Abstract
Dietary restriction (DR) extends lifespan and promotes metabolic health in evolutionary distinct species. DR is widely believed to promote longevity by causing an energy deficit leading to increased mitochondrial respiration. We here show that inhibitors of mitochondrial complex I promote physical activity, stress resistance as well as lifespan of Caenorhabditis elegans despite normal food uptake, i.e. in the absence of DR. However, complex I inhibition does not further extend lifespan in dietarily restricted nematodes, indicating that impaired complex I activity mimics DR. Promotion of longevity due to complex I inhibition occurs independently of known energy sensors, including DAF-16/FoxO, as well as AAK-2/AMPK and SIR-2.1/sirtuins, or both. Consistent with the concept of mitohormesis, complex I inhibition transiently increases mitochondrial formation of reactive oxygen species (ROS) that activate PMK-1/p38 MAP kinase and SKN-1/NRF-2. Interference with this retrograde redox signal as well as ablation of two redox-sensitive neurons in the head of the worm similarly prevents extension of lifespan. These findings unexpectedly indicate that DR extends organismal lifespan through transient neuronal ROS signaling rather than sensing of energy depletion, providing unexpected pharmacological options to promote exercise capacity and healthspan despite unaltered eating habits.
Abbreviations: DAF, abnormal dauer formation; AAK, C. elegans AMP-activated protein kinase; SIR, yeast silent information regulator related; SKN, skinhead; PMK, C. elegans p38 map kinase family; FoxO, forkhead box protein O; NRF, nuclear factor erythroid 2-related factor; EAT, eating: abnormal pharyngeal pumping; MPTP, 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine; BHA, butylated hydroxyl-anisole
Keywords: Caenorhabditis elegans, Aging, Dietary restriction, Mitochondria, Mitohormesis, Redox signaling, Reactive oxygen species
1. Introduction
Dietary restriction (DR) is a well-established intervention known to promote longevity of various model organisms, including Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, different rodent models, and possibly monkeys, as previously summarized elsewhere [1]. By definition, DR causes energy deprivation which has been previously shown to increase mitochondrial respiration in yeast [2] and nematodes [3,4]. Such deprivation of available energy has been shown to be sensed by either sirtuins [5] or AMP-activated protein kinase (AMPK) [3,6]. Activation of either sirtuins or AMPK is believed to promote respiration which is necessary for lifespan extension [3,5].
Increased respiration has been linked to lifespan extension due to increased formation of mitochondrial reactive oxygen species (ROS) which are essential for DR-mediated lifespan extension [3] based on the concept of mitohormesis, as exemplified [3,7–12] and reviewed [1] elsewhere. Consistently, genetic impairment of mitochondrial function [13] and in particular impairment of mitochondrial complex I [14,15] has been shown to extend C. elegans lifespan. Independently, inhibitor-mediated inhibition of complex I is known to promote ROS formation for many decades [16]. Consistent with the mitohormesis concept, genetic impairment of the complex I subunit nuo-6, an orthologue of mammalian nudfb4, not only extends C. elegans lifespan but also promotes mitochondrial ROS formation which appears to be necessary to promote longevity [17].
While DR causes energy deprivation by reduced calorie intake, it is interesting to note that impairment of mitochondrial complex I impairs oxidative phosphorylation (OXPHOS) culminating in reduced ATP availability, i.e. intracellular energy deprivation. Nevertheless the published body of evidence does not preclude the possibility that complex I-derived ROS signaling promotes longevity independently of known energy sensors which, however, has not been tested experimentally. This hypothesis is of significant interest since it would anticipate pharmacological approaches to mimic DR despite normal food uptake, including approaches to ameliorate DR-preventable diseases like diabetes mellitus, obesity and possibly cancer without the necessity for individual changes in lifestyle and/or eating habits.
By using different chemical inhibitors, we here analyze both the acute and long-term effects of complex I inhibition on fitness and lifespan in C. elegans to determine the respective roles of energy sensing and ROS signaling in regards to DR-mediated lifespan extension.
2. Materials and methods
2.1. Chemicals
All chemicals were obtained from Sigma-Aldrich (Munich, Germany) unless stated otherwise.
2.2. Nematode strains and maintenance
C. elegans strains used for this publication were provided by the Caenorhabditis Genetics Centre (Univ. of Minnesota, USA) except for the SKN-1::GFP reporter strain [18] which was a kind gift from T Keith Blackwell. MIR13 (aak-2(ok524); sir-2.1(ok434)) double mutants were obtained as described below, and have been deposited at CGC. Maintenance was performed as previously published [12].
2.3. Compound treatment
Treatment of C. elegans was carried out on NGM agar plates containing respective complex I inhibitors. All agar plates were prepared from the same batch of NGM agar, whereas treatment plates were supplemented with the respective compound and control plates with the respective solvent as described [19]. After plates were poured and dried for about 30 min, they were sealed and stored at 4 °C.
Complex I inhibitors treatment was performed using heat inactivated bacteria (OP50 HIT) to avoid interference by the xenobiotic-metabolizing activity of Escherichia coli [19,20]. To obtain heat-inactivated bacteria, an overnight liquid culture of E. coli was treated 30 min at 65 °C. The bacteria suspension was then concentrated 20-fold by centrifugation (30 min at 3200×g) and resuspended within S-buffer containing 10 mM MgSO4 and 5 μg/ml cholesterol as described [19,21]. NGM plates used for compound treatment were additionally supplemented with 100 μg/ml ampicillin. It should be noted that this treatment causes an inactivation of metabolic activity of E. coli rather than a complete lethality. The antioxidant butylated hydroxyanisole (BHA) was dissolved in DMSO and applied at a final concentration of 10 μM, as previously described [12].
Incubations of a synchronized population with complex I inhibitors started after L4 by washing the synchronized, young adult worms and transferring them to the respective treatment plates using S-buffer.
2.4. Lifespan assays
All lifespan assays were performed at 20 °C according to standard protocols and as previously described [3]. Briefly, a C. elegans population was treated with hypochlorite/NaOH solution to synchronize the population at day zero of the lifespan. Eggs were transferred to freshly spotted plates to allow hatching and development. After L4 around 120 nematodes were manually transferred to fresh incubation plates containing the respective compounds. Experiments were conducted in triplicates. For the first 10–14 days, worms were transferred every day and afterwards every second day. Nematodes that show no reaction to gentle stimulation were scored as death. Those animals that crawled off the plates or displayed non-natural death particularly due to internal hatching were censored.
2.5. Dietary restriction by bacterial dilution
Dietary restriction lifespans were performed as previously described [22]. Briefly, nematodes were synchronized as described above. After L4, 40 worms were transferred to DR plates (50 mm diameter) containing 100 μg/ml ampicillin and 25 μg/ml 5-fluoro-2′-deoxyuridine spotted with 300 μl of the respective concentrations of E. coli suspensions. In addition, treatment plates contained 100 nM rotenone whereas control plates were poured with the respective solvent as described above. Worms were transferred daily.
2.6. Paraquat stress resistance
Resistance to lethal oxidative stress derived from paraquat was determined with minor modifications as previously described [3]. Briefly, nematodes were pre-treated with the respective complex I inhibitor for 6 days. Thereafter, worms were transferred manually to fresh NGM plates containing 10 mM paraquat (Sigma-Aldrich) spotted with a lawn of heat-inactivated OP50 followed by daily determining the survival rate until all nematodes were death. As described for lifespan analysis, worms were counted as censored in case of internal hatching, crawling off and bursting.
2.7. Immunoblotting
Nematodes were washed three times with ice-cold S-Buffer and pellets were shock frozen in liquid nitrogen. Frozen pellets were grinded in a nitrogen-chilled mortar and suspended in phosphate buffer containing protease and phosphatase inhibitors (Complete protease inhibitor cocktail [Roche, Penzberg, Germany] and additionally 2 mM sodiumfluoride, 2 mM sodium orthovanadate, 1 mM PMSF, and 2 mM EDTA). Extracts were sonicated three times and centrifuged for 7 min at 12,000×g. Supernatants were used for protein quantification, and an aliquiot was boiled in Laemmli buffer and applied for SDS-PAGE. Antibodies against phosphorylated p38 MAP kinase (Cell Signaling, Beverly, MA, USA) and α-tubulin (clone DM1A, Sigma-Aldrich) were used.
2.8. Oxygen consumption
Respiration was quantified using a DW1/AD Clark-type electrode (Hansatech, King's Lynn, England) as previously described [3]. After individual incubation period, worms were harvested, washed and transferred into the DW1 chamber. Oxygen consumption was monitored for at least 10 min. Afterwards, worms were carefully removed from the chamber and collected for a subsequent protein determination. Therefore, worms were sonicated 3 times and centrifuged for 10 min at 12,000×g. Supernatant was used for protein determination.
2.9. ATP determination
A worm lysate-based method [23] with minor changes was used. Worms were harvested and immediately shock frozen in liquid nitrogen. The frozen worm pellet was grinded in a nitrogen chilled mortar to yield a well disrupted worm powder. Four molar guanidinium-HCL was prepared, heated to 100 °C and then mixed with the frozen worm powder to destroy ATPase activity and to further lyse worms. The mixture was boiled for 15 min at 100 °C with a subsequent centrifugation step (30 min at 13,200×g and 4 °C). The supernatant was diluted with ddH2O 1:200 and analyzed using a commercially available kit (CellTiter Glo; Promega, Fitchburg, WI, USA) according to the manufacturer's instructions. For normalization of the luminescence signal, protein values were determined.
2.10. Laser ablation of ASI neurons
For focussed laser ablation the output laser beam of a UV pulsed laser (diode-pumped, Q-Switched Frequency-Tripled Laser System: Triton; TEM00, 349 nm; maximum power 1 W, pulsed; repetition rate: 1–1000 Hz; pulse width: <15 ns; pulse energy: adjustable from 1 to 200 μJ; Spectra Physics, Darmstadt, Germany) was expanded by a telescope system and was coupled into a confocal laser scanning microscope (LSM 510) via epifluorescence illumination path. The laser beam was focused into the object plane by a Zeiss Plan-Neofluar 100/1.30 oil objective (spot diameter <500 nm) after reflection by a dielectric mirror (Laser Optik, Germany). The dielectric mirror is placed on the empty laser scanning position of the fluorescence reflector slider and transmits the scanning lasers as well as the emitted fluorescence and reflects the pulsed laser beam of Triton. Thus the imaging functions of the LSM are not reduced. Before entering the microscope, laser pulse energy was reduced by a gradient position dependent attenuator (Laser Optik) to 80%.
The laser ablation of ASI neurons was done as described [4]. Briefly, paralyzed post-L4 nematodes that expresses GFP from a ASI-specific str-3 promoter (kyIs128[str-3::gfp]) were irradiated by several laser pulses at laser energy of 4 μJ. The live cell damage was recorded and controlled by time series function of the Zeiss laser scanning microscope which is equipped with an Argon ion laser and emission filter sets for the detection of EGFP signals (BP530/20) and the Zeiss LSM software version 3.2. Immediately after ablation, worms were transferred to the corresponding treatment plates and analyzed for life expectancy according to the described lifespan protocol above.
2.11. Protein determination
Protein content in nematodes was determined by standard methods as previously described [3].
2.12. Quantification of ROS using Mitotracker Red CM-H2X ROS
Quantification of mtROS was done as described [12,19]. Briefly, prior to ROS measurement MitoTracker Red CM-H2X ROS (Invitrogen, Carlsbad, CA, USA) incubation plates were prepared as following: For each treatment 500 μl heat inactivated OP50 (65 °C, 30 min) were mixed with 100 μl MitoTracker Red CM-H2X stock solution (100 μM) and spotted on a large NGM agar plate which was allowed to dry for approx. 20 min. Nematodes were incubated with corresponding compounds, then washed off the plates with S-Basal and allowed to settle by gravitation to remove offspring. Worms were washed two additional times with S-Basal and centrifuged (300×g, 30 s). The worm pellet was transferred to freshly prepared MitoTracker Red CM-H2X solution and incubated for 2 h at 20 °C. To remove excess dye from the gut, worms were transferred to NGM agar plates spotted with the corresponding compounds. Aliquots of 100 μl worm suspension were distributed into 96-well FLUOTRAC™ plate (Greiner Bio-One, Frickenhausen, Germany). Fluorescence intensity was measured in a microplate reader (FLUOstar Optima, BMG Labtech, Offenburg, Germany) using well-scanning mode (ex: 570 nm, em: 610 nm). To normalize fluorescence signal, remaining worm suspension was used for protein determination.
2.13. Amplex Red-based quantification of supernatant hydrogen peroxide
Quantification of hydrogen peroxide was performed as described [12]. Worms were removed from plates with 50 mM sodium-phosphate buffer pH 7.4, washed twice and transferred into an upright plexiglas cylinder (1.5 ml volume) with continuous stirring at low speed (100 rpm) at 20 °C. Firstly determination of fluorescence was done without horse radish peroxidase (HRP) only in the presence of 1 μM Amplex Red (Invitrogen, Carlsbad, CA, USA) to detect possible unspecific increase in fluorescence (which was not observed). Next, 0.01 U/ml HRP was added and changes of fluorescence were recorded with a fluorescence detector (LF402 ProLine, IOM, Berlin, Germany) for at least 15 min at excitation and emission wavelengths of 571 nm and 585 nm, respectively. Immediately afterwards, worms were removed and collected for protein determination to normalize fluorescence values.
2.14. Locomotion assay
Single worm movements within a liquid system were recorded using a digital CCD camera (Moticam 2300, Motic, St. Ingbert, Germany) coupled microscope (SMZ 168, Motic, St. Ingbert, Germany) with a subsequent analysis using the program DanioTrack (Loligo Systems, Tjele, Denmark). Worms (n=15) at the age of 7 days of adult age were transferred from agar plates to S-buffer and immediately afterwards a 20-second video sequence was recorded. During the subsequent video analysis, the DanioTrack software subtracted the background and determined the center of gravity of all object pixels in contrast with the background. Finally, the moving distance of this worm gravity center was tracked and calculated. Thus, this locomotion assay can be considered as a quantitative analysis of the maximum movement capacity of a single worm.
2.15. Superoxide dismutase activity assay
SOD activity in nematodes was determined by a standard photometric assay as previously described [12].
2.16. Extraction of RNA
Total RNA was isolated using QIAzol (Qiagen, Hilden, Germany) based on the phenol/chloroform extraction method. Afterwards the RNA was quantified photometrically with a NanoDrop 1000 (PeqLab, Erlangen, Germany) and stored at −80 °C until use.
2.17. Illumina next-generation sequencing (RNAseq)
Total RNA was inspected for degradation using Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). For library preparation an amount of 2 μg of total RNA per sample was processed using Illumina's TruSeq™ RNA Sample Prep Kit (Illumina; San Diego; CA, USA) following the manufacturer's instruction. Each library includes its own index sequencing to allow multiplexing. The libraries were sequenced using v2 sequencing chemistry and a HiSeq2000 (Illumina, San Diego, CA, USA) in a single read approach with 50 cycles resulting in reads with length of 50 nucleotides. Libraries were sequenced in a multiplex manner pooling four libraries per lane. Sequencing ends up with around 30–40 mio reads per sample. Sequence data were extracted in FASTQ format and used for mapping as described next.
2.18. Bioinformatics of RNA expression data
The FASTQ files were mapped using the TopHat read mapper [24] vs. the C. elegans genome assembly WS220 (Ensembl release 66). Gene counts were obtained using the Python package HTSeq and the respective Ensembl annotation. Raw counts for the genes were analyzed using the R Statistical Computing Environment [25] and the Bioconductor packages edgeR [26]. EdgeR provides statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting p-values were adjusted using the Benjamini and Hochberg's approach for controlling the false discovery rate (FDR) [27]. Transcripts with an adjusted p-value smaller 0.05 were assigned as differentially expressed. The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE46051 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE46051).
2.19. FunCat analyses
For analyses of functional annotations of C. elegans genes we used the FunCat categorization method included in the FungiFun tool as previously described [28].
2.20. Generation of double mutants
In order to generate double mutants, we crossed male nematodes to hermaphrodites using standard techniques [29] briefly by placing 2–3 aak-2 hermaphrodites on a small plate together with 10–12 sir-2.1 males (generated by heat shock treatment; 6 h, 30 °C). Subsequently single F1 progenies were isolated and genotyped using PCR. Once a heterozygous progeny was identified, the corresponding F2 progenies were screened for homozygous double mutants. Therefore, the genotype of isolated progenies was again examined using single worm PCR. The following primers were used for genotyping: aak-2 5′-TCAGTTGGAATCCATGAGAC-3′ and 5′-ATGACTCCACACGACCATAC-3′ and sir-2.1 5′-AACTTCTGGTCGTCTTCTTC-3′ and 5′-GATGAACATCACGAACCAGT-3′ for forward and reverse primers, respectively.
2.21. Fluorescence microscopy
Translocation and accumulation of SKN-1 within the ASI neurons was analyzed as described [18,30]. Briefly, worms were treated with 100 nM rotenone. Afterwards, individual worms were placed on agarose pads and paralyzed with 1 mg/ml tetramisole. Pictures were taken using a Zeiss LSM 410 confocal unit (excitation wavelength 488 nm; emission filter 510–525 nm).
2.22. Food uptake quantification
To analyze the total amount of incorporated food, i.e. OP50 E. coli, nematodes were pretreated for 5 days under standard conditions in the presence and absence of 100 nM rotenone as described above. Next, worms were transferred to assay plates that were spotted with a defined volume of heat inactivated OP50. Worms were allowed to consume bacteria for 6 h. Afterwards, the remaining bacteria and worms were thoroughly removed and transferred in an reaction tube. Worms were spun down at low speed and an aliquot of the supernatant was removed for a subsequent optical density (OD) determination using a microplate reader (FLUOstar Optima, BMG Labtech, Offenburg, Germany, at λ=600 nm). An empty reference plate, i.e. w/o worms but with the same amount of bacteria, was equally handled. ΔOD was calculated by subtracting the OD of a worm plate by the OD value derived from the reference plate. The remaining worms were used for protein determination in order to normalize the OD 600 values.
2.23. Statistical analyses
Data are expressed as means±SD unless otherwise indicated. Statistical analyses were performed by Student's t-test (unpaired, two-tailed) after testing for equal distribution of the data and equal variances within the data set. For comparing significant distributions between different groups in the life-span assays and stress resistance assays, statistical calculations were performed using JMP software version 9.0 (SAS Institute Inc., Cary, NC, USA) applying the log-rank test. All other calculations were performed using Excel 2007 (Microsoft, Albuquerque, NM, USA) and SPSS version 13.0 (IBM, Armonk, NY, USA). A P-value below 0.05 was considered as statistically significant.
3. Results and discussion
3.1. Compound-based inhibition of mitochondrial complex I increases lifespan, stress resistance and physical activity
We have used three well-established inhibitors of mitochondrial complex I, namely rotenone, piericidin A and 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) [31] to block electron transport from iron–sulfur clusters to the ubiquinone pool causing reduced ATP production (OXPHOS) and elevated mtROS formation [32]. These were analyzed in regards to their putative effects on resistance against lethal paraquat stress, on lifespan, and on physical activity.
We first tested whether low doses of the complex I inhibitors rotenone (100 nM), piericidin A (100 nM) and MPTP (10 nM) are capable to improve stress resistance against paraquat-derived lethal oxidative stress which is considered to be an indicator not only for potentially beneficial lifespan outcomes but also for an elevated ROS defense capacity. As depicted in Figure 1 A, D and G, we found that all three compounds increase stress resistance against lethal oxidative stress. We subsequently analyzed the influence of a compound based inhibition of mitochondrial complex I on the life expectancy of C. elegans. By applying rotenone, piericidin A or MPTP to nematodes increased median lifespan by 10 percent (P<0.00001), 5 percent (P=0.00005) and 8 percent (P<0.00001), respectively (Figure 1 B, E and H) (pls. see Suppl. Table 1 for lifespan details, applies to all subsequent lifespan assays). Consistent with previous findings indicating that physical activity is associated with longevity in C. elegans [33,34] and humans [35], we found that worms treated with complex I inhibitors are characterized by increased activity as determined by quantifying locomotion at an age of 7 adult days (Figures 1C, F and I).
Figure 1.
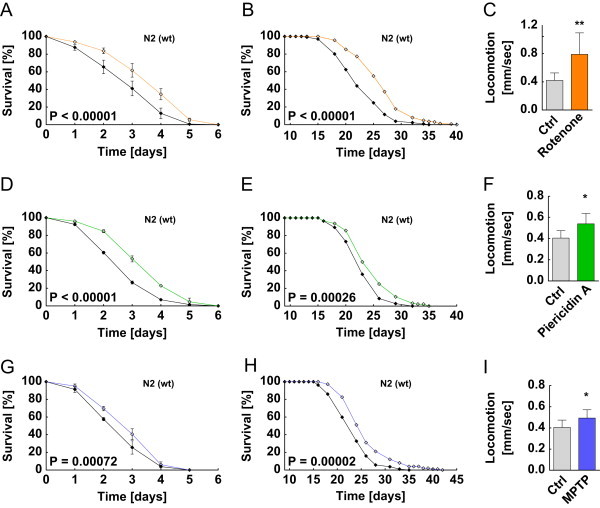
Increased stress resistance, lifespan and locomotion due to compound-based inhibition of mitochondrial complex I. (A, D, and G) Stress assay for N2 wild type nematodes, i.e. survival during 10 mM paraquat exposure after pretreatment with complex I inhibitors; control (black, applies to all panels) vs. (A) 100 nM rotenone (orange), (D) 100 nM piericidin A (green), or (G) 10 nM MPTP (blue), respectively. (B, E, and H) Lifespan assay for N2 wild type nematodes in the presence of complex I inhibitors; control vs. (B) 100 nM rotenone (orange), (C) 100 nM piericidin A (green), or (H) 10 nM MPTP (blue), respectively. (C, F, and I) Locomotion quantification for N2 wild type nematodes after 7 d exposition to complex I inhibitors; control bars (gray) compared with (C) 100 nM rotenone, (F) 100 nM piericidin A and (I) 10 nM MPTP treatment according to lifespan assay setup.
3.2. Compound-based inhibition of mitochondrial complex I causes a characteristic transcriptomic signature in early adult life that is sufficient to extend lifespan
Complex I inhibition causes increased formation of superoxide, a component of the ROS family, which is neutralized by superoxide dismutase (SOD) [32]. To test whether the previously described increase in resistance against ROS stress (Figures 1 A, D and G) is caused by changes in ROS defense enzymes, we quantified activity of SOD and found it to be increased (Figures 2A).
Figure 2.
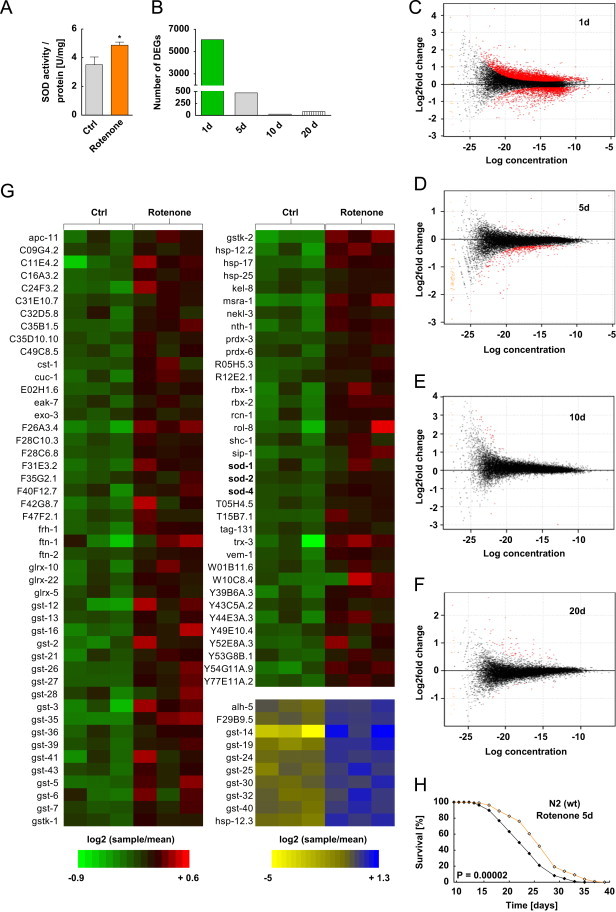
Compound-based inhibition of mitochondrial complex I induces an orchestrated adaptive response. (A) SOD activity after 48 h treatment with 100 nM rotenone. (B) Absolute number of differentially expressed genes (DEGs) after 1, 5, 10 and 20 days rotenone treatment (according to edgeR, adj. P<0.05). (C–F) MA plot depicting DEG as quantified by deep sequencing technology following 1 day (C), 5 days (D), 10 days (E) and 20 days (F) rotenone treatment. Not differentially regulated genes are depicted as black dots. Red dots indicate significantly regulated DEGs. (G) Relative expression level of up-regulated DEGs after 1 day rotenone treatment, annotated to oxidative stress response (n=3 for control and treatment). (H) Lifespan assay for N2 wild type nematodes in the presence of rotenone for 5 days only; control (black) vs. 100 nM rotenone (orange).
To identify additional molecular mechanism responsible for transducing complex I inhibition into extended longevity, we performed so-called deep sequencing (RNAseq) of RNA samples extracted from nematodes that were treated with rotenone for 1, 5, 10 or 20 days, respectively, and compared them to solvent-supplemented worms of the same ages. We observed the maximal number of differentially expressed genes (n=6064) already after 24 h of treatment with rotenone (Figures 2B and C and Suppl. Table 3), whereas at later time points the number of regulated genes was strikingly reduced (Figures 2B and D–F and Supp. Tables 4–6). Analyzing all genes upregulated at 24 h by clustering them into functional groups revealed a significant overrepresented group (p=0.000072; 93 genes) annotated to oxidative stress response (Figure 2G and Suppl. Table 2) as well as additional groups linked to mitochondrial function (Suppl. Table 2). Notably, different subtypes of SOD were also contained in this list (Figure 2G) consistent with the fact that SOD activity was found to be increased following rotenone treatment, as discussed above (Figure 2A).
Given the fact that most differentially expressed genes were detected before an adult age of 5 days (Figures 2B and C), we questioned whether exposure of nematodes to rotenone for 5 days would be sufficient to extend lifespan which was indeed the case (Figure 2H) (8 percent), indicating that short-term complex I inhibition is sufficient to extend C. elegans lifespan since the vast majority of transcriptional regulations has occurred at this stage (Figures 2B and C).
3.3. Lifespan-extending effects following both dietary restriction and complex I inhibition share a common denominator
Next we questioned whether inhibitor treatment would affect food uptake of C. elegans. We observed no effect of rotenone on consumption of the nematodal food source E. coli OP50 (Figure 3A and Suppl. Figure 1A) whereas a C. elegans model of impaired food uptake, eat-2, that is characterized by defect in pharyngeal pumping [36], served as a positive control (Figure 3A and Suppl. Figure 1A).
Figure 3.
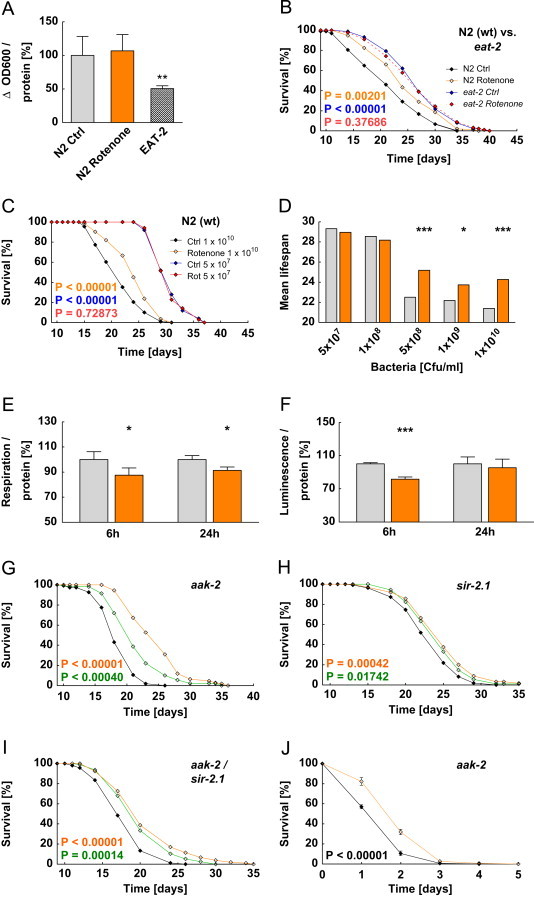
Inhibition of mitochondrial complex I extends lifespan in a dietary restriction (DR) manner and causes a short-term energy deficit but does not require AMPK or sirtuin signaling to extend lifespan. (A) Food uptake after 120 h treatment with 100 nM rotenone in N2 wild type nematodes and in untreated eat-2 (ad453) mutants. (B) Lifespan assay for N2 wild type and eat-2 (ad453) mutants in the presence of 100 nM rotenone; N2 control (black) vs. N2+100 nM rotenone (orange) vs. eat-2 control (blue) vs. eat-2+100 nM rotenone (dotted red). (C) Lifespan assay for N2 wild type under DR. Ad libitum fed (black) vs. ad libitum+100 nM rotenone fed (orange) vs. DR (5×107 c.f.u. ml−1; blue) vs. DR+100 nM rotenone (5×107 c.f.u. ml−1; red). (D) Summary of lifespan experiments during various bacterial dilutions. (E) Oxygen consumption for different incubation periods (control: gray bars; 100 nM rotenone: orange bars). (F) ATP content for various rotenone incubation periods (control: gray bars; 100 nM rotenone: orange bars). (G) Lifespan assay for aak-2 (ok524) mutants; control (black) vs. 100 nM rotenone (orange) vs. 100 nM piericidin A (green). (H) Lifespan assay for sir-2.1 (ok434) mutants; control (black) vs. 100 nM rotenone (orange) vs. 100 nM piericidin A (green). (I) Lifespan assay for MIR13 (aak-2(ok524); sir-2.1(ok434)) mutants; control (black) vs. 100 nM rotenone (orange) vs. 100 nM piericidin A (green). (J) Stress assay for aak-2 (ok524) deficient mutants; control (black) vs. 100 nM rotenone (orange).
We then exposed eat-2 worms to rotenone to test whether the inhibitor would be capable of extending lifespan further in a long-lived model of DR. Rotenone had no effect on eat-2 lifespan (Figure 3B) indicating that reduced food uptake (eat-2) and complex I inhibition share a common mechanistic denominator in regards to lifespan extension. To further support this we used a different model of DR in C. elegans, so-called bacterial dilution [22], and analyzed lifespan in the presence and absence of rotenone. While the inhibitor was capable of extending lifespan in states of unlimited food availability (1×1010 c.f.u./ml) (Figures 3C and D, see also Figure 1B), in states of limited food availability (1×108 c.f.u./ml and below) we confirmed [22] a pronounced effect of DR on lifespan per se (Figures 3C and D). However, rotenone did not further extend lifespan of worms on bacterial DR (Figures 3C and D). These findings indicate that, in two models of DR, rotenone has no additional effect on lifespan, indicating that DR and complex I inhibition share the same mechanistic denominator. Inversely, this indicates that complex I inhibitor mimic DR and may hence be considered calorie restriction mimetics (CRMs) [37].
3.4. Compound-based inhibition of mitochondrial complex I induces metabolic stress but does not require AMPK or sirtuin signaling to extend lifespan
Dietary restriction causes changes in OXPHOS and hence ATP availability [3,38]. Similarly, inhibition of the electron transport chain (ETC) at complex I compromises oxidative phosphorylation (OXPHOS) and thus evokes depletion of ATP [15,32]. We consistently observed a significant decrease in oxygen consumption (12%) and ATP content (18%) after short-term rotenone incubation, whereas prolonged exposure periods results in a compensatory increase, however for ATP only (Figures 3E and F, Suppl. Figures 1B and C).
ATP depletion has been shown to activate important cellular energy sensors such as AMPK [6] and sirtuins [5]. Accordingly, activation of either AMPK or sirtuins has been shown to mediate longevity-promoting effects of DR [3,5,21,39,40]. To address the question whether activation of AMPK or sirtuins is required for the lifespan-extending effects of complex I inhibition we performed lifespan analyses using nematodes deficient for the corresponding C. elegans orthologues, AAK-2 and SIR-2.1, respectively. Surprisingly, we found that rotenone and piericidin A were still capable of extending life expectancy of those mutants (Figure 3G and H), unexpectedly suggesting a mechanism independent of AMPK or sirtuins. Moreover, we generated a double mutant deficient for both AMPK and SIR-2.1. Both rotenone as well as piericidin A were still capable of extending lifespan in this double mutant (Figure 3I). In addition, the induction of stress resistance observed in wild-type worms (Figure 1A) was also unaffected by impaired AAK-2 expression (Figure 3J). These finding indicate that complex I inhibition does deplete ATP in a transient manner, however sensing of this ATP depletion by the two energy sensors, AMPK and sirtuins, is dispensable for lifespan extension in nematodes. Moreover, these findings suggest that ATP depletion is not the cause of lifespan extension mediated by complex I inhibition.
3.5. Compound-based inhibition of mitochondrial complex I causes mtROS formation that requires PMK-1/p38 to promote longevity
As stated above, complex I inhibition is known to promote formation of mitochondrial ROS (mtROS). To test whether this is likewise the case in nematodes, we quantified mtROS formation using a ROS-sensitive fluorophore targeted to the mitochondrial compartment. When we exposed wild-type nematodes to rotenone for 4 h, we observed a transient increase in mtROS formation, which was undetectable at 6 or 24 h after initiation of exposure (Figure 4A and Suppl. Figure 2A). Interestingly, at 48 h and beyond, i.e. in the steady-state, a persistent reduction of mtROS levels was observed following exposure to either compound (Figure 4A and Suppl. Figure 2A). As previously shown for other mtROS-inducing interventions [3,10,12,17] this is due to a compensatory induction of ROS-detoxifying enzymes, including SOD which was found to be increased in rotenone-treated worms, as shown above (Figure 2A). We additionally quantified ROS formation by an independent method based on AmplexRed fluorescence that is proportional to accumulation of hydrogen peroxide in the supernatant of worm suspensions. These data similarly show that rotenone induces formation of hydrogen peroxide 4 h after initiation of exposure (Figure 4B and Suppl. Figure 2B).
Figure 4.
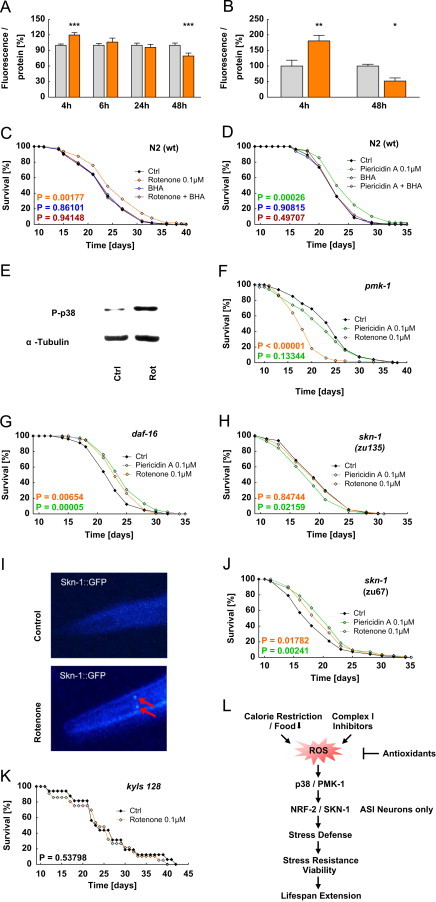
Compound-based inhibition of mitochondrial complex I causes mtROS formation that mediates lifespan extension dependent on PMK-1/p38 and ASI specific neuronal SKN-1/NRF-2 signaling. (A) mtROS production, depicted for different rotenone incubation periods (control: gray bars; 100 nM rotenone: orange bars). (B) H2O2 production after 4 and 48 h rotenone treatment (control: gray bars; 100 nM rotenone: orange bars). (C) Lifespan assay for N2 wild type nematodes in the presence of rotenone co-applied with an antioxidant; control (black line) vs. 100 nM rotenone (orange line) vs. 10 μM BHA (blue line) vs. 100 nM rotenone in combination with 10 μM BHA (purple line). (D) Lifespan assay for N2 wild type nematodes in the presence of piericidin A co-treated with an antioxidant; control (black line) vs. 100 nM piericidin A (orange line) vs. 10 μM BHA (blue line) vs. 100 nM piericidin A in combination with 10 μM BHA (purple line). (E) Western blot analysis of the phosphorylated form of PMK-1/p38 after 24 h rotenone exposure. (F) Lifespan assay for pmk-1 (km25) mutants; control (black line) vs. 100 nM rotenone (orange line) vs. 100 nM piericidin A (green line). (G) Lifespan assay for daf-16 (mgDf47) mutants; control (black line) vs. 100 nM rotenone (orange line) vs. 100 nM piericidin A (green line). (H) Lifespan assay for skn-1 (zu135) mutants; control (black line) vs. 100 nM rotenone (orange line) vs. 100 nM piericidin A (green line). (I) Rotenone induced nuclear translocation and accumulation within ASI neurons (arrows) in young adult C. elegans after 6 h short-term incubation, determined by using SKN-1::GFP. (J) Lifespan assay for skn-1 (zu67) mutants; control (black line) vs. 100 nM rotenone (orange line) vs. 100 nM piericidin A (green line). (K) Lifespan assay for ASI ablated kyIs128 nematodes (expresses GFP from ASI-specific str-3 promoter); control (black line) vs. 100 nM rotenone (orange line). (L) Graphical summary.
We next exposed nematodes to a ROS scavenger to impair formation of mtROS following exposure to rotenone or piericidin. As previously shown [12], the scavenger butylated hydroxyl-anisole (BHA) had no effect on C. elegans lifespan per se (Figures 4C and D). However, the lifespan-extending capabilities of both rotenone and piericidin A were completely abolished in the presence of BHA (Figures 4C and D). These findings indicate that the transient induction of ROS is essential for the extension of C. elegans lifespan mediated by complex I inhibitors.
Several pathways were described to be involved in oxidative stress response, including the mitogen-activated protein kinase signaling pathway [41] also in C. elegans [42,43]. In particular, the C. elegans orthologue of mammalian p38 MAP kinase, PMK-1 [44], exerts similar stress related functions in particular following compound-mediated mtROS formation [19]. Consistently, we here observed that exposure to rotenone promotes phosphorylation, reflecting activation, of PMK-1 (Figure 4E). Moreover, complex I inhibitors were incapable of extending lifespan in nematodes deficient for PMK-1 (Figure 4F) but rather exert lifespan-shortening effects on these mutants (Figure 4F).
Taken together, these findings indicate that energy sensors like AMPK or sirtuins or both are dispensable for ROS-mediated lifespan extension (Figures 3G–I) whereas ROS sensors like p38/PMK-1 are essential for this phenotype (Figures 4E and F).
3.6. SKN-1-dependent redox signaling in ASI neurons only promotes longevity
Oxidative stress response on transcriptional level is significantly mediated by the Keap-1-associated transcription factor NRF-2 as well as the forkhead box (Fox) containing transcription factor family FoxO. Both transcription factors are capable of regulating gene expression of a variety of defense mechanisms including oxidative stress [45,46] or phase II xenobiotic detoxifying enzymes [18,43] in order to counteract the initial redox signal. We first analyzed the C. elegans daf-16 mutant being deficient for the FoxO pathway which also has been involved into energy sensing [47]. Unexpectedly, both rotenone and piericidin A were capable of extending lifespan in daf-16 mutants (Figures 4G) indicating that FoxO signaling is dispensable for mtROS-mediated lifespan extension.
We next analyzed the effects of complex I inhibitors on the NRF-2 signaling pathway by analyzing nematodes deficient for the C. elegans orthologue SKN-1 [48]. The loss-of-function allele skn-1(zu135) is deficient for all three isoforms of SKN-1 [30]. Neither rotenone nor piericidin A promoted longevity in this mutant (Figure 4H), indicating that redox-sensitive NRF-2 signaling [18,43] is essential for mtROS-mediated lifespan extension, consistent with previously published findings [12]. Moreover, these findings indicate that NRF-2 mediates a starvation response, as previously suggested [4,12] and as further extended while this manuscript was in preparation [49].
Next, we used a skn-1 GFP reporter strain [18] to study the effects of complex I inhibition. Rotenone treatment preferentially induced fluorescence in the two nematodal ASI neurons (Figure 4I) which interestingly have been previously linked to lifespan extension in states of DR [4]. Our findings suggest that neuronal ROS signaling may promote lifespan following complex I inhibition and mtROS induction. To further support this hypothesis we analyzed worms that were deficient for only two of the three SKN-1 isoforms, named skn-1(zu67) [30]. Notably, the neuronal SKN-1 isoform is functional in these worms [50]. Consistent with neuronal ROS signaling both rotenone as well as piericidin A extended lifespan in this isoform-specific mutant (Figure 4J). To further elucidate which neuronal subsets may be involved into mtROS-mediated lifespan extension we laser-ablated selectively ASI neurons in wild-type nematodes, consistent with previous findings [4], and subsequently tested the ability of rotenone to extend lifespan in the absence of the two ASI neurons. Notably, rotenone did not extend lifespan in the absence of ASI neurons (Figure 4K) altogether indicating that these serve as mtROS sensors in a SKN-1-dependent fashion (Figure 4L).
4. Conclusions
Taken together, these data suggest that a compound-based inhibition of mitochondrial complex I induces a mtROS signal that is sufficient to extend lifespan in the absence of known energy sensors like AMPK/AAK-2 and sirtuins. The mtROS signal initiates SKN-1-mediated redox response to increase stress resistance and fitness that finally culminates in lifespan extension. These findings not only extend the role of SKN-1/NRF-2 as mediator of transcriptional starvation response [4,12,49] but also indicate that activators of SKN-1/NRF-2, and hence complex I inhibitors, may be considered as a promising pharmacological approach to extend lifespan independent of impaired food uptake. In this regard it is interesting to note that a significant number of pharmaceutically effective compounds, including phytochemicals like resveratrol [51], sulforaphane [52], niacin [53] and berberine [54], as well as anti-diabetics like metformin [55] and PPARγ-activating thiazolidinediones [56,57], and lastly cholesterol-lowering HMG-CoA-synthase inhibitors (“statins”) [57] and PPARα-activating fibrates [57,58], have been found to inhibit mitochondrial complex I of the ETC, opposing the disease-promoting effects of high-dose complex I inhibition, as reviewed elsewhere [32]. These findings indicate that a limited inhibition of complex I and hence limited ROS production, as exerted by these before-mentioned compounds [51–58], promote metabolic health and lifespan, while high-dose complex I inhibition promotes neurodegenerative disorders [32], providing a prime example for non-linear dose-response characteristics, i.e. hormesis [59,60] and in particular mitohormesis [1], in regards to longevity regulation.
5. Conflict of interest
None declared.
Acknowledgments
C. elegans strains used in this work were provided by the Caenorhabditis Genetics Centre (Univ. of Minnesota), which is funded by the NIH National Center for Research Resources (NCRR). We thank T. Keith Blackwell for a skn-1::GFP reporter strain [18]. The excellent technical assistance of Ivonne Heinze, Beate Laube, Annett Müller, Susann Richter and Waltraud Scheiding, as well as the excellent secretarial assistance of Mandy Schalowski are gratefully acknowledged. This work is in part supported by the research program of the Jena Centre for Systems Biology of Ageing (JenAge) funded by the German Ministry for Education and Research (Bundesministerium für Bildung und Forschung; support code BMBF 0315581). Funding for this project was denied by the German Research Association (Deutsche Forschungsgemeinschaft, DFG), grant application number RI 1976/3-1.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.molmet.2013.02.002.
Appendix A. Supporting information
Figure S1 (A) Absolute values for food uptake after 120 hrs treatment with 100nM Rotenone in N2 wild type nematodes and in untreated eat-2 (ad453) mutants. (B) Absolute values of oxygen consumption for different incubation periods with 100nM rotenone (orange bars) vs. control (grey bars). (C) Absolute values of ATP content for various rotenone incubation periods. Figure S2 (A) Absolute values of mtROS production, depicted for different rotenone incubation periods (control: grey bars; rotenone: orange bars). (B) Absolute values of H2O2 production after 4 and 48 hrs rotenone treatment (control: grey bars; rotenone: orange bars).
Supplementary Tables
References
- 1.Ristow M., Schmeisser S. Extending life span by increasing oxidative stress. Free Radical Biology and Medicine. 2011;51:327–336. doi: 10.1016/j.freeradbiomed.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 2.Lin S.J., Kaeberlein M., Andalis A.A., Sturtz L.A., Defossez P.A., Culotta V.C. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 3.Schulz T.J., Zarse K., Voigt A., Urban N., Birringer M., Ristow M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metabolism. 2007;6:280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 4.Bishop N.A., Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- 5.Lin S.J., Defossez P.A., Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 6.Hardie D.G. Sensing of energy and nutrients by AMP-activated protein kinase. American Journal of Clinical Nutrition. 2011;93:891S–896. doi: 10.3945/ajcn.110.001925. [DOI] [PubMed] [Google Scholar]
- 7.Doonan R., McElwee J.J., Matthijssens F., Walker G.A., Houthoofd K., Back P. Against the oxidative damage theory of aging: superoxide dismutases protect against oxidative stress but have little or no effect on life span in Caenorhabditis elegans. Genes and Development. 2008;22:3236–3241. doi: 10.1101/gad.504808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ristow M., Zarse K., Oberbach A., Klöting N., Birringer M., Kiehntopf M. Antioxidants prevent health-promoting effects of physical exercise in humans. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8665–8670. doi: 10.1073/pnas.0903485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Raamsdonk J.M., Hekimi S. Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans. PLoS Genetics. 2009;5:e1000361. doi: 10.1371/journal.pgen.1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mesquita A., Weinberger M., Silva A., Sampaio-Marques B., Almeida B., Leao C. Caloric restriction or catalase inactivation extends yeast chronological lifespan by inducing H2O2 and superoxide dismutase activity. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15123–15128. doi: 10.1073/pnas.1004432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan Y., Schroeder E.A., Ocampo A., Barrientos A., Shadel G.S. Regulation of yeast chronological life span by TORC1 via adaptive mitochondrial ROS signaling. Cell Metabolism. 2011;13:668–678. doi: 10.1016/j.cmet.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zarse K., Schmeisser S., Groth M., Priebe S., Beuster G., Guthke R. Impaired insulin/IGF1-signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell Metabolism. 2012;15:451–465. doi: 10.1016/j.cmet.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dillin A., Hsu A.L., Arantes-Oliveira N., Lehrer-Graiwer J., Hsin H., Fraser A.G. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- 14.Rea S.L., Ventura N., Johnson T.E. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in caenorhabditis elegans. Plos Biology. 2007;5:e259. doi: 10.1371/journal.pbio.0050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang W., Hekimi S. Two modes of mitochondrial dysfunction lead independently to lifespan extension in Caenorhabditis elegans. Aging Cell. 2010;9:433–447. doi: 10.1111/j.1474-9726.2010.00571.x. [DOI] [PubMed] [Google Scholar]
- 16.Gutman M., Singer T.P., Casida J.E. Studies on the respiratory chain-linked reduced nicotinamide adenine dinucleotide dehydrogenase. XVII. Reaction sites of piericidin A and rotenone. Journal of Biological Chemistry. 1970;245:1992–1997. [PubMed] [Google Scholar]
- 17.Yang W., Hekimi S. A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans. PLoS Biology. 2010;8:e1000556. doi: 10.1371/journal.pbio.1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.An J.H., Blackwell T.K. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes and Development. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmeisser S., Zarse K., Ristow M. Lonidamine extends lifespan of adult Caenorhabditis elegans by increasing the formation of mitochondrial reactive oxygen species. Hormone and Metabolic Research. 2011;43:687–692. doi: 10.1055/s-0031-1286308. [DOI] [PubMed] [Google Scholar]
- 20.Gruber J., Ng L.F., Poovathingal S.K., Halliwell B. Deceptively simple but simply deceptive-Caenorhabditis elegans lifespan studies: considerations for aging and antioxidant effects. FEBS Letters. 2009;583:3377–3387. doi: 10.1016/j.febslet.2009.09.051. [DOI] [PubMed] [Google Scholar]
- 21.Wood J.G., Rogina B., Lavu S., Howitz K., Helfand S.L., Tatar M. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 22.Park S.K., Link C.D., Johnson T.E. Life-span extension by dietary restriction is mediated by NLP-7 signaling and coelomocyte endocytosis in C. elegans. Faseb Journal. 2010;24:383–392. doi: 10.1096/fj.09-142984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumsta C., Thamsen M., Jakob U. Effects of oxidative stress on behavior, physiology, and the redox thiol proteome of Caenorhabditis elegans. Antioxidants and Redox Signaling. 2011;14:1023–1037. doi: 10.1089/ars.2010.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.R Development Core Team. R: a language and environment for statistical computing. Vienna; 2008.
- 26.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B. 1995;57:289–300. [Google Scholar]
- 28.Priebe S., Linde J., Albrecht D., Guthke R., Brakhage A.A. FungiFun: a web-based application for functional categorization of fungal genes and proteins. Fungal Genetics and Biology. 2011;48:353–358. doi: 10.1016/j.fgb.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 29.Hope I.A. Oxford University Press, New York; 1999. C. elegans: a practical approach. [Google Scholar]
- 30.Tullet J.M., Hertweck M., An J.H., Baker J., Hwang J.Y., Liu S. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Degli Esposti M. Inhibitors of NADH-ubiquinone reductase: an overview. Biochimica et Biophysica Acta. 1998;1364:222–235. doi: 10.1016/s0005-2728(98)00029-2. [DOI] [PubMed] [Google Scholar]
- 32.Schapira A.H. Complex I: inhibitors, inhibition and neurodegeneration. Experimental Neurology. 2010;224:331–335. doi: 10.1016/j.expneurol.2010.03.028. [DOI] [PubMed] [Google Scholar]
- 33.Hosono R., Sato Y., Aizawa S.I., Mitsui Y. Age-dependent changes in mobility and separation of the nematode Caenorhabditis elegans. Experimental Gerontology. 1980;15:285–289. doi: 10.1016/0531-5565(80)90032-7. [DOI] [PubMed] [Google Scholar]
- 34.Johnson T.E., Conley W.L., Keller M.L. Long-lived lines of Caenorhabditis elegans can be used to establish predictive biomarkers of aging. Experimental Gerontology. 1988;23:281–295. doi: 10.1016/0531-5565(88)90031-9. [DOI] [PubMed] [Google Scholar]
- 35.Warburton D.E., Nicol C.W., Bredin S.S. Health benefits of physical activity: the evidence. Canadian Medical Association Journal (CMAJ) 2006;174:801–809. doi: 10.1503/cmaj.051351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lakowski B., Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ingram D.K., Zhu M., Mamczarz J., Zou S., Lane M.A., Roth G.S. Calorie restriction mimetics: an emerging research field. Aging Cell. 2006;5:97–108. doi: 10.1111/j.1474-9726.2006.00202.x. [DOI] [PubMed] [Google Scholar]
- 38.Houthoofd K., Braeckman B.P., Lenaerts I., Brys K., Matthijssens F., De Vreese A. DAF-2 pathway mutations and food restriction in aging Caenorhabditis elegans differentially affect metabolism. Neurobiology of Aging. 2005;26:689–696. doi: 10.1016/j.neurobiolaging.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y., Tissenbaum H.A. Overlapping and distinct functions for a Caenorhabditis elegans SIR2 and DAF-16/FOXO. Mechanisms of Ageing and Development. 2006;127:48–56. doi: 10.1016/j.mad.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 40.Greer E.L., Dowlatshahi D., Banko M.R., Villen J., Hoang K., Blanchard D. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Current Biology. 2007;17:1646–1656. doi: 10.1016/j.cub.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emerling B.M., Platanias L.C., Black E., Nebreda A.R., Davis R.J., Chandel N.S. Mitochondrial reactive oxygen species activation of p38 mitogen-activated protein kinase is required for hypoxia signaling. Molecular Cell Biology. 2005;25:4853–4862. doi: 10.1128/MCB.25.12.4853-4862.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kondo M., Yanase S., Ishii T., Hartman P.S., Matsumoto K., Ishii N. The p38 signal transduction pathway participates in the oxidative stress-mediated translocation of DAF-16 to Caenorhabditis elegans nuclei. Mechanisms of Ageing and Development. 2005;126:642–647. doi: 10.1016/j.mad.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 43.Inoue H., Hisamoto N., An J.H., Oliveira R.P., Nishida E., Blackwell T.K. The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes and Development. 2005;19:2278–2283. doi: 10.1101/gad.1324805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berman K., McKay J., Avery L., Cobb M. Isolation and characterization of pmk-(1-3): three p38 homologs in Caenorhabditis elegans. Molecular Cell Biology Research Communications. 2001;4:337–344. doi: 10.1006/mcbr.2001.0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Henderson S.T., Johnson T.E. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Current Biology. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- 46.Yanase S., Yasuda K., Ishii N. Adaptive responses to oxidative damage in three mutants of Caenorhabditis elegans (age-1, mev-1 and daf-16) that affect life span. Mechanisms of Ageing and Development. 2002;123:1579–1587. doi: 10.1016/s0047-6374(02)00093-3. [DOI] [PubMed] [Google Scholar]
- 47.Greer E.L., Banko M.R., Brunet A. AMP-activated protein kinase and FoxO transcription factors in dietary restriction-induced longevity. Annals of the New York Academy of Sciences. 2009;1170:688–692. doi: 10.1111/j.1749-6632.2009.04019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blackwell T.K., Bowerman B., Priess J.R., Weintraub H. Formation of a monomeric DNA binding domain by Skn-1 bZIP and homeodomain elements. Science. 1994;266:621–628. doi: 10.1126/science.7939715. [DOI] [PubMed] [Google Scholar]
- 49.Paek J., Lo J.Y., Narasimhan S.D., Nguyen T.N., Glover-Cutter K., Robida-Stubbs S. Mitochondrial SKN-1/Nrf mediates a conserved starvation response. Cell Metabolism. 2012;16:526–537. doi: 10.1016/j.cmet.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ventura N., Rea S.L., Schiavi A., Torgovnick A., Testi R., Johnson T.E. p53/CEP-1 increases or decreases lifespan, depending on level of mitochondrial bioenergetic stress. Aging Cell. 2009;8:380–393. doi: 10.1111/j.1474-9726.2009.00482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zini R., Morin C., Bertelli A., Bertelli A.A., Tillement J.P. Effects of resveratrol on the rat brain respiratory chain. Drugs Experimental Clinical Research. 1999;25:87–97. [PubMed] [Google Scholar]
- 52.Singh S.V., Srivastava S.K., Choi S., Lew K.L., Antosiewicz J., Xiao D. Sulforaphane-induced cell death in human prostate cancer cells is initiated by reactive oxygen species. Journal of Biological Chemistry. 2005;280:19911–19924. doi: 10.1074/jbc.M412443200. [DOI] [PubMed] [Google Scholar]
- 53.Fukushima T. Niacin metabolism and Parkinson's disease. Environmental Health and Preventive Medicine. 2005;10:3–8. doi: 10.1265/ehpm.10.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turner N., Li J.Y., Gosby A., Cheng S.W., Miyoshi Z., Taketo H. Berberine and its more biologically available derivative, dihydroberberine, inhibit mitochondrial respiratory complex I: a mechanism for the action of berberine to activate AMP-activated protein kinase and improve insulin action. Diabetes. 2008;57:1414–1418. doi: 10.2337/db07-1552. [DOI] [PubMed] [Google Scholar]
- 55.El-Mir M.Y., Nogueira V., Fontaine E., Averet N., Rigoulet M., Leverve X. Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. Journal of Biological Chemistry. 2000;275:223–228. doi: 10.1074/jbc.275.1.223. [DOI] [PubMed] [Google Scholar]
- 56.Brunmair B., Staniek K., Gras F., Scharf N., Althaym A., Clara R. Thiazolidinediones, like metformin, inhibit respiratory complex I: a common mechanism contributing to their antidiabetic actions? Diabetes. 2004;53:1052–1059. doi: 10.2337/diabetes.53.4.1052. [DOI] [PubMed] [Google Scholar]
- 57.Nadanaciva S., Dykens J.A., Bernal A., Capaldi R.A., Will Y. Mitochondrial impairment by PPAR agonists and statins identified via immunocaptured OXPHOS complex activities and respiration. Toxicology and Applied Pharmacology. 2007;223:277–287. doi: 10.1016/j.taap.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 58.Brunmair B., Lest A., Staniek K., Gras F., Scharf N., Roden M. Fenofibrate impairs rat mitochondrial function by inhibition of respiratory complex I. Journal of Pharmacology and Experimental Therapeutics. 2004;311:109–114. doi: 10.1124/jpet.104.068312. [DOI] [PubMed] [Google Scholar]
- 59.Calabrese E.J., Baldwin L.A. Toxicology rethinks its central belief. Nature. 2003;421:691–692. doi: 10.1038/421691a. [DOI] [PubMed] [Google Scholar]
- 60.Gems D., Partridge L. Stress-response hormesis and aging: that which does not kill us makes us stronger. Cell Metabolism. 2008;7:200–203. doi: 10.1016/j.cmet.2008.01.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 (A) Absolute values for food uptake after 120 hrs treatment with 100nM Rotenone in N2 wild type nematodes and in untreated eat-2 (ad453) mutants. (B) Absolute values of oxygen consumption for different incubation periods with 100nM rotenone (orange bars) vs. control (grey bars). (C) Absolute values of ATP content for various rotenone incubation periods. Figure S2 (A) Absolute values of mtROS production, depicted for different rotenone incubation periods (control: grey bars; rotenone: orange bars). (B) Absolute values of H2O2 production after 4 and 48 hrs rotenone treatment (control: grey bars; rotenone: orange bars).
Supplementary Tables


