, Jarjoura D, Marsh CB. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Kaise M, Miwa J, Fujimoto A, Tashiro J, Tagami D, Sano H, Ohmoto Y. Influence of Helicobacter pylori status and eradication on the serum levels of trefoil factors and pepsinogen test: serum trefoil factor 3 is a stable biomarker. Gastric Cancer. 2013;16:329–337. doi: 10.1007/s10120-012-0185-y. [DOI] [PubMed] [Google Scholar]
- Kamada T, Kurose H, Yamanaka Y, Manabe N, Kusunoki H, Shiotani A, Inoue K, Hata J, Matsumoto H, Akiyama T, Hirai T, Sadahira Y, Haruma K. Relationship between gastroesophageal junction adenocarcinoma and Helicobacter pylori infection in Japan. Digestion. 2012;85:256–260. doi: 10.1159/000336352. [DOI] [PubMed] [Google Scholar]
- Kawai T, Miki K, Ichinose M, Kenji Y, Miyazaki I, Kawakami K, Kataoka M, Yamagishi T, Sofuni A, Itoi T, Moriyasu F, Takagi Y, Aoki T, Matsubayashi J, Mukai K. Changes in evaluation of the pepsinogen test result following Helicobacter pylori eradication therapy in Japan. Inflammopharmacology. 2007;15:31–35. doi: 10.1007/s10787-006-0009-y. [DOI] [PubMed] [Google Scholar]
- Kim YK, Yu J, Han TS, Park SY, Namkoong B, Kim DH, Hur K, Yoo MW, Lee HJ, Yang HK, Kim VN. Functional links between clustered microRNAs: suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 2009;37:1672–1681. doi: 10.1093/nar/gkp002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitahara F, Kobayashi K, Sato T, Kojima Y, Araki T, Fujino MA. Accuracy of screening for gastric cancer using serum pepsinogen concentrations. Gut. 1999;44:693–697. doi: 10.1136/gut.44.5.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo T, Kakizaki S, Sohara N, Onozato Y, Okamura S, Inui Y, Mori M. Analysis of ABC (D) stratification for screening patients with gastric cancer. World J Gastroenterol. 2011;17:4793–4798. doi: 10.3748/wjg.v17.i43.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, Hatton CS, Harris AL. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- Liu H, Zhu L, Liu B, Yang L, Meng X, Zhang W, Ma Y, Xiao H. Genome-wide microRNA profiles identify miR-378 as a serum biomarker for early detection of gastric cancer. Cancer Lett. 2012;316:196–203. doi: 10.1016/j.canlet.2011.10.034. [DOI] [PubMed] [Google Scholar]
- Liu R, Zhang C, Hu Z, Li G, Wang C, Yang C, Huang D, Chen X, Zhang H, Zhuang R, Deng T, Liu H, Yin J, Wang S, Zen K, Ba Y, Zhang CY. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer. 2011;47:784–791. doi: 10.1016/j.ejca.2010.10.025. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Maehata Y, Nakamura S, Fujisawa K, Esaki M, Moriyama T, Asano K, Fuyuno Y, Yamaguchi K, Egashira I, Kim H, Kanda M, Hirahashi M, Matsumoto T. Long-term effect of Helicobacter pylori eradication on the development of metachronous gastric cancer after endoscopic resection of early gastric cancer. Gastrointest Endosc. 2012;75:39–46. doi: 10.1016/j.gie.2011.08.030. [DOI] [PubMed] [Google Scholar]
- Matsushima K, Isomoto H, Inoue N, Nakayama T, Hayashi T, Nakayama M, Nakao K, Hirayama T, Kohno S. MicroRNA signatures in Helicobacter pylori-infected gastric mucosa. Int J Cancer. 2011;128:361–370. doi: 10.1002/ijc.25348. [DOI] [PubMed] [Google Scholar]
- Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton SD, Genta RM, Souza RF. Biomarkers and molecular diagnosis of gastrointestinal and pancreatic neoplasms. Nat Rev Gastroenterol Hepatol. 2010;7:620–628. doi: 10.1038/nrgastro.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki K. Gastric cancer screening by combined assay for serum anti-Helicobacter pylori IgG antibody and serum pepsinogen levels - "ABC method". Proc Jpn Acad Ser B Phys Biol Sci. 2011;87:405–414. doi: 10.2183/pjab.87.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki K, Ichinose M, Shimizu A, Huang SC, Oka H, Furihata C, Matsushima T, Takahashi K. Serum pepsinogens as a screening test of extensive chronic gastritis. Gastroenterol Jpn. 1987;22:133–141. doi: 10.1007/BF02774209. [DOI] [PubMed] [Google Scholar]
- Miki K, Morita M, Sasajima M, Hoshina R, Kanda E, Urita Y. Usefulness of gastric cancer screening using the serum pepsinogen test method. Am J Gastroenterol. 2003;98:735–739. doi: 10.1111/j.1572-0241.2003.07410.x. [DOI] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58:1375–1381. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- Okubo M, Tahara T, Shibata T, Yamashita H, Nakamura M, Yoshioka D, Yonemura J, Ishizuka T, Arisawa T, Hirata I. Association between common genetic variants in pre-microRNAs and gastric cancer risk in Japanese population. Helicobacter. 2010;15:524–531. doi: 10.1111/j.1523-5378.2010.00806.x. [DOI] [PubMed] [Google Scholar]
- Peter ME. Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle. 2009;8:843–852. doi: 10.4161/cc.8.6.7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, Cavazzini L, Volinia S, Alder H, Ruco LP, Baldassarre G, Croce CM, Vecchione A. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Resnick KE, Alder H, Hagan JP, Richardson DL, Croce CM, Cohn DE. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol. 2009;112:55–59. doi: 10.1016/j.ygyno.2008.08.036. [DOI] [PubMed] [Google Scholar]
- Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Samloff IM. Cellular localization of group I pepsinogens in human gastric mucosa by immunofluorescence. Gastroenterology. 1971a;61:185–188. [PubMed] [Google Scholar]
- Samloff IM. Pepsinogens, pepsins, and pepsin inhibitors. Gastroenterology. 1971b;60:586–604. [PubMed] [Google Scholar]
- Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- Sano T, Sasako M, Kinoshita T, Maruyama K. Recurrence of early gastric cancer. Follow-up of 1475 patients and review of the Japanese literature. Cancer. 1993;72:3174–3178. doi: 10.1002/1097-0142(19931201)72:11<3174::aid-cncr2820721107>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Shiotani A, Cen P, Graham DY.2013Eradication of gastric cancer is now both possible and practical Semin Cancer Biole-pub ahead of print 19 July 2013;doi: 10.1016/j.semcancer.2013.07.004 [DOI] [PubMed]
- Shiotani A, Murao T, Uedo N, Iishi H, Yamanaka Y, Kamada T, Kusunoki H, Inoue K, Haruma K. Eradication of H. pylori did not improve abnormal sonic hedgehog expression in the high risk group for gastric cancer. Dig Dis Sci. 2012a;57:643–649. doi: 10.1007/s10620-011-1916-3. [DOI] [PubMed] [Google Scholar]
- Shiotani A, Uedo N, Iishi H, Murao T, Kanzaki T, Kimura Y, Kamada T, Kusunoki H, Inoue K, Haruma K. H. pylori eradication did not improve dysregulation of specific oncogenic miRNAs in intestinal metaplastic glands. J Gastroenterol. 2012b;47:988–998. doi: 10.1007/s00535-012-0562-7. [DOI] [PubMed] [Google Scholar]
- Shiotani A, Uedo N, Iishi H, Yoshiyuki Y, Ishii M, Manabe N, Kamada T, Kusunoki H, Hata J, Haruma K. Predictive factors for metachronous gastric cancer in high-risk patients after successful Helicobacter pylori eradication. Digestion. 2008;78:113–119. doi: 10.1159/000173719. [DOI] [PubMed] [Google Scholar]
- Song J, Bai Z, Han W, Zhang J, Meng H, Bi J, Ma X, Han S, Zhang Z. Identification of suitable reference genes for qPCR analysis of serum microRNA in gastric cancer patients. Dig Dis Sci. 2012a;57:897–904. doi: 10.1007/s10620-011-1981-7. [DOI] [PubMed] [Google Scholar]
- Song MY, Pan KF, Su HJ, Zhang L, Ma JL, Li JY, Yuasa Y, Kang D, Kim YS, You WC. Identification of serum microRNAs as novel non-invasive biomarkers for early detection of gastric cancer. PLoS One. 2012b;7:e33608. doi: 10.1371/journal.pone.0033608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, Kosuga T, Konishi H, Morimura R, Deguchi K, Fujiwara H, Okamoto K, Otsuji E. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer. 2010;102:1174–1179. doi: 10.1038/sj.bjc.6605608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang Q, Liu H, Hu B, Zhou W, Cheng Y. MicroRNA expression and its implication for the diagnosis and therapeutic strategies of gastric cancer. Cancer Lett. 2010;297:137–143. doi: 10.1016/j.canlet.2010.07.018. [DOI] [PubMed] [Google Scholar]
- Wittmann J, Jack HM. Serum microRNAs as powerful cancer biomarkers. Biochim Biophys Acta. 2010;1806:200–207. doi: 10.1016/j.bbcan.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Wong TS, Liu XB, Chung-Wai HoA, Po-Wing Yuen A, Wai-Man NgR, Ignace Wei W. Identification of pyruvate kinase type M2 as potential oncoprotein in squamous cell carcinoma of tongue through microRNA profiling. Int J Cancer. 2008;123:251–257. doi: 10.1002/ijc.23583. [DOI] [PubMed] [Google Scholar]
- Wu WK, Lee CW, Cho CH, Fan D, Wu K, Yu J, Sung JJ. MicroRNA dysregulation in gastric cancer: a new player enters the game. Oncogene. 2010;28:5761–5771. doi: 10.1038/onc.2010.352. [DOI] [PubMed] [Google Scholar]
- Yanaoka K, Oka M, Ohata H, Yoshimura N, Deguchi H, Mukoubayashi C, Enomoto S, Inoue I, Iguchi M, Maekita T, Ueda K, Utsunomiya H, Tamai H, Fujishiro M, Iwane M, Takeshita T, Mohara O, Ichinose M. Eradication of Helicobacter pylori prevents cancer development in subjects with mild gastric atrophy identified by serum pepsinogen levels. Int J Cancer. 2009;125:2697–2703. doi: 10.1002/ijc.24591. [DOI] [PubMed] [Google Scholar]
- Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]