Abstract
A hierarchical control of fimbrial gene expression limits laboratory grown cultures of Salmonella enterica serovar typhimurium (S. typhimurium) to the production of type I fimbriae encoded by the fimAICDHF operon. Here we show that an unlikely culprit, namely the 5′-untranslated region (5′-UTR) of a messenger (m)RNA, coordinated the regulation. Binding of CsrA to the 5′-UTR of the pefACDEF transcript was required for expression of plasmid-encoded fimbriae. The 5′-UTR of the fimAICDHF transcript cooperated with two small untranslated RNAs, termed CsrB and CsrC, in antagonizing the activity of the RNA binding protein CsrA. Through this post-transcriptional mechanism, the 5′-UTR of the fimAICDHF transcript prevented production of PefA, the major structural subunit of plasmid-encoded fimbriae. This regulatory mechanism limits the costly expression of plasmid-encoded fimbriae to host environments in a mouse model. Collectively, our data suggest that the 5′-UTR of an mRNA coordinates a hierarchical control of fimbrial gene expression in S. typhimurium.
Keywords: fimbriae, gene regulation, Salmonella
Introduction
The genomes of Escherichia coli and Salmonella enterica contain a large number of fimbrial gene clusters belonging to the chaperone/usher assembly class, a group defined by sequence homology of the encoded periplasmic chaperone and outer membrane usher assembly proteins (Nuccio and Bäumler, 2007; Yue et al, 2012). Many of these operons are required for intestinal colonization and/or the pathogenesis of urinary tract infection (Bäumler et al, 1997; Bergsten et al, 2005; Nielubowicz and Mobley, 2010; Wagner and Hensel, 2011). Each operon encodes structural subunits that are assembled into a fimbrial filament on the cell surface by a periplasmic chaperone and an outer membrane usher protein (Hung and Hultgren, 1998; Proft and Baker, 2009; Waksman and Hultgren, 2009). Fimbriated bacteria can express >200 fimbrial filaments on their surface, each composed of up to 1000 copies of the major structural subunit (Klemm, 1994; Proft and Baker, 2009). Thus, upon expression of a fimbrial gene cluster, the respective major structural subunit becomes one of the most abundant proteins in the bacterial cell.
The costly expression of these surface structures is tightly controlled by regulatory mechanisms that prevent their elaboration in vitro. For example, laboratory-grown cultures of S. enterica serovar typhimurium (S. typhimurium) commonly elaborate only type 1 fimbriae encoded by the fimAICDHF operon (Duguid et al, 1966; Clegg et al, 1987), while expression of the remaining 11 chaperone/usher (C/U)-type fimbrial operons that are present in its genome cannot be detected by western blotting (Humphries et al, 2005) or flow cytometry (Humphries et al, 2003). Similarly, 9 of the 13 C/U-type fimbrial operons present in the genome of enterohaemorrhagic E. coli are not expressed under in vitro growth conditions (Low et al, 2006).
One of the reasons why only a selected few C/U-type fimbrial operons are expressed under standard laboratory conditions is the hierarchical control of fimbrial gene expression in E. coli and S. enterica (Xia et al, 2000; Snyder et al, 2005; Holden et al, 2006; Nuccio et al, 2007). For example, the elaboration of type I fimbriae by uropathogenic E. coli strain CFT073 suppresses the expression of pyelonephritis-associated fimbriae, which in turn suppresses expression of F1C fimbriae (Snyder et al, 2005). Similarly, expression of PefA, the major structural subunit of plasmid-encoded fimbriae, can be detected by western blotting in S. typhimurium after the biosynthesis genes for type 1 fimbriae (fimAICDHF) have been deleted (Nuccio et al, 2007). The S. typhimurium fimAICDHF operon encodes a periplasmic chaperone (FimC), an outer membrane usher (FimD), a major structural subunit (FimA), three minor structural subunits (FimI, FimF and FimH), but not regulatory proteins. It is thus not obvious by which mechanism deletion of the fimAICDHF operon induces expression of plasmid-encoded fimbriae.
Here, we investigated the mechanism by which the presence of the fimAICDHF operon prevents expression of plasmid-encoded fimbriae. Our results identify a novel mechanism of bacterial gene regulation that ensures hierarchical expression of fimbrial biosynthesis genes.
Results
SirA and the fimAICDHF operon synergize in repressing plasmid-encoded fimbriae
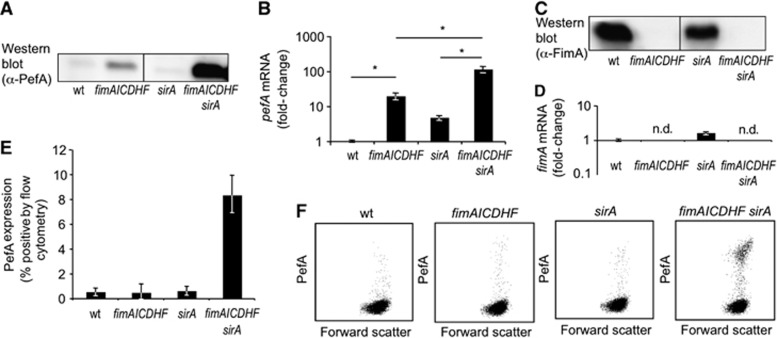
The goal of this study was to determine the mechanism by which the presence of type I fimbrial biosynthesis genes interferes with expression of plasmid-encoded fimbriae in S. typhimurium. Consistent with a previous report (Nuccio et al, 2007), deletion of the fimAICDHF genes induced expression of PefA as detected by western blotting (Figure 1A). Deletion of type I fimbrial biosynthesis genes was accompanied by an increase in pefA transcript levels, as determined by quantitative real-time PCR (Figure 1B). Two possible mechanisms could account for the observation that the fimAICDHF genes reduce pefA transcript levels. The first possibility was that the fimAICDHF messenger RNA (mRNA) interfered with pefA transcription or with pefA transcript stability. This scenario seemed unlikely, since there was no precedent for such a regulatory mechanism. The second possible scenario was that expression of the fimbrial proteins encoded by the fimAICDHF operon reduced pefA transcript levels by an unknown mechanism. We reasoned that this mechanism might require signal transduction across the cytoplasmic membrane, because the periplasmic chaperone (FimC), the outer membrane usher (FimD), and the structural subunits (FimA, FimI, FimF and FimH) encoded by fimAICDHF operon are located in the cell envelope.
Figure 1.
SirA and the fimAICDHF operon synergize in repressing PefA expression. (A) Expression of PefA was detected in cell lysates of the indicated strains using western blot. (B) Relative expression of pefA-transcripts was assessed in RNA isolated from the indicated bacterial strains by real-time PCR. Bars represent the average of four independent experiments ±standard error. *P<0.05 (Student’s t-test). (C) Expression of FimA was detected in cell lysates of the indicated strains using western blot. (D) Relative expression of fimA-transcripts was assessed in RNA isolated from the indicated bacterial strains by real-time PCR. Bars represent the average of four independent experiments ±standard error. n.d., not detected. (E) Surface expression of PefA was detected by flow cytometry in the indicated bacterial strains. Bars represent the average of four independent experiments ±standard error. (F) Representative images of PefA expression detected by flow cytometry. wt, S. typhimurium wild type (SR-11); sirA, S. typhimurium sirA mutant (TS23); fimAICDHF, S. typhimurium ΔfimAICDHF mutant (SPN342); fimAICDHF sirA, S. typhimurium sirA ΔfimAICDHF mutant (TS24).
Source data for this figure is available on the online supplementary information page.
When we investigated the possible contribution of two component regulatory systems to this process, we noted that inactivation of sirA had little effect on pefA transcript levels or PefA expression on its own, but a marked increase in PefA expression was observed in a sirA ΔfimAICDHF mutant (TS24) compared to a ΔfimAICDHF mutant (SPN342) (Figure 1A and B). Inactivation of sirA did not substantively alter expression of FimA (Figure 1C and D), suggesting that chromosomally encoded SirA did not exert its effect on pefA expression by altering expression of type I fimbriae. A marked increase in surface assembly of plasmid-encoded fimbriae was detected by flow cytometry with anti-PefA antiserum in the S. typhimurium sirA ΔfimAICDHF mutant (TS24) compared to the sirA mutant (TS23), the ΔfimAICDHF mutant (SPN342) or wild type (SR-11) (Figure 1E and F). These data suggested that suppression of plasmid-encoded fimbriae involved some synergistic interaction between SirA and the presence of the fimAICDHF genes, which provided our first lead for investigating the mechanism by which the presence of type I fimbrial biosynthesis genes interferes with pef expression.
The fim operon cooperates with CsrB and CsrC to repress PefA expression
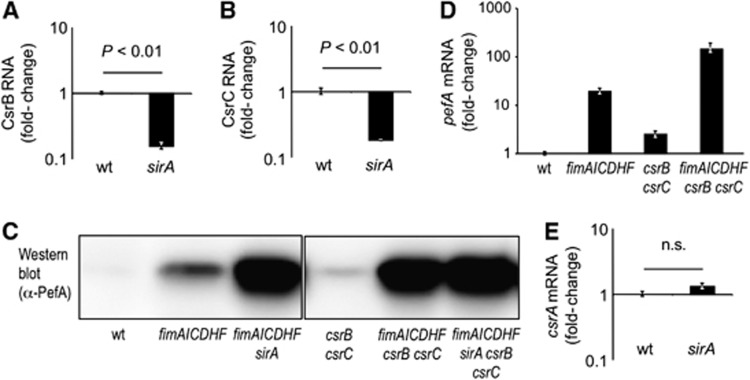
SirA is the response regulator of the BarA/SirA two-component regulatory system in S. typhimurium (Ahmer et al, 1999), which is also known as BarA/UvrY in E. coli. In E. coli and S. typhimurium, SirA is a positive regulator of csrB and csrC, two genes encoding small untranslated RNAs termed CsrB and CsrC (Suzuki et al, 2002; Teplitski et al, 2003; Weilbacher et al, 2003). Consistent with these reports, we found that levels of CsrB and CsrC RNA were significantly reduced in a S. typhimurium sirA mutant (TS23) (Figure 2A and B). We next investigated whether increased PefA expression observed in a sirA ΔfimAICDHF mutant (TS24) was due to reduced expression of CsrB and CsrC RNA. Similar to the enhanced PefA expression observed in a sirA ΔfimAICDHF mutant (TS24) (Figure 1B), we found a profound increase in PefA expression in a csrB csrC ΔfimAICDHF mutant (TS131), while only marginal expression of PefA was detected by western blotting in a csrB csrC mutant (TS130) (Figure 2C). A similar trend was observed when pefA transcripts were quantified by real-time PCR (Figure 2D). No further increase in PefA expression was apparent in a sirA csrB csrC ΔfimAICDHF mutant (TS133) (Figure 2C), thus the lack of SirA or inactivation of the SirA-regulated genes csrB and csrC produced similar effects on PefA expression. These data suggested that the fimAICDHF genes and the small regulatory RNAs CsrB and CsrC synergized in suppressing expression of PefA.
Figure 2.
SirA represses expression of PefA via downregulation of CsrB and CsrC. Expression of CsrB RNA (A) and CsrC RNA (B) was quantified by real-time PCR using RNA isolated from S. typhimurium wild type (wt) and the sirA mutant (sirA). Bars represent the average of four independent experiments ±standard error. (C) Expression of PefA was detected in cell lysates of the indicated strains using western blot. (D) Relative expression of pefA transcripts was assessed in RNA isolated from the indicated bacterial strains by real-time PCR. Bars represent the average of three independent experiments ±standard error. (E) Expression of csrA mRNA was quantified by real-time PCR using RNA isolated from the indicated strains. Bars represent the average of four independent experiments ±standard error. Statistical analysis was performed by a Student’s t-test.
Source data for this figure is available on the online supplementary information page.
CsrA is a positive regulator of plasmid-encoded fimbriae
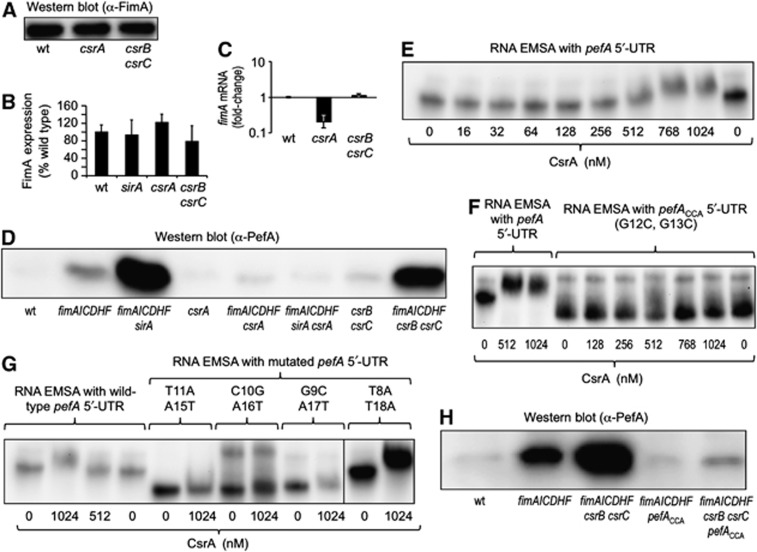
CsrB and CsrC contain multiple high-affinity binding sites for CsrA (carbon storage regulator), a post-transcriptional regulator that binds the 5′-untranslated region (UTR) of mRNAs, thereby either reducing translation (by preventing ribosome binding or enhancing mRNA decay) or enhancing translation (by increasing translation and/or RNA stability) (Wei et al, 2001; Jonas et al, 2008; Bhatt et al, 2009). Binding of CsrB and CsrC sequesters CsrA, thereby repressing the activity of this RNA-binding protein (Liu et al, 1997; Weilbacher et al, 2003). In light of this connection, we next investigated whether CsrA was involved in regulating fimbrial expression in S. typhimurium. Expression of FimA was not markedly changed in a csrA mutant (TS112) or a csrB csrC mutant (TS130) compared to the S. typhimurium wild type (SR-11) (Figure 3A and B), although inactivation of csrA reduced fimA transcript levels modestly (Figure 3C). Interestingly, expression of PefA was abrogated in a csrA ΔfimAICDHF mutant (TS113) compared to the ΔfimAICDHF mutant (SPN342) (Figure 3D). Similarly, the profound PefA expression observed in a sirA ΔfimAICDHF mutant (TS24) was abrogated in a csrA sirA ΔfimAICDHF mutant (TS115). These data suggested that inactivation of sirA and deletion of the fimAICDHF genes increased PefA expression through a mechanism that was fully dependent on the positive regulator CsrA.
Figure 3.
CsrA regulates expression of PefA. (A) Expression of FimA was detected in cell lysates of the indicated S. typhimurium strains using western blot. (B) Quantification of FimA levels in western blots (N=3) by densitometry. (C) Relative expression of fimA mRNA was quantified in RNA isolated from the indicated bacterial strains by real-time PCR. Bars represent the average of three independent experiments ±standard error. (D) Expression of PefA was detected in cell lysates of the indicated S. typhimurium strains using western blot. wt, S. typhimurium wild type (SR-11). (E–G) Electrophoretic mobility shift assays (EMSAs). 3′-end biotinylated pefA 5′-UTR RNA (E), pefACCA 5′-UTR RNA (F) or pefA 5′-UTR RNA mutated at the indicated positions (G) was incubated with the indicated concentrations of CsrA-6xHis dimers. RNA protein complexes were separated on a native 5% TBE gel to perform EMSA. (H) Expression of PefA was detected in cell lysates of the indicated S. typhimurium strains using western blot.
Source data for this figure is available on the online supplementary information page.
CsrA induces expression of plasmid-encoded fimbriae by binding the pefA 5′-UTR
Since the substantial PefA expression observed in a sirA ΔfimAICDHF mutant was CsrA dependent (Figure 3D), we wanted to study the mechanism by which CsrA activates expression of plasmid-encoded fimbriae. We reasoned that this regulatory effect could be either indirect or a direct consequence of CsrA binding to the 5′-UTR of the pefA transcript. We performed 5′-rapid amplification of cDNA ends (5′-RACE) to determine the transcriptional start site of the pefACDEF transcript, which was located 76 bp upstream of the pefA start codon (RefSeq accession NC_003277.1 coordinate 14163). We next synthesized a biotinylated transcript containing nucleotides +1 to +148 relative to the transcriptional start site using in vitro transcription and investigated binding of CsrA to this transcript employing an RNA electrophoretic mobility shift assay (EMSA). When the biotinylated pefA 5′-UTR transcript was incubated with increasing concentrations of CsrA-6xHis, we observed a mobility shift at a CsrA-6xHis dimer concentration of 512 nM (Figure 3E). CsrA binds to a conserved sequence present in CsrB RNA, CsrC RNA and in the 5′-UTR of its target mRNAs, which contains a central GGA motif that is 100% conserved (Dubey et al, 2005; Mercante et al, 2009). Inspection of the pefA 5′-UTR nucleotide sequence revealed a single site (nucleotides 9–16, GCTGGAAA) with conserved GGA motif but otherwise marginal similarity to the proposed CsrA consensus sequence (ACAGGATG).
To determine whether this potential CsrA-binding site was responsible for the observed mobility shift, we generated a transcript (pefACCA 5′-UTR) in which the conserved GGA motif was mutagenized to CCA. No mobility shift was observed when the biotinylated pefACCA 5′-UTR transcript was incubated with increasing concentrations of CsrA-6xHis (Figure 3F). These data suggested that CsrA bound specifically to a single GGA binding site in the pefA 5′-UTR. To further map the CsrA-binding site in the pefA 5′-UTR, we mutated nucleotides directly adjacent to the GGA motif (T11A, A15T) or in a distance of one nucleotide (C10G, A16T) or two nucleotides (G9C, A17T) from the GGA motif. An altered mobility suggested that the respective mutations altered the secondary structure of the resulting biotinylated constructs. Importantly, no electrophoretic shift was observed when these biotinylated constructs were incubated with increasing concentrations of CsrA-6xHis (Figure 3G). In contrast, mutation of nucleotides located in a distance of three bases from the GGA motif (T8A, T18A) did no longer prevent an electrophoretic shift with CsrA-6xHis.
To further investigate whether binding of CsrA to the GGA motif in the pefA 5′-UTR was required for expression of plasmid-encoded fimbriae, we introduced point mutations into the S. typhimurium genome to change the central GGA motif in the pefA 5′-UTR to CCA. The resulting pefACCA mutation was introduced into a ΔfimAICDHF mutant and a ΔfimAICDHF csrB csrC mutant and PefA expression was determined by western blotting. Introduction of a pefACCA mutation into a ΔfimAICDHF mutant (TS139) or a ΔfimAICDHF csrB csrC mutant (TS 140) markedly lowered PefA protein levels (Figure 3H). Collectively, these data suggested that CsrA activates expression of plasmid-encoded fimbriae by binding a GGA motif in the 5′-UTR of the pefACDEF transcript.
The 5′-UTR of the fimAICDHF transcript is sufficient for suppressing PefA expression
We next investigated the mechanism by which SirA and the fimAICDHF genes suppressed the CsrA-mediated activation of PefA expression. Inactivation of sirA did not alter the transcript levels of csrA (Figure 2E), but lowered the levels of CsrB RNA (Figure 2A) and CsrC RNA (Figure 2B), two known inhibitors of CsrA activity (Suzuki et al, 2002; Weilbacher et al, 2003). These data were consistent with the idea that the mechanism by which SirA suppressed CsrA activity was by increasing the levels of the small untranslated RNAs CsrB and CsrC, which contain multiple high-affinity binding sites for CsrA and sequester this RNA binding protein (Suzuki et al, 2002; Weilbacher et al, 2003). However, the mechanism by which the fimAICDHF genes suppressed CsrA activity remained obscure.
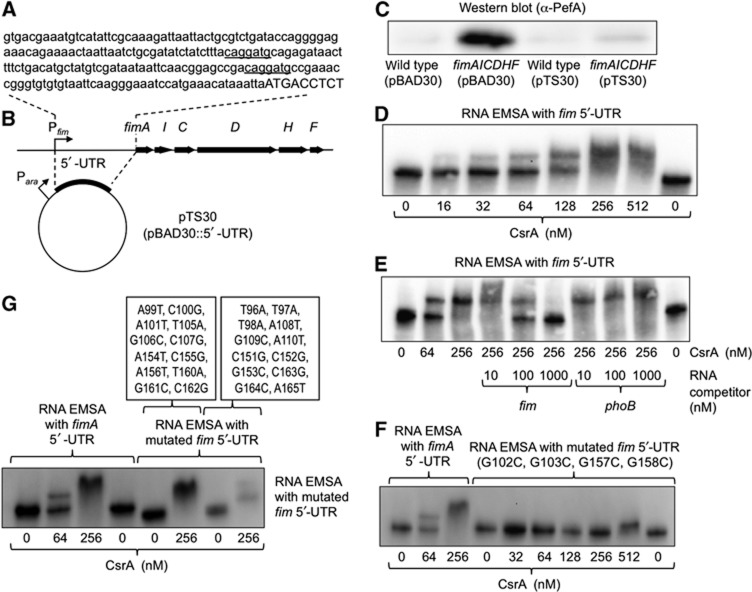
As mentioned above, one possibility was that the fimAICDHF mRNA was responsible for the observed suppression of PefA expression (Figure 1A). Interestingly, we noticed that the 5′-UTR of the fimAICDHF transcript contained two potential CsrA-binding sites that matched the proposed consensus sequence (ACAGGAUG, Figure 4A). To investigate whether the 5′-UTR of the fimAICDHF transcript was sufficient for preventing PefA expression, we cloned the corresponding DNA region in the vector pBAD30 under the control of the E. coli arabinose operon promoter (Figure 4B). The resulting plasmid (pTS30) or the vector control (pBAD30) was introduced into wild-type S. typhimurium (SR-11) or the ΔfimAICDHF mutant (SPN342) and expression of PefA detected by western blotting after growing bacteria in the presence of the inducer arabinose. Expression of the 5′-UTR of the fimAICDHF transcript fully suppressed PefA expression in the S. typhimurium ΔfimAICDHF mutant (SPN342) (Figure 4C). In contrast, introduction of the vector control (pBAD30) did not prevent expression of PefA in the ΔfimAICDHF mutant (SPN342). These data provided compelling evidence that proteins encoded by the fimAICDHF operon were not required for suppression of PefA expression. Instead, our data raised the surprising possibility that the 5′-UTR of a chromosomally encoded mRNA (i.e., the fimAICDHF transcript) could regulate the expression of a gene product (i.e., PefA) encoded on the virulence plasmid.
Figure 4.
The 5′-UTR of the fimA transcript suppresses PefA expression and binds to CsrA. (A) Sequence of the 5′-UTR of the fimA transcript according to Kroger et al (2012). Predicted CsrA-binding sites are underlined. Capital letters indicate the start of the pefA open reading frame. (B) Schematic depiction of the fimAICDHF operon and the cloning strategy for overexpressing the 5′-UTR of the fimA transcript. (C) Expression of PefA was detected in cell lysates of the indicated strains using western blot. Bacteria were grown for 48 h statically in the presence of 0.2% arabinose and carbenicillin. (D–G) Electrophoretic mobility shift assays (EMSAs). RNA protein complexes were separated on a native 5% TBE gel to perform EMSAs. 3′-end biotinylated fimA 5′-UTR RNA was incubated with the indicated concentrations of CsrA-6xHis dimers in the absence (D) or presence (E) of unlabelled competitor RNA. The concentrations of specific (fim) or non-specific (phoB) competitor RNA are indicated on the bottom of each lane. (F, G) 3′-end biotinylated fimA 5′-UTR RNA mutated at the indicated positions was incubated with the indicated concentrations of CsrA-6xHis dimers.
Source data for this figure is available on the online supplementary information page.
CsrA binds the 5′-UTR of the fimAICDHF transcript
To further investigate the mechanism by which the fimAICDHF genes suppress PefA expression, we determined whether CsrA can bind to the 5′-UTR of the fimAICDHF transcript by an EMSA. A biotinylated transcript containing nucleotides +1 to +264 relative to the published start site of the fimAICDHF transcript (Kroger et al, 2012) was synthesized by in vitro transcription. When this biotinylated 5′-UTR transcript was incubated with increasing concentrations of purified His-tagged CsrA protein (CsrA-6xHis) (Mercante et al, 2006), we observed a partial mobility shift starting at a CsrA-6xHis dimer concentration of 16 nM, whereas a complete mobility shift was observed at a CsrA-6xHis dimer concentration of 256 nM (Figure 4D). To verify the specificity of the binding reaction, 256 nM CsrA-6xHis dimer was incubated with biotinylated in vitro transcribed 5′-UTR of the fimAICDHF transcript in the presence of increasing concentrations of unlabelled in vitro transcribed 5′-UTR of the fimAICDHF transcript. The unlabelled in vitro transcribed 5′-UTR of the fimAICDHF transcript was able to compete for CsrA binding as indicated by the disappearance of the mobility shift at higher concentrations of the competitor (Figure 4E). In contrast, competition for CsrA binding with a non-specific competitor (an in vitro transcribed fragment of the phoB gene) (Martinez et al, 2011) did not result in the disappearance of the mobility shift. These results suggested that CsrA specifically binds the 5′-UTR of the fimAICDHF transcript.
To determine whether the potential CsrA-binding sites were responsible for the observed mobility shift, we generated a transcript in which both GGA motifs were mutagenized to CCA (G102C, G103C, G157C, G158C). No mobility shift was observed when the mutated (G102C, G103C, G157C, G158C) biotinylated 5′-UTR transcript of the fimAICDHF transcript was incubated with increasing concentrations of CsrA-6xHis (Figure 4F). These data suggested that CsrA bound specifically to GGA binding sites in the 5′-UTR of the fimAICDHF transcript. To further map the CsrA-binding sites in the 5′-UTR of the fimAICDHF transcript, we mutated nucleotides directly adjacent to each GGA motif (A99T, C100G, A101T, T105A, G106C, C107G, A154T, C155G, A156T, T160A, G161C, C162G). An electrophoretic shift was observed when the resulting biotinylated construct was incubated with increasing concentrations of CsrA-6xHis (Figure 4G). Finally, we mutated nucleotides in a distance of three nucleotides from each GGA motif (T96A, T97A, T98A, A108T, G109C, A110T, C151G, C152G, G153C, C163G, G164C, A165T). Mutation of nucleotides located in a distance of three bases from the GGA motif did not prevent an electrophoretic shift with CsrA-6xHis (Figure 4G).
The 5′-UTR of the fimAICDHF transcript antagonizes the regulatory effects of CsrA
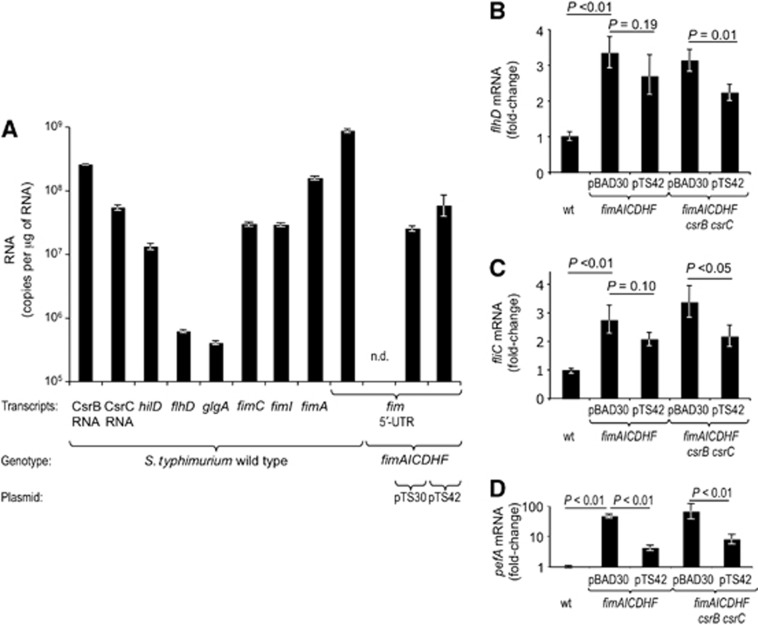
The small untranslated RNAs CsrB and CsrC antagonize the regulatory effects of CsrA presumably by sequestering this RNA binding protein. Our finding that CsrA binds the 5′-UTR of the fimAICDHF transcript (Figure 4D and E) raised the possibility that the fimAICDHF genes antagonized the regulatory effects of CsrA by a similar mechanism. CsrB and CsrC are highly abundant in the cell, which explains in part why these small untranslated RNAs can antagonize CsrA activity. However, no antagonistic activity has been ascribed to mRNA targets of CsrA, presumably because these mRNAs are expressed at considerably lower levels than CsrB RNA and CsrC RNA. We thus compared expression levels of the 5′-UTR of the fimAICDHF transcript with those of CsrB RNA, CsrC RNA and the mRNAs of known CsrA targets (hilD, flhD and glgA) by quantitative real-time PCR (Yang et al, 1996; Altier et al, 2000; Jackson et al, 2002; Teplitski et al, 2003; Teplitski et al, 2006; Jonas et al, 2010; Yakhnin et al, 2013). As expected, the levels of CsrB RNA and CsrC RNA were ∼10- to 100-fold higher than transcript levels of hilD, flhD or glgA (Figure 5A). However, the levels of the fimAICDHF 5′-UTR were not only higher than those of other target mRNAs (i.e., hilD, flhD or glgA), but exceeded even the levels of the highly abundant CsrB RNA and CsrC RNA. Transcript levels of fimA, fimI and fimC were lower than that of the fimAICDHF 5′-UTR.
Figure 5.
The 5′-UTR of the fimA transcript regulates gene expression. RNA was isolated from the indicated bacterial strains grown in LB5.5. (A) Absolute copy numbers of the indicated transcript per microgram of total RNA were determined by quantitative real-time PCR for the indicated transcripts. (B–D) Relative changes in transcript levels were determined by quantitative real-time PCR for the indicated transcripts. Bars represent geometric means±standard error from three (A) or six (B–D) different experiments. Statistical analysis was performed by a Student’s t-test and statistical significance of differences is indicated by brackets. ND, none detected.
We next investigated whether expression of the fimAICDHF transcript could alter expression of other genes whose transcripts are controlled by CsrA. CsrA stabilizes the flhDC transcript (Jonas et al, 2010), which encodes a master regulator (FlhDC) of the flagellar regulon. FlhDC controls expression of over 90 genes in S. typhimurium (Frye et al, 2006), including fliC, which encodes flagellin. Compared to the S. typhimurium wild type, flhD and fliC transcript levels were significantly increased in a S. typhimurium ΔfimAICDHF mutant (Figure 5B and C). However, introduction of a plasmid encoding the fim 5′-UTR (pTS30) did not lead to a reduction in flhD and fliC transcript levels (data not shown). We noted that expression levels of the fimA 5′-UTR were considerably higher in the S. typhimurium wild type than in a S. typhimurium ΔfimAICDHF mutant expressing the fimA 5′-UTR from plasmid pTS30 (Figure 5A), which might explain the lack of complementation. As a follow-up to this experiment, we constructed a second plasmid (pTS42) in which the E. coli arabinose operon promoter drove expression of the fimA and fimC genes in addition to the fimA 5′-UTR, as the UTR may be less stable when expressed alone. Although introducing pTS42 into a S. typhimurium ΔfimAICDHF mutant increased the fimA 5′-UTR transcript levels compared to a strain carrying pST30, they still remained below those observed in the S. typhimurium wild type (Figure 5A). Introduction of pTS42 into the ΔfimAICDHF mutant did not significantly alter the flhD and fliC transcript levels (Figure 5B and C). In contrast, introduction of pTS42 significantly (P<0.01) reduced expression of pefA in a ΔfimAICDHF mutant (Figure 5D), suggesting that expression levels of the fimA 5′-UTR below that of the wild type were sufficient to partially restore repression of plasmid-encoded fimbriae (Figure 5D).
Assuming the fim message functions in sequestering CsrA, we reasoned that a partial complementation of flhD and/or fliC repression might be observed even with plasmids yielding relatively low levels of fimA 5′-UTR provided that small RNAs with redundant function would be removed. This is particularly relevant, because CsrA activates transcription of csrB/C and represses CsrB/C turnover through a negative feedback loop (Weilbacher et al, 2003; Suzuki et al, 2006), a mechanism that might also partly compensate for loss of the fim message. We thus introduced plasmid pTS42 or a vector control (pBAD30) into a S. typhimurium ΔfimAICDHF csrB csrC mutant and determined flhD and fliC transcript levels. Introduction of pTS42 significantly (P<0.05) reduced flhD and fliC transcript levels in the ΔfimAICDHF csrB csrC mutant (Figure 5B and C). These data suggested that the increased flhD and fliC expression resulting from deletion of the fimAICDHF operon could be restored, at least in part, by restoring intermediate expression levels of fimA 5′-UTR in the cell.
The CsrA-binding site in the pefA 5′-UTR controls expression of plasmid-encoded fimbriae in vivo
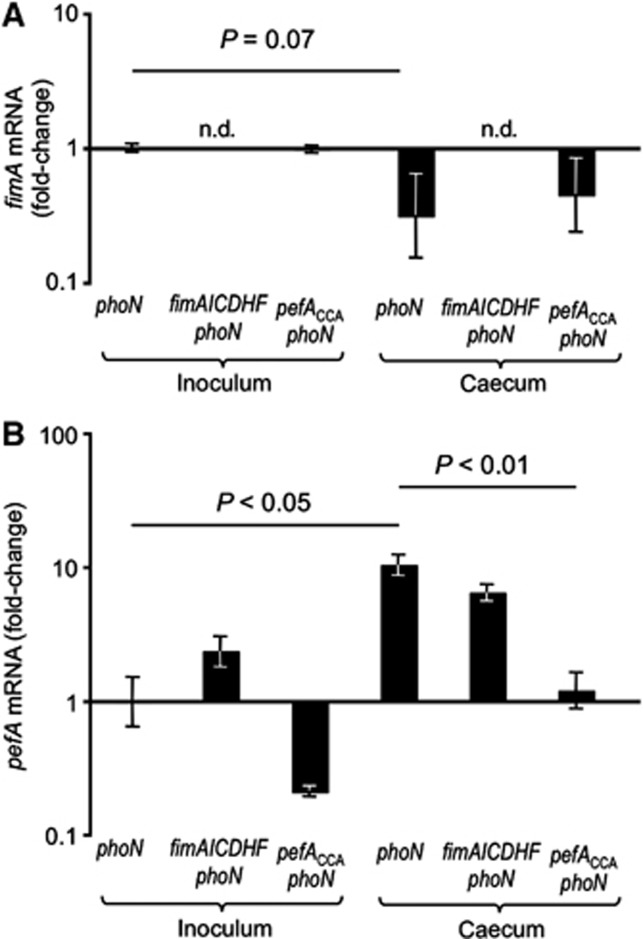
A possible biological function for the hierarchical control of fimbrial expression would be to ensure that S. typhimurium does not express plasmid-encoded fimbriae while residing outside a host. To test this assumption, we investigated expression of the fim and pef operons in the mouse colitis model. Mice were preconditioned with streptomycin and inoculated with a S. typhimurium wild-type strain that was marked by a phoN::Cm insertion to facilitate recovery (TS141). At 72 h after infection, organs were collected to determine bacterial gene expression in the caecum by quantitative real-time PCR. Similar numbers of bacteria were recovered from caecal contents of all groups (data not shown). Compared to expression levels in the inoculum, mRNA levels of fimA were reduced in the phoN mutant (TS141) recovered from the caecum (Figure 6A). In contrast, pefA transcript levels were significantly (P<0.05) higher in RNA recovered from caecal contents than from the inoculum of the phoN mutant (TS141) (Figure 6B). As expected, the transcript levels of pefA were elevated in an in vitro culture of a ΔfimAICDHF phoN mutant (TS142) compared to a phoN mutant. However, both strains exhibited similar pefA transcript levels in vivo.
Figure 6.
The CsrA-binding site in the 5′-UTR of the pefA transcript limits expression to host environments. Streptomycin pretreated mice were inoculated with the indicated S. typhimurium mutants and RNA was isolated from the inoculum cultures (inoculum) and from caecal washes collected 72 h after infection (caecum). Transcript levels of fimA (A) and pefA (B) relative to the respective mRNA levels present in the inoculum culture of the S. typhimurium phoN mutant were determined by quantitative real-time PCR. Bars represent geometric means±standard error from at least four different animals. Statistical analysis was performed by a Student’s t-test. n.d., no transcripts detected.
To determine whether increased pefA expression in vivo was dependent on the CsrA-binding site in the pefA 5′-UTR, mice were infected with a pefACCA phoN mutant (TS147). Importantly, pefA expression in the caecum was significantly (P<0.01) lower in the pefACCA phoN mutant compared to a phoN mutant, supporting the idea that the regulatory mechanism proposed here was operational in vivo and contributed to increased expression of plasmid-encoded fimbriae in the murine caecum. In summary, these data support a model in which CsrA-mediated hierarchical control of fimbrial expression limits expression of plasmid-encoded fimbriae to host environments (Figure 7).
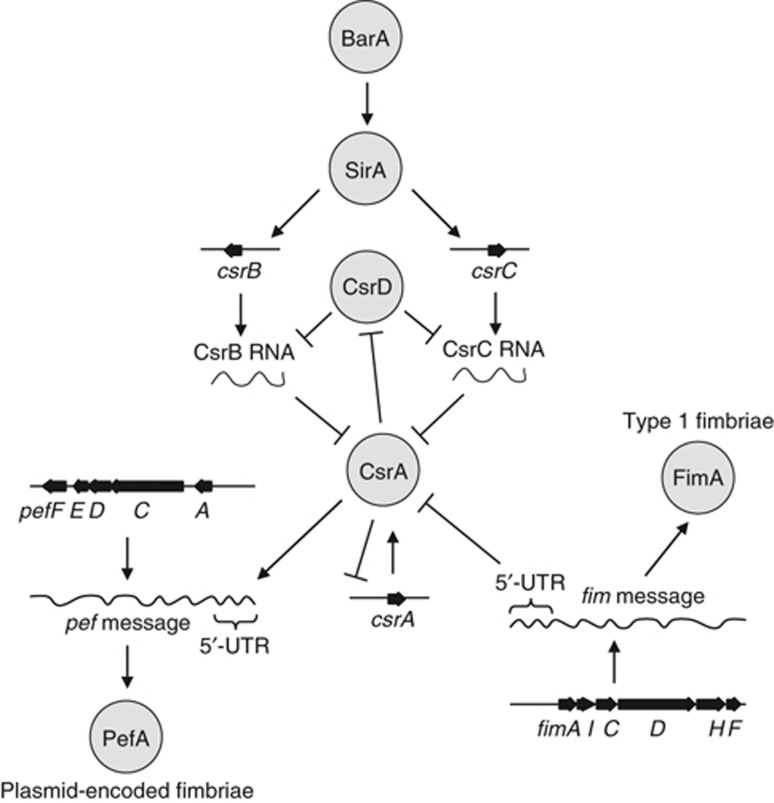
Figure 7.
Proposed model for the hierarchical control of fimbrial expression in S. typhimurium mediated by CsrA. The exceptionally high transcript levels of the fimAICDHF 5′-UTR can antagonize the regulatory effects of CsrA by binding and sequestering this RNA binding protein through a mechanism similar to that proposed for CsrB RNA and CsrC RNA. Via this mechanism, the fimAICDHF mRNA synergizes with CsrB RNA and CsrC RNA in antagonizing the activity of CsrA, a regulatory protein that activates expression of plasmid-encoded fimbriae by binding a GGA motif in the 5′-UTR of pefACDEF transcript. This mechanism is ultimately responsible for the hierarchical control of type I fimbriae and plasmid-encoded fimbriae in S. typhimurium.
Discussion
Small untranslated RNAs are important regulators of bacterial gene expression (Romeo, 1998; Vogel and Luisi, 2011; Romeo et al, 2012; Heroven et al, 2012). The activity of CsrA, a global post-transcriptional regulator, is antagonized by two small untranslated RNAs, termed CsrB and CsrC. CsrB and CsrC possess high-affinity CsrA-binding sites (Altier et al, 2000; Fortune et al, 2006; Martinez et al, 2011) that enable these small untranslated RNAs to bind to CsrA, thereby repressing its activity (Liu et al, 1997; Weilbacher et al, 2003). CsrA molecules that are not sequestered by CsrB or CsrC can post-transcriptionally regulate gene expression by binding to their mRNA targets, thereby preventing ribosome binding, enhancing mRNA decay or increasing mRNA stability (Wei et al, 2001; Jonas et al, 2008; Bhatt et al, 2009). CsrA was initially identified as a negative regulator of glycogen synthesis encoded by the glgCAY operon in E. coli (Romeo et al, 1993), but it is now clear that this post-transcriptional regulator controls a wide range of properties related to metabolism and virulence (Edwards et al, 2011; Romeo et al, 2012). For example, in S. typhimurium and E. coli, CsrA is a positive regulator of motility because it stabilizes the transcript encoding FlhC and FlhD, the two subunits of the master regulator of flagella expression (Wei et al, 2001; Lawhon et al, 2003). Furthermore, CsrA regulates expression of the invasion-associated type III secretion system of S. typhimurium via post-transcriptional regulation of HilD (Altier et al, 2000; Fortune et al, 2006). CsrA also regulates expression of eight diguanylate cyclases (GGDEF-domain containing proteins) and phosphodiesterases (EAL-domain containing proteins) in S. typhimurium, which control the switch between a motile and a sessile state by regulating cyclic di-guanylate (c-di-GMP) metabolism (Jonas et al, 2010). Here, we show that CsrA is a positive regulator of PefA expression in S. typhimurium by binding a GGA motif in the 5′-UTR of the pefACDEF transcript (Figure 3).
It is uncommon that an mRNA molecule, independently of its encoded gene product, can alter the activity of a global regulatory protein, thereby changing protein expression in bacteria. While the binding of small untranslated RNAs to CsrA can regulate the activity of this RNA-binding protein, binding of CsrA to mRNAs generally functions in regulating expression of the encoded proteins rather than regulating CsrA activity (Romeo, 1998). However, a regulatory effect of an mRNA has recently been described that is based on the competition for binding of a small untranslated RNA (Figueroa-Bossi et al, 2009). Based on this observation, it has been proposed that some mRNAs are regulated by small untranslated RNAs but have no effect on their availability while other mRNAs may act only as decoys that regulate the availability of a small untranslated RNA but are not subject to regulation by this small untranslated RNA (Mandin and Gottesman, 2009). Similarly, binding of CsrA to the 5′-UTR of the fimAICDHF transcript had little effect on expression of FimA (Figure 3A and B), suggesting that the main function for the binding of CsrA to the 5′-UTR of the fimAICDHF transcript was not to regulate expression of the encoded type I fimbrial proteins. Instead, our data raised the intriguing possibility that binding of the fimAICDHF mRNA to CsrA functioned mainly in regulating CsrA activity.
The surprising regulatory activity of the 5′-UTR of the fimAICDHF transcript might be explained in part by its high abundance in the cell. For example, the fimA 5′-UTR was present in ∼100-fold to 1000-fold greater abundance than hilD, flhD and glgA mRNAs (Figure 5), which are known targets of CsrA (Yang et al, 1996; Altier et al, 2000; Jackson et al, 2002; Teplitski et al, 2003; Teplitski et al, 2006; Jonas et al, 2010). The high abundance of the 5′-UTR of the fimAICDHF transcript was a feature shared with CsrB RNA and CsrC RNA (Figure 5). It thus seems likely that the 5′-UTR of the fimAICDHF transcript sequesters CsrA by a mechanism similar to that proposed for CsrB RNA and CsrC RNA (Liu et al, 1997; Weilbacher et al, 2003). Our model for the hierarchical control of fimbrial gene expression mediated by CsrA (Figure 7) provides a plausible explanation for the observation that the presence of the fimAICDHF transcript synergized with CsrB RNA and CsrC RNA to suppress expression of plasmid-encoded fimbriae in S. typhimurium (Figure 2). Transcript levels of pefA were more sensitive to sequestration of CsrA by the 5′-UTR of the fimAICDHF transcript (Figure 5D) than transcript levels of flhD (Figure 5B). This may be explained by the fact that the 5′-UTR of the fimAICDHF transcript binds CsrA with a higher affinity than the 5′-UTR of the pefACDEF transcript. As a result, the fimAICDHF transcript is very effective in sequestering CsrA away from the pefACDEF transcript. In contrast, CsrA binding of mRNAs that have a similar or higher affinity than the 5′-UTR of the fimAICDHF transcript (e.g., the flhD mRNA) will be affected to a lesser extent by fluctuations in fimAICDHF-transcript levels. Thus, differences in the affinities of different mRNA targets for CsrA are predicted to determine the magnitude by which expression of each target is regulated by the 5′-UTR of the fimAICDHF transcript.
Materials and methods
Bacterial strains and growth conditions
The bacterial strains used in this study are listed in Supplementary Table S1. Unless otherwise noted, cultures of S. typhimurium and E. coli were routinely incubated aerobically at 37°C in Lysogeny broth (LB) (per liter: 10 g tryptone, 5 g yeast extract, 10 g NaCl) or on LB agar plates (LB containing 1.5% Difco agar) overnight. Sucrose selection was performed aerobically on 5% sucrose agar (SA) (per liter: 50 g sucrose, 8 g nutrient broth, 15 g Bacto agar) at 30°C (Lawes and Maloy, 1995). Antibiotics were added at the following concentrations (mg/l) as needed: chloramphenicol (Cm), 30; kanamycin (Km), 100; nalidixic acid (Nal), 50; tetracycline (Tc), 20; gentamicin (Gm), 10; spectinomycin (Spec), 100. For fimbrial expression, strains were grown statically in 50 ml conical tubes in 5 ml LB buffered to pH 5.5 with 100 mM 2-(N-morpholino)ethanesulphonic acid (MES) (LB5.5). A sterile pipette tip was routinely added to standing cultures to support Pef expression as previously observed (Nuccio et al, 2007).
Plasmid construction
Chemically competent E. coli was prepared as previously described (Maniatis et al, 1989). To generate unmarked fim operon deletions, the unmarked Δfim cassette within pSF2 was relocated within pRDH10 to the tetC gene as pSF2 did not confer a sucrose-sensitive phenotype to S. typhimurium cells harbouring it. To achieve this, pSF2 was digested with EcoRI (New England Biolabs), the ΔfimAICDHF cassette gel purified (Qiagen Qiaex II kit), blunted (NEB Quick Blunting kit), purified (Qiagen MinElute kit) and then ligated (NEB Quick Ligation kit) to BamHI-linearized, blunted, purified and phosphatase-treated (NEB Antarctic Phosphatase) pRDH10. The ligation was transformed into chemically competent CC118 λpir cells and the cells then plated on LB+Cm agar. Colonies were screened for Tc sensitivity on LB+Tc agar. A plasmid containing a single ΔfimAICDHF cassette (determined by plasmid linearization with PstI) in a blunted BamHI site (negative for BamHI digestion and negative for Tc resistance) was labelled as pSPN22.
A standard for measuring fliC mRNA expression was generated by PCR amplifying (Supplementary Table S2) the fliC open reading frame of ATCC 14028 and then cloning it into pCR2.1 with the TOPO TA cloning kit (Invitrogen). The ligation mixture was transformed into TOP10 cells and plated on LB+Km agar. Clones were sequenced with M13 universal primers by SeqWright (Houston, TX) and a plasmid with an accurate fliC sequence was designated as pSPN31. Standards for hilD, csrA, csrB, csrC, glgA and flhD were generated in a similar fashion by PCR amplifying (Supplementary Table S2) fragments of the respective genes and cloning into pCR2.1 using the TOPO TA cloning kit to yield plasmids pTS33 through pTS38, respectively. Primers incorporating a T7 promoter (Supplementary Table S2) were similarly used to generate pCR2.1-cloned RNA transcription templates for the fimA 5′-UTR (pTS31), the wild-type pefA 5′-UTR (pTS32), pefACCA 5′-UTR (pTS48) and other mutations in the pefA 5′-UTR (pTS54-57). To generate pCR2.1-cloned RNA transcription templates for the fimA 5′-UTR containing mutations (pTS51-51), overlapping primers containing the desired mutation were used for PCR together with the distal primers used for the construction of pTS31. The two resulting PCR products were mixed in an equimolar ratio and used as a PCR template with the two distal primers. The resulting PCR product was then cloned into pCR2.1 or, to introduce additional mutations, used as a PCR template as described above.
The 5′-UTR of the fimAICDHF operon was PCR amplified (Supplementary Table S2) and the resulting PCR product was cloned into pCR2.1 using the TOPO TA cloning kit. After sequencing with M13 universal primers, the insert was subcloned into pBAD30 using the unique EcoRI and XbaI restriction sites. The resulting plasmid was designated as pTS30.
To generate a construct expressing the 5′-UTR of the fimAICDHF operon along with FimA and FimC, a DNA region encompassing the fimA 5′-UTR, the fimA gene as well as the fimC gene were PCR amplified (Supplementary Table S2). The resulting PCR products were cloned into pCR2.1 using the TOPO TA cloning kit to generate pTS39 and pTS40, respectively. After verification of the sequence, the insert of pTS39 was subcloned into pBAD30 using the unique EcoRI and XbaI restriction sites to generate pTS41. The insert of pTS40 was then subcloned into pTS41 using the XbaI and SalI restriction sites to generate pTS42.
To generate a mutant containing an altered CsrA-binding site in the 5′-UTR of the pefA transcript, a region upstream and a region downstream of the predicted CsrA-binding site were PCR amplified (Supplementary Table S2). The resulting PCR products were gel purified, digested with SacI and ligated. The resulting ligation mixture served as a template for a PCR with the distal primer of each region’s amplification relative to the CsrA-binding site. The resulting PCR was cloned into pCR2.1 using the TOPO TA cloning kit, yielding pTS43. Following sequence confirmation, the insert was subcloned into pGP704 using the unique XbaI and EcoRI sites, giving rise to pTS44. pTS45 was then generated by introducing the kanamycin-resistance cassette from pKSAC (Pharmacia) into the SacI site of pTS44.
To generate pTS47, which contains the mutated CsrA-binding site in the 5′-UTR of the pefA transcript and the same flanking regions as pTS45, the fragments were amplified employing the primers listed in Supplementary Table S2. The overlapping primers proximal to the CsrA-binding site contained point mutations that yielded the desired mutation. The resulting PCR products were gel purified and an equimolar mixture served as a template for PCR with the distal primer from each region. The resulting PCR product was cloned into pCR2.1 with the TOPO TA cloning kit and sequenced to verify the mutation, yielding pTS46. The insert was then subcloned into pRDH10 using the unique BamHI and SacI restriction sites to generate pTS47.
Construction of S. typhimurium mutants
To generate an unmarked deletion of the fimAICDHF operon, E. coli S17-1 λpir was transformed with the suicide plasmid pSPN22 and then conjugated with EHW2 (IR715 ΔfimAICDHF::KSAC). S. typhimurium transconjugants containing pSPN22 integrated into the chromosome were selected for by plating on LB+Cm+Nal agar plates. Colonies from these plates were then plated on SA to select for bacteria that had recombined out and titrated pSPN22. Sucrose-resistant colonies were then screened on LB+Cm and LB+Km agar plates. A colony sensitive to both antibiotics and confirmed by PCR (Supplementary Table S2) to contain an unmarked ΔfimAICDHF locus and to be missing fimA was dubbed SPN192. To transduce the unmarked fim deletion of SPN192 into SR-11, a method similar to that of Kang et al (2002) was employed. Integration of pSPN22 into SPN192 was achieved by conjugation as described above, resulting in a merodiploid ΔfimAICDHF state flanking the integrated plasmid pSPN22, which is maintained by selection for the plasmid’s Cm-resistance cassette. A Cm-resistant transconjugant negative for the PCR product from unmarked ΔfimAICDHF amplification was labelled as SPN227 (ΔfimAICDHF::pSPN22). The ΔfimAICDHF::pSPN22 locus was then transduced into SR-11 with phage P22 HT105/1 int-201, selecting for Cm resistance. As SR-11 is resistant to P22-mediated lysis, purification from phage was not required and transductants were plated on SA to select for the loss of pSPN22. A Cm-sensitive colony positive for unmarked ΔfimAICDHF amplification and negative for fimA amplification was called SPN342 (SR-11 ΔfimAICDHF).
Insertion mutants in sirA were generated by transducing the kanamycin-resistance cassette from BA736 into the respective target strains by P22-mediated transduction. Insertion mutants of csrA, csrB and csrC were constructed by linear transformation as described previously (Penrod and Roth, 2006). PCR products were amplified with primers listed in Supplementary Table S2. The primers contained a stretch of ∼40 bp homologous to the intended insertion site in the genome of S. typhimurium and a universal linker sequence used to amplify the respective resistance cassette (chloramphenicol-resistance cassette for csrA, spectinomycin-resistance cassette for csrB, and gentamicin-resistance cassette for csrC). The resulting PCR products were recombined into S. typhimurium LT2 using linear transformation mediated by the Red functions of phage lambda (Datsenko and Wanner, 2000). The constructed mutations were transduced by phage P22 into strain TT10674. The respective mutations in csrA, csrB or csrC were then transduced into the respective strains as indicated in Supplementary Table S1 by phage P22 and selection for the respective antibiotic resistance. The presence or absence of the respective resistance cassettes in csrA, csrB or csrC was confirmed by PCR with primers listed in Supplementary Table S2.
To generate a point mutant in the CsrA binding sequence of the pefA 5′-UTR, E. coli S17-1 λpir was transformed with pTS45 and then conjugated with S. typhimurium IR715. Transconjugants were selected for by plating on LB+Nal+Kan agar. Colonies were screened on LB+Carb for being Carb sensitive to locate transconjugants that were the product of a double crossover. The resulting locus containing the kanamycin-resistance cassette was then transduced into S. typhimurium SR11, generating TS137. E. coli S17-1 λpir was then transformed with pTS47 and conjugated with TS137. Single-crossover transconjugants were selected for by plating on LB+Kan+Cm agar plates. Colonies were then grown on SA plates to select for the loss of pTS45, resulting in colonies that had either reverted to the same genotype as TS137 (evidenced by retention of the kanamycin-resistance cassette), or had incorporated the pTS45 region with the pefA 5′-UTR CsrA-binding site mutation (loss of kanamycin-resistance cassette). To isolate cells having undergone the latter event, colonies were screened for sensitivity to both kanamycin and chloramphenicol. The pefA 5′-UTR was then amplified with primers (Supplementary Table S2) and subcloned into pCR2.1 using the TOPO TA cloning kit. The insert was sequenced to verify that the CsrA-binding site of the pefA 5′-UTR contained the mutated sequence.
Western blotting
Polyclonal rabbit anti-PefA and anti-FimA antiserum (Humphries et al, 2003) were diluted 1:5 in phosphate-buffered saline (PBS) supplemented with 0.2% sodium azide and preabsorbed (Gruber and Zingales, 1995) eight times with ORN172 carrying plasmid pGEX-4T-2 and four times with ADH19 (anti-PefA) or SPN342 (anti-FimA). Unless otherwise noted, cultures were resuspended to 2 × 108 CFU/20 μl in sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) loading buffer containing 0.1% glycine/HCl pH 2.2 and boiled for 5 min. Afterwards, samples were neutralized by adding 1 M Tris–HCl pH 7.1 to a final concentration of 100 mM. In all, 20 μl was loaded onto a 15% SDS–PAGE gel, and, after electrophoresis, proteins were transferred onto an Immobilon-P membrane (Millipore) using a Trans-Blot SD semi-dry electrophoretic transfer unit (Bio-Rad). Blots were incubated with preabsorbed antiserum diluted 1:500, detected with a goat anti-rabbit IgG antibody conjugated to horseradish peroxidase and the Immun Star chemiluminescent substrate (Bio-Rad), followed by visualization with a BiospectrumAC Imaging System (UVP). Blot density quantification was performed with the Labworks Image Acquisition and Analysis Software (UVP version 4.6.00.0).
Bacterial RNA isolation and real-time PCR
Bacterial RNA from in vitro cultures was isolated using the Aurum Total RNA Kit (Bio-Rad) according to the manufacturer’s recommendations. An additional DNase treatment was performed using the Ambion DNA removal and inactivation kit. RNA from caecal content was essentially isolated as described previously (Lopez et al, 2012) except that purified RNA was subjected to a second purification using the RNeasy MinElute Cleanup kit (Qiagen).
Reverse-transcription PCR and real-time PCR were performed as described previously (Thiennimitr et al, 2011) with primers listed in Supplementary Table S2. Transcription of the target genes in each sample originating from in vitro cultures was normalized to the respective levels of guanyl nucleotide kinase mRNA, encoded by the gmk gene. RNA originating from caecal contents was normalized to the respective levels of S. typhimurium-specific 16S rRNA using oligonucleotides specific to the gene encoding for S. typhimurium 16S rRNA. Absolute transcript numbers for the fimA 5′-UTR were obtained by amplification of pISF101 (Clegg et al, 1987) as a control template, pTS19 for the pefA 5′-UTR, pSPN31 for fliC, or a plasmid from pTS33 through pTS38 for the remaining genes.
5′-RACE
To determine the exact start site of the pefACDEF transcript, we used a slightly modified version of a 5′-RACE protocol (2005). In all, 1 μg of total RNA isolated from a S. typhimurium SR11 ΔfimAICDHG sirA mutant was reverse transcribed as described previously using primer GSP1-PefA (Supplementary Table S2) instead of random hexamers. The reaction was then purified using a PCR purification kit (Qiagen). A 3′ polyA tail was added by incubating purified cDNA in 1 × terminal transferase buffer in the presence of 0.1 mM dATPs, 0.25 mM CoCl2 and 40 units of terminal transferase (NEB) for 30 min at 37°C. The reaction was terminated by incubating the reaction mixture for 10 min at 70°C. In all, 1 μl of 1:10 diluted cDNA was then amplified by PCR with primers T17-adaptor, adaptor and GSP2-PefA. The resulting PCR product was gel purified and cloned into pCR2.1 using the Topo TA cloning kit (Invitrogen). The insert was sequenced using M13 universal primers and the transcriptional start site determined.
RNA EMSAs
CsrA-6xHis protein was purified as described previously (Mercante et al, 2006). DNA templates for generating phoB RNA transcript were produced by annealing primers (Martinez et al, 2011), also described in Supplementary Table S2. The T7 promoter-driven wild-type, mutated fimA 5′-UTRs, and pefA 5′-UTRs were cut out of their respective vectors (pTS31, pTS3, pTS48 and pTS51-57) with EcoRI, gel purified and then used as templates for in vitro transcription. RNA was synthesized with the MEGAShort script kit (Ambion, Austin, TX) according to the manufacturer’s instructions and the resulting RNA was gel purified. In all, 50 pmol of purified RNA was 3′-biotinylated using the RNA 3′ End Biotinylation kit (Pierce, Rockford, IL) according to the manufacturer’s protocols.
Biotinylated RNA was boiled 3 min and reconstituted by incubating 30 min at room temperature. Increasing concentrations of purified CsrA-6xHis protein were added to the biotinylated RNA in a total volume of 10 μl in CsrA-binding buffer (10 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 100 mM KCl, 3.25 ng yeast tRNA, 20 mM DTT, 7.5% glycerol, 2 U/μl RNAse inhibitor) and incubated for 30 min at room temperature. Afterwards, 2.5 μl of REMSA loading buffer was added and 3 μl of sample was run on a 5% native TBE gel (Bio-Rad). Blots were transferred onto a positively charged nylon membrane using a semidry transfer apparatus and biotin was detected using the chemiluminescent nucleic acid detection kit (Ambion).
Flow cytometry
Flow cytometry was done as described previously (Chessa et al, 2008) with the following modification. Instead of fixing the bacteria in 4% paraformaldehyde, bacteria were fixed in 0.2% sodium azide. For gating ADH19 (SR-11 ΔpefBACDHF::KmR) was used as a negative control.
Animal experiments
All mouse experiments were approved by the Institutional Animal Care and Use Committees at the University of California at Davis. Female 8- to 12-week-old wild-type C57BL/6 mice were obtained from the Jackson Laboratory (Bar Harbor). The bacterial strains for the inoculum were grown statically for 16 h in LB5.5 in a volume of 50 ml. Animals were pretreated with 20 mg streptomycin and intragastrically infected 24 h later with 1 × 109 CFU of S. typhimurium in 0.1 ml LB as described previously (Godinez et al, 2008). Animals were euthanized 72 h after infection, caecal content was collected for bacteriology and caecal washes were collected for RNA purification as described previously (Lopez et al, 2012).
Statistical tests
Statistical analysis was performed by a two-sample Student’s t-test with unequal variance.
Supplementary Material
Acknowledgments
We would like to thank Cyril Bussiere and John R Roth for providing bacterial strains. We would like to acknowledge support by Public Health Service Grant AI040124 to AJB, GM059969 to TR, AI097116 to TR and AI074693 to SC. TS was supported by a stipend from the Deutsche Forschungsgemeinschaft.
Author contributions: TS, KTN, MGW, SPN and CV performed experiments. TS, SC, TR and AJB conceived the study and designed experiments. AJB, TS and SPN wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- (2005) Rapid amplification of 5′ complementary DNA ends (5′ RACE). Nat Methods 2: 629–630 [DOI] [PubMed] [Google Scholar]
- Ahmer BM, van Reeuwijk J, Watson PR, Wallis TS, Heffron F (1999) Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol Microbiol 31: 971–982 [DOI] [PubMed] [Google Scholar]
- Altier C, Suyemoto M, Lawhon SD (2000) Regulation of Salmonella enterica serovar typhimurium invasion genes by csrA. Infect Immun 68: 6790–6797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäumler AJ, Tsolis RM, Heffron F (1997) Fimbrial adhesins of Salmonella typhimurium. Role in bacterial interactions with epithelial cells. Adv Exp Med Biol 412: 149–158 [PubMed] [Google Scholar]
- Bergsten G, Wullt B, Svanborg C (2005) Escherichia coli, fimbriae, bacterial persistence and host response induction in the human urinary tract. Int J Med Microbiol 295: 487–502 [DOI] [PubMed] [Google Scholar]
- Bhatt S, Edwards AN, Nguyen HT, Merlin D, Romeo T, Kalman D (2009) The RNA binding protein CsrA is a pleiotropic regulator of the locus of enterocyte effacement pathogenicity island of enteropathogenic Escherichia coli. Infect Immun 77: 3552–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chessa D, Dorsey CW, Winter M, Bäumler AJ (2008) Binding specificity of Salmonella plasmid-encoded fimbriae assessed by glycomics. J Biol Chem 283: 8118–8124 [DOI] [PubMed] [Google Scholar]
- Clegg S, Purcell BK, Pruckler J (1987) Characterization of genes encoding type 1 fimbriae of Klebsiella pneumoniae, Salmonella typhimurium, and Serratia marcescens. Infect Immun 55: 281–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey AK, Baker CS, Romeo T, Babitzke P (2005) RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA 11: 1579–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duguid JP, Anderson ES, Campbell I (1966) Fimbriae and adhesive properties in Salmonellae. J Pathol Bacteriol 92: 107–138 [DOI] [PubMed] [Google Scholar]
- Edwards AN, Patterson-Fortin LM, Vakulskas CA, Mercante JW, Potrykus K, Vinella D, Camacho MI, Fields JA, Thompson SA, Georgellis D, Cashel M, Babitzke P, Romeo T (2011) Circuitry linking the Csr and stringent response global regulatory systems. Mol Microbiol 80: 1561–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Bossi N, Valentini M, Malleret L, Fiorini F, Bossi L (2009) Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev 23: 2004–2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune DR, Suyemoto M, Altier C (2006) Identification of CsrC and characterization of its role in epithelial cell invasion in Salmonella enterica serovar Typhimurium. Infect Immun 74: 331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye J, Karlinsey JE, Felise HR, Marzolf B, Dowidar N, McClelland M, Hughes KT (2006) Identification of new flagellar genes of Salmonella enterica serovar Typhimurium. J Bacteriol 188: 2233–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinez I, Haneda T, Raffatellu M, George MD, Paixao TA, Rolan HG, Santos RL, Dandekar S, Tsolis RM, Bäumler AJ (2008) T cells help to amplify inflammatory responses induced by Salmonella enterica serotype Typhimurium in the intestinal mucosa. Infect Immun 76: 2008–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A, Zingales B (1995) Alternative method to remove antibacterial antibodies from antisera used for screening of expression libraries. Biotechniques 19: 30. [PubMed] [Google Scholar]
- Heroven AK, Bohme K, Dersch P (2012) The Csr/Rsm system of Yersinia and related pathogens: a post-transcriptional strategy for managing virulence. RNA Biol 9: 379–391 [DOI] [PubMed] [Google Scholar]
- Holden NJ, Totsika M, Mahler E, Roe AJ, Catherwood K, Lindner K, Dobrindt U, Gally DL (2006) Demonstration of regulatory cross-talk between P fimbriae and type 1 fimbriae in uropathogenic Escherichia coli. Microbiology 152: 1143–1153 [DOI] [PubMed] [Google Scholar]
- Humphries A, Deridder S, Bäumler AJ (2005) Salmonella enterica serotype Typhimurium fimbrial proteins serve as antigens during infection of mice. Infect Immun 73: 5329–5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries AD, Raffatellu M, Winter S, Weening EH, Kingsley RA, Droleskey R, Zhang S, Figueiredo J, Khare S, Nunes J, Adams LG, Tsolis RM, Bäumler AJ (2003) The use of flow cytometry to detect expression of subunits encoded by 11 Salmonella enterica serotype Typhimurium fimbrial operons. Mol Microbiol 48: 1357–1376 [DOI] [PubMed] [Google Scholar]
- Hung DL, Hultgren SJ (1998) Pilus biogenesis via the chaperone/usher pathway: an integration of structure and function. J Struct Biol 124: 201–220 [DOI] [PubMed] [Google Scholar]
- Jackson DW, Suzuki K, Oakford L, Simecka JW, Hart ME, Romeo T (2002) Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J Bacteriol 184: 290–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas K, Edwards AN, Ahmad I, Romeo T, Romling U, Melefors O (2010) Complex regulatory network encompassing the Csr, c-di-GMP and motility systems of Salmonella Typhimurium. Environ Microbiol 12: 524–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas K, Edwards AN, Simm R, Romeo T, Romling U, Melefors O (2008) The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol Microbiol 70: 236–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HY, Dozois CM, Tinge SA, Lee TH, Curtiss R 3rd (2002) Transduction-mediated transfer of unmarked deletion and point mutations through use of counterselectable suicide vectors. J Bacteriol 184: 307–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm P (1994)) Fimbriae: Adhesion, Genetics, Biogenesis, and Vaccines. CRC Press: Boca Raton, FL,
- Kroger C, Dillon SC, Cameron AD, Papenfort K, Sivasankaran SK, Hokamp K, Chao Y, Sittka A, Hebrard M, Handler K, Colgan A, Leekitcharoenphon P, Langridge GC, Lohan AJ, Loftus B, Lucchini S, Ussery DW, Dorman CJ, Thomson NR, Vogel J et al. (2012) The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci USA 109: E1277–E1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawes M, Maloy S (1995) MudSacI, a transposon with strong selectable and counterselectable markers: use for rapid mapping of chromosomal mutations in Salmonella typhimurium. J Bacteriol 177: 1383–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawhon SD, Frye JG, Suyemoto M, Porwollik S, McClelland M, Altier C (2003) Global regulation by CsrA in Salmonella typhimurium. Mol Microbiol 48: 1633–1645 [DOI] [PubMed] [Google Scholar]
- Liu MY, Gui G, Wei B, Preston JF 3rd, Oakford L, Yuksel U, Giedroc DP, Romeo T (1997) The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J Biol Chem 272: 17502–17510 [DOI] [PubMed] [Google Scholar]
- Lopez CA, Winter SE, Rivera-Chavez F, Xavier MN, Poon V, Nuccio SP, Tsolis RM, Bäumler AJ (2012) Phage-mediated acquisition of a type III secreted effector protein boosts growth of salmonella by nitrate respiration. MBio 3: pii: e00143–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low AS, Holden N, Rosser T, Roe AJ, Constantinidou C, Hobman JL, Smith DG, Low JC, Gally DL (2006) Analysis of fimbrial gene clusters and their expression in enterohaemorrhagic Escherichia coli O157:H7. Environ Microbiol 8: 1033–1047 [DOI] [PubMed] [Google Scholar]
- Mandin P, Gottesman S (2009) Regulating the regulator: an RNA decoy acts as an OFF switch for the regulation of an sRNA. Genes Dev 23: 1981–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Sambrook J, Fritsch EF (1989) Molecular Cloning 2nd edn. New York: Cold Spring Harbor Laboratory Press, [Google Scholar]
- Martinez LC, Yakhnin H, Camacho MI, Georgellis D, Babitzke P, Puente JL, Bustamante VH (2011) Integration of a complex regulatory cascade involving the SirA/BarA and Csr global regulatory systems that controls expression of the Salmonella SPI-1 and SPI-2 virulence regulons through HilD. Mol Microbiol 80: 1637–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercante J, Edwards AN, Dubey AK, Babitzke P, Romeo T (2009) Molecular geometry of CsrA (RsmA) binding to RNA and its implications for regulated expression. J Mol Biol 392: 511–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercante J, Suzuki K, Cheng X, Babitzke P, Romeo T (2006) Comprehensive alanine-scanning mutagenesis of Escherichia coli CsrA defines two subdomains of critical functional importance. J Biol Chem 281: 31832–31842 [DOI] [PubMed] [Google Scholar]
- Nielubowicz GR, Mobley HL (2010) Host-pathogen interactions in urinary tract infection. Nat Rev Urol 7: 430–441 [DOI] [PubMed] [Google Scholar]
- Nuccio SP, Bäumler AJ (2007) Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol Mol Biol Rev 71: 551–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuccio SP, Chessa D, Weening EH, Raffatellu M, Clegg S, Bäumler AJ (2007) SIMPLE approach for isolating mutants expressing fimbriae. Appl Environ Microbiol 73: 4455–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penrod JT, Roth JR (2006) Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica. J Bacteriol 188: 2865–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft T, Baker EN (2009) Pili in Gram-negative and Gram-positive bacteria - structure, assembly and their role in disease. Cell Mol Life Sci 66: 613–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo T (1998) Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol Microbiol 29: 1321–1330 [DOI] [PubMed] [Google Scholar]
- Romeo T, Gong M, Liu MY, Brun-Zinkernagel AM (1993) Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J Bacteriol 175: 4744–4755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo T, Vakulskas CA, Babitzke P (2012) Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems. Environ Microbiol 15: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder JA, Haugen BJ, Lockatell CV, Maroncle N, Hagan EC, Johnson DE, Welch RA, Mobley HL (2005) Coordinate expression of fimbriae in uropathogenic Escherichia coli. Infect Immun 73: 7588–7596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Babitzke P, Kushner SR, Romeo T (2006) Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev 20: 2605–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Wang X, Weilbacher T, Pernestig AK, Melefors O, Georgellis D, Babitzke P, Romeo T (2002) Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J Bacteriol 184: 5130–5140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplitski M, Al-Agely A, Ahmer BM (2006) Contribution of the SirA regulon to biofilm formation in Salmonella enterica serovar Typhimurium. Microbiology 152: 3411–3424 [DOI] [PubMed] [Google Scholar]
- Teplitski M, Goodier RI, Ahmer BM (2003) Pathways leading from BarA/SirA to motility and virulence gene expression in Salmonella. J Bacteriol 185: 7257–7265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Bäumler AJ (2011) Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci USA 108: 17480–17485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Luisi BF (2011) Hfq and its constellation of RNA. Nat Rev Microbiol 9: 578–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner C, Hensel M (2011) Adhesive mechanisms of Salmonella enterica. Adv Exp Med Biol 715: 17–34 [DOI] [PubMed] [Google Scholar]
- Waksman G, Hultgren SJ (2009) Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat Rev Microbiol 7: 765–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei BL, Brun-Zinkernagel AM, Simecka JW, Pruss BM, Babitzke P, Romeo T (2001) Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol Microbiol 40: 245–256 [DOI] [PubMed] [Google Scholar]
- Weilbacher T, Suzuki K, Dubey AK, Wang X, Gudapaty S, Morozov I, Baker CS, Georgellis D, Babitzke P, Romeo T (2003) A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol Microbiol 48: 657–670 [DOI] [PubMed] [Google Scholar]
- Xia Y, Gally D, Forsman-Semb K, Uhlin BE (2000) Regulatory cross-talk between adhesin operons in Escherichia coli: inhibition of type 1 fimbriae expression by the PapB protein. EMBO J 19: 1450–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin AV, Baker CS, Vakulskas CA, Yakhnin H, Berezin I, Romeo T, Babitzke P (2013) CsrA activates flhDC expression by protecting flhDC mRNA from RNase E-mediated cleavage. Mol Microbiol 87: 851–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Liu MY, Romeo T (1996) Coordinate genetic regulation of glycogen catabolism and biosynthesis in Escherichia coli via the CsrA gene product. J Bacteriol 178: 1012–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue M, Rankin SC, Blanchet RT, Nulton JD, Edwards RA, Schifferli DM (2012) Diversification of the salmonella fimbriae: a model of macro- and microevolution. PLoS One 7: e38596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.