Abstract
The EMBO J 32: 2804–2818 ; DOI: 10.1038/emboj.2013.198; published online September 03 2013
In his 1543 monumental work De humanis corpori fabrica, Andreas Vesalius used rigorous dissection practices and a mechanistic view of the organ’s function to provide the first accurate anatomical description of a human heart. Guided by similar principles of meticulous structural probing and mechanistic explanatory potential, Anokhina and colleagues in this issue of The EMBO Journal dissect the molecular topology of the RNA heart of the Spliceosome, the ribonucleoprotein machinery in charge of intron removal. Their findings reveal key structural features with important implications for understanding the mechanisms of pre-mRNA splicing catalysis.
Most eukaryotic genes display a counter-intuitive organization, in which primary transcripts require the removal of internal (intronic) sequences and splicing together of the remaining parts (exons) to generate functional mRNAs. The mystery is further spiced by the limited conservation of sequences at exon–intron boundaries (splice sites) and the striking complexity of the Spliceosome, the enzymatic machinery in charge of intron removal, which with over 200 proteins and five small nuclear ribonucleoprotein particles (snRNPs) is among the most complex molecular machineries in the cell (Wahl et al, 2009). Whatever their origins, the actual existence of introns, the variety of sequence elements at their boundaries and the complexity of the Spliceosome have set the stage for the evolution of alternative splice site selection as a prevalent mechanism for regulation of gene expression in multicellular organisms (Braunschweig et al, 2013).
More than 30 years of intensive research have worked out, in broad outline, the stepwise process of Spliceosome assembly and the chemical mechanism of the splicing reaction. Two general principles have emerged. The first is that dynamic formation and melting of RNA:RNA interactions involving splice sites and snRNAs (the RNA components of snRNPs), as well as within and between snRNAs, are essential to identify exon/intron boundaries and to bring together the chemical groups that undergo the two steps of the splicing reaction (Figure 1). In the first step, the phosphodiester bond between the last nucleotide of the exon and the first of the intron is excised, concomitant with formation of a 2′–5′ bond between the 5′ end of the intron and an internal adenosine (the branch point) usually located near the 3′ end of the intron (Figure 1). In the second step, a new concerted excision and formation of phosphodiester bonds leads to ligation of the two exons and release of the intron in a branched (lariat) configuration. This chemical mechanism of intron removal from pre-mRNAs is identical to that of self-catalytic group II introns. There are in fact profound similarities between elements of the highly elaborate three-dimensional structure that group II introns adopt to mediate self-catalysis and key pre-mRNA:snRNA and snRNA:snRNA interactions in the Spliceosome, implying that intron removal from pre-mRNAs can indeed be catalysed by RNA, as is the case for peptide bond formation catalysed by the Ribosome, another complex ribonucleoprotein machine (Staley and Woolford, 2009).
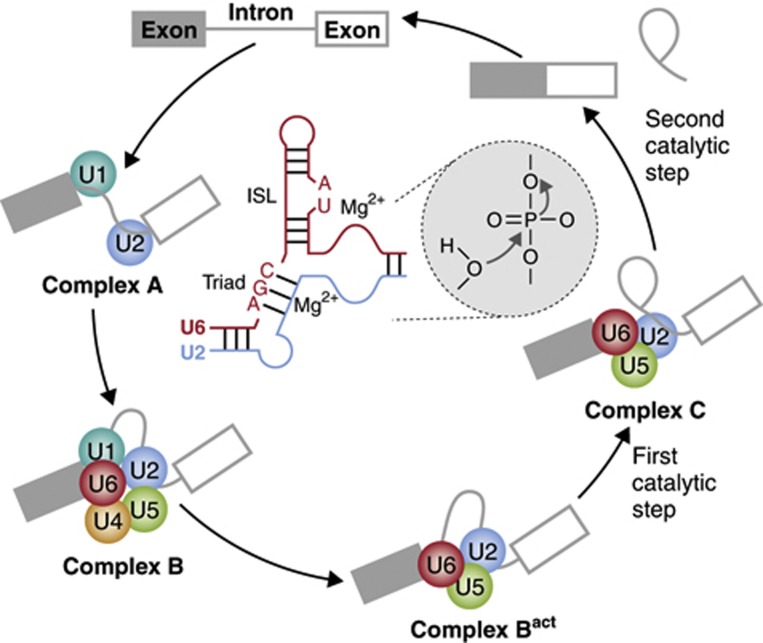
Figure 1.
Schematic representation of the process of Spliceosome assembly and of key RNA interactions at its core. Recognition of the 5’ splice site by U1 snRNP and/or the branch point region by U2 snRNP form complex A, which serves as a platform for assembly of the U4/5/6 tri-snRNP in complex B. Conformational rearrangements result in release of U1 and U4 and formation of the catalytically active Bact complex, in which the first step of the splicing reaction takes place, producing the free first exon and a lariat intron–exon intermediate. New conformational changes generate complex C, where exons 1 and 2 get ligated and the intron lariat released after the second catalytic step. Base pairing interactions between U2 and U6 snRNAs are critical for catalysis (central part). In particular, the AGC catalytic triad and an ISL in U6 help to coordinate magnesium ions that are believed to stabilize the nucleophile and leaving groups of the phosphates involved in each catalytic step.
The second general principle is that substantial changes in the protein composition of Spliceosomal complexes accompany the dynamic transitions in RNA:RNA interactions that characterize the different steps of assembly and catalysis. This flux of proteins and their associated RNA–protein, protein–protein and enzymatic activities facilitates splice site recognition, remodelling of RNA:RNA contacts and the establishment of proofreading mechanisms that discard illegitimate interactions, thus ensuring accurate intron removal even in the face of the multiplicity of sequences present at intron boundaries (Wahl et al, 2009).
Despite these major advances, key outstanding questions remain. First, what is the nature of the catalyst molecule: is it just RNA, is it a protein or is the catalytic centre contributed by the concerted action of chemical groups from both types of molecules? Second, is there a single catalytic centre for the two steps of the splicing reaction or are there separate catalytic centres for each step? Third, is the network of RNA–RNA interactions conserved or are there variant configurations in different organisms? The work of Anokhina et al (2013) in this issue of The EMBO Journal provides substantial insights into these questions.
Major advances in biochemical purification allowed Anokhina et al (2013) to obtain significant amounts of highly homogeneous Spliceosomes from mammalian nuclear extracts at different stages of assembly and catalysis. This provided a unique opportunity to carry out detailed structural probing of its RNA components at each step, using reagents that either modify exposed bases (thus revealing their non-involvement in contacts with other RNAs or with proteins) or that detect base pairing interactions by forming covalent bonds between the interacting strands. Comprehensive mapping confirmed well-known interactions, further validating them as well as validating the approach itself, and revealed novel features, particularly helping to decide between previously proposed alternative RNA folds. To illustrate the value of the approach, we will focus here on insights obtained about the structure of two important regions of U6 snRNA that have been involved in the positioning of magnesium ions at the core of the Spliceosome. These ions play key role in catalysis of group II introns and have also been proposed to function similarly in the Spliceosome: one magnesium stabilizes the nucleophile, while the other stabilizes the leaving group in each of the two phosphotransesterification splicing steps (Steitz and Steitz, 1993; Villa et al, 2002).
One of the magnesium binding sites includes an AGC sequence in U6 snRNA known as the catalytic triad. Two alternative base pairing schemes between U2 and U6 in the triad region have been proposed to form just before catalysis. In one of the conformations, previously inferred from work showing that purified U2 and U6 snRNAs can assemble in vitro to catalyse two-step splicing reactions on short model substrates (Valadkhan and Manley, 2001; Jaladat et al, 2011), U2 and U6 adopt a four-way junction structure. In contrast, previous work in yeast Spliceosomes suggested a model in which U2/U6 adopt a three-way junction structure in the same region (Madhani and Guthrie, 1992). The data from Anokhina et al (2013) indicate that residues predicted to base pair with two of the triad nucleotides in the four-way junction structure (but not in the three-way junction structure) are exposed through splicing catalysis. This result favours the idea that the three-way junction is the functional conformation conserved between yeast and mammalian Spliceosomes. While the four-way junction remains a valid option for an RNA-only ribozyme, the Spliceosome appears to have adopted the alternative structural solution that, in concert with other components, including proteins, may improve features of the catalytic process. Because the structure persists through catalytic step 1 and contains metal ligands that function in both steps, a single reactive center for the two steps of the splicing reaction seems to be the most likely scenario, as proposed before (Steitz and Steitz, 1993).
Having mapped in detail the status of individual nucleotides, Anokhina et al (2013) then took the challenge of developing a highly educated three-dimensional model of the RNA structure of the catalytic core. The model assembled available structural and biochemical information from catalytic complexes, high-resolution structures from group II introns or from RNA folds found in other molecules (e.g., ribosomal or transfer RNAs), with final touches of structural intuition to adjust three-dimensional orientations in ways that could better explain catalysis. In particular, enforcing a two nucleotide bulge in the so-called internal stem loop (ISL) of U6, another point of magnesium coordination, led to an overall structure that not only fit best structural probing data and three-dimensional constraints but also resembled the configuration of the equivalent structure present in group II introns (Villa et al, 2002).
Full validation of this model will require the determination of the structure of Spliceosomal complexes at high resolution, which remains a formidable challenge. Meanwhile, Anokhina et al (2013) made the courageous prediction that if the model would be true, it might fit in the structure of a particular surface of the Prp8 protein. Prp8 is the most conserved component of the Spliceosome, which has been shown to be crosslinked (and therefore in close proximity) to many of the chemical groups in the pre-mRNA or snRNAs engaged in the two catalytic steps (Grainger and Beggs, 2005). The excellent docking between key domains of Prp8 and the model RNA built by Anokhina et al (2013) argues in favour of the validity of the predicted RNA structure. Furthermore, the intimate structural fit suggests that, even if catalysis is fundamentally driven by the two-metal ion coordination of RNAs, Prp8 and perhaps other proteins play a key role in optimizing the RNA fold to mediate efficient catalysis. In the same tradition that allowed Vesalius to dismount the 1400-year-old belief that the left and right ventricles were physically interconnected, further meticulous dissection of the Rnatomy of the Spliceosome’s heart, combined with intuition about the function of its parts, will help to unveil more secrets of this fascinating molecular device.
Footnotes
The authors declare that they have no conflict of interest.
References
- Anokhina M, Bessonov S, Miao Z, Westhof E, Hartmuth K, Lührmann R (2013) RNA structure analysis of human spliceosomes reveals a 3D arrangement of snRNAs at the catalytic core. The EMBO J 32: 2804–2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig U, Gueroussov S, Plocik AM, Graveley BR, Blencowe BJ (2013) Dynamic integration of splicing within gene regulatory pathways. Cell 152: 1252–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger RJ, Beggs JD (2005) Prp8 protein: at the heart of the spliceosome. RNA 11: 533–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaladat Y, Zhang B, Mohammadi A, Valadkhan S (2011) Splicing of an intervening sequence by protein-free human snRNAs. RNA Biol 8: 372–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C (1992) A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell 71: 803–817 [DOI] [PubMed] [Google Scholar]
- Staley JP, Woolford JL Jr (2009) Assembly of ribosomes and spliceosomes: complex ribonucleoprotein machines. Curr Opin Cell Biol 21: 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz TA, Steitz JA (1993) A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci USA 90: 6498–6502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadkhan S, Manley JL (2001) Splicing-related catalysis by protein-free snRNAs. Nature 413: 701–707 [DOI] [PubMed] [Google Scholar]
- Villa T, Pleiss JA, Guthrie C (2002) Spliceosomal snRNAs: Mg(2+)-dependent chemistry at the catalytic core? Cell 109: 149–152 [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R (2009) The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]