Abstract
Elucidating molecular links between cell-fate regulatory networks and dynamic patterning modules is a key for understanding development. Auxin is important for plant patterning, particularly in roots, where it establishes positional information for cell-fate decisions. PIN genes encode plasma membrane proteins that serve as auxin efflux transporters; mutations in members of this gene family exhibit smaller roots with altered root meristems and stem-cell patterning. Direct regulators of PIN transcription have remained elusive. Here, we establish that a MADS-box gene (XAANTAL2, XAL2/AGL14) controls auxin transport via PIN transcriptional regulation during Arabidopsis root development; mutations in this gene exhibit altered stem-cell patterning, root meristem size, and root growth. XAL2 is necessary for normal shootward and rootward auxin transport, as well as for maintaining normal auxin distribution within the root. Furthermore, this MADS-domain transcription factor upregulates PIN1 and PIN4 by direct binding to regulatory regions and it is required for PIN4-dependent auxin response. In turn, XAL2 expression is regulated by auxin levels thus establishing a positive feedback loop between auxin levels and PIN regulation that is likely to be important for robust root patterning.
Keywords: MADS proteins, root development, PIN regulation, stem cell niches, auxin
Introduction
Plant and animal development is guided by transcriptional networks that control cell-fate decisions in conjunction with dynamic patterning modules, such as those that regulate the differential distribution (gradients) of hormones (Davidson and Erwin, 2006; Newman et al, 2009). Establishing the molecular links between such networks and hormone or nutrient distribution is a key for understanding cell patterning. Our current comprehension of how these two types of processes are linked spatially and temporally in vivo is limited (Newman et al, 2009).
The root of Arabidopsis thaliana has become an exemplar for in vivo studies of molecular developmental mechanisms, particularly of the molecular basis of stem-cell niche patterning and dynamics (van den Berg et al, 1997; Aida et al, 2004; Sarkar et al, 2007; Fulcher and Sablowski, 2009). The root stem-cell niche is a part of the cell proliferation domain (Ivanov and Dubrovsky, 2013) and, like that of other multicellular organisms, has in the centre an organizer, called the Quiescent Centre (QC) in roots. Tissue-specific progenitor cells, multipotent stem cells or initial cells, surround the QC. The derivatives of the initial cells exhibit a brief period of rapid cell proliferation shootward of the root apical region. After 4–6 rounds of division, cells begin elongation and ultimately differentiation (Dolan et al, 1993; van den Berg et al, 1998).
Expression of GRAS family SCARECROW and SHORTROOT (SCR/SHR) and AP2 family PLETHORA (PLT) transcription factors is necessary for the formation and maintenance of the stem-cell niche in Arabidopsis roots (Sabatini et al, 1999, 2003; Helariutta et al, 2000; Aida et al, 2004). Auxins are fundamental plant hormones in embryonic development (Möller and Weijers, 2009), organogenesis (Vanneste and Friml, 2009), and root cell patterning (Friml et al, 2003). An auxin gradient with a maximum level at the QC is required for correct specification of the stem-cell niche. Moreover, the cellular levels of auxin define root cell fate: intermediate auxin concentrations are found in the cell proliferation zone and low auxin levels characterize the zones of elongation and differentiation along the root longitudinal axis (Burstrom, 1957; Friml and Palme, 2002; Petersson et al, 2009; Jurado et al, 2010). Directional auxin transport between cells, partially mediated by PIN auxin-efflux carriers, is crucial for generating these auxin gradients (Blilou et al, 2005; Vieten et al, 2005; Wisniewska et al, 2006). Differential PIN expression has been explored extensively (Aida et al, 2004; Vieten et al, 2005; Ruzicka et al, 2009), but little is known about the factors that directly regulate PIN gene transcription or PIN expression in response to auxins, as well as how this regulation impacts PIN-dependent developmental processes.
Initial studies suggested that the PLT and SHR/SCR genes constituted the main components of root stem-cell niche patterning; based on the recent gene discoveries and a new gene regulatory network (GRN) model it is hypothesized that additional components remain undiscovered (Sarkar et al, 2007; Welch et al, 2007; Azpeitia et al, 2010). Plant MADS-box genes have been extensively characterized as regulators of reproductive development, flower transition, and organ identity (Coen and Meyerowitz, 1991; Álvarez-Buylla et al, 2000a; Álvarez-Buylla et al, 2000b; Burgeff et al, 2002), and recent studies suggest that auxin-regulated MADS-box genes, such as XAANTAL1/AGL12 (XAANTAL is the Mayan word for ‘go slower’ in recognition of the retarded root growth phenotypes of xaantal mutants), also regulate root development (Gan et al, 2005; Tapia-López et al, 2008). We report here that the Arabidopsis MADS-box gene, AGL14/XAANTAL2, closely related to the flowering gene, SUPPRESOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) (Martinez-Castilla and Alvarez-Buylla, 2003) is required for root stem-cell niche and meristem patterning. Furthermore, AGL14/XAL2 regulates auxin transport and gradients in the root via direct regulation of PIN transcription.
Results
XAANTAL2 (XAL2/AGL14) controls root stem-cell niche delimitation and cell proliferation
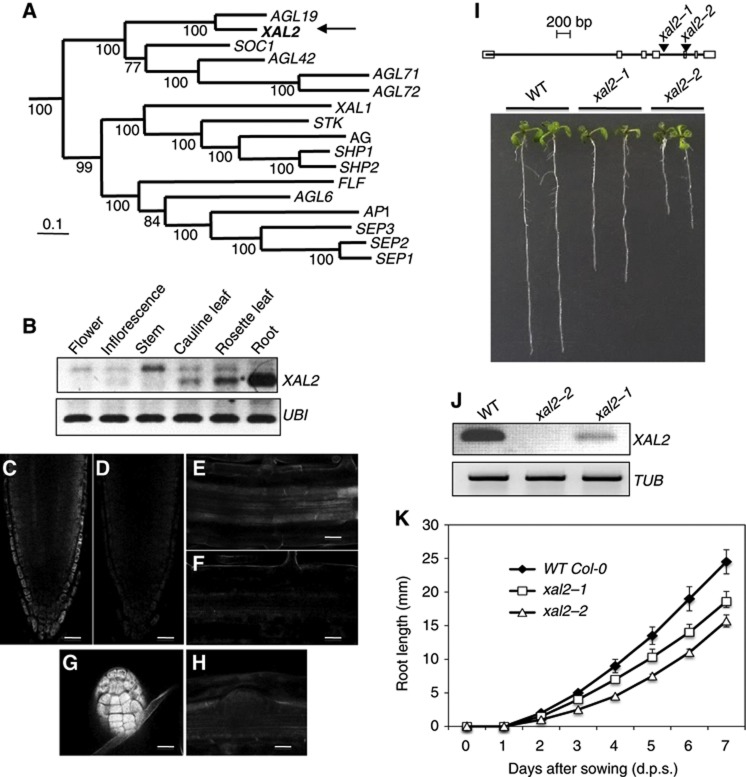
We sought to test the role of MADS-box gene XAL2 in Arabidopsis root development. We obtained lines with loss-of-function mutations in XAL2, a type II MADS-box gene that belongs to the SOC1 clade (Figure 1A) and is highly expressed in roots (Figure 1B). ‘In situ’ data of RNA expression derived from both ‘in situ’ PCR and dig-labelled ‘in situ’ experiments demonstrated that mRNA is found in lateral root cap, epidermis, endodermis, and columella of the root meristematic region, as well as in the vascular cylinder in differentiated zones of the primary root and in emerged lateral root primordia (Figures 1C–H; Supplementary Figures S1B–H and S2). In addition, we generated a 1-kb XAL2 promoter that also recovers the expression of the gene promoter in the vascular tissue and in the lateral roots. This line recovers only a scanty and light expression in the root meristematic tissues after a few hours of GUS staining, maybe because the cloned promoter fraction misses some important enhancer sequences. This genetic marker is strongly expressed in the root meristem only after several days of staining (data not shown). Nonetheless, both this and the ‘in situ’ data show clear and strong expression of XAL2 in the central cylinder and the emerging lateral roots. These data parallel the reports by Birnbaum et al (2003) and Nawy et al (2005) for XAL2; who report expression in the meristematic tissues as our ‘in situ’ assays, and in the QC (see Supplementary Figure S1A). We isolated two loss-of-function alleles for this XAL2 (called hereafter xaantal2-1 and xaantal2-2). Both mutants develop shorter wild-type roots in the Columbia background, similar in length to XAL1 mutants (Figure 1I; Tapia-López et al, 2008). The xal2-2 allele exhibited the most pronounced retarded root growth and altered cellular structure at the stem-cell niche, and concordantly lacked detectable levels of XAL2 mRNA, whereas the xal2-1 allele exhibited intermediate phenotypes between wild type and xal2-2, and exhibited residual XAL2 expression (Figures 1I–K). The root growth and stem-cell niche phenotypes, as well as the correlated levels of expression of XAL2 in these two independent alleles demonstrate that the observed phenotype can be attributed to the loss of XAL2 function (Figure 2A). For the rest of the functional analyses, we therefore focused on the strong allele xal2-2.
Figure 1.
XAL2, a MADS-box gene, is highly expressed in roots and affects root growth. (A) Bayesian reconstruction of the phylogenetic relationships among selected type II A. thaliana MADS-box genes, with XAL2 position indicated by an arrow. (B) RT–PCR used to visualize XAL2 expression in different tissues from 7 days post sowing (d.p.s.) in wild-type. Ubiquitin (UBI) was used as an internal control. (C–H) Whole-mount ‘in situ’ RT–PCR hybridization (see Materials and methods for details of procedures used) of XAL2 in columella cells and lateral root cap (C), in the vascular bundle in differentiated cells (E) and in the primordia of lateral roots (G). (D, F, and H) Negative sense controls (Bar=50 μm). (I) XAL2 gene structure schematic model with the sites of transposon insertions. Squares correspond to exons while lines represent introns. Root length phenotypes of seedlings at 7 d.p.s. of Col-0 wild type, xal2-1, and xal2-2 (n=60). Tubulin (TUB) was used as an internal control. (J) RT–PCR of XAL2 at 7 d.p.s. in wild type, xal2-1, and xal2-2. (K) Root growth curves of wild-type, xal2-1, and xal2-2 plants grown in medium supplemented with 2% sucrose for 10 days (n=30). Average and s.d. are shown.
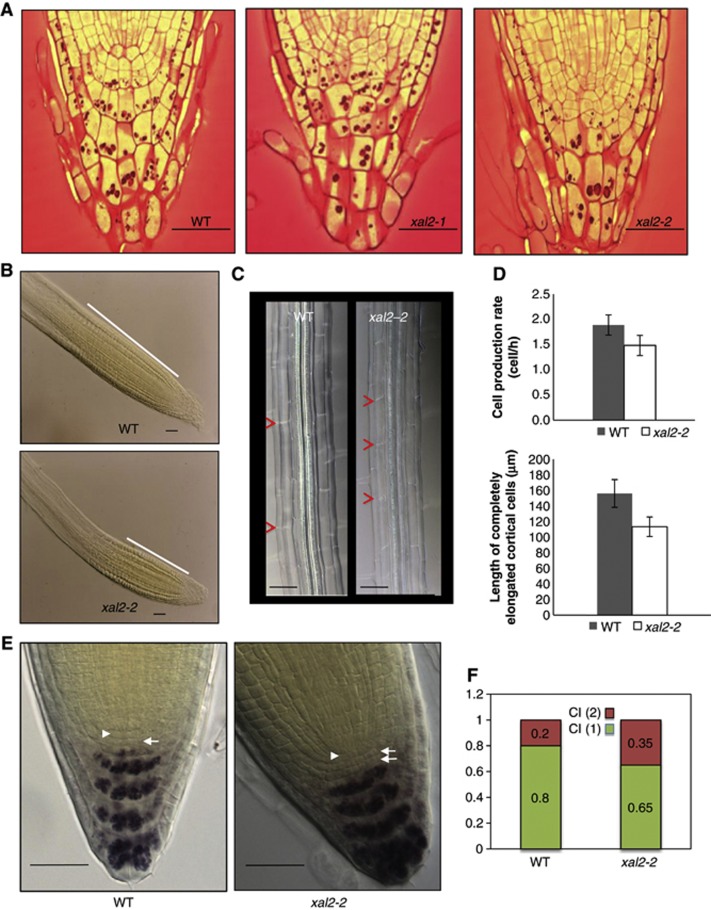
Figure 2.
XAL2 controls meristem size and patterning of the root apical meristem. (A) Organization of the quiescent center and initial cells in the stem-cell niche from representative phenotypes for wild-type and XAL2 mutants in longitudinal root sections (100%; n=30) (Bar=50 μm). (B) Meristem length of wild-type and xal2-2 plants at 7 d.p.s.; the white line indicates the size of the meristematic region (Bar=50 μm). (C) Cell length of fully elongated cells (see red arrowheads) at 7 d.p.s. (n=30) in cleared roots observed with Nomarski optics (Bar=50 μm). (D) Cell production rate and length of completely elongated cortical differentiated cells; quantifications are described in detail in Materials and methods. All data were analysed with the JMP 5.1.1 version statistical package. Measurements were obtained directly by measuring 10 cortical cells from 10 plants. Average and s.d. are shown. (E) Nomarski photographs of wild-type and xal2-2 plants. The position of the QC is indicated (see white arrowhead) and white arrows indicate position of columella initial tiers. Lugol staining marks starch granules observed in mature wild-type and xal2-2 columella-differentiated cells (Bar=50 μm). (F) Percentage of one or two tiers of columella initials (CI) in wild-type plants and xal2-2 mutants (n=56 for wild type; n=69 for xal2-2).
Quantitative growth analyses of xal2-2 roots (Table I) showed that the mutant displays a significantly lower number of meristematic cells and lower rate of cell production, fewer cells in the elongation zone and shorter fully elongated cells compared with wild-type roots (Figures 2B–D; Table I) and the xal2-2 line exhibits normal cell-cycle duration and pCYCB1;1DB::GUS expression (Supplementary Figures S3A and B). Consequently, decreased root growth in plants with this strong loss-of-function xal2-2 allele results from a combination of decreased cell production and shorter cell length in the differentiation zone. The diminished size of the completely elongated cells observed in xal2-2 accounts for up to 86% of the decrease in root length in the mutant, while only 14% of such length decrease could be attributed to changes in a number of cells in the root apical meristem (RAM) proliferation domain (Table I). In addition, we documented that the diameter of the stele (provascular tissues) of wild-type plants is not larger (see Supplementary Figures S4A–E), in the root meristem compared to that of xal2-2 plants. In wild type, a greater number of pericycle cells are present in the transverse plane of the wild-type roots in comparison to the mutant ones (14.8 cells in wild type and 12.0 cells in xal2-2, Supplementary Figures S4C, D, and F). It is important to note, that wild-type and xal2-2 mutant lines had roots and steles with the same width also in the differentiation zone (see Supplementary Figure S4G). We can conclude that xal2-2 loss-of-function mutation significantly affects the proliferation of the pericycle cells, particularly the radial anticlinal divisions at the fifth cortical cell.
Table 1. Quantitative cellular analysis of root development for wild-type and xal2-2 seedlings (7 d.p.s.).
| Root growth rate (μm h−1) | Proliferation domain (PD) length (μm) | Cortical cell number within PD | Cortical cell length within PD (μm) | Combined length of the transition domain and the elongation zone (μm) | |
|---|---|---|---|---|---|
| ****** | * | **** | ** | ||
| WT | 265±11 | 173±12 | 33.3±2.14 | 5.2±0.1 | 510±32 |
| xal2-2 | 163±3 | 142±5 | 26.5±1.43 | 5.4±0.1 | 385±22 |
Average (marked with bold letters)±s.d. with α<0.1, α<0.05, α<0.02, α<0.005, α<0.002, and α<0.001 indicated as *, **, ***, ****, *****, and ******, respectively; n=30.
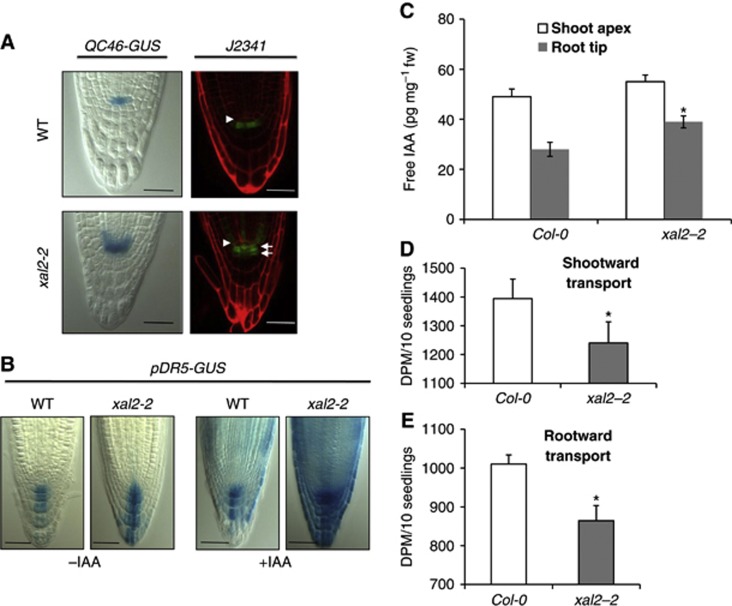
To determine whether XAL2 is necessary for QC identity and stem-cell niche patterning, we analysed whole mount optical and thin plastic sections of roots for each allele. We identified an altered stem-cell niche phenotype characterized by altered shapes and distributions of the QC, initial, and columella cells. XAL2 does not seem to be necessary for QC identity, as WOX5, SCR, and PLT1 (COL148 (plt1-1-GUS); Aida et al, 2004) were properly expressed in the xal2-2 mutants (Supplementary Figure S5) and xal2-2 roots continued growth until day 10, although at a lower rate than wild-type roots (Figure 1K). The domains of expression of the QC-specific markers QC25 and QC46 were abnormally expanded in xal2-2 mutants towards the columella cells and provascular initials, respectively (Figure 3A; Supplementary Figure S5), suggesting that XAL2 is necessary for restriction of the QC to its normal spatial domain. Expression of the columella initial marker J2341 was expanded as well in 25% of the xal2-2 mutant roots (Figure 3A), and in these plants a supernumerary columella layer lacking starch granules was observed (Figures 2E and F); the rest of the mutant roots did not show this phenotype. We interpret this as indicating a delayed transition to columellar cell differentiation. The above results suggest that XAL2 seems to be required for normal spatial organization and patterning of the root stem-cell niche, as well as for maintaining meristem homeostasis.
Figure 3.
XAL2 is necessary to restrict QC and auxin distribution to its normal spatial domain in Arabidopsis root tip. (A) Two different markers in wild-type and xal2-2 mutant plants: QC46 expression (95%; n=50) and specific enhancer trap J2341 expression (25%; n=30). In the latter marker, two tiers of columella initials were observed in xal2-2 (white arrows) and the position of the QC is indicated (white arrowhead). For this line, red signal is emitted by propidium iodide that was used as a counterstain. 7 d.p.s. plants were used for the experiment with QC46 transgenic lines and 5 d.p.s. plants for the J2341 marker (Bar=50 μm). (B) pDR5-GUS (90%; n=80) expression of wild-type and mutant xal2-2 plants with or without an auxin treatment (IAA 1 μM, 4 h) (Bar=50 μm). (C) Quantification of free IAA levels in the root of wild-type and xal2-2 plants (P<0.01, n=50). (D) Direct measurements of 3H-IAA transport in shootward transport capacity from the root apex to the first 2 mm section (P=0.052, n=10). (E) Direct measurements of 3H-IAA transport in rootward transport capacity in the hypocotyl to root–shoot transition zone (P<0.001, n=10). In (C–E), bars represent standard errors (s.e.) and the * indicates significant differences (P<0.01, n=50).
XAL2 controls auxin transport and concentration
Auxin is required for normal cell proliferation and elongation: intermediate auxin levels are associated with highest cell proliferation, while lower levels with cell elongation and differentiation along the root (Burstrom, 1957; Grieneisen et al, 2007; Jurado et al, 2010). Given the growth defects of xal2-2 mutant roots, we hypothesized that XAL2 could be important for maintaining auxin gradients along the root longitudinal axis. Visualization of the auxin-responsive DR5 promoter (Sabatini et al, 1999; Friml et al, 2003) reporter suggests that auxin levels or responses are elevated in the QC, provascular initials, and columella cells of xal2-2 mutants compared with normal siblings (Figure 3B). Moreover, exogeneous indole acetic acid (IAA) treatment of xal2-2 roots resulted in increased pDR5:GUS expression throughout the root (Figure 3B; Supplementary Figure S6A) compared with wild-type treated roots. We performed several experiments to distinguish whether these effects reflected alterations in downstream signalling, free auxin levels, or auxin transport. First, we tested several auxin responsive markers in the wild-type and xal2-2 plants with or without auxin treatment and found that all these markers were similarly induced by auxin treatment in both genotypes (Supplementary Figure S6B), implying that auxin response is not altered in the mutant.
Next, direct quantification of free IAA levels in xal2-2 seedlings confirmed that auxin levels are significantly increased in the xal2-2 root compared with wild type (Figure 3C). A reduction in rootward 3H-IAA transport, similar to levels seen in pin1 mutants (Blakeslee et al, 2007), was also observed (Figure 3E). Diminished shootward transport from the root apex (Figure 3D) is consistent with the increased pDR5::GUS signal observed in xal2-2. These results indicated that XAL2 is a positive regulator of auxin transport towards and within the root. We then addressed whether such alterations were a consequence, at least in part, of the misregulation of the transcription of PIN auxin transporters, in the xal2-2 background.
XAL2 regulates the transcript accumulation of several PIN genes
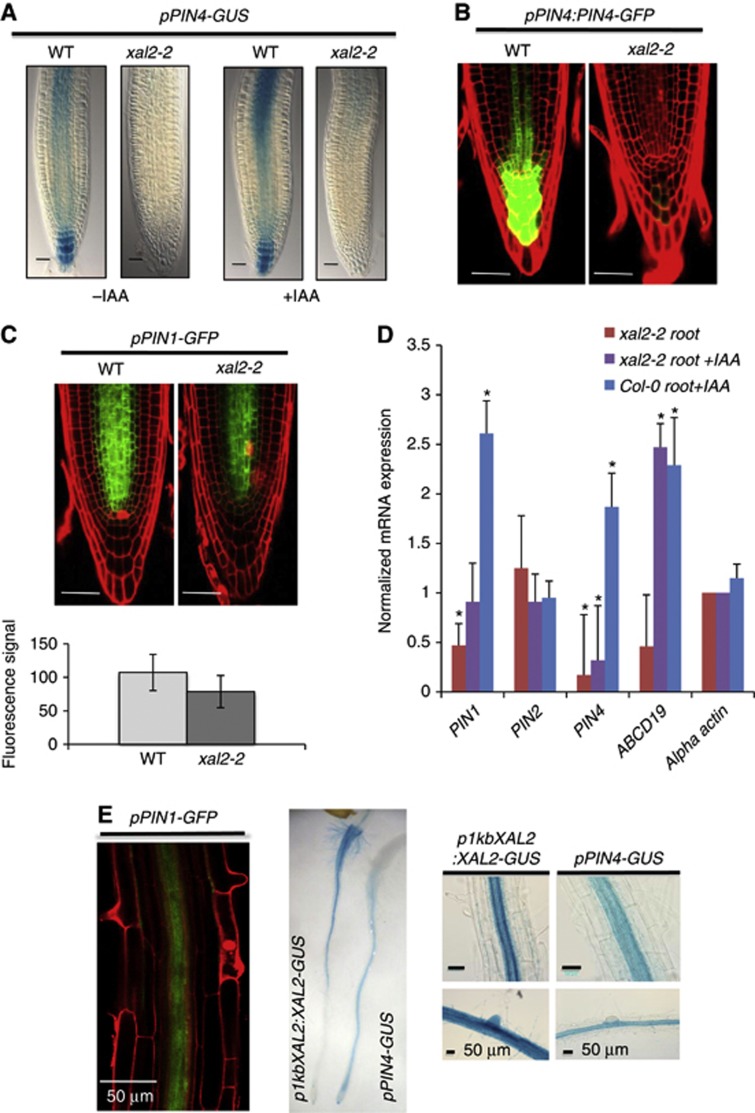
Despite functional redundancy, each PIN protein has been implicated in particular developmental processes (Gälweiler et al, 1998; Luschnig et al, 1998; Friml et al, 2002a, 2002b, 2003; Mravec et al, 2009). For example, PIN4 has been associated with root meristem activity and patterning (Friml et al, 2002b). Interestingly, roots with pin4 loss-of-function alleles are strikingly similar to xal2-2 roots, with altered stem-cell niches, two files of columella initials, expanded expression domains of QC markers, and altered auxin gradients (Friml et al, 2002b). Hence, we addressed whether XAL2 is involved in regulation of PIN4 expression. We crossed xal2-2 with a pPIN4:GUS line to assay an impact on RNA accumulation and a pPIN4:PIN4-GFP translational fusion line. The progeny of these crosses displayed clearly diminished GUS and GFP signals, respectively (Figures 4A and B). Consistent with this result, PIN4 transcript abundance in response to auxin was also decreased by this visual assay in xal2-2 mutants compared to wild type (Figures 4A and D).
Figure 4.
XAL2 upregulates PIN1 and PIN4 gene expression and controls PIN4 auxin response. (A) pPIN4-GUS (100%; n=60) expression of wild-type and mutant xal2-2 plants with or without an auxin treatment (IAA 1 μM, 4 h) (Bar=50 μm). (B) pPIN4:PIN4-GFP (30%; n=65) expression in wild-type and xal2-2 mutant plants. In these lines, confocal image of longitudinal optical sections of root meristem and red signal is emitted by propidium iodide that was used as a counterstain (Bar=50 μm). (C) pPIN1-GFP (50%: n=80) expression in wild-type and xal2-2 mutant plants (Bar=50 μm). Quantification of the fluorescent signal of the pPIN1-GFP reporter (shown above) for wild-type and xal2-2 plants (n=57 for wild type and n=52 for xal2-2 plants) Average and s.d. are shown. (D) qRT-PCR analysis of PIN1, PIN2, PIN4, and ABCB19 normalized to actin expression in 5 d.p.s. wild-type and xal2-2 plants. Bars represent standard deviation (s.d.). *Marks that the change is significant at P<0.001 by Student’s t-test followed by Neuman Keuls post hoc ANOVA. (E) Co-localization of expression of PIN1, PIN4, and XAL2 in different tissues of the root: pPIN1-GFP, pPIN4-GUS, and p1kbXAL2:XAL2-GUS.
Lower expression of PIN4 in xal2-2 mutants is insufficient to explain the shorter roots and the rootward and shootward auxin transport alterations observed in this mutant. Hence, we decided to analyse the expression of other PIN genes. xal2-2 exhibits similar root growth alterations observed in double and triple pin mutants (Blilou et al, 2005). PIN1 is preferentially expressed in the stele and is fundamental for rootward auxin transport (Blilou et al, 2005; Vieten et al, 2005; Blakeslee et al, 2007); pPIN1:GFP signals in xal2-2 are lower in comparison with the wild-type roots (Figures 4C and D; Supplementary Figure S7). To distinguish whether the observed differences reflected anatomical differences in stele width between wild-type plants and the xal2-2 mutant, transverse sections were analysed by light and confocal microscopy. As can be seen in Supplementary Figures S4F and G, there were no differences between these plants in terms of width of the root and the stele in the differentiation zone although we can see more rounded cortical cells in some xal2-2 mutant roots (Supplementary Figure S4F). We also observed that there were a significantly lower number of pericycle cells at the level of the fifth cortical cell from the QC in xal2-2 compared to wild-type plants (see Supplementary Figures S4A–E). Nonetheless, the magnitude of the difference is insufficient to explain the decrease found in the expression of PIN1 in xal2-2 mutant. Moreover, we did not find that XAL2 regulates PIN7 expression despite the latter is expressed also in the vascular tissue (see Supplementary Figure S8A) but we found that PIN7 has two overlapping phenotypes (see Supplementary Figure S8A); in one of them, the expression of PIN7 was expanded to the xylem root axis in both wild-type and xal2-2 plants, thus suggesting that the mutation in XAL2 is not the causal factor of such patterns. The decrease in PIN1 and PIN4 expression was further confirmed quantitatively by qRT-PCR measurements. Confirming the qualitative assessment with visual PIN reporter lines, transcripts of PIN1 and PIN4 genes were reduced in xal2-2 compared with wild-type plants (Figure 4D). As can be seen in Figure 4E, PIN1, PIN4, and XAL2 are strongly expressed and their expression overlaps in the vascular tissue in the differentiation zone of the root; PIN4 and XAL2 expression also overlaps in the columella and the three genes seem to have coincident patterns of expression in the root meristem (Blilou et al, 2005; Vieten et al, 2005; Figures 1C and 4E). XAL2 is not a general regulator of PIN genes, because we found that this MADS transcription factor does not regulate PIN2 or PIN7 (Supplementary Figure S8). We conclude that XAL2 is a positive regulator of the mRNA expression of PIN1 and PIN4 in the root. It is important to note that xal2-2 loss-of-function allele has several phenotypes that cannot be explained by the loss of function of PIN1 and PIN4, thus suggesting that XAL2 is a regulator of several additional genes involved in the networks important for root development.
XAL2 directly binds to PIN regulatory sequences
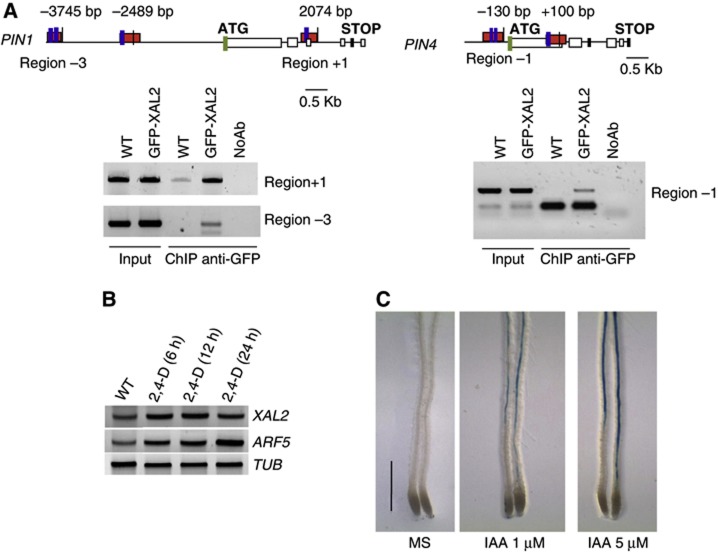
Both PIN1 and PIN4 have several MADS DNA binding motifs (CArG boxes; Riechmann et al, 1996) in their regulatory regions and hence we tested whether XAL2 is able to directly bind to any of such CArG boxes. Chromatin immunoprecipitation (ChIP) assays with 35S:GFP-XAL2 were performed (Supplementary Figure S9). Three CArG box-containing fragments, one in the promoter region of PIN1, another one in the second intron of PIN1, and the third in the promoter region of PIN4 were consistently enriched in three different biological experiments in the IP, indicating that XAL2 directly binds to and presumably regulates the expression of both PIN1 and PIN4 (Figure 5A). We also tested whether XAL2 recognizes additional CArG boxes present in upstream regions of these genes (two for PIN1 and one for PIN4) and confirmed that XAL2 does not bind to all CArG boxes found in these regions (see −2489, bp in the PIN1 promoter and +100 pb in the PIN4 promoter in Figure 5A).
Figure 5.
XAL2 directly regulates PIN1 and PIN4 gene expression and is induced by auxin treatment. (A) Scheme of PIN1 and PIN4 genes; blue boxes indicate putative CArG-boxes, red boxes indicate the XAL2 binding sites, and green box indicates the translation start site. PCR on total DNA (Input) and on DNA recovered by immunoprecipitation with (ChIP anti-GFP) or without (NoAb) GFP antibody from 5 d.p.s. roots of wild-type and 35S-GFP:XAL2 lines, using primers to amplify region −3, −1 and +1. (B) RT–PCR of XAL2, ARF5, and TUB2. RNA was isolated from 4 d.p.s. seedlings subject to 96 hours of 1-N-naphthylphthalamic acid (NPA) treatment and then transferred to 10 μM 2,4-D for 6, 12, or 24 h as indicated. (C) p1KbXAL2:XAL2-GUS line induced by different auxin treatments. Plants were grown for 6 d.p.s. in hormone-free medium and then transferred to growth media supplemented with 1 and 5 μM IAA (indole acetic acid) (n=4) (Bar=0.5 mm).
Do the observed xal2-2 phenotypes result from the downregulation of PIN1 and PIN4 that, in turn, alter auxin distribution and transport in the root? The extent of decrease (1010±23.51 in wild type and 864±39 in xal2-2) in rootward transport in xal2-2 mutant (Figure 3E) is approximately what we would expect for the loss of PIN1 and PIN4 activity (20–40% in Blakeslee et al, 2007). Ectopic auxin accumulation observed in xal2-2 also appears to induce compensatory auxin efflux mechanisms, as expression of the auxin-inducible ABCB19 auxin transporter gene (Noh et al, 2001) was significantly increased in auxin-treated roots compared with the wild-type roots (Figure 4D; Supplementary Figure S10); this result parallels what is observed in the weak pin1-5 allele (Supplementary Figure S10). In conclusion, decreased PIN1 expression (and by implication, function) in xal2-2 appears to increase local auxin concentrations, which, in turn, increases ABCB19 abundance to compensate for the partial PIN1 loss-of-function in the xal2-2 background.
XAL2 gene responds to exogeneous auxins
According to TGA (http://bioinformatics.psb.ugent.be/webtools/plantcare/html), there are several auxin response elements (AREs) in the regulatory regions of XAL2. Does this gene respond to auxin resulting in increased transcript abundance, similar to XAANTAL1? (Tapia-López et al, 2008). Expression analysis using RT–PCR after 2,4-Dichlorophenoxyacetic acid (2,4-D) (Figure 5B) or IAA induction of p1Kb::XAL2::gXAL2-GUS reporter lines (Figure 5C) indicates that this XAL2 is positively regulated by auxins. Therefore, there is a positive feedback loop between auxin levels and PIN regulation via XAL2, which in turn, affects auxin levels and distribution in the root meristem (Figure 6).
Figure 6.
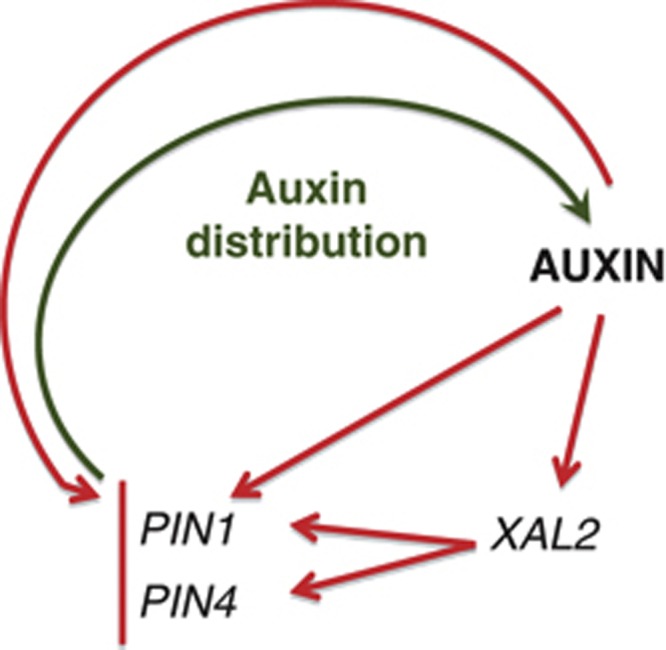
Positive feedback loop between XAL2 gene transcription and auxin distribution. Auxin upregulates XAL2 gene transcription and this MADS regulates efflux auxin carriers controlling auxin distribution/participation in the proliferation of the root meristematic cells. It is shown that while the response to auxin of PIN4 is mediated by XAL2, the auxin response of PIN1 does not depend on XAL2. Red arrows indicate transcript regulation based on the experimental data and green arrow indicates proteins involved in auxin distribution.
Discussion
The focus of this study is XAL2, a MADS-box gene belonging to the SOC clade (Martinez-Castilla and Alvarez-Buylla, 2003). We establish that XAL2 is a component of the Arabidopsis GRNs underlying root stem-cell and meristem patterning and that XAL2 can control auxin transport via direct regulation of PIN1 and PIN4 genes. XAL2 directly binds to discrete motifs within the regulatory regions of PIN1 and PIN4 and results in increased transcript abundance; this activity is selective, because XAL2 does not affect PIN2 and PIN7. Furthermore, PIN4 response to auxin depends on XAL2. Thus, we have uncovered a direct molecular link between auxin transport dynamics, which is a key plant hormone, and transcriptional MADS-box GRNs that underpin cell-fate decisions.
xal2-2 roots showed an increased free auxin accumulation in comparison to wild-type roots. Although this could appear to contradict the observation of lower PIN1 and PIN4 expression in the mutant, redundant auxin efflux mechanisms must compensate, which in turn could explain augmented ABCB19 expression levels. Such compensatory mechanisms are expected from the reported complexity of phenotypic effects with multiple PIN mutations (Blilou et al, 2005). The magnitude of ABCB19 overexpression in xal2-2 mutant could be important for auxin accumulation in the root tip, especially when the mutants are treated with IAA. The fact that xal2-2 roots showed increased free auxin levels along the entire length of the Arabidopsis seedling root could explain the decreased size of completely elongated and differentiated cells in the mutant in comparison to wild-type cells.
Because both XAL2 and XAL1 (Tapia-López et al, 2008) are auxin-responsive genes, we hypothesize that a complex MADS network functions to regulate auxin transport, distribution patterns, and auxin levels within root cells. Auxin levels in turn modulate PIN expression via at least one of these MADS factors. Such feedback may contribute to the establishment of a robust auxin and morphogenetic pattern along the root axis, because it has been shown that positive feedback loops amplify a received signal or stimulus and can generate robust and precise patterns as it has been suggested by theoretical dynamical systems (Kitano, 2004; Jaeger et al, 2008; Mitrophanov and Groisman, 2008). In Arabidopsis roots, the complete mechanism likely involves XAL1 and XAL2 plus additional MADS-box proteins that form heterodimeric transcriptional activators and repressors (Honma and Goto, 2001). MADS proteins already identified as important regulators of root development include AGL16, AGL20, and AGL21; XAL1 can interact by yeast two and three hybrid, with all three of these AGLs and XAL2 with AGL16 and AGL20 (de Folter et al, 2005; Immink et al, 2009). We propose that both MADS-domain heteromultimeric complexes and yet to be identified transcription factors are involved in the control and readout mechanisms of auxin gradients along the root. Those with maximum impact on auxin distribution should be factors expressed primarily at the root stem-cell niche.
The PLETHORA genes have been reported to be crucial genes to root patterning and auxin readout mechanisms (Aida et al, 2004; Galinha et al, 2007). Future studies should search directly for biochemical evidence of MADS–PLT interactions combined with tests of functional relevance in stem-cell patterning and auxin control and readout mechanisms. In fact, XAL and PLT mutants are quite similar in root length and organization of stem-cell niche (Aida et al, 2004; Galinha et al, 2009) although the loss-of function or the gain-of-function of PLT genes shows more drastic effects than those observed in the MADS mutant lines.
A recent GRN model of root stem-cell niche patterning suggests that additional components of the mechanisms involved in root stem-cell patterning remain undiscovered (Azpeitia et al, 2010). Here, we have provided data for XAL2 mutants, which have altered cellular patterns in the stem-cell niche and apical meristem of Arabidopsis roots. We also showed that this MADS factor is an important regulator of transport and distribution of auxin, a hormone required for stem-cell patterning. Future models should incorporate the role of these and other MADS factors utilizing ChIP-Seq data to define their target genes and detailed phenotypic quantification to clarify their roles in controlling cell patterning and proliferation in the root apex. Indeed, the relatively more drastic alterations in the stem-cell and meristem cellular patterns of xal2-2, in comparison to pin mutants (Blilou et al, 2005), suggest that XAL2 regulates other genes in addition to PIN1 and PIN4.
Materials and methods
Plant materials, DNA constructs, and growth conditions
Arabidopsis thaliana wild-type, xal2-1, and xal2-2 plants, pPIN1-GFP, pPIN2:PIN2-GFP, pPIN4:PIN4-GFP, pPIN4-GUS, pWOX-GFP, and pSCR-GFP constructs and DR5-GUS auxin reporter lines are in a Col-0 genetic background. QC25-GUS, QC46-GUS, and PLT1-GUS are in the Ws ecotype, and J2341 is in C24 ecotype. All lines were homozygous. The xal2-1 and xal2-2 alleles were identified by screening for En-I insertions among a collection of Arabidopsis plants carrying ∼50 000 independent insertions of the autonomous maize transposable element (En; Baumann et al, 1998). The collection was screened in pools using the En-I transposon primer En8130 and the internal XAL2 primers: XAL2F and XAL2R (see Supplementary Table S1 for primer sequences) and single insertion lines were confirmed with Southern blot analyses and propagated for further analysis. For the 35S:GFP-XAL2 construction, an (AGL14) XAL2 cDNA entry clone (de Folter et al, 2005) was recombined using the Gateway system (Invitrogen) in the pK7WG2 destination vector (Karimi et al, 2002). In the case of the 35S:XAL2 construction, an (AGL14) XAL2 cDNA entry clone (de Folter et al, 2005) was recombined using the Gateway system (Invitrogen) in the pH7WG2 destination vector (Karimi et al, 2007). For the pXAL2-GUS promoter construction, Arabidopsis (Col-0 ecotype) genomic DNA was used as a template to amplify a 1-kb fragment upstream of the XAL2 start codon. The oligonucleotides used were up-p14-FW and Atg-14-RV (+NcoI; see Supplementary Table S1 for primer sequences). The PCR product was purified by using a kit (Qiagen; Cat. no. 28104) and cloned in pENTR/D, followed by sequencing and recombination with pBGWFS7 (Karimi et al, 2007). Arabidopsis thaliana plants were transformed using the floral dip method (Clough and Bent, 1998), the transgenic lines were selected based on their antibiotic resistance (different for each line), and expression analysis was carried out on T3 homozygous lines. All seeds were sown on vertical plates with 0.2X MS salts, 1% sucrose and 1% agar (unless otherwise indicated). Plants were grown under long-day (LD; 16 h light/8 h dark) conditions in growth chambers at 22oC and with a 16-h photo/8-h dark period (110 μE m−2 s−1).
Seed carrying pPIN1-GFP was obtained from J Friml; pPIN4:PIN4-GFP and pPIN4:GUS were obtained from E Benková; QC25-GUS, pPIN2:PIN2-GFP, pPIN7:PIN7-GFP pDR5-GUS, pWOX5-GFP, plt1-1-GUS, and QC46-GUS were obtained from B Scheres; pSCR-GFP was obtained from P Benfey, pCYCB1;1DB::GUS was obtained from P Doerner and J2341 was obtained from the Arabidopsis Stock Center.
Phylogenetic analysis
We performed a Bayesian reconstruction of the phylogenetic relationships among selected type II Arabidopsis thaliana MADS-box genes using the complete cDNA sequences from Martinez-Castilla and Alvarez-Buylla (2003). Bayesian methods with Mr Bayes (Huelsenbeck and Ronquist, 2001) were used with a Markov chain Monte Carlo exploration of the tree likelihood surface. Four independent Markov chains (three heated) were used according to the Metropolis coupled scheme. The codon substitution model used was that of Goldman and Yang (1994). Four independent runs of 2 500 000 generations each were performed, and every 100th tree was saved. After checking for Markov chain convergence, we discarded the first 15 000 trees and used the remaining trees to calculate Bayesian posterior probabilities of the clades. Results from every independent run were similar.
Histological analysis
Roots were fixed, embedded in Historesin (Leica Instruments GmbH) as described previously (Dubrovsky et al, 2000) and 2.5 μm sections obtained on a Leica RM2155 microtome (Cambridge Instruments, Nussloch, Germany). Sections were mounted on gelatin-coated slides (Baum and Rost, 1996), and after 30 min of hydrolysis in 5N of HCl at room temperature, were stained by the Feulgen technique (De Tomasi, 1936) for 1 h. The same material subsequently was stained by periodic Acid-Schiff reaction (Baum and Rost, 1996).
‘In situ’ hybridization
For whole-mount ‘in situ’ RT–PCR hybridization, we used the protocol of Ruiz-Medrano et al (1999); probes were chosen to avoid the presence of the MADS-box sequence and to have an average length of 200 bp. Sense probes were used as negative controls for XAL2 in all experiments performed for different tissues.
GUS staining
Plant material for light microscopy and GUS staining was prepared as previously described by Malamy and Benfey (1997) and visualized microscopically (see below). Seedlings were subjected to GUS reaction in the dark during 40 min at room temperature for pDR5-GUS and plt1-1-GUS, 1 h at 37oC for pPIN4-GUS, 5 h at 37oC for QC25-GUS, overnight at 37oC for QC46-GUS, 24 or 48 h, 4 or 6 days at 37oC for p1KbXAL2:XAL2-GUS. To restrict the diffusion of GUS blue staining, 2 mM of K3FE(CN)6 and K4FE(CN)6 was added in the GUS solution at the beginning.
GFP quantification
Fluorescence intensity of green channel was measured using the FV10-ASW 1.7 Software (Olympus, USA). To measure the intensity of GFP, we used the same region shown in Figure 4C of WT and xal2-2 plants. The figure shows the mean and standard deviation.
Microscopy
Plant material for light microscopy was cleared using a modified Malamy and Benfey protocol (1997). Roots were visualized using Nomarski optics under an Olympus BX60 microscope and photographed with a Leica camera. Confocal images were acquired using an inverted Zeiss LSM 510 Meta microscope (Carl Zeiss, Oberkochen, Germany) with a dry × 40 or a × 63 water immersion C-Apochromat objectives after root tissue was stained with 5 or 10 gμl−1 propidium iodide. For starch staining, live roots were stained for 5 min in Lugol (Sigma Aldrich) solution 1 × , washed in 30% glycerol for 1 min, mounted into the NaI solution without DMSO as described previously (Dubrovsky et al, 2009) and immediately analysed using Nomarski optics.
Quantitative analysis of cellular parameters of root growth
Length of the cell proliferation domain of the RAM was determined for the cortex cells as a point where the distances between nuclei in neighbouring cells in a cell file became greater than the diameter of the nuclei (Dubrovsky et al, 1998). The combined length of the elongation zone and the transition domain defined as in Ivanov and Dubrovsky (2013) was measured as the distance between the shootward border of the RAM proliferation domain and the location of the most distal root-hair bulge. The number of cortical cells in the cell proliferation domain in 7-day and 8-day plants (a period during which the rate of the root growth was estimated) was similar in wild-type and mutant plants, which enabled us to consider root growth to be at steady state and apply the rate of cell production method (Ivanov and Dubrovsky, 1997) for estimation of average cell-cycle duration in the RAM of 8-day plants. Cell-cycle duration (T, hours) was calculated as T=(NPD le ln2)V−1 where NPD is the number of cells in the cell proliferation domain, le (μm) is the average length of 10 fully elongated cells and V (μm h−1) is the rate of root growth during last day of growth (Ivanov and Dubrovsky, 1997). As the transition domain of the RAM previously has not been defined, the number of meristematic cells in the cited work corresponds to the NPD in the current study. The rate of cell production was estimated as V(le)−1 (Baskin, 2000). Student’s t-test or the Tukey-Kramer test (depending on the sample size) was calculated by the JMP program version 5.1.1 at different confidence level (see Table I).
Expression analysis by RT–PCR
Total RNA was isolated from seedlings using the Trizol reagent (Invitrogen). Columbia wild-type, xal2-1, and xal2-2 seedlings were grown for 7 days post sowing (d.p.s.); for PIN1 expression analysis Columbia wild-type, xal2-2, and 35S:GFP-XAL2 plants were grown for 5 d.p.s.; in both cases, the seedlings were grown in MS plates. RT–PCR was done starting from 1 μg of total RNA to obtain cDNA using Super Script II (Invitrogen). TUBULIN or UBIQUITIN was amplified as an internal positive control; primer sequences of PCR primers are included in Supplementary Table S1. The primers for XAL2 were XAL2F1 and XAL2R2, for PIN1 were PIN1F and PIN1R, for ARF5 were ARF5F1 and ARF2R2; for TUB2 were TUB2F and TUB2R and for UBI were UBIF and UBIR; primer sequences are described in Supplementary Table S1. For auxin induction assays, seeds were germinated in plates with 10 μM 1-N-naphthylphthalamic acid (NPA) for 96 h and then were transferred to fresh plates with or without 10 μM 2,4-D and incubated for 6, 12, and 24 h, as indicated.
Real-time PCR
Seedlings were grown as described by Murphy and Taiz (1995). RNA extraction and PCR conditions were as described by Blakeslee and collaborators (2004). See http://www.psla.umd.edu/faculty/Murphy/index_files/Page530.htm for complete details and Supplementary Table S1 for primer sequences. Primer sequences for quantitative PCR are marked with a q at the beginning of the primer name. The qRT-PCR data were analysed using the value of the wild-type amplification cycle divided by the internal standard compared to the value of xal2-2 against the same standard for PIN1, PIN2, PIN4, ABCB19, and alpha actin. The standard deviations are the sum of standard deviations for N biological replicates with N technical replicates of each sample. Quantification and data acquisition was done using the iCycler Real Time system (Bio-Rad) using Syber Green I as a fluorophor. Real-time PCR was run using 1:50 dilution of first strand at 95°C for 40 s (denaturation), 54°C for 40 s (annealing), and 72°C for 20 s (extension) for 35 cycles. The ABI program was setup with an s.d. threshold for three replicates and was set to <0.25. If data are ⩾0.25, then they are not significant.
Chromatin immunoprecipitation
ChIP assays were carried out as previously described (Sanchez and Gutierrez, 2009) with several modifications. For immunoprecipitation, 4 μl of an anti-GFP antibody (sc-8334; Santa Cruz Biotechnology) was incubated in phosphate-buffered saline (PBS) with Protein A agarose; 500 ng of root fixed-chromatin extracts from 5-day-old 35S:GFP-XAL2 complementation plants or wild-type plants was added to the IP reaction. The immunoprecipitated DNA was resuspended in 30 μl of TE and 1 μl of this DNA and a 1/5 dilution of INPUT samples were used for each PCR. The primers sequences are described in Supplementary Table S1.
Hormone treatments
Plants of the lines harbouring the 1KbXAL2:XAL2-GUS or PIN4-GUS constructs were grown for 7 days in hormone-free medium plates and then transferred to growth media supplemented with 1 or 5 μM of IAA for 1 day. For the RT–PCR experiment, plants were grown in 2,4-D for 4 days in NPA and then transferred to 10 μM of 2,4-D for 6, 12, and 24 h as indicated. After IAA treatment, some seedlings were subjected to GUS staining, pDR5-GUS (40 min at room temperature) and pPIN4-GUS (1 h at 37oC). In parallel, duplicate samples were subjected to RNA extraction for RT–PCR experiments. T3 seeds of two representative independent p1KbXAL2:XAL2-GUS transformants were surface sterilized and sown in plates containing 0.2XMS medium with 1% sucrose and 1% micropropagation grade agar. After three nights at 4°C, the plates were placed in a growth cabinet (Percival) at 16 h photoperiod at 22°C.
Quantification of endogeneous-free auxin
Seedlings were grown as described by Murphy and Taiz (1995). The protocol for quantification of free IAA is described in Kim et al (2007).
Auxin transport assays
Seedlings were grown as described by Murphy and Taiz (1995). Protocols for quantifying auxin transport are provided in Blakeslee and collaborators (2007) and in Peer and Murphy (2007). A detailed protocol can be found at http://www.psla.umd.edu/faculty/Murphy/index_files/Page530.htm.
Supplementary Material
Acknowledgments
We thank M Yanofsky for his help, guidance, and support during the early stages of this work. We also thank C Gutiérrez for support and advice during the final phase of this study. Thanks to B Scheres, V Ivanov, E Azpeitia, and M Benítez who provided insightful comments on the manuscript, and to R Pérez, G Coello, D Romo, T Romero, S Napsucialy-Mendivil, A Saralegui, X Alvarado-Affantranger, JA Pimentel-Cabrera, JM Hurtado-Ramírez, K Jiménez-Durán and L Rodríguez who helped with logistical and laboratory tasks. We also thank J Lin for assistance with the preliminary set of transport assays, Professor Wendy Peer for statistical evaluation and validation of qRT-PCR results and A Chase for preparing the materials and plants for auxin transport assays. We thank Dr Virginia Walbot for her careful editing and overall revision of the paper. Any remaining mistake is our responsibility. This work was supported by grants from CONACYT, México: (1) Red Temática de Investigación: ‘Complejidad, Ciencia y Sociedad’ (124909; ERAB; BGP; AGA) and (2) 81542 and 105678 (ERAB), 167705 (AGA), 152649 (MPS), 81433 (BGP), 177739 (SF) and 127957 (JGD), from PAPIIT-UNAM, IN204011-3 (BGP), IN229009-3 (ERAB), IN226510-3 (AGA), IB201212 (MPS), and IN204312 (JGD), from the Spanish Government BFU2012-33746 (SP) and from the National Science Foundation (NSF-IOS) 0820648 (ASM). ERAB acknowledges the support of the Miller Institute for Basic Research in Science, University of California, Berkeley while spending a sabbatical leave in the laboratory of Chelsea Specht at UC-B.
Author contributions: ERA-B designed and coordinated this study, designed many experiments, performed the initial experiments, analysed all experimental results, and wrote the paper; AG-A conceptualized and performed most of the genetic experiments and some of the qRT-PCR assays, analysed data, and wrote the paper; EO-M conceptualized and performed the mutant line characterizations and some of the crosses, made whole-mount ‘in situ’ hybridization experiments, performed root histology and assisted with data analysis. AC-P participated in the genetic and molecular biology experiments and analysis. MPS, NM-M, and SF conceptualized, performed, and analysed ChIP experiments and made the overexpression constructs and the prom-GUS fusion construct. NM-M also performed the auxin induction experiments for long periods of time. ASM conceptualized, performed, and analysed auxin transport, response, and level experiments, as well as qRT-PCR assays for PINs and auxin responsive markers and participated in writing the manuscript. BGP conceptualized genetic and molecular biology experiments, helped analyse them and participated in writing the paper. FJ-M participated in genetic and molecular experiments and MAP-E participated in the visualization of roots and analysis of results. JGD established and helped coordinate the quantitative cellular, histological, and confocal microscopy analyses of wild-type and mutant roots, participated in analysis of genetic experiments, and participated in manuscript preparation. SP performed some of the first experiments and helped in the isolation of the transposon mutants of XAL2.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120 [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla ER, Pelaz S, Liljegren SJ, Gold SE, Burgeff C, Ditta GS, Ribas de Pouplana L, Martínez-Castilla L, Yanofsky MF (2000a) An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc Natl Acad Sci 97: 5328–5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Buylla ER, Liljegren SJ, Pelaz S, Gold SE, Burgeff C, Ditta GS, Vergara-Silva L, Yanofsky MF (2000b) MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. Plant J 24: 457–466 [DOI] [PubMed] [Google Scholar]
- Azpeitia E, Benítez M, Vega I, Villarreal C, Alvarez-Buylla E (2010) Single-cell and coupled GRN models of cell patterning in the Arabidopsis thaliana root stem-cell niche. BMC Syst Biol 4: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin TI (2000) On the constancy of cell division rate in the root meristem. Plant Mol Biol 43: 545–554 [DOI] [PubMed] [Google Scholar]
- Baum SF, Rost TL (1996) Root apical organization in Arabidopsis thaliana: 1. Root cap and protoderm. Protoplasma 192: 178–188 [Google Scholar]
- Baumann E, Lewald J, Saedler H, Schultz B, Wisman E (1998) Successful PCR-based reverse genetic screen using an En-1 mutagenised Arabidopsis thaliana population generated via single-seed descent. Theor Appl Genet 97: 729–734 [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Blakeslee JJ, Bandyopadhyay A, Lee OR, Mravec J, Titapiwatanakun B, Sauer M, Makam SN, Cheng Y, Bouchard R, Adamec J, Geisler M, Nagashima A, Sakai T, Martinoia E, Friml J, Peer WA, Murphy AS (2007) Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 19: 131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeslee JJ, Bandyopadhyay A, Peer WA, Makam SN, Murphy AS (2004) Relocalization of the PIN1 auxin efflux facilitator plays a role in phototropic responses. Plant Physiol 134: 28–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Burgeff C, Liljegren SJ, Tapia-Lopez R, Yanofsky MF, Alvarez-Buylla ER (2002) MADS-box gene expression in lateral primordia, meristems and differentiated tissues of Arabidopsis thaliana roots. Planta 214: 365–372 [DOI] [PubMed] [Google Scholar]
- Burstrom H (1957) Auxin and the mechanism of root growth. Symp Soc Exp Biol 11: 44–62 [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Davidson EH, Erwin DH (2006) Gene regulatory networks and the evolution of animal body plans. Science 311: 796–800 [DOI] [PubMed] [Google Scholar]
- de Folter S, Immink RG, Kieffer M, Parenicová L, Henz SR, Weigel D, Busscher M, Kooiker M, Colombo L, Kater MM, Davies B, Angenent GC (2005) Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell 17: 1424–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Tomasi JA (1936) Improving the technic of the Feulgen stain. Stain Technol 11: 137–144 [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B (1993) Cellular organization of the Arabidopsis thaliana root. Development 119: 71–84 [DOI] [PubMed] [Google Scholar]
- Dubrovsky JG, Doerner PW, Colón-Carmona A, Rost TL (2000) Pericycle cell proliferation and lateral root initiation in Arabidopsis. Plant Physiol 124: 1648–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, North GB, Nobel PS (1998) Root growth, developmental changes in the apex, and hydraulic conductivity for Opuntia ficus-indica during drought. New Phytol 138: 75–82 [Google Scholar]
- Dubrovsky JG, Soukup A, Napsucialy-Mendivil S, Jeknic Z, Ivanchenko MG (2009) The lateral root initiation index: an integrative measure of primordium formation. Ann Bot 103: 807–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Benková E, Blilou I, Wisniewska J, Hamann T, Ljung K, Woody S, Sandberg G, Scheres B, Jürgens G, Palme K (2002b) AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108: 661–673 [DOI] [PubMed] [Google Scholar]
- Friml J, Palme K (2002) Polar auxin transport--old questions and new concepts? Plant Mol Biol 49: 273–284 [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Friml J, Wiśniewska J, Benková E, Mendgen K, Palme K (2002a) Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809 [DOI] [PubMed] [Google Scholar]
- Fulcher N, Sablowski R (2009) Hypersensitivity to DNA damage in plant stem cell niches. Proc Natl Acad Sci USA 106: 20984–20988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B (2007) PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Gan Y, Filleur S, Rahman A, Gotensparre S, Forde BG (2005) Nutritional regulation of ANR1 and other root-expressed MADS-box genes in Arabidopsis thaliana. Planta 222: 730–742 [DOI] [PubMed] [Google Scholar]
- Goldman N, Yang Z (1994) A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol 11: 725–736 [DOI] [PubMed] [Google Scholar]
- Grieneisen VA, Xu J, Marée AF, Hogeweg P, Scheres B (2007) Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449: 1008–1013 [DOI] [PubMed] [Google Scholar]
- Gälweiler L, Guan C, Müller A, Wisman E, Mendgen K, Yephremov A, Palme K (1998) Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282: 2226–2230 [DOI] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN (2000) The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567 [DOI] [PubMed] [Google Scholar]
- Honma T, Goto K (2001) Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 525–529 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755 [DOI] [PubMed] [Google Scholar]
- Immink RG, Tonaco IA, de Folter S, Shchennikova A, van Dijk AD, Busscher-Lange J, Borst JW, Angenent GC (2009) SEPALLATA3: the 'glue' for MADS box transcription factor complex formation. Genome Biol 10: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov VB, Dubrovsky JG (1997) Estimation of the cell-cycle duration in the root meristem: a model of linkage between cell-cycle duration, rate of cell production, and rate of root growth. Int. J. Plant Sci 158: 757–763 [Google Scholar]
- Ivanov VB, Dubrovsky JG (2013) Longitudinal zonation pattern in plant roots: conflicts and solutions. Trends Plant Sci 18: 237–243 [DOI] [PubMed] [Google Scholar]
- Jaeger J, Irons D, Monk N (2008) Regulative feedback in pattern formation: towards a general relativistic theory of positional information. Development 135: 3175–3183 [DOI] [PubMed] [Google Scholar]
- Jurado S, Abraham Z, Manzano C, López-Torrejón G, Pacios LF, Del Pozo JC (2010) The Arabidopsis cell cycle F-box protein SKP2A binds to auxin. Plant Cell 22: 3891–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Depicker A, Hilson P (2007) Recombinational cloning with plant gateway vectors. Plant Physiol 145: 1144–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kim JI, Sharkhuu A, Jin JB, Li P, Jeong JC, Baek D, Lee SY, Blakeslee JJ, Murphy AS, Bohnert HJ, Hasegawa PM, Yun DJ, Bressan RA (2007) yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol 145: 722–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H (2004) Biological robustness. Nat Rev Genet 5: 826–837 [DOI] [PubMed] [Google Scholar]
- Luschnig C, Gaxiola RA, Grisafi P, Fink GR (1998) EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev 12: 2175–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Martinez-Castilla LP, Alvarez-Buylla ER (2003) Adaptive evolution in the Arabidopsis MADS-box gene family inferred from its complete resolved phylogeny. Proc Natl Acad Sci USA 100: 13407–13412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrophanov AY, Groisman EA (2008) Positive feedback in cellular control systems. Bioessays 30: 542–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravec J, Skůpa P, Bailly A, Hoyerová K, Krecek P, Bielach A, Petrásek J, Zhang J, Gaykova V, Stierhof YD, Dobrev PI, Schwarzerová K, Rolcík J, Seifertová D, Luschnig C, Benková E, Zazímalová E, Geisler M, Friml J (2009) Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459: 1136–1140 [DOI] [PubMed] [Google Scholar]
- Murphy A, Taiz L (1995) A new vertical mesh transfer technique for metal-tolerance studies in Arabidopsis-ecotypic variation and copper-sensitive mutants. Plant Physiol 108: 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller B, Weijers D (2009) Auxin control of embryo patterning. Cold Spring Harb Perspect Biol 1: a001545 Review [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawy T, Lee JY, Colinas J, Wang JY, Thongrod SC, Malamy JE, Birnbaum K, Benfey PN (2005) Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell 17: 1908–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman SA, Bhat R, Mezentseva NV (2009) Cell state switching factors and dynamical patterning modules: complementary mediators of plasticity in development and evolution. J Biosci 34: 553–572 [DOI] [PubMed] [Google Scholar]
- Noh B, Murphy AS, Spalding EP (2001) Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13: 2441–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer WA, Murphy AS (2007) Flavonoids and auxin transport: modulators or regulators? Trends Plant Sci 12: 556–563 Review [DOI] [PubMed] [Google Scholar]
- Petersson SV, Johansson AI, Kowalczyk M, Makoveychuk A, Wang JY, Moritz T, Grebe M, Benfey PN, Sandberg G, Ljung K (2009) An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell 21: 1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Krizek BA, Meyerowitz EM (1996) Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc Natl Acad Sci USA 93: 4793–4798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Medrano R, Xoconostle-Cázares B, Lucas WJ (1999) Phloem long-distance transport of CmNACP mRNA: implications for supracellular regulation in plants. Development 126: 4405–4419 [DOI] [PubMed] [Google Scholar]
- Ruzicka K, Simásková M, Duclercq J, Petrásek J, Zazímalová E, Simon S, Friml J, Van Montagu MC, Benková E (2009) Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc Natl Acad Sci USA 106: 4284–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, Scheres B (1999) An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472 [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B (2003) SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev 17: 354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez MP, Gutierrez C (2009) Arabidopsis ORC1 is a novel PHD-containing H3K4me3 effector that regulates transcription. Proc Natl Acad Sci USA 106: 2065–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T (2007) Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814 [DOI] [PubMed] [Google Scholar]
- Tapia-López R, García-Ponce B, Dubrovsky JG, Garay-Arroyo A, Pérez-Ruíz RV, Kim SH, Acevedo F, Pelaz S, Alvarez-Buylla ER (2008) An AGAMOUS-related MADS-box gene, XAL1 (AGL12), regulates root meristem cell proliferation and flowering transition in Arabidopsis. Plant Physiol 146: 1182–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg C, Weisbeek P, Scheres B (1998) Cell fate and cell differentiation status in the Arabidopsis root. Planta 205: 483–491 [DOI] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B (1997) Short-range control of cell differentiation in the Arabidopsis root meristem. Nature 390: 287–289 [DOI] [PubMed] [Google Scholar]
- Vanneste S, Friml J (2009) Auxin: a trigger for change in plant development. Cell 136: 1005–1016 Review [DOI] [PubMed] [Google Scholar]
- Vieten A, Vanneste S, Wisniewska J, Benková E, Benjamins R, Beeckman T, Luschnig C, Friml J (2005) Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132: 4521–4531 [DOI] [PubMed] [Google Scholar]
- Welch D, Hassan H, Blilou I, Immink R, Heidstra R, Scheres B (2007) Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev 21: 2196–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska J, Xu J, Seifertová D, Brewer PB, Ruzicka K, Blilou I, Rouquié D, Benková E, Scheres B, Friml J (2006) Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.