Abstract
Visualization is a powerful mechanism for extracting information from data. ggplot2 is a contributed visualization package in the R programming language, which creates publication-quality statistical graphics in an efficient, elegant, and systematic manner. This article summarizes key features of the package with examples from pharmacometrics and pointers to available resources for learning ggplot2.
The ggplot2 package, authored by Hadley Wickham,1 is an implementation of the theory described in “The Grammar of Graphics” by Leland Wilkinson.2 In a nutshell, the grammar defines a set of rules by which components of a statistical graphic are organized, coordinated, and rendered. The package is programmed entirely in the R statistical programming environment3 using the grid graphics system,4 extending Wilkinson's theory to a “layered” grammar of graphics.5
The grammar implemented in ggplot2 provides an infrastructure for composing a graphic from multiple elements:
An input data object, usually an R data frame.
Aesthetics (abbreviated “aes”), which refer to visual attributes that affect how data are displayed in a graphic, e.g., color, point size, or line type.
Geometric objects (“geoms” for short), such as points, lines, polygons, box plots, error bars, etc.
Scale transformations, which map aesthetics to unique values of variables, in addition to mathematical transformations to produce positional axes (e.g., logarithms). This process entails a conversion from “data units” to “graphical units.” The inverse mapping of a scale transformation is rendered in the graphic as a guide, either a positional scale or a legend, in the original data units.
Statistical transformations (“stats”), which refer to some type of data summarization such as a five-number summary for a box plot (stat_boxplot) or counts of observations by bin (stat_bin). The objective of statistical transformation is to supply the inputs necessary to produce a geom; for example, stat_bin sets up the data structure necessary for geom_bar and geom_histogram. Many stat_functions can be invoked directly to generate ggplot layers.
Coordinate transformations (“coord”), which specify how a coordinate system is specified in a graphic. The default is the Cartesian coordinate system, but several others are built in, such as polar coordinates (coord_polar) or flips of the x and y coordinates (coord_flip).
Faceting or conditioning, which applies the same type of graph to each defined subset of the data, usually indicated by the unique values of a categorical variable or factor.
A theming system, which controls the nondata aspects of a statistical graphic, such as the size and font of axis labels, legends, and titles or the appearance of the plot background.
Annotation, which allows you to add text and/or external graphics to a ggplot.
Positional adjustments, such as point jittering to reduce overplotting of points or various ways to maneuver bar segments associated with different groups in a bar chart.
The creation of a ggplot involves a stepwise process that takes the defined component pieces, called layers, and coordinates them through a sequence of transformations to produce the final graph.
In ggplot2, two functions can be used to create a graphic: qplot() and ggplot(). The former is shorthand for “quick plot” and is particularly useful when you want to create relatively simple graphs. Its original purpose was to provide a transition from R base graphics to ggplot2 graphics. For more complex graphics, you should use ggplot(), which is the function used for all of the examples in this article. The chapter in the ggplot2 book1 corresponding to qplot() is available on the book's web page: http://ggplot2.org/book/. Click on the link to “Getting started with qplot.”
R version 2.15.3 and ggplot2 version 0.9.3.1 were used to create all the plots in this article. It helps if the reader is familiar with base and lattice graphics in R, but it is not a prerequisite. The R code and data sets to reproduce all plots shown in this article can be downloaded online (see Supplementary Material online). Some topics cannot be fully illustrated in the text due to space constraints, so the annotated code will be supplied in the supplemental materials for you to try on your own. A useful companion is the online help pages for ggplot2 (http://docs.ggplot2.org/current), which contains a series of illustrative examples under each help page.
Basic Syntax in ggplot2
Two concepts at the core of ggplot2 are essential for its flexibility and efficiency: layers and aesthetic mappings. A ggplot object is composed of one or more layers, where each layer contains a different graphical object, or grob for short. The ggplot() function defines the base layer of a ggplot, indicating the name of the input data frame and establishing the association between a certain subset of its variables and their corresponding roles in the graph.
More specifically, the ggplot() function has two optional arguments: (i) data (an input data frame), and (ii) aes, which defines the “x” and “y” variables in addition to any other variables to be associated with specific aesthetics, such as color, point shape, or line type. A standard call to define the base layer of a ggplot is:
ggplot(data = Data, aes(x, y))
This is sufficient to define a ggplot object, but it does not produce any plotted output until at least one layer is added that specifies a geometry (geom). For example, to create a scatter plot, type:
ggplot(data = Data, aes(x, y)) +
geom_point()
In this case, no argument is supplied to geom_point. The required information (data and aesthetic mappings) is taken from the default set up in the base layer by the ggplot() call.
A powerful feature of ggplot() is that it can use different data frames to produce separate layers. For example, suppose we have two data frames d1 and d2 with variables x1,y1 and x1,y2, respectively, where x1 is common to both data frames and y1 and y2 are distinct variables. The following code creates two separate scatter plot layers, one from each data frame:
ggplot(data = d1, aes(x = x1, y = y1)) +
geom_point() +
geom_point(data = d2, aes(x = x1, y = y2))
Two equivalent ways to code this plot are
ggplot(data = d1, aes(x = x1)) +
geom_point(aes(y = y1)) +
geom_point(data = d2, aes(y = y2))
and
ggplot() +
geom_point(data = d1, aes(x = x1, y = y1)) +
geom_point(data = d2, aes(x = x1, y = y2))
This example illustrates a few important points about coding a ggplot graphic:
A base layer can be empty, with the data and aesthetic definitions passed to individual geom_ or stat_ layers, as shown in the last code block above.
The data argument in a ggplot() call must either be a data frame or NULL (the latter implicit in the third code block above).
Any aesthetic defined in a base layer by ggplot() is passed on to all subsequent layers. In the first code block above, it is expected that every layer will have an x variable named x1 and a y variable named y1, but it is possible to override the defaults in a specific layer, as shown in the second geom_point call. In the second code block, we took advantage of the fact that x1 is common to both geom_point calls; therefore, for efficiency, we can define it in the base layer as the default x variable and set up separate aes() calls for y in the two geom_point layers.
In the same manner, one can sequentially add layers to a ggplot, as in the following:
ggplot(Data, aes(x, y)) +
geom_point()+
geom_line() +
geom_smooth()+ ...
How It Works
Let us look at a few examples, saving certain technical details for later sections. Below is a snapshot of the R code for each example in Figure 1. The ggplot() call defines the “data” (i.e., data frame) that are to be input and the variable aesthetic mappings to be passed to all layers. The “+” operator adds layers to the ggplot object and must be at the end of a line of code if an additional layer is to be added (see Supplementary Material online for the code).
Figure 1.
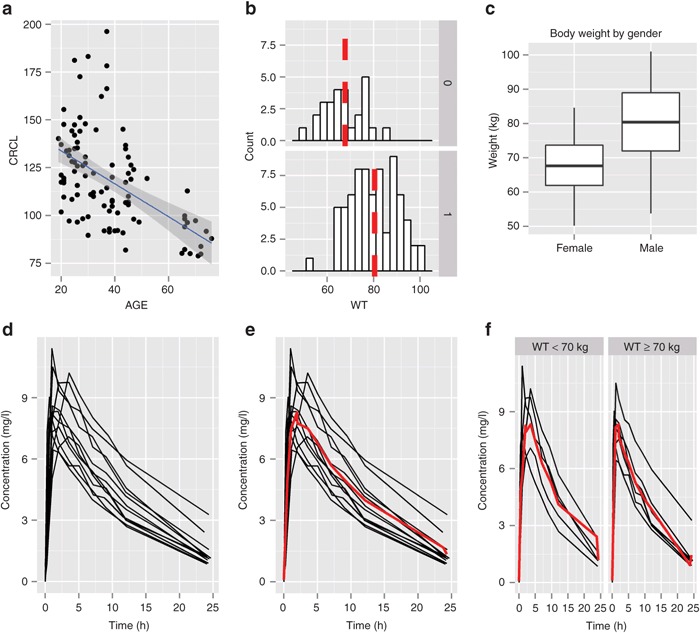
Example plots using ggplot2. (a) Scatter plot adding a layer of a linear regression line. (b) A histogram by gender (using facet_grid) adding a layer for median value for each panel. (c) A box plot conditioned by gender (using aesthetic mapping) with a customized title and x and y labels. (d) A spaghetti plot for Theoph data (nlme package). (e) Add a mean line to the spaghetti plot. (f) A spaghetti plot by covariate (body weight category) using facet_grid().
# Figure 1(a)
ggplot(data=d1, aes(x=AGE, y=CRCL)) +
geom_point() +
geom_smooth(method = “lm”)
# Figure 1(b)
ggplot(d1, aes(x=WT)) +
geom_histogram(binwidth = 3, color = “black”, fill = “white”) +
facet_grid(GEN ~.) +
geom_vline(data=tabWT, aes(xintercept=WT.median),
linetype=“dashed”, size=2, color=“red”)
# Figure 1(c)
ggplot(d1, aes(x= GEN, y=WT)) +
geom_boxplot() +
labs(x = “”, y = “Weight (kg)”) +
scale_x_discrete(labels=c(“Female”, “Male”)) +
ggtitle(“Body weight by gender”) +
theme(axis.text.x = element_text(angle=90, hjust=1, size=12))
In Figure 1a, geom_point() adds a scatter plot to the base layer, followed by a fitted least-squares linear regression line in a separate layer with geom_smooth() using method = “lm” as an optional argument (The default method for geom_smooth is “loess” if the sample size is less than 1,000 and “gam” (from the mgcv package) otherwise.).
In Figure 1b, geom_histogram() creates a histogram layer. Because no y aesthetic was defined in the base layer, stat_bin is called first to transform WT into a set of class intervals of binwidth 3, with corresponding counts saved in a derived variable ..count.. before the geom is rendered. The output from stat_bin is then passed to geom_histogram, yielding a frequency histogram.
Arguments exist in each geom function to control appearance; in geom_histogram(), these include binwidth (control the width of each bin), color (color for outline of each histogram bar), and fill (fill color for histogram). Because these are not defined inside aes(), the color and fill aesthetics are set to constant values rather than mapped. Pairings of aesthetics with variables take place inside an aes() call, which defines a mapping between values of aesthetics and values of the defined variable. The facet_grid() call creates a histogram for each level of GEN (Male = 0, Female = 1) in separate panels. Note that facet_grid(GEN~.) creates a panel plot by row, whereas facet_grid(.~ GEN) creates a panel plot by column (see “Faceting” section).
Finally, geom_vline()adds a new layer consisting of a vertical line in each panel, using another data frame (data=tabWT) that contains precomputed median values for each gender.
> tabWT
GEN WT.median
1 0 67.7
2 1 80.4
Several more aesthetics are set in this code chunk. To get individual lines for each panel of Figure 1b, it is necessary for the input data frame to contain a GEN variable so that ggplot2 knows which vertical line to associate with each panel. Without this information, the default action would be to plot both lines in each panel.
In Figure 1c, the x variable GEN is a factor and the y variable WT is numeric. geom_boxplot() produces separate box plots of WT for both females and males. The remaining code is used to customize the plot by adding a title, changing the axis labels, etc.
Next, let us use Theoph (data from an experiment on the pharmacokinetics of theophylline) from the nlme package to illustrate another feature of ggplot2. We often want to be able to plot lines for different groups of data without mapping a variable to an aesthetic. The group = aesthetic in ggplot2 allows you to do this easily. We consider two common examples: (i) spaghetti plots and (ii) plotting a line connecting mean values across levels of a factor.
The first example can be coded as follows:
library(nlme)
head(Theoph)
Grouped Data: conc ~ Time | Subject
Subject Wt Dose Time conc
1 1 79.6 4.02 0.00 0.74
2 1 79.6 4.02 0.25 2.84
3 1 79.6 4.02 0.57 6.57
4 1 79.6 4.02 1.12 10.50
5 1 79.6 4.02 2.02 9.66
6 1 79.6 4.02 3.82 8.58
# Figure 1(d)
ggplot(data=Theoph, aes(x=Time, y=conc, group=Subject)) + geom_line() + labs(x=“Time (hr)”, y=“Concentration (mg/L)”)
This produces separate lines for each level of Subject without having to define an aesthetic.
For case (ii), consider the problem of adding a line that connects the means of each group (note: for this case, we need a nominal time postdose (ntpd) to calculate mean values, and the R-code to calculate “ntpd” is provided in the Supplementary Material online). The default behavior in ggplot2 is to ignore a call to geom_line() when the x variable is a factor; to get around this, we use the group = 1 aesthetic, which is used to plot an overall “average” line across the levels of the factor.
# Figure 1(e)
p <- ggplot(data=Theoph, aes(x=Time, y=conc, group=Subject)) +
geom_line() +
labs(x=“Time (hr)”, y=“Concentration (mg/L)”) +
stat_summary(fun.y=median, geom=“line”
,aes(x=ntpd, y=conc, group=1), color=“red”, size=1)
print(p) # “p” is a ggplot object
Notice this plot (plot object) is saved as “p” (you can name it whatever you like, e.g., plot1, my.plot, etc); we will call it “p” in the following code.
Once you have created a plot (plot object) as above, it is easy to create a multipanel plot conditioned by a covariate, such as DOSE or SEX (see “Faceting” section).
# create a flag for body weight
Theoph$WT <- ifelse(Theoph$Wt<70, “WT < 70kg”, “WT >= 70kg”)
# Figure 1(f)
p + facet_grid(.~WT)
Let us save this plot as “plot1” for later use (for “Multiple Plots on One Page” section).
plot1 <- p + facet_grid(.~WT)
Core Elements of ggplot2
Generating a ggplot graphic entails an ordered sequence of transformations from data units to graphical units and back. Three types of transformations occur in the process of rendering a graph, in the following order: (i) scale transformations, which convert from data units to graphical units used by the computer; (ii) statistical transformations, which reduce input data to a form required by a geom, and (iii) coordinate transformations, which manipulate the coordinate system of a graph. The system underlying ggplot2 enacts a training process that coordinates all of the layers and transformations before rendering the graphic. The final graph applies inverse transformations on positional axes and legends so that the labels are expressed in the original data units. Each element of the system described below contributes in some way to the training process.
geoms and stats
In ggplot2, geoms are functions that convert transformed numeric data to some type of geometric object, such as points, lines, bars, or box plots. The functions that transform the input data into a form that can be used by geoms are called stats. Strictly speaking, stats and geoms are independent of one another; however, every geom in ggplot2 has a default stat. For example, both geom_bar and geom_histogram use stat_bin as their core stat function, whereas geom_contour() has stat_contour() as its default stat. Usually, a geom is called to produce a ggplot layer, but it is possible to call a stat function directly to perform both the necessary statistical transformation and the rendering of the geometry through the geom = argument of the stat function. Typical usage of a stat_ function within a ggplot() call entails some type of data reduction, followed by a call to the geom = argument, which triggers the visualization. An example of this feature is shown in the code that generates Figure 1, but many more examples are shown in the online help pages for the stat_* functions of ggplot2 cited in the references.
Mapping and setting aesthetics
To visualize data conditioned by the values of one or more grouping variables, e.g., SEX or DOSE, we can (i) associate (or map) individual values of a grouping variable to corresponding values of an aesthetic, such as color, size, or shape; or (ii) create separate panels for each value/level of a grouping variable, which is covered in the “Faceting” section. For illustration, suppose we want to map SEX to color (“colour” is used in the example code here following Hadley Wickham's ggplot web page (http://had.co.nz/ggplot2) and his book, but “color” works as a substitute for “colour.”); to do this, use aes()to define the mapping:
ggplot(Data, aes(x, y, colour = SEX))
This reveals a few more properties of aesthetics in ggplot2:
Aesthetics fall into two general groups: positional and non-positional.
By default, each mapped aesthetic defined inside aes() produces a guide to aid interpretation of the graph: for positional aesthetics, the guide is a scaled axis, whereas for non-positional aesthetics, the default guide is typically a legend.
All mappings inside aes() are actually transformations in the mathematical sense (i.e., 1–1 and onto) so that the inverse mappings are uniquely defined.
In the above example, the default behavior would be to set up positional axes for both x and y, and to produce a legend for color.
Another important feature of aesthetics is that they can either be mapped inside aes()or set outside of aes() inside the call to a geom. A mapping uniquely associates values of a variable with default values of an aesthetic; by contrast, setting refers to assigning a single value to an aesthetic. Here is an illustration of the difference between mapping and setting aesthetics:
Data <- data.frame(x1 = 1:12,
y1 = 0.5*1:12 + rnorm(12),
drug = rep(factor(c(“A”,“B”,“C”)), each=4))
# Figure 2(a)
ggplot(Data, aes(x = x1, y = y1, size=3, color=“darkblue”)) +
geom_point()
Figure 2a maps x1 and y1 to the positional aesthetics x and y, and maps “3” and “darkblue” to size and color, respectively, creating two new factors named “3” and “darkblue,” each with one level. The first color in the default color palette is assigned to the points (pink). Because new factors (“3” and “darkblue”) are created on the fly and mapped inside the ggplot() call (regardless of whether you wanted it or not), ggplot() automatically creates legends for size and color, as seen in Figure 2a.
Figure 2.
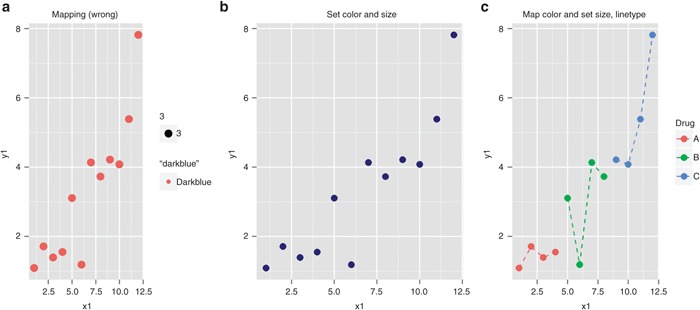
Mapping and setting aesthetics in ggplot2. (a) Variables that are not in the data set are mapped to color and size inside aes, which creates new factor variables on the fly and produce a plot with unwanted legends. (b) Correct settings for color and size. (c) Map the levels of the “drug” variable to color and set size and line type in separate geom calls.
To avoid this “feature,” you need to set size and color outside of aes() in the layer call:
# Figure 2(b)
ggplot(Data, aes(x=x1,y=y1)) +
geom_point(size=3, color=“darkblue”)
For another example of the difference between mapping and setting aesthetics, we can map color to levels of a grouping variable for all layers and set size, line type, etc. separately in distinct layers:
# Figure 2(c)
ggplot(Data, aes(x=x1, y=y1, color=drug)) +
geom_point(size=3) +
geom_line(size=0.5, linetype=2)
The above code creates Figure 2c. Factor “drug” is mapped to color, which assigns the default ordering of colors in ggplot2 to levels (“A,”, “B,” and “C”) in its defined order. Points and lines are drawn accordingly, and a corresponding legend is created by default to associate colors with drug names.
Guides
In the grammar of graphics, a guide is a graphical object that aids in the interpretation of a statistical graphic. There are two classes of guides: positional and nonpositional. A positional guide is an axis, a reference (i) to the range of values in a single direction if the variable to which it is mapped is continuous or (ii) to the levels of a factor, if discrete. A nonpositional guide is usually a legend, which illustrates the relationship between individual values of an aesthetic and its corresponding variable values.
Point shape and line type are discrete aesthetics, which means that they cannot be mapped to numeric variables (must be either factor or character variables). Point size is continuous, and therefore, it must be mapped to a numeric variable. Color and fill aesthetics, on the contrary, may be mapped to either discrete (e.g., “red,” “blue,” etc.) or continuous variables (color gradient). In a legend guide, continuous values are discretized into bins whose number can be controlled by the user. To get a smooth range of (fill) colors, a color bar guide is available.
The guides() function in ggplot2 allows a user to manipulate various aspects of a legend or color bar guide, typically in conjunction with the guide_legend and guide_colorbar functions. The online help page for guide_legend6 has a fairly comprehensive set of useful examples that go beyond the intended scope of this article.
Positional guides can assume several forms. In scatter plots, for example, both positional guides are continuous. In the special case of a time-related scatter plot, one of the axes (usually horizontal) may represent a date or a date–time object. In a strip chart or a Cleveland dot chart, one of the axes is discrete, whereas the other is continuous. You may want to represent a continuous axis in a suitably transformed metric such as a logarithmic or square root scale.
Scale functions
Each mapped aesthetic produces a default guide. Scale functions allow a user to control the rendering of a guide, whether positional or nonpositional.
The scale functions in ggplot2 have the following common arguments:
breaks: the set of values that are used to define the tick locations in an axis guide or the unique values of a legend guide;
values: the desired values of the aesthetic in a legend guide;
labels: the desired set of labels in an axis or legend guide.
Other arguments may be present in individual functions to handle specific properties of a scale.
Positional axes tend to have the form scale_dir_type, e.g., scale_x_discrete, scale_x_date, or scale_y_continuous (e.g., Figures 5 and 7). Certain scale transformations are built in, such as scale_x_sqrt or scale_y_log10, but most scale transformations now need to be defined through the scales package, which is beyond the scope of this article. In a positional axis scale, breaks would generally represent the desired locations of tick marks, and labels would represent the tick labels to be associated with the breaks. For dates or transformed continuous scales, manual specification of the labels is a common practice.
Figure 5.
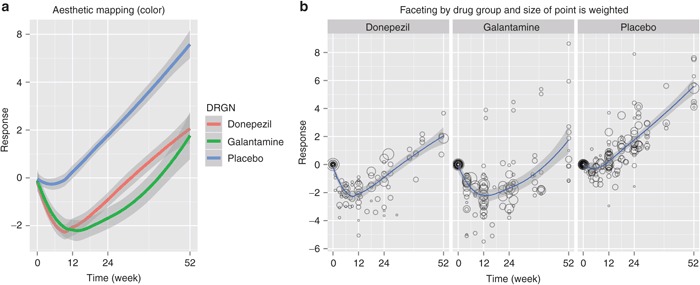
Visualize clinical data with weighted information. (a) Drug groups are differentiated using aesthetic mapping (color), (b) multipanels using facet_grid, and size of shape visually represents the number of patients in each treatment arm.
Figure 7.
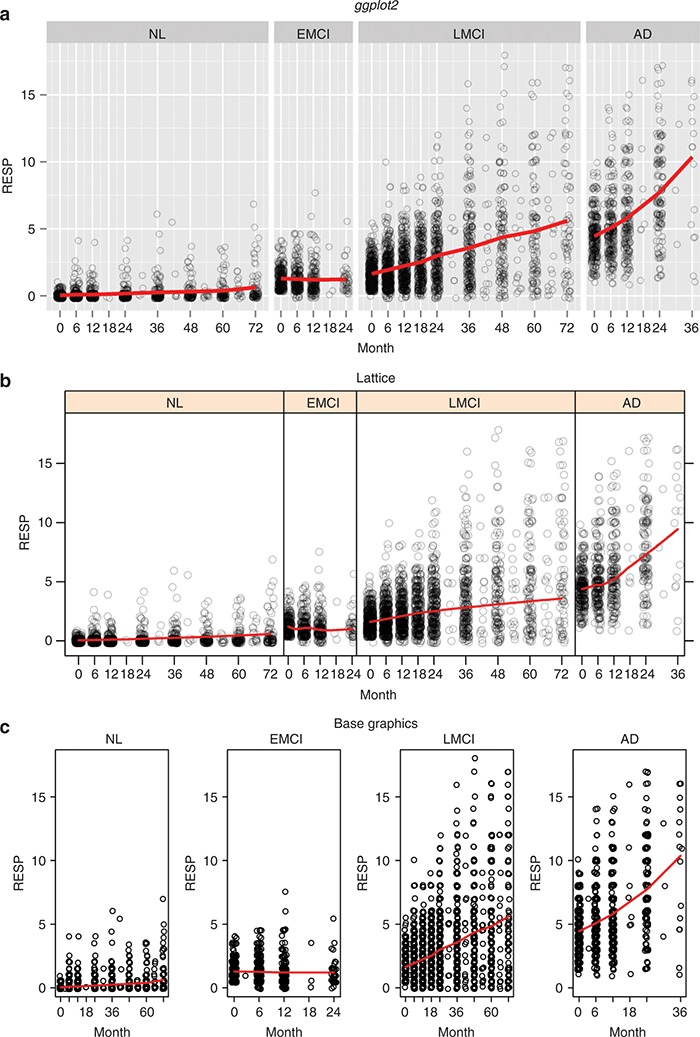
Same plots using ggplot2, lattice, and base graphics. (a) ggplot2, (b) lattice, and (c) base graphics. AD, Alzheimer's disease; EMCI, early mild cognitive impairment; LMCI, late mild cognitive impairment; NL, normal elderly; RESP, response.
Legend-related scale functions are modified through scale functions of the form scale_aes_type, such as scale_fill_gradient, scale_colour_identity, or scale_shape_manual. The way in which these arguments are applied depends somewhat on the type of scale selected. For legend guides associated with discrete variables such as factors, breaks typically represent unique values of the variable (e.g., factor levels), values represent the values of the aesthetic to be mapped to breaks, and labels represent the desired labels in the legend guide. For continuous aesthetics, the corresponding scale functions have different sets of arguments, e.g., range substitutes for breaks and/or values.
The aesthetics for which scale functions exist are color, fill, shape, size, linetype, and alpha, the last of which applies when alpha transparency is mapped to a variable. Two types of scale functions that apply to all aesthetics are identity and manual: an identity scale is appropriate when the desired values of the aesthetic are defined in the input data frame; on the contrary, a manual scale is appropriate when you want to customize the breaks, values, and/or labels used in a legend. Additional scale types specific to color and fill aesthetics include brewer, gradient, gradient2, gradientn, grey, and hue.
ggplot2 has a predefined set of default values for aesthetics. For example, the first default shape is a closed circle (pch=16 in base graphics), the next is a closed triangle (pch=17), and so on. A list of values for the primary aesthetics is given in Appendix B of the ggplot2 book,1 found at http://ggplot2.org/book/appendices.pdf.
We use the data set d3 (which contains mean CONC by TIME conditioned by DOSE) to illustrate how to change default shapes and colors using scale_ functions.
p <- ggplot(d3, aes(x=TIME, y=CONC, shape=DOSE)) +
geom_line() + geom_point(size=3) +
geom_errorbar(width=.1, aes(ymin=CONC-ci, ymax=CONC+ci))
#Figure 3(a)-left panel
print(p)
To change the shape of points:
#Figure 3(a)-middle panel
p + scale_shape_manual(values=c(1,2,7))
Observe that you need to have mapped DOSE to shape in the original ggplot() call before using scale_shape_manual. Note that the defined values in scale_shape_manual are pch (point character) values from par() in base graphics.
Similarly, if you want to change the color for each dose (Figure 3a, right panel), the DOSE variable needs to be mapped to color in the ggplot() call, e.g.,
Figure 3.
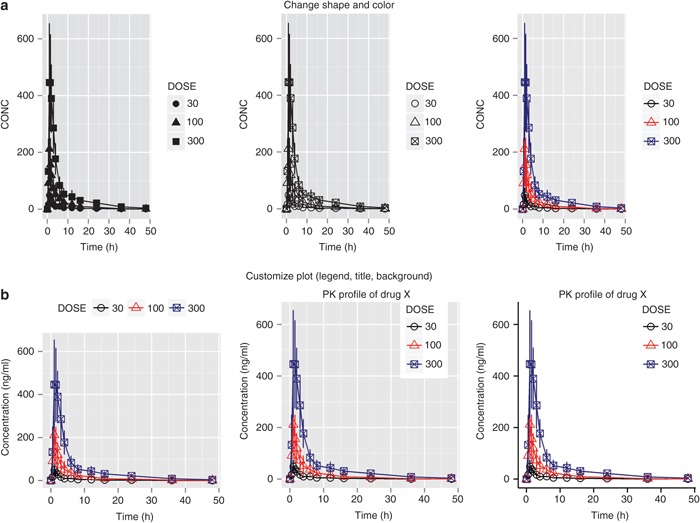
Controlling appearance and customizing plot. (a) Changing color and shape of points. (b) Adding legend and title, changing background.
p <- ggplot(d3, aes(x=TIME, y=CONC, shape=DOSE, color=DOSE)) +
geom_line() + geom_point(size=3) +
geom_errorbar(width=.1, aes(ymin=CONC-ci, ymax=CONC+ci)) +
scale_shape_manual(values=c(1,2,7)) +
scale_colour_manual(values=c(“black”,”red”,”darkblue”))
#Figure 3(a)-right panel
print(p)
Because the same variable is mapped to both shape and color, the scales are merged if they have the same title, breaks, and labels.
Themes
The theming system in ggplot2 controls the nondata aspects of a ggplot, primarily its general appearance. The system was overhauled in version 0.9.2 of the package so that it now supports relative sizing and inheritance of theme elements.
The key concept in the theming system is the theme, or more precisely, theme function. Every ggplot is controlled by a theme function that controls the general appearance of a ggplot. The default is theme_grey, which produces a light gray background with white gridlines. There are several reasons1 why it was selected as the default theme: (i) the white gridlines aid in the judgment of position but have little visual impact and can easily be “tuned out” (ii) the gray background gives the plot a similar color (in a typographical sense) as the remainder of the text, ensuring that the graphics fit in with the flow of the text without standing out over a bright white background; (iii) the gray background creates a continuous field of color, which ensures that the plot is perceived as a single visual entity. The default theme function can be changed globally either in an R session with theme_set() or within a ggplot() call by invoking the desired theme function. We will consider the latter in this section.
A theme function is composed of theme elements, which are individual nondata components of a graphic, such as axis.text, axis.ticks, legend.key, legend.position, and panel.background. To alter specific theme elements in a ggplot() call, we use the theme() function. Each argument of theme() is a pairing of a specific theme element with a function call that modifies one or more properties of the theme element. There are four element functions: element_text(), element_rect(), element_line(), and element_blank(). Most theme elements are associated with one of the first three element_*() functions; element_blank() sets all properties associated with a theme element to NULL.
You can specify/change the position of the legends as shown below (Figure 3b, left panel):
#Figure 3(b)-left
p + theme(legend.position=“top”)
You can also specify the x and y location as proportions of the graphics page (not the graphics region) and add a title using ggtitle (Figure 3b, middle panel):
#Figure 3(b)-middle
p + theme(legend.position=c(0.8,0.8)) +
ggtitle(“pharmacokinetic Profile of Drug X”)
#save this plot for later use (section “Multiple Plots on One Page”)
plot2 <- p + theme(legend.position=c(0.8,0.8))
To remove the gridlines:
p + theme(panel.grid.minor=element_blank(),
panel.grid.major=element_blank())
To change the background color:
p + theme(panel.background = element_rect(fill = “#003DF5”))
In the new system, there are 38 theme elements, some of which are nested within others. The three primary theme elements are text, line, and rect, and most (but not all) elements are associated with these. Theme elements contain one or more properties arranged in a list structure. The default values are typically defined in the primary elements, and changes are defined as needed in the nested elements. To see what a complete theme function looks like in the new theming system, type theme_grey() at the R prompt. A complete list of available arguments and definitions of theme elements can be found in the online help pages (http://docs.ggplot2.org/current/theme.html).
Several theme elements are associated with measurement units, defined in terms of the unit() function from the grid package. These elements can be modified directly, but the grid package needs to be loaded first or an error is thrown. Some examples include axis.ticks.length, plot.margin, and legend.key.size. For example,
theme(axis.ticks.length = unit(0.1, “in”))
ggplot2 also natively supports two additional built-in theme functions: theme_bw() and theme_classic(). All of the graphs produced thus far in this article have used theme_grey; theme_bw is a slight variation that replaces the gray panel background with a white one, whereas theme_classic is designed to mimic a base graphics plot. A number of other theme functions for use in ggplot2 can be accessed from the ggthemes package authored by Jeff Arnold.7 Below is an example that replaces the default theme function with theme_classic() (Figure 3b, right panel):
#Figure 3(b)-right panel
p + theme_classic() +
theme (legend.position=c(0.8,0.8),
legend.key = element_rect(fill=NA, color=NA))+ ggtitle(“pharmacokinetic Profile of Drug X”)
Axis and Main Titles
ggplot2 lets you define titles in several ways. The simplest method is to use separate functions to define x, y, and main titles: xlab(), ylab(), and ggtitle(), respectively. Each takes a character string as its sole argument. A more general labeling function is labs(), which lets you define not only x, y, and title but also the text of legend titles named by aesthetic, e.g.,
labs(x = “Type”, y = “Concentration”, title = “Main title”, colour = “Gender”, linetype = “Method”)
This provides a cleaner mechanism for titling in ggplot2 and is recommended when you want to specify several titles for axes, legends, and/or main titles.
Positional Adjustments
Positional adjustments are used to overcome two types of problems that occur in the process of rendering graphics: overplotting of points at the same or nearby locations and arrangement of a collection of graphical objects, particularly in the case of bar charts. In the former case, a common remedy is to jitter points, which is implemented by either geom_jitter() or by use of the position_jitter() function within geom_point(). Other positional adjustments include stack, dodge, fill, and identity. In the case of a bar chart with multiple groups associated with a fill aesthetic, any one of these can be taken as the value of the position = argument. The default, stack, produces a vertically stacked bar chart; dodge produces side-by-side bar charts, fill is a special case of stack where, in each group, the sum of the bar values is 100%, and identity stacks bars according to the ordering of the levels of the stacking variable.
Two examples of jittering points in ggplot2 are shown in the code chunk below:
ggplot(subset(d7a, group == “AD”), aes(x = Month, y = RESP)) + geom_jitter()
ggplot(subset(d7a, group == “AD”), aes(x = Month, y = RESP)) + geom_point(position = position_jitter(width = 0.5))
Coordinate Systems
ggplot2 contains six functions that specify or modify coordinate systems in two-dimensional (2D) plots: Cartesian (coord_cartesian, default), equal-scale Cartesian coordinates (coord_equal), interchange of x and y Cartesian coordinates (coord_flip), transformed Cartesian coordinates (coord_trans), map projections (coord_map), and polar coordinates (coord_polar).
For example, suppose we want to ascertain the relative impact of covariates on certain parameters such as clearance (CL) in population pharmacokinetic analysis and the relative effects (magnitude) on area under the concentration–time curve or the peak plasma concentration (Cmax) with concomitant medications such as Cytochrome P450 3A4 (CYP3A4) inhibitors. In these situations, a forest plot (also known as a tornado plot) is helpful as a visual guide to interpret the results. We can create one easily using coord_flip().
Assume that we have a data set “d4” as shown below, where x is the name of a covariate in the population pharmacokinetic analysis, y is the estimated relative change from its reference value, and ylo and yhi are the 2.5th and 97.5th percentiles of a bootstrap confidence interval.
>d4
x y ylo yhi
1 age 0.942 0.899 1.020
2 body weight 1.442 1.220 1.752
3 gender 1.123 1.059 1.197
4 CYP2D6 0.995 0.867 1.123
5 medication 0.981 0.940 1.025
6 diabetes 1.064 1.011 1.117
We start by creating an error bar plot and use geom_hline() to add reference lines (horizontal line) at 0.8, 1.0, and 1.25 to visualize a range of 80–125% (Figure 4a). In this example, we assign the output of the call on the right to an object “p”, which is of class gg and ggplot; it is common practice to assign the output of a ggplot()call to an object and add layers to it (with optional intermediate assignments):
Figure 4.
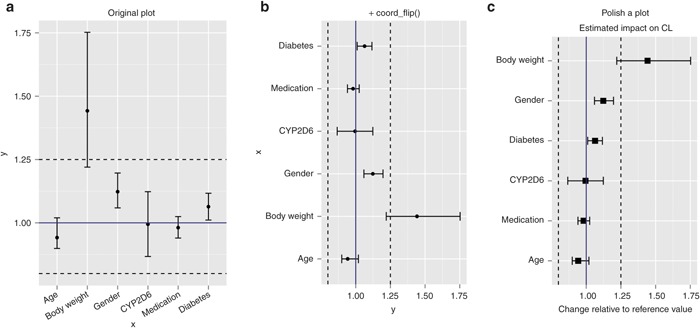
Coordinate system (coord_flip). (a) First attempt to create a forest plot before swapping the x and y axes. (b) Flip a plot using coord_flip(). (c) Reorder the variable based on the relative effect and add a title, axis labels, etc. to polish a plot.
#Figure 4(a)
p <- ggplot(d4, aes(x=x, y=y)) +
geom_point() +
geom_errorbar(aes(ymin=ylo,
ymax=yhi),width=0.2) +
geom_hline(yintercept=1, col=“darkblue”)
+geom_hline(yintercept=c(0.8,1.25),
linetype=2)
print(p) # “p” is a ggplot object
To flip the axis, use coord_flip()(Figure 4b).
#Figure 4(b)
p + coord_flip()
Observe that all mapped geometries (points, error bars, and lines) are flipped, and the x and y axes are swapped.
To polish this plot, let us change the size and shape of points, add a main title and axis label, and sort the levels of the covariate by the magnitude of the relative effect (Figure 4c).
#Figure 4(c)
ggplot(d4, aes(x=reorder(x,y), y=y)) +
geom_point(size=3, shape=15)+
geom_errorbar(aes(ymin=ylo,
ymax=yhi),width=0.2)+
geom_hline(yintercept=1, col=“darkblue”) +
geom_hline(yintercept=c(0.8,1.25), linetype=2) +
coord_flip() +
labs(title=“Estimated Impact of Covariates on CL”
,x=“”,y=“Change Relative to Reference Value”)
If you want to create a forest plot with a box plot, the same logic can be applied. Therefore, the R-code looks as shown below (and save it as “plot3” for later use):
plot3 <- ggplot(d4, aes(x=reorder(x,y))) +
geom_boxplot(aes(ymin=ylo, lower = ylo, middle = y,
upper = yhi, ymax= yhi), stat = “identity”) +
geom_hline(yintercept=1, col=“darkblue”) +
geom_hline(yintercept=c(0.8,1.25), linetype=2) +
coord_flip() +
labs(title=“Estimated Impact on CL”
,x=“”,y=“Change Relative to Reference Value”)
print(plot3)
Faceting
The concept of conditioning plots by the levels of one or more factors is called faceting in ggplot2. There are two faceting functions: facet_wrap() and facet_grid(). Both use a formula to determine the layout, and both share a common argument scales =, but otherwise, they are separate entities. facet_grid() is capable of generating a 2D grid of graphics panels―by column and/or by row. For example, facet_grid(.~ group) generates multipanel plots per level of a factor group by column, but facet_grid(group ~.) will generate plots by row. The dot (.) indicates that no variable is specified for that side of the formula. By specifying both sides, such as facet_grid(group ~ sex), a 2D grid of multipanel plots by group and by sex will be produced. In contrast, facet_wrap() reshapes a 1D ribbon of plots into a 2D arrangement. Instead of a 2D grid of panels associated with the level combinations of two variables, facet_wrap() creates a string of panels and “wraps” them into the graphics region like a ribbon, patterned on how the lattice behaves. You can specify ncol and/or nrow to control the arrangement of panels. You could write facet_wrap(~ group, ncol=4)to create a graph with four panels per row. The default (if you do not specify ncol or nrow) action is to attempt to lay out the panels as close to a square as possible. The default action is to start in the top left corner, moving left to right and then down, starting each new row from the left. The argument as.table = TRUE controls this ordering. Setting it to FALSE initiates wrapping from the bottom left corner of the graphics region and upward (see Supplementary Material online for the example code for facet_wrap()).
Here is an illustration to visualize complex clinical data (e.g., multiple dose levels, different drugs, etc.). You can choose aesthetic mappings to distinguish behavior among subgroups in a single graphics region or use faceting to separate group-wise behavior into multiple subpanels. In the former case, choose one or more of (fill) color, point shape, line type, and/or size as aesthetics to map to variables. A legend is automatically created for each defined nonpositional mapping.
Data set d5 contains summary information from several clinical studies (mean response value at each time point, number of patients for each treatment arm, etc.). To visualize the trend for each drug using smooth lines, map the color aesthetic to levels of the factor DRGN (drug name) and use the weight argument to influence the shape of a locally weighted smooth curve that weights Response (RESP) with reference to the number of patients (NTRT) in the data set (Figure 5a).
#Figure 5(a)
ggplot(d5,aes(x=WEEK, y=RESP, colour=DRGN)) +
geom_smooth(aes(weight=NTRT), size=1.5) +
scale_x_continuous(“Time (week)”, breaks=c(0,12,24,52))+
scale_y_continuous(“Response”, breaks=c(-6,-4,-2,0,2,4,6,8))
Instead of using color to differentiate each drug group, we can use facet_grid() to create multiple panels, one per group. This would be useful for visualizing the data by taking observation weights into account. For example, in Figure 5b, with mean values taken from the literature (summary data), the sizes of the data points represents their “weight”—i.e., greater size of the point represents more patients in the arm of the study making up the data point. This type of display is easily created with geom_point() using the size argument in aes (Figure 5b).
#Figure 5(b)
p <- ggplot(d5, aes(x=WEEK, y=RESP)) +
geom_point(aes(size=NTRT), shape=1, alpha=0.4) +
geom_smooth(aes(weight=NTRT)) +
scale_x_continuous(“Time (week)”, breaks=c(0,12,24,52))+
scale_y_continuous(“Response”, breaks=c(-6,-4,-2,0,2,4,6,8)) +
facet_grid(.~DRGN)
p + theme(legend.position=“none”)
facet_grid()uses common (same range) scales in all plots by default, but one can produce different ranges for each plot by using scales=“free”. To limit this freedom to one direction, one can specify “free_x” or “free_y”. ggplot2 supports an additional argument, space=“free”, to adjust the width or height of each panel in proportion to the maximum extent of the x and/or y scales; the free_x and free_y options also apply here. For example, in Figure 7, in the “Miscellaneous” section, the EMCI group (early mild cognitive impairment) only has data up to 24 months; therefore, this plot only takes one-third of the space of the width of the normal elderly (NL) group (where data is available up to 72 months).
Annotation
Annotation refers to textual or graphical embellishments of a ggplot object, including such things as text labels, fitted equations, P values, tables, pictures, or inset graphics. It is possible to tailor annotations on a facet-by-facet basis with a bit of extra work.
The primary way to add text data to a ggplot is through geom_text. Its basic syntax is as follows:
geom_text(data, aes(x, y, label), size, hjust, vjust, ...)
The x and y arguments define the locations where the centers of text strings are located, and label designates the variable name associated with the text strings. The arguments outside aes() correspond to the (constant) size of the text string along with its horizontal and vertical justifications relative to its location. If any of these are mapped to a variable, then they should be placed inside aes().
It is often the case that only a few text strings are desired in a graph (e.g., outlier identification); therefore, a common practice is to create an external data frame that contains (at least) variables corresponding to the (x, y) locations and labels. If the strings are to be distributed among facets, then you should include a variable with the same name as the faceting variable. This data frame can then be passed into the geom_text() call as its data = argument. If the variable names are not the same as those of corresponding aesthetics in the ggplot() call, then they need to be defined as aesthetics in geom_text().
To illustrate this, let us create a WRES (weighted residuals) vs. PRED (predicted) plot using a data set “d6a.” First, create a scatter plot using geom_point(), and then add a layer of horizontal lines using geom_hline() as follows:
p <- ggplot(data=d6a, aes(x=PRED, y=WRES)) + geom_point(shape=1) +
geom_hline(yintercept = 0, color=“red”) +
geom_hline(yintercept = c(-5,5),linetype = 2)
Next, create a separate data frame (subset of “d6a”), which only contains outlier subjects:
outlier <- d6a[abs(d6a$WRES)> 5,]
Add a layer to p using geom_text() with outlier as the input data frame. Use label=ID to place the subject ID (identity) numbers in the plot along with the x and y locations inside aes(), which could be omitted because these are the same as in the original ggplot() call. Note also that vjust (vertical justification) is used to adjust the location of the labels (Figure 6a).
Figure 6.
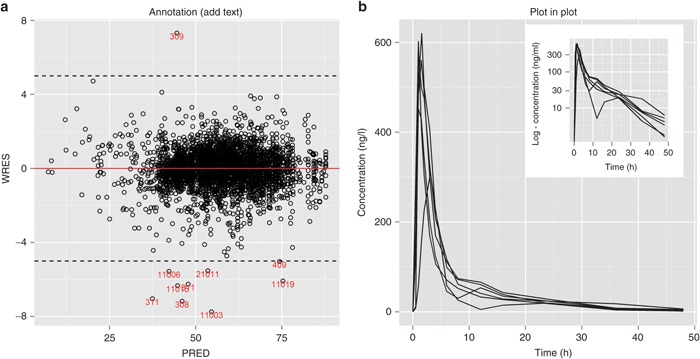
Annotations in plots. (a) Add text (labels) layer for outlier points; (b) new annotation function annotation_custom(): insert an image (semilog scale plot) into the original plot (normal scale).
#Figure 6(a)
p + geom_text(data=outlier, aes(x=PRED, y=WRES, label=ID)
,vjust=1, size=3, color=“red”)
#saved for later use (section “Multiple Plots on One Page”)
plot4 <- p + geom_text(data=outlier, aes(x=PRED, y=WRES, label=ID)
, vjust=1, size=3, colour=“red”)
In contrast, the annotate() function is typically used to add single features to a ggplot object such as a single text string, a rectangle, a segment, or any geom that can be defined in terms of a vector. No mapping is allowed in an annotate() call. Assuming you have already a plot saved in an object “p”, an example usage is:
#add text “hello”
p + annotate(“text”, x=20, y=7, label=“hello”, col=“red”, size=12) +
#add arrow (requires the grid package to be loaded for unit())
p + annotate(“segment”, x=20, y=5, xend=48, yend=7
,col=“red”, size=2
,arrow = arrow(angle=30, length=unit(0.2, “in”)))
#add shaded area
p + annotate(“rect”, xmin=25, xmax=30, ymin=-Inf, ymax=Inf,
fill=“blue”, alpha=0.2)
Plot in Plot (annotation_custom)
In more recent versions of ggplot2 (v-0.9.0 or later), a function called annotation_custom() was introduced to allow insertion of an image (graph, table, or other graphical object of interest) into a plot.
Assume that we want to create a pharmacokinetic concentration profile plot from a data set named “d6b” and add a log-scale version of the same plot in a corner. The code below illustrates how to create such plots step by step.
Step 1: Create the initial plot (normal scale) and save it as an object p1:
p1 <- ggplot(d6b, aes(x=TIME, y=CONC, group=ID)) +
geom_line() +
labs(x=“Time (hr)”, y=“Concentration (ng/mL)”)
Step 2: Create a semilog plot as the second plot and save it as p2:
p2 <- p1 +
scale_y_log10(“log - Concentration (ng/mL)” ,
breaks = c(1,10,30,100,300))
Step 3: To add this plot as a image into the first plot, “read” the plot information using ggplotGrob(). This requires loading the grid and gridExtra packages.
library(grid)
library(gridExtra)
g <- ggplotGrob(p2) #image information is saved as “g”
We can now insert the second plot into the first with annotation_custom() by specifying its bounding box locations xmin, xmax, ymin, and ymax (Figure 6b):
#Figure 6(b)
p1 + annotation_custom(grob = g,
xmin = 20, xmax = 48, ymin = 300, ymax = 640)
You can also insert pictures, maps, or a (raster) image into a plot. See more details in Section 4 of the ggplot2-0.9.0 transition guide8 or the online help.
Miscellaneous Topics
ggplot2 vs. other R graphics
There are four primary ways of creating a graph in R: (i) traditional (base) graphics, (ii) grid graphics (iii) lattice, and (iv) ggplot2. Each has its own strengths and weaknesses, but once you get over the learning curve, ggplot2 has a lot of elegance and power. Traditional (base) graphics are easy to start with and very flexible, but when it comes to creating more complex plots, the code quickly becomes cumbersome. By contrast, both lattice and ggplot2 are programmed in the grid graphics system developed by Paul Murrell through the grid package.4 Both are designed to be more user-friendly when plotting multivariate data, which means that certain design decisions are hard coded; however, each package has its own philosophy of how to produce a graphic. The most noticeable difference is that the code to produce a lattice graphic is contained in a single-function call, whereas in a ggplot2 graphic, several function calls are strung together, separated by a “+” operator. Both packages have a set of core functions that perform the bulk of the work (panel functions in lattice; stats and geoms in ggplot2). Panel functions are typically called within a high-level lattice function (e.g., xyplot, histogram), whereas geoms and stats are added to an existing ggplot object. Moreover, each package is an implementation of a particular theory of graphics: lattice was originally a port of the Trellis graphics system9 in S-PLUS to R by Deepayan Sarkar, whereas ggplot2 is an extension of the aforementioned Grammar of Graphics.2 The biggest advantages of lattice over ggplot2 are as follows: (i) it has several functions for 3D graphics; (ii) you can write your own panel functions and pass them into a high-level lattice function call; (iii) it is faster than ggplot2; and (iv) in conjunction with the latticeExtra package, one can interactively edit a lattice object and add (superimpose) two compatible lattice objects together. However, some advantages of ggplot2 over lattice include the following:
Panel functions in lattice can be tricky to write, especially for conditioning plots;
It is much easier to tailor legends in ggplot2 than in lattice;
Usually, modifying panels in a conditioning plot is much easier in ggplot2;
ggplot2 accepts different input data frames in different panels—lattice does not;
ggplot2 gives the user a simpler way to control the nondata features of a graphic through its theming system;
The code to produce a ggplot is usually easier to read, write, and modify than the corresponding lattice code.
Figure 7 shows the differences in plot appearance using (i) ggplot2, (ii) lattice, and (iii) base graphics when creating a similar type of conditioning plot. The R code snippets to create each plot are shown below.
#--ggplot2 version--#
ggplot(d7a, aes(Month, RESP)) +
geom_jitter(shape=1, alpha=0.2) +
geom_line(data=d7b, aes(Month, RESP), col=“red”, size=1.2) +
facet_grid(.~ group, scale=“free”, space=“free”) +
scale_x_continuous(breaks=c(0,6,12,18,24,36,48,60,72))
#--lattice version--#
library (lattice)
library (latticeExtra) ## re-size panels in a trellis object post-hoc
trellis.par.set(plot.symbol = list(pch=1, col=“black”, alpha=0.2))
lt <- xyplot(RESP ~ Month|group, data=d7a, layout=c(4,1)
,panel = function(x,y,...){
panel.xyplot(x,y, col=“black”, cex=0.8,
jitter.x = TRUE, jitter.y = TRUE, factor=2,...)
panel.loess(x, y, col=“red”, lwd=2,...)
}
,scales = list(x = list(relation = “free”, at=c(0,6,12,18,24,36,48,60,72),cex=0.7))
)
resizePanels(ls, w=c(6,2,6,3))
#--base graphics version--#
par(mfrow=c(1,4), mex=0.7, mai=c(0.7,0.4,0.3,0.2))
group <- c(“NL”, “EMCI”, “LMCI”, “AD”)
for (i in group){
temp1 <- d7a[d7a$group==i,]
temp2 <- d7b[d7b$group==i,]
plot(jitter(temp1$Month), jitter(temp1$RESP)
,xlab=“Month”, ylab=“RESP”, ylim=c(0,18), axes=FALSE)
axis(1, at=c(0,6,12,18,24,36,48,60,72))
axis(2)
box()
lines(temp2$Month, temp2$RESP, col=“red”, lwd=2)
title(i)
}
Let us examine the differences and similarities among the three methods step by step. First, it is easy to use different data sets in separate layers of a ggplot() call. In this example, geom_line() uses data=d7b instead of the one from the base layer (data=d7a). d7b is a summary data frame (see Supplementary Material online) containing mean values of RESP by Month; because it contains variables named Month and RESP, there is no need to modify aes() from the base layer, although no harm is done by including it. d7b is also used in the base graphics code with lines() to add mean lines to the existing plot, and temp2 is a subset of d7b created in each iteration of the for loop to select the data used to produce each subplot.
The lattice call can produce average lines internally with panel_average(); however, panel_smooth() is used in this example to maintain comparability with the other two graphs.
The equivalent of panel_average() in lattice is produced in ggplot2 with the stat_summary() function:
#--replace--#
geom_line(data=d7b, aes(Month, RESP), col=“red”, size=1.2)
#--with--#
stat_summary(fun.y = mean, geom = “line”,
colour = “red”, size = 1.2)
facet_grid(. ~ z) in ggplot2 is the analogue of the right-hand side of the conditioning formula (y ~ x | z) in lattice graphics. Base graphics have no built-in mechanism to create multipanel plots. Instead, par() or layout() is used to apportion the graphics region into subregions that accommodate multiple plots. for loops or apply family functions can be used to subset of the data by condition and create each plot, which are rendered in successive order according to the plot layout.
The most apparent difference among the three plots is how ggplot2 allocates space for the individual panels with a space = argument in a faceted plot, as described in the Faceting section. The only way one could replicate the behavior of the ggplot2 graph in Figure 7 with base graphics would be to (i) carefully define how to allocate the graphics space for each panel and (ii) write separate graphics code for each panel. In lattice, you can define such space as a post processing using resizePanels function in conjunction with latticeExtra package, however, there is no single argument in lattice to mimic ggplot2's behavior.
Two other features of Figure 7 are of note: “alpha transparency” and “jittering.” The alpha aesthetic ranges between zero (completely transparent) and one (completely opaque). Similar to jittering, alpha transparency is useful for handling overplotted data, making it easier to see where the majority of points lie in a scatter plot. lattice also supports alpha transparency, but it needs to be set with trellis.par.set() and remains set as long as the trellis device is open (i.e., you need to close the current graphic device by dev.off() to undo the altered settings).
Multiple Plots on One Page
The gridExtra package is a convenient tool for positioning multiple plots on a page without having to learn the underlying graphics system (grid).
Once each plot is saved as a separate R object as shown in the previous example, it is easy to combine them on one page using grid.arrange(). We have already created four plots and saved them as plot1–plot4; you can place them in one page as follows:
library(gridExtra)
grid.arrange(plot1, plot2, plot3, plot4, ncol=2)
This creates a 2 × 2 plot (Figure 8). You can specify how many plots you want to display by column or by row using the ncol or nrow arguments, respectively.
Figure 8.
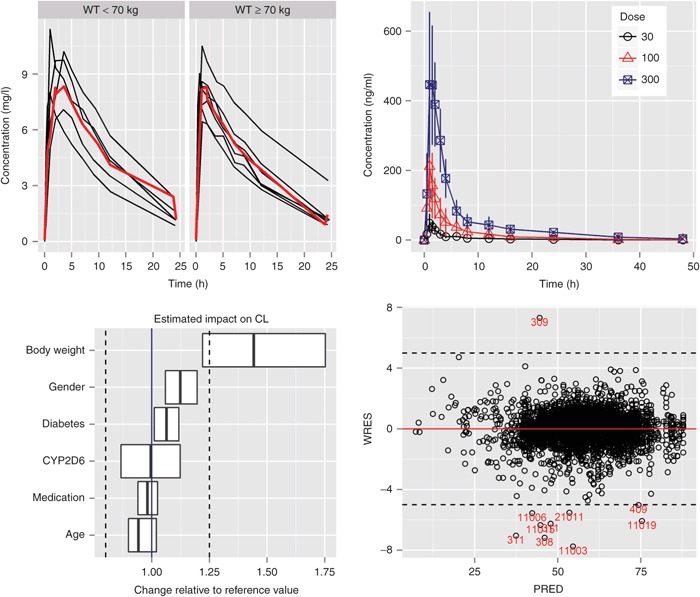
Multiple plots (row × column) in one page using grid.arrange (gridExtra package).
Saving Plots
A ggplot object is a list composed of data components, mappings, layers, scales, etc. A ggplot object can be rendered in a graphics window or device with print(). The user may also save it to disk with ggsave(), a special function in ggplot2 that saves the current ggplot. You can also use the typical R method to save a (gg)plot. For example,
ggsave(“myplot.pdf”)
is the same as:
pdf(“myplot.pdf”)
print(p) # p is a ggplot object
dev.off()
ggsave() outputs to various formats, such as png, pdf, etc. Grid graphics objects (including ggplots and trellis objects) need to be wrapped inside a print() statement when a graphics device is open; see R FAQ 7.22 for details.
Note that each method has different default settings for width and height. pdf() defaults to 7 inches for width and height, whereas ggsave() defaults to the dimensions of the current graphics device. It can be easily modified (set) by specifying width and height in the function call.
Resources
There are many ggplot2 learning resources available online, in books, and in commercially available training courses. The list below provides several examples of material available to learn ggplot2, from beginner to more advanced levels.
Hadley Wickham's web page for ggplot2
ggplot2 help documentation
http://docs.ggplot2.org/current/
ggplot2 book “ggplot2: Elegant Graphics for Data Analysis” by Hadley Wickham
(all codes are available from the author's website)
Cookbook for common graphics by Winston Chang10
http://wiki.stdout.org/rcookbook/Graphs/
ggplot2 (0.9.0) transition guide8
http://cloud.github.com/downloads/hadley/ggplots2/guide-col.pdf
ggplot2 mailing list
http://groups.google.com/group/ggplot2
stackoverflow
http://stackoverflow.com/tags/ggplot2
ggplot2 Wiki
https://github.com/hadley/ggplot2/wiki
lattice to ggplot2 conversion
Conclusion
ggplot2 combines the advantages of both base and lattice graphics and is able to create “publication-ready” plots. Conditioning and shared axes are handled automatically, and you can build up a plot step by step from multiple data sources. It also implements a sophisticated multidimensional conditioning system and a consistent interface to map data to aesthetic attributes.
There are many advanced features in ggplot2, which are not covered in this article because it focuses on the core elements of ggplot2 with examples from pharmacometrics. Note that ggplot2 has been rapidly evolving over the past several years; therefore, refer to the most current help files (http://docs.ggplot2.org/current/) as the canonical reference.
Conflict of Interest
The authors declared no conflict of interest.
References
- Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer; Berlin; 2009. [Google Scholar]
- Wilkinson L. The Grammar of Graphics, 2nd edition. Springer; Berlin; 2005. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria; 2013 ). < http://www.R-project.org/ >. [Google Scholar]
- Murrell P. R Graphics, 2nd edition. Chapman and Hall; Boca Raton, FL; 2011. [Google Scholar]
- Wickham H. A layered grammar of graphics. J. Comput. Graph. Stat. 2010;19 1:3–28. [Google Scholar]
- < http://docs.ggplot2.org/current/guide_legend.html >
- Arnold J.B.ggthemes: Extra themes, scales and geoms for ggplot. R package version 1.3.3. 2013 ) < http://CRAN.R-project.org/package=ggthemes >
- < http://cloud.github.com/downloads/hadley/ggplot2/guide-col.pdf >
- Cleveland W.S. Visualizing Data. Hobart Press; Hobart, Australia; 1993. [Google Scholar]
- Chang W. R Graphics Cookbook. O'Reilly; Sebastopol, CA; 2013. [Google Scholar]


