Abstract
Background:
Five-year survival after resection of colorectal cancer liver metastasis (CRLCM) is <30%. We recently found that aurora kinase A (AURKA) drives 20q gain-associated tumour progression and is associated with disease recurrence. This study evaluates the prognostic value of AURKA expression in CRCLM of patients who underwent liver resection.
Methods:
Tissue microarrays (TMAs) were generated using formalin-fixed paraffin-embedded CRCLM and matched primary tumour from a multi-institutional cohort of patients with CRCLM who underwent liver resection between 1990 and 2010. Tissue microarrays were stained for AURKA using immunohistochemistry, and a hazard rate ratio (HRR) for the association between overall survival (OS) and nuclear AURKA expression in CRCLM was calculated. Results were validated by 500-fold cross-validation.
Results:
The expression of AURKA was evaluated in CRCLM of 343 patients. High AURKA expression was associated with poor OS (HRR 1.55, P<0.01), with a cross-validated average HRR of 1.57 (P=0.02). Average HRR was adjusted for the established prognostic clinicopathological variables in a multivariate analysis (average HRR 1.66; P=0.02). The expression of AURKA in CRCLM was correlated to its expression in corresponding primary tumour (P<0.01).
Conclusion:
The expression of AURKA protein is a molecular biomarker with prognostic value for patients with CRCLM, independent of established clinicopathological variables.
Keywords: colorectal cancer, liver metastasis, prognosis, biomarker, AURKA
Annually, >1.2 million people are diagnosed with colorectal cancer (CRC) worldwide and almost half of them die as a consequence of disseminated disease (Ferlay et al, 2010). The majority of haematogenous CRC metastases are located in the liver, followed by the lung and the peritoneum (Welch and Donaldson, 1979). Liver resection, occasionally in combination with radiofrequency ablation (RFA), is currently the only option with curative intent for CRC patients with liver metastases (CRCLM) (Fong et al, 1997; Abdalla et al, 2004). Patients eligible for liver resection are selected based on well-established prognostic clinicopathological variables that have been combined in a clinical risk score as defined by Fong et al (1999), which was supported by others in independent study populations (Nordlinger et al, 1996; Rees et al, 2008; Yamaguchi et al, 2008). Application of molecular imaging using 18F-labelled fluorodeoxyglucose (18FDG) PET in combination with computed tomography (CT) further reduced the proportion of futile surgeries by more than one-third (Ruers et al, 2009). Nevertheless, only ∼30% of patients survive for >5 years after surgery. Considering the significant morbidity and mortality risk associated with liver resections (Leporrier et al, 2006; Ruers et al, 2009), there is a clear need for better prognostic biomarkers.
Aurora kinase A (AURKA) is a cell cycle-regulated kinase involved in spindle formation and chromosome segregation (Lens et al, 2010). AURKA is located on chromosome 20q, a genomic region that is frequently amplified in CRC, that has been associated with adenoma-to-carcinoma progression and that is an indicator of poor prognosis (Bischoff et al, 1998; Hermsen et al, 2002; Aust et al, 2004; Nakao et al, 2004; Postma et al, 2007; Carvalho et al, 2009; Sillars-Hardebol et al, 2010). We recently demonstrated that AURKA, together with TPX2, drives 20q gain-associated adenoma-to-carcinoma progression based on the correlation of AURKA DNA copy number status to mRNA and protein expression levels combined with its functional effects on cell viability, anchorage-independent growth and invasion (Sillars-Hardebol et al, 2012). Furthermore, we demonstrated that high protein levels of AURKA in stage III colon cancer – that is, patients with disseminated cells present in lymph nodes – were associated with increased disease recurrence (Belt et al, 2012). These data indicate that AURKA overexpression drives malignant behaviour and imply that AURKA may be a prognostic biomarker for CRC. In the present retrospective study, we aimed to investigate whether AURKA is a prognostic molecular biomarker with added value to clinicopathological variables for CRCLM patients selected for liver resection with curative intent.
Materials and Methods
Patient study population
Patients who underwent liver resection with curative intent, sometimes with the addition of RFA, in the seven Dutch hospitals affiliated with the DeCoDe PET group were identified by cross-referencing surgery and pathology databases using Dutch MeSH terms for ‘colon', ‘rectum', ‘carcinoma', ‘adenocarcinoma', ‘colorectal neoplasms', ‘liver', ‘neoplasm metastasis' and ‘(hemi)hepatectomy'. Clinicopathological data from patients who were operated on between 1990 and 2010 were extracted from these databases. Formalin-fixed paraffin-embedded (FFPE) tissue specimens were collected from one CRCLM sample and an adjacent control liver sample. When available, also the corresponding primary tumour and adjacent control colon tissue were collected. Only specimens of patients with histologically confirmed CRCLM were included in the study, whereas tissue samples of patients with multiple primary tumours were excluded. Collection, storage and use of clinicopathological data and tissue specimens were performed in compliance with the ‘Code for Proper Secondary Use of Human Tissue in The Netherlands' and approved in protocol 2011–03 of our Department of Pathology (Stichting FMWV Rotterdam, 2011).
Tissue microarrays
A total of 21 TMAs were generated using methodology as described previously (Simon et al, 2004; Belt et al, 2011). In brief, three tissue core biopsies of 0.6 mm in diameter were punched from morphologically representative areas of all FFPE donor blocks and transferred into TMA-recipient paraffin blocks using the 3DHISTECH TMA Master (v1.14; 3DHISTECH Ltd., Budapest, Hungary).
Evaluation of AURKA protein expression
Sections of TMAs (4 μm) were mounted on glass slides and immunohistochemically stained for AURKA (mouse monoclonal, NCL-L-AK2, Novocastra Laboratories, Newcastle, UK) as described previously (Carvalho et al, 2009). Immunohistochemical stainings were digitally captured using the Mirax slide scanner system equipped with a × 20 objective with a numerical aperture of 0.75 (Carl Zeiss B.V., Sliedrecht, The Netherlands) and a Sony DFW-X710 Fire Wire 1/3' type progressive SCAN IT CCD (pixel size 4.65 × 4.65 μm). Actual scan resolution at × 20 was 0.23 μm. Computer monitors were calibrated using the Spyder2PRO software (v1.0–16, Pantone Colorvision, Regensdorf, Switzerland). Tissue microarray core biopsies were scored for an intensity of AURKA protein expression in the nuclei of neoplastic epithelial cells (categories negative, weak, moderate, strong) using the dedicated TMA-scoring software (v1.14.25.1; 3DHISTECH Ltd.). For facilitating scoring, a chart with visual analogue scales of staining patterns was used. Tissue samples were evaluated unaware of corresponding clinicopathological information at the time of assessment. Tissue samples were independently evaluated by a second investigator in a blinded manner with high interobserver agreement (Kw=0.75).
Statistical analysis
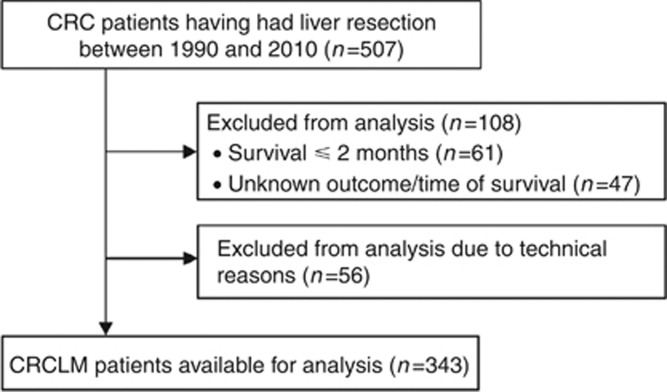
Overall survival (OS) was defined as the time in months after surgery until death in a follow-up period of 10 years. Patients were excluded from the analysis if OS⩽2 months, if OS or survival status were unknown or if tissue cores could not be evaluated for technical reasons (Figure 1). The prognostic value of AURKA expression was tested in a cross-validation procedure, repeated 500 times. In each cross-validation round the study, population was randomly subdivided into a training set (50%) and a validation set (50%). The training set was used to determine the optimal cutoff for dichotomising intensity scores into ‘low AURKA' and ‘high AURKA' staining intensities. This was performed using the receiver operating characteristic (ROC) curve analysis for survival data, with 3-year OS as the outcome of interest (Heagerty et al, 2000; Heagerty and Zheng, 2005; Zlobec et al, 2007). The optimal cutoff was defined as the point on the ROC curve, giving the smallest distance to the point (1-specificity, sensitivity)=(0,1) (Supplementary Figure S1A). Using this cutoff, AURKA intensity scores in the validation set were dichotomised, and the crude hazard rate ratio (HRR) for AURKA expression was calculated in a Cox regression analysis with OS as outcome. Additionally, established prognostic clinicopathological variables combined in the clinical risk score defined by Fong et al (1999) were included in a multivariate Cox regression analysis. The average cross-validated HRR (HRRav) of the validation sets was calculated, and the P-value of the cross-validation procedure was defined as the percentage of average cross-validated HRRav<1. To visualise the relation between OS and AURKA expression in the total study cohort and a number of cohort subgroups, Kaplan–Meier curves were obtained using the most frequently selected cutoff in the training sets as the optimal cutoff and the HRR was calculated using the Cox regression analysis. The level of concordance of protein expression in primary CRC and CRCLM was calculated using the Pearson's χ2 test. All statistical tests were two-sided and executed using IBM SPSS Statistics 20.0 software (SPSS Inc., IL, USA) or R Statistics 14.0 software (RStudio Inc., Boston, MA, USA). P-values <0.05 were considered significant. All data reported were REMARK-compliant (McShane et al, 2005).
Figure 1.
Flow diagram of the total study population.
A power calculation provided an estimate of the required size of the study population. Hazard rate ratio was the primary outcome measure for OS difference between patients with 'low AURKA' and 'high AURKA' expression. Ten-year OS of the study population was estimated as 30% and, therefore, the proportion of long-term and short-term survivors after liver resection as 30% and 70%, respectively (Leporrier et al, 2006). Assuming similar proportions of patients with low and high AURKA expression, 227 events were required to detect an OS difference between both subgroups with an HRR of 1.5. With a follow-up period of 10 years, this required an estimated total study population of 361 patients.
Results
Demographic characteristics
A study cohort of 507 consecutive CRCLM patients who underwent liver resection with curative intent was assembled. CRCLM samples were obtained from all patients, and from 234 patients the corresponding primary CRC samples were also collected. Demographic characteristics of the cohort are summarised in Table 1. OS after liver resection is depicted in Figure 2A. The median survival of the cohort was 27 months (range, 3–212 months); 5-year OS was 41.3%. The majority of patients were male (63.1%). The median age at the time of liver resection was 64.3 years (range, 27.6–83.9 years). The primary tumour was of colonic origin in 67.0% of patients and of rectal origin in 29.8% of patients. In 3.2% of patients, the origin of the primary tumour was unknown. No positive lymph nodes were detected at the time of primary tumour resection in 30.4% of patients. Metastases presented within 12 months after diagnosis of the primary tumour in 43.6% of patients. The median number of liver metastases was 2 (range, 1–12), with a median size of 3.5 cm (range, 0.2–22.0 cm). Extrahepatic disease was present in 7.1% of patients. Chemo- and/or antibody-based therapy during the months prior to or following liver resection was received by 33.3% of patients (Supplementary Table S1). Additional perioperative RFA of liver metastases was performed in 21.5% of patients.
Table 1. Patient characteristics.
| |
Total population |
|
|---|---|---|
| Clinicopathological variable | n/mean/median | %/s.d./range |
|
General characteristics | ||
| All |
507 |
|
|
Gender | ||
| Male (%) | 320 | 63.1 |
| Female (%) |
187 |
36.9 |
|
OS after liver resection in months | ||
| Mean (s.d.) | 37.2 | 1.7 |
| Median (range) |
27 |
3.0–212.0 |
|
Primary CRC characteristics | ||
|
Age at primary CRC resection | ||
| Mean (s.d.) | 61.9 | 0.5 |
| Median (range) |
62.8 |
24.4–83.3 |
|
Primary tumour location | ||
| Left colon (%) | 245 | 48.3 |
| Right colon (%) | 95 | 18.7 |
| Rectum (%) | 151 | 29.8 |
| Unknown (%) |
16 |
3.2 |
|
Histological grade | ||
| Poorly differentiated (%) | 38 | 7.5 |
| Moderately differentiated (%) | 288 | 56.8 |
| Well-differentiated (%) | 14 | 2.8 |
| Unknown (%) |
167 |
32.9 |
|
Maximal CRC diameter in cm | ||
| Mean (s.d.) | 4.4 | 0.1 |
| Median (range) |
4 |
0.2–12.0 |
|
Positive lymph nodes detecteda | ||
| Yes (%)a | 190 | 37.5 |
| No (%)a | 154 | 30.4 |
| Unknown (%)a |
163 |
32.1 |
|
Systemic treatment of primary CRC: colon cancer | ||
| Preoperative (%) | 9 | 2.6 |
| Perioperative (%) | 0 | 0 |
| Postoperative (%) | 91 | 26.8 |
| None (%) | 226 | 66.5 |
| Unknown (%) |
14 |
4.1 |
|
Systemic treatment of primary CRC: rectal cancer | ||
| Preoperative (%) | 25 | 16.5 |
| Perioperative (%) | 1 | 0.7 |
| Postoperative (%) | 24 | 15.9 |
| None (%) | 98 | 64.9 |
| Unknown (%) |
3 |
2 |
|
Radiotherapeutic treatment of primary CRC | ||
| Yes (%) | 113 | 22.3 |
| No (%) | 392 | 77.3 |
| Unknown (%) |
2 |
0.4 |
|
Liver metastasis characteristics | ||
|
Age at liver resection | ||
| Mean (s.d.) | 63.3 | 0.5 |
| Median (range) |
64.3 |
27.6–83.9 |
|
Synchronous liver metastasisa | ||
| Yes (%)a | 221 | 43.6 |
| No (%)a | 261 | 51.5 |
| Unknown (%)a |
25 |
4.9 |
|
Distribution liver metastases | ||
| Left half | 90 | 17.8 |
| Segment 1 (%) | 18 | 3.6 |
| Segment 2 (%) | 116 | 22.9 |
| Segment 3 (%) | 135 | 26.6 |
| Segment 4 (%) | 134 | 26.4 |
| Right half | 259 | 51 |
| Segment 5 (%) | 171 | 33.7 |
| Segment 6 (%) | 212 | 41.8 |
| Segment 7 (%) | 211 | 41.6 |
| Segment 8 (%) | 182 | 35.9 |
| Both left and right half | 146 | 28.8 |
| Unknown |
12 |
2.4 |
|
Maximal CRCLM diameter in cma | ||
| Mean (s.d.) | 4.1 | 0.1 |
| Median (range) | 3.5 | 0.2–22.0 |
| Maximal diameter ⩽5.0 cm (%)a | 372 | 73.4 |
| Maximal diameter >5.0 cm (%)a | 126 | 24.8 |
| Maximal diameter unknown (%)a |
9 |
1.8 |
|
Number of CRCLMsa | ||
| Mean (s.d.) | 2 | 0.1 |
| Median (range) | 2 | 1–12 |
| Nr of crclms=1 (%)a | 248 | 48.9 |
| No. of crclms >1 (%)a | 253 | 49.9 |
| No. of crclms unknown (%)a |
6 |
1.2 |
|
Serum CEA levela | ||
| Mean (s.d.) | 101.6 | 38.5 |
| Median (range) | 15 | 0.5–6625.0 |
| Serum CEA level ⩽200 ng ml−1 (%)a | 166 | 32.7 |
| Serum CEA level >200 ng ml−1 (%)a | 13 | 2.6 |
| Serum CEA level unknown (%)a |
328 |
64.7 |
|
Extrahepatic metastases | ||
| Yes (%) | 36 | 7.1 |
| No (%) | 423 | 83.4 |
| Unknown (%) |
48 |
9.5 |
|
Resection in combination with RFA | ||
| Yes (%) | 109 | 21.5 |
| No (%) | 357 | 70.4 |
| Unknown (%) |
41 |
8.1 |
|
Systemic treatment of liver metastases | ||
| Preoperative (%) | 60 | 11.8 |
| Perioperative (%) | 12 | 2.4 |
| Postoperative (%) | 97 | 19.1 |
| None (%) | 321 | 63.3 |
| Unknown (%) | 17 | 3.4 |
Abbreviations: CEA=carcino-embryonic antigen; OS=overall survival; RFA=radiofrequency ablation.
Clinical risk score variable (Fong et al, 1999).
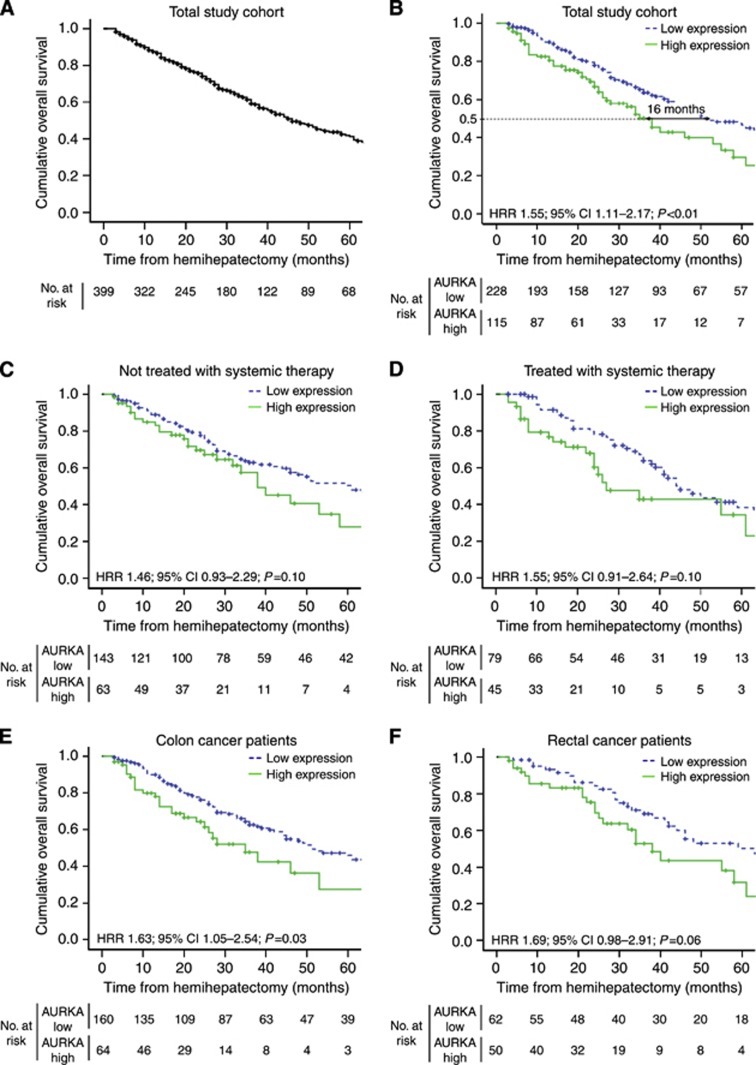
Figure 2.
(A) Kaplan–Meier graph depicting OS in months of the total study population. (B–F) Kaplan–Meier graphs depicting OS in months stratified by intensity of AURKA expression in hepatic metastases (B) of the total population, (C) of patients in whom liver metastases were not treated with systemic therapy (n=206), (D) of patients in whom liver metastases were treated with systemic therapy (n=124), (E) originating from colon cancer (n=224) and (F) originating from rectal cancer (n=112). Information of systemic therapy was unavailable for n=13 patients. Origin of primary tumour (colon or rectum) was unkown for n=7 patients.
Presence of more than one liver metastasis was inversely and significantly related to OS (HRR 1.38; 95% CI 1.04–1.83; P=0.03). Positive lymph nodes at the time of primary tumour resection and presentation of metastases within 12 months after diagnosis of the primary tumour both tended to be associated with decreased OS (HRR 1.39; 95% CI 0.99–1.96; P=0.06 and HRR 1.24; 95% CI 0.93–1.66; P=0.14, respectively). Maximal diameter of the metastasis and serum CEA levels were not associated with OS (HRR 1.05; 95% CI 0.76–1.46; P=0.75 and HRR 0.94; 95% CI 0.37–2.36; P=0.90, respectively). No significant differences in OS were observed between patients having had surgery between 1990 and 1999 vs patients who were operated on after 1999 (HRR 1.09; 95% CI 0.80–1.50; P=0.58; Supplementary Figure S2).
AURKA expression is associated with poor prognosis
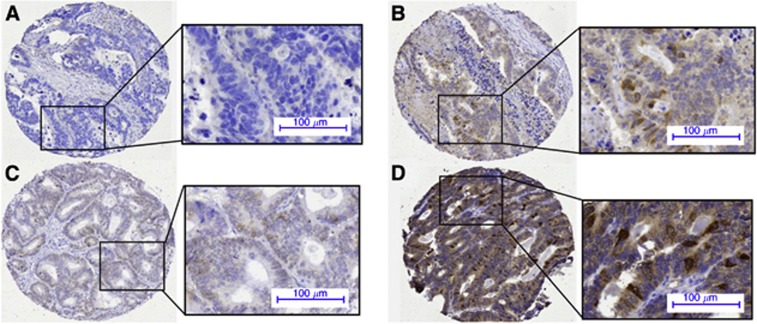
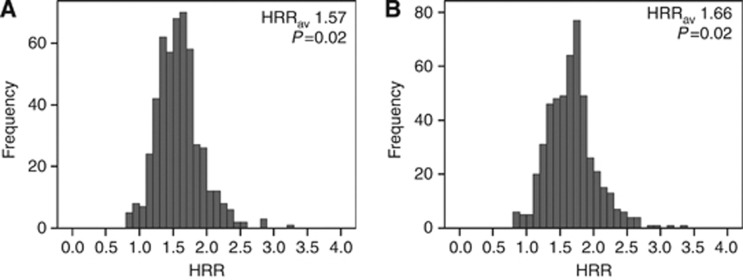
Immunohistochemical staining of AURKA expression could be evaluated for 343 patients (Figure 1), and AURKA nuclear staining intensity of neoplastic CRCLM epithelium was scored (Figure 3 and Supplementary Table S2). The cross-validated HRRav for AURKA expression was 1.57 (P=0.02; Figure 4A), indicating a significant association between high levels of AURKA in CRCLM and decreased OS. The cutoff between the categories negative, weak and moderate on one hand and strong on the other emerged in >95% of cross-validation cycles as most optimal (Supplementary Figure S1B). When dichotomising the total study population based on this cutoff, a difference of 16 months in the median OS was observed (HRR 1.55; 95% CI 1.11–2.17; P<0.01; Figure 2B) between patients with low AURKA expression (median OS 51 months) and high AURKA expression (median OS 35 months).
Figure 3.
Expression pattern of AURKA in the epithelium of CRCLM. Staining intensity of the nuclei was evaluated as (A) negative, (B) weak, (C) moderate or (D) strong.
Figure 4.
Distribution of the cross-validated HRRs after (A) univariate analysis and (B) multivariate analysis together with established prognostic clinicpathological variables.
Next, a multivariate Cox regression analysis was performed to evaluate whether the prognostic value of AURKA protein expression was independent of the established prognostic clinicopathological variables, that is, primary tumour-to-liver metastasis interval ⩽12 months, number of liver metastases >1, maximal tumour diameter >5.0 cm and lymph node positivity at the time of diagnosis of the primary tumour. The HRRav after multivariate analysis was 1.66 (P=0.02; Figure 4B), indicating an autonomous association between AURKA expression and OS.
Prognostic value of AURKA expression in patient subgroups
Preoperative systemic therapy can affect tissue characteristics at the time of surgery and may bias immunohistochemical analysis of AURKA expression. When excluding patients who had received systemic treatment within 6 months preoperative to liver resection (n=281), results were similar to those of the total study population (HRR 1.52; 95% CI 1.03–2.23; P=0.03; Supplementary Figure S3), indicating the lack of significant bias.
Next, the effects of systemic therapy regardless of administration time point (that is, pre-, peri- or postoperative to liver resection) were analysed. Overexpression of AURKA tended to be associated with poor survival, both in the subgroup of patients that did not receive systemic treatment (HRR 1.46; 95% CI 0.93–2.29; P=0.10; Figure 2C) and in the subgroup of patients who were treated with systemic therapy (HRR 1.55; 95% CI 0.91–2.64; P=0.10; Figure 2D). Furthermore, the prognostic value was evaluated separately for CRCLM patients whose primary tumour originated in the colon (HRR 1.63; 95% CI 1.05–2.54; P=0.03; Figure 2E) and the rectum (HRR 1.69; 95% CI 0.98–2.91; P=0.06; Figure 2F). Comparable results were obtained for both tumour sites.
AURKA expression in primary CRC and corresponding CRCLM
Primary CRC tissue material is more readily obtained for pathological examination than CRCLM tissue material. Therefore, we investigated whether AURKA expression in the primary tumour was correlated to expression in its corresponding liver metastasis. For 152 patients, both CRCLM and matched primary CRC tissue could be evaluated for AURKA staining intensity (Supplementary Figure S4), revealing 63.8% concordant and 36.2% discordant pairs of intensity scores (P<0.01; Supplementary Table S3). Similar level of concordance was found when patients treated with systemic therapy preoperative to primary CRC resection and/or CRCLM resection were excluded from analysis, with a level of concordance of 60.4%, and 39.6% disconcordant pairs (P<0.01; Supplementary Table S4).
Discussion
Our data demonstrate that high expression of AURKA in liver metastases of CRC patients who underwent liver resection was associated with poor survival (HRR 1.55; P<0.01), which was validated in a 500-fold cross-validation procedure (HRRav 1.57; P=0.02). The difference in the median OS time between patients with high- vs low AURKA-expressing CRCLM was 16 months. This difference in the median OS time is substantial, considering a median survival after liver resection of 27 months in our total study population. Multivariate analysis revealed that the prognostic value of AURKA expression was independent of prognostic clinicopathological variables, such as presence of more than one liver metastasis, positive lymph nodes at time of primary tumour resection, maximal CRCLM diameter >5 cm and presentation of metastases within 12 months after diagnosis of the primary tumour.
The CRCLM cohort used for the current study lacked prognostic value for two established clinicopathological risk factors – that is, liver metastasis diameter >5.0 cm and serum CEA >200 ng ml−1. Some prognostic effects for liver metastasis diameter >5.0 cm could be observed in our study population within 30 months follow-up after liver resection (HRR 1.48; 95% CI 0.99–2.20; P=0.06), an effect that diminished when considering a longer follow-up period of 10 years. For CEA serum levels, data availability was limited (n=179) and the number of patients that exceeded the preset threshold of serum CEA levels >200 ng ml−1 within this group was small (n=13). Therefore, we excluded this variable from the multivariate analysis. Resection margin status and presence of extrahepatic metastases are two additional clinicopathological prognostic variables regularly used in clinical practice. On resection margin status, no data were available for current study population; hence, this parameter could not be included in the multivariate analysis. Including presence of extrahepatic metastases as additional prognostic variable in the multivariate Cox regression analysis merely altered the average HRR (HRRav 1.61; P=0.02).
The OS of patients having had liver resection in the period from 2000 to 2010 was not improved compared with patients operated on in the period from 1990 to 2000 (Supplementary Figure S2). This observation was somewhat unexpected, although could be explained by an increase in patients having been operated on since 2000 who presented with affected lymph nodes (P=0.05) and/or extrahepatic metastases (P=0.04).
Although a clinical risk score is a useful and easily applicable method to select patients for liver resection, it is insufficient to represent the complexity of CRC biology. Molecular biomarkers have the potential to reveal differences in biological behaviour among apparently similar tumour samples. They are indicators of changes in cancer-related biological processes, such as proliferation, differentiation, invasion and angiogenesis, because of their direct or indirect involvement in these processes. AURKA has been described to function in proliferation, chromosomal instability, anchorage-independent growth and invasion (Lee et al, 2008; Wang et al, 2009; Moore et al, 2010; Zhang et al, 2011; Sillars-Hardebol et al, 2012). Importantly, changes in AURKA expression appear to cause rather than to follow differences in these biological processes, implying that AURKA is a driver rather than a passenger of tumour progression and thereby a direct indicator of tumour biology (Sillars-Hardebol et al, 2012). The present study demonstrates that differences in AURKA protein expression in CRCLM are also associated with clinical consequences – that is, overall survival, analogous to our previously observed association with disease recurrence in stage III colon cancer patients (Belt et al, 2012). The causal relationship between AURKA overexpression and tumour progression, and its association with reduced patient survival in different CRC stages, may explain why AURKA is a strong and independent prognostic biomarker for CRC. Similar to our observations, association of high AURKA protein expression with poor survival has been reported for stage III CRC (Lam et al, 2008) and for triple-negative breast cancer (Xu et al, 2013). However, no such association was found in another study of AURKA protein expression in stage-I- to stage-IV CRC samples (Baba et al, 2009). In addition, contradictory findings have been reported by studies that determined AURKA DNA copy number status, one indicating association with tumour progression (Zhang et al, 2010) and another association with prolonged survival (Dotan et al, 2012). Differences among these and our studies may be explained by variation in size and composition of patient cohorts, methodology and reagents to determine AURKA expression levels as well as methodology to determine its cutoff values for low vs high expression.
The primary tumour of CRCLM is located in either the colon or the rectum, the two organs that are often combined for experimental analyses. Although adenocarcinomas of colon and rectum are hardly distinguishable at the genomic level, it remains debatable whether colon and rectal cancers should be considered as one or two distinct entities (Frattini et al, 2004; Cancer Genome Atlas Network, 2012). Our data demonstrated prognostic value of AURKA expression in CRCLM, irrespective of the intestinal origin of the primary tumour (Figure 2E and F) – that is, colon or rectum – which renders AURKA a candidate biomarker for both colon and rectal cancer patients. Next, we examined whether the expression of AURKA in CRCLM was correlated to its expression in the corresponding patient-matched primary tumour, as molecular alterations that are acquired during early stages of tumour development are likely to be present in the metastases (Stange et al, 2010; Knijn et al, 2011). Conform our expectations, a significant correlation was found (P<0.01), indicating that the level of AURKA expression in CRCLM is to a certain extent predetermined by molecular alterations in the primary tumour. Although this correlation suggests that one may predict the level of AURKA expression in CRCLM based on analysis of its corresponding primary tumour, for example, when CRCLM specimens are not (yet) available, it should be noted that AURKA expression was concordant in only 63.8% of CRC–CRCLM pairs.
In addition to principally curative liver resection, about one-third of all patients also received chemotherapy, sometimes combined with antibody-based systemic therapy. Commonly used chemotherapeutic agents, such as Fluorouracil (5-FU) or its prodrug Capecitabine target mitosis, are one of the hallmarks of cancer cells. As AURKA is involved in spindle formation and chromosome segregation, we hypothesised that treatment with antiproliferative agents might influence its prognostic value as determined in this retrospective study. This turned out not to be the case, as similar prognostic effects of AURKA were observed for the groups of patients who did and did not receive systemic treatment (Figure 2C and D). One explanation may be that the biological effects of AURKA are not restricted to a role in proliferation. In vitro studies demonstrated that AURKA activation can switch cells from a pro- to an anti-apoptotic transcriptional programme through the regulation of mRNA splicing (Moore et al, 2010), suggesting that AURKA is a master regulator that affects the function of many genes. Other studies showed that AURKA affects anchorage-independent growth and invasion, properties that match characteristics that are essential for malignant metastasising cells and therefore may highly influence OS (Zhang et al, 2011; Ratushny et al, 2012; Sillars-Hardebol et al, 2012). As such, although targeting mitosis is currently being challenged as a flawed rationale to treat cancer (Komlodi-Pasztor et al, 2012), targeting of AURKA kinase activity may affect cancer-driving properties other than proliferation.
Several small molecule inhibitors have been developed specifically inhibiting AURKA rather than the other aurora kinase isoforms. These inhibitors have demonstrated anticancer activity in various preclinical cancer models and some have entered clinical trials (Katayama and Sen, 2010; Lens et al, 2010; Cervantes et al, 2012). Our data suggest that targeted treatment of CRC patients with AURKA inhibitors may improve OS, provided that within a given tumour AURKA functions as a driver of the carcinogenic process rather than as a passenger of the mitotic machinery. Further research is required to establish and validate the potential of AURKA as a molecular biomarker to predict patient responsiveness to AURKA inhibitors – for example, at the DNA level through determination of chromosome 20q AURKA amplification or at the protein level using immunohistochemical evaluation of tumour tissue. Alternatively, AURKA inhibitors may be used for the development of novel PET tracers to directly visualise and quantify AURKA as a molecular drug target within a given patient in vivo and to monitor efficiency of treatment. Therefore, we conclude that AURKA expression can serve both as a promising prognostic and potentially also as a predictive biomarker for CRC, which fits current trends towards personalised medicine.
Acknowledgments
Funding was supported by the Center for Translational Molecular Medicine, DeCoDe project (grant 03O-101).
Appendix
DeCoDe PET group collaborators: All collaborators of the DeCoDe PET group have contributed to data acquisition and approved the final paper after critical revision. Collaborators of the DeCoDe PET group are listed in alphabetical order per medical center: N.C.T. van Grieken, L.R. Perk, E.A. te Velde, A.D. Windhorst (VU University/VU University Medical Center – Amsterdam); J. Baas, A.M. Rijken (Amphia Medical Center – Breda); M.W. van Beek, H.J. Pijpers (Catharina Medical Center/PAMM Foundation – Eindhoven); H. Bril, H.B.A.C. Stockmann, A. Zwijnenburg (Kennemer Gasthuis/Spaarne Medical Center – Haarlem); K. Bosscha, A.J. van den Brule, C.J. Hoekstra, J.C. van der Linden (Jeroen Bosch Medical Center‘s – Hertogenbosch); I.H. Borel Rinkes, P.J. van Diest, R. van Hillegersberg, O. Kranenburg, M.G. Lam, N. Snoeren (UMCU – Utrecht); I.H. Liem, R.M. Roumen, W. Vening (MMC – Veldhoven).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Abdalla EK, Vauthey JN, Ellis LM, Ellis V, Pollock R, Broglio KR, Hess K, Curley SA. Recurrence and outcomes following hepatic resection, radiofrequency ablation, and combined resection/ablation for colorectal liver metastases. Ann Surg. 2004;239:818–825. doi: 10.1097/01.sla.0000128305.90650.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aust DE, Muders M, Kohler A, Schmidt M, Diebold J, Muller C, Lohrs U, Waldman FM, Baretton GB. Prognostic relevance of 20q13 gains in sporadic colorectal cancers: a FISH analysis. Scand J Gastroenterol. 2004;39:766–772. doi: 10.1080/00365520410003191. [DOI] [PubMed] [Google Scholar]
- Baba Y, Nosho K, Shima K, Irahara N, Kure S, Toyoda S, Kirkner GJ, Goel A, Fuchs CS, Ogino S. Aurora-A expression is independently associated with chromosomal instability in colorectal cancer. Neoplasia. 2009;11:418–425. doi: 10.1593/neo.09154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belt EJT, Brosens RPM, Delis-van Diemen P, Bril H, Tijssen M, van Essen DF, Heymans MW, Belien JAM, Stockmann HBAC, Meijer S, Meijer GA. Cell cycle proteins predict recurrence in stage II and III colon cancer. Ann Surg Oncol. 2012;19 (Suppl 3:S682–S692. doi: 10.1245/s10434-012-2216-7. [DOI] [PubMed] [Google Scholar]
- Belt EJT, Fijneman RJA, van den Berg EG, Bril H, is-van Diemen PM, Tijssen M, van Essen HF, de Lange-de Klerk ESM, Belien JAM, Stockmann HBAC, Meijer S, Meijer GA. Loss of lamin A/C expression in stage II and III colon cancer is associated with disease recurrence. Eur J Cancer. 2011;47:1837–1845. doi: 10.1016/j.ejca.2011.04.025. [DOI] [PubMed] [Google Scholar]
- Bischoff JR, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, Schryver B, Flanagan P, Clairvoyant F, Ginther C, Chan CS, Novotny M, Slamon DJ, Plowman GD. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17:3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho B, Postma C, Mongera S, Hopmans E, Diskin S, van de Wiel MA, van Criekinge W, Thas O, Matthai A, Cuesta MA, Terhaar Sive Droste JS, Craanen M, Schrock E, Ylstra B, Meijer GA. Multiple putative oncogenes at the chromosome 20q amplicon contribute to colorectal adenoma to carcinoma progression. Gut. 2009;58:79–89. doi: 10.1136/gut.2007.143065. [DOI] [PubMed] [Google Scholar]
- Cervantes A, Elez E, Roda D, Ecsedy J, Macarulla T, Venkatakrishnan K, Rosello S, Andreu J, Jung J, Sanchis-Garcia JM, Piera A, Blasco I, Manos L, Perez-Fidalgo JA, Fingert H, Baselga J, Tabernero J. Phase I pharmacokinetic/pharmacodynamic study of MLN8237, an investigational, oral, selective aurora a kinase inhibitor, in patients with advanced solid tumors. Clin Cancer Res. 2012;18:4764–4774. doi: 10.1158/1078-0432.CCR-12-0571. [DOI] [PubMed] [Google Scholar]
- Dotan E, Meropol NJ, Zhu F, Zambito F, Bove B, Cai KQ, Godwin AK, Golemis EA, Astsaturov I, Cohen SJ. Relationship of increased aurora kinase A gene copy number, prognosis and response to chemotherapy in patients with metastatic colorectal cancer. Br J Cancer. 2012;106:748–755. doi: 10.1038/bjc.2011.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. GLOBOCAN 2008, Cancer Incidence and Mortality Worldwide. IARC Cancer Base No. 2010;10 [Google Scholar]
- Fong Y, Cohen AM, Fortner JG, Enker WE, Turnbull AD, Coit DG, Marrero AM, Prasad M, Blumgart LH, Brennan MF. Liver resection for colorectal metastases. J Clin Oncol. 1997;15:938–946. doi: 10.1200/JCO.1997.15.3.938. [DOI] [PubMed] [Google Scholar]
- Fong Y, Fortner J, Sun RL, Brennan MF, Blumgart LH. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: analysis of 1001 consecutive cases. Ann Surg. 1999;230:309–318. doi: 10.1097/00000658-199909000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frattini M, Balestra D, Suardi S, Oggionni M, Alberici P, Radice P, Costa A, Daidone MG, Leo E, Pilotti S, Bertario L, Pierotti MA. Different genetic features associated with colon and rectal carcinogenesis. Clin Cancer Res. 2004;10:4015–4021. doi: 10.1158/1078-0432.CCR-04-0031. [DOI] [PubMed] [Google Scholar]
- Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337–344. doi: 10.1111/j.0006-341x.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- Heagerty PJ, Zheng Y. Survival model predictive accuracy and ROC curves. Biometrics. 2005;61:92–105. doi: 10.1111/j.0006-341X.2005.030814.x. [DOI] [PubMed] [Google Scholar]
- Hermsen M, Postma C, Baak J, Weiss M, Rapallo A, Sciutto A, Roemen G, Arends JW, Williams R, Giaretti W, De Goeij A, Meijer G. Colorectal adenoma to carcinoma progression follows multiple pathways of chromosomal instability. Gastroenterology. 2002;123:1109–1119. doi: 10.1053/gast.2002.36051. [DOI] [PubMed] [Google Scholar]
- Katayama H, Sen S. Aurora kinase inhibitors as anticancer molecules. Biochim Biophys Acta. 2010;1799:829–839. doi: 10.1016/j.bbagrm.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knijn N, Mekenkamp LJM, Klomp M, Vink-Borger ME, Tol J, Teerenstra S, Meijer JWR, Tebar M, Riemersma S, van Krieken JHJM, Punt CJA, Nagtegaal ID. KRAS mutation analysis: a comparison between primary tumours and matched liver metastases in 305 colorectal cancer patients. Br J Cancer. 2011;104:1020–1026. doi: 10.1038/bjc.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komlodi-Pasztor E, Sackett DL, Fojo AT. Inhibitors targeting mitosis: tales of how great drugs against a promising target were brought down by a flawed rationale. Clin Cancer Res. 2012;18:51–63. doi: 10.1158/1078-0432.CCR-11-0999. [DOI] [PubMed] [Google Scholar]
- Lam AK-Y, Ong K, Ho YH. Aurora kinase expression in colorectal adenocarcinoma: correlations with clinicopathological features, p16 expression, and telomerase activity. Hum Pathol. 2008;39:599–604. doi: 10.1016/j.humpath.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Lee HH, Zhu Y, Govindasamy KM, Gopalan G. Downregulation of Aurora-A overrides estrogen-mediated growth and chemoresistance in breast cancer cells. Endocr Relat Cancer. 2008;15:765–775. doi: 10.1677/ERC-07-0213. [DOI] [PubMed] [Google Scholar]
- Lens SMA, Voest EE, Medema RH. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat Rev Cancer. 2010;10:825–841. doi: 10.1038/nrc2964. [DOI] [PubMed] [Google Scholar]
- Leporrier J, Maurel J, Chiche L, Bara S, Segol P, Launoy G. A population-based study of the incidence, management and prognosis of hepatic metastases from colorectal cancer. Br J Surg. 2006;93:465–474. doi: 10.1002/bjs.5278. [DOI] [PubMed] [Google Scholar]
- McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies. J Clin Oncol. 2005;23:9067–9072. doi: 10.1200/JCO.2004.01.0454. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Wang Q, Kennedy CJ, Silver PA. An alternative splicing network links cell-cycle control to apoptosis. Cell. 2010;142:625–636. doi: 10.1016/j.cell.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao K, Mehta KR, Fridlyand J, Moore DH, Jain AN, Lafuente A, Wiencke JW, Terdiman JP, Waldman FM. High-resolution analysis of DNA copy number alterations in colorectal cancer by array-based comparative genomic hybridization. Carcinogenesis. 2004;25:1345–1357. doi: 10.1093/carcin/bgh134. [DOI] [PubMed] [Google Scholar]
- Nordlinger B, Guiguet M, Vaillant JC, Balladur P, Boudjema K, Bachellier P, Jaeck D, Association Francaise de Chirurgie Surgical resection of colorectal carcinoma metastases to the liver. A prognostic scoring system to improve case selection, based on 1568 patients. Cancer. 1996;77:1254–1262. [PubMed] [Google Scholar]
- Postma C, Terwischa S, MAJA Hermsen, van der Sijp JRM, Meijer GA. Gain of chromosome 20q is an indicator of poor prognosis in colorectal cancer. Cell Oncol. 2007;29:73–75. doi: 10.1155/2007/137592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratushny V, Pathak HB, Beeharry N, Tikhmyanova N, Xiao F, Li T, Litwin S, Connolly DC, Yen TJ, Weiner LM, Godwin AK, Golemis EA. Dual inhibition of SRC and Aurora kinases induces postmitotic attachment defects and cell death. Oncogene. 2012;31:1217–1227. doi: 10.1038/onc.2011.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees M, Tekkis PP, Welsh FKS, O'Rourke T, John TG. Evaluation of long-term survival after hepatic resection for metastatic colorectal cancer: a multifactorial model of 929 patients. Ann Surg. 2008;247:125–135. doi: 10.1097/SLA.0b013e31815aa2c2. [DOI] [PubMed] [Google Scholar]
- Ruers TJM, Wiering B, van der Sijp JRM, Roumen RM, de Jong KP, Comans EFI, Pruim J, Dekker HM, Krabbe PFM, Oyen WJG. Improved selection of patients for hepatic surgery of colorectal liver metastases with (18)F-FDG PET: a randomized study. J Nucl Med. 2009;50:1036–1041. doi: 10.2967/jnumed.109.063040. [DOI] [PubMed] [Google Scholar]
- Sillars-Hardebol AH, Carvalho B, de Wit M, Postma C, is-van Diemen PM, Mongera S, Ylstra B, van de Wiel MA, Meijer GA, RJA Fijneman. Identification of key genes for carcinogenic pathways associated with colorectal adenoma-to-carcinoma progression. Tumour Biol. 2010;31:89–96. doi: 10.1007/s13277-009-0012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillars-Hardebol AH, Carvalho B, Tijssen M, Belien JAM, de Wit M, is-van Diemen PM, Ponten F, van de Wiel MA, Fijneman RJA, Meijer GA. TPX2 and AURKA promote 20q amplicon-driven colorectal adenoma to carcinoma progression. Gut. 2012;61:1568–1575. doi: 10.1136/gutjnl-2011-301153. [DOI] [PubMed] [Google Scholar]
- Simon R, Mirlacher M, Sauter G. Tissue microarrays. Biotechniques. 2004;36:98–105. doi: 10.2144/04361RV01. [DOI] [PubMed] [Google Scholar]
- Stange DE, Engel F, Longerich T, Koo BK, Koch M, Delhomme N, Aigner M, Toedt G, Schirmacher P, Lichter P, Weitz J, Radlwimmer B. Expression of an ASCL2 related stem cell signature and IGF2 in colorectal cancer liver metastases with 11p15.5 gain. Gut. 2010;59:1236–1244. doi: 10.1136/gut.2009.195701. [DOI] [PubMed] [Google Scholar]
- Stichting FMWV Rotterdam . Code Goed Gebruik van lichaamsmateriaal. Stichting FMWV Rotterdam: Rotterdam; 2011. [Google Scholar]
- Wang X, Dong L, Xie J, Tong T, Zhan Q. Stable knockdown of Aurora-A by vector-based RNA interference in human esophageal squamous cell carcinoma cell line inhibits tumor cell proliferation, invasion and enhances apoptosis. Cancer Biol Ther. 2009;8:1852–1859. doi: 10.4161/cbt.8.19.9550. [DOI] [PubMed] [Google Scholar]
- Welch JP, Donaldson GA. The clinical correlation of an autopsy study of recurrent colorectal cancer. Ann Surg. 1979;189:496–502. doi: 10.1097/00000658-197904000-00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wu X, Wh Zhou, Aw Liu, Jb Wu, Jy Deng, Cf Yue, Sb Yang, Wang J, Zy Yuan, Liu Q. Aurora-A identifies early recurrence and poor prognosis and promises a potential therapeutic target in triple negative breast cancer. PLoS One. 2013;8:e56919. doi: 10.1371/journal.pone.0056919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Mori T, Takahashi K, Matsumoto H, Miyamoto H, Kato T. A new classification system for liver metastases from colorectal cancer in Japanese multicenter analysis. Hepatogastroenterology. 2008;55:173–178. [PubMed] [Google Scholar]
- Zhang C, Fang Z, Xiong Y, Li J, Liu L, Li M, Zhang W, Wan J. Copy number increase of aurora kinase A in colorectal cancers: a correlation with tumor progression. Acta Biochim Biophys Sin (Shanghai) 2010;42:834–838. doi: 10.1093/abbs/gmq088. [DOI] [PubMed] [Google Scholar]
- Zhang H, Chen X, Liu B, Zhou L. Effects of stable knockdown of Aurora kinase A on proliferation, migration, chromosomal instability, and expression of focal adhesion kinase and matrix metalloproteinase-2 in HEp-2 cells. Mol Cell Biochem. 2011;357:95–106. doi: 10.1007/s11010-011-0879-1. [DOI] [PubMed] [Google Scholar]
- Zlobec I, Steele R, Terracciano L, Jass JR, Lugli A. Selecting immunohistochemical cut-off scores for novel biomarkers of progression and survival in colorectal cancer. J Clin Pathol. 2007;60:1112–1116. doi: 10.1136/jcp.2006.044537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.