Abstract
The sirtuins are a family of highly conserved NAD+-dependent lysine deacylases with important roles in metabolic regulation. Of the seven mammalian sirtuins, three localize to the mitochondria: SIRT3, SIRT4, and SIRT5. Mitochondrial sirtuins are crucial regulators of the metabolic network that controls energy homeostasis and impacts cancer, obesity, diabetes, mitochondrial diseases, metabolic disorders, and many other human diseases of aging. To best study the mitochondrial function of the sirtuins, we have employed an oxygen flux analyzer as a tool to track and record the extracellular oxygen consumption rate and acidification rate that reflects mitochondrial respiration and glycolysis, respectfully. Here we described the methods using this assay to study the substrate utilization and mitochondrial function in a human hepato-cellular carcinoma cell line, Huh7. Additionally, we have generated a stable SIRT4 knocked-down Huh7 cell line. With this cell line, we evaluated how the absence of SIRT4 affects mitochondrial function, glucose utilization, glutamine oxidation, and fatty acid oxidation in these cells.
Keywords: Mitochondrial sirtuins, Seahorse XF extracellular flux analyzer, Oxygen consumption rate, Seahorse assay, Substrate utilization, Mitochondrial function, SIRT4
1 Introduction
The major energy-producing pathway in cells when oxygen is present is oxidative phosphorylation. Nutrients derived from different metabolic pathways are oxidized to produce cellular energy in the form of ATP and macromolecules for cellular function. The mitochondrion is the central organelle where all of these processes take place.
Currently, the most common method to study substrate oxidation in vitro is to provide the cells, or isolated mitochondria, with radiolabeled substrate and then track and quantify the radiolabeled intermediates and end-products. This method has several challenges including the use of toxic radioactive materials, as well as limited sensitivity. For example, the end-products of some reactions are the substrates of others, and therefore will be challenging to monitor pathway flux.
Alternatively, oxygen dissolved in the reaction vessel of liquid-phase systems, or that accumulates in the sample chamber of gas-phase systems, can be directly detected polarographically by Clark type electrodes [1]. Isolated mitochondria are often used for this measurement. Although this system has had some success with a suspension of cultured cells, it does not work for adherent cells due to low oxygen diffusion. Additionally, setup of the Clark electrode system is tedious and requires considerable experience.
Seahorse XF extracellular flux analyzer is an alternative to the Clark type electrode. It was first introduced in 2006. By using the optical sensors, it simultaneously measures the proton and oxygen level in a very small volume of media above a monolayer of cultured cells. Valuable insight into the physiological state of cells, and the alteration of the state of those cells, can be gained through measuring the rate of oxygen consumed by the cells (OCR), an indicator of mitochondrial respiration. The cells also generate ATP through glycolysis, the conversion of glucose to lactate, independent of oxygen. The measurement of lactic acid produced is indirectly via the protons released into the extracellular medium surrounding the cells. Therefore, the extracellular acidification rate (ECAR) obtained from a “seahorse assay” reflects the glycolytic function of the cells [2]. Together, OCR and ECAR can provide important insight into the metabolic role of mitochondrial proteins.
The mitochondrial sirtuins remove acyl groups from lysine residues on proteins and control the levels of these posttranslational modifications in the mitochondria [3]. SIRT3 is the best studied of the three mitochondrial sirtuins and functions to remove acetyl groups from target lysine residues. Acetylation is emerging as an important regulatory mechanism in mitochondria where over one third of proteins contain acetylation sites and metabolic enzymes are preferentially modified [4]. Furthermore, acetylation levels and sirtuin activity change depending on the nutritional status of the cell [5–9]. In the case of SIRT3 a pattern is emerging where nutrient starvation leads to activation of the sirtuin followed by deacetylation of substrates and return to metabolic homeostasis. This is illustrated by the regulation of LCAD, an enzyme important in fatty acid oxidation, by SIRT3 [5]. During metabolic stress brought on by fasting acetylation levels of LCAD increase leading to decreased activity of the enzyme and lower levels of fatty acid oxidation. However, when SIRT3 is present LCAD is deacetylated during fasting leading to increased activity of the enzyme and increased fatty acid oxidation, an important source of fuel during fasting. Thus, SIRT3 contributes to shifts in substrate use during times of metabolic stress.
Less is known about the functions of the other mitochondrial sirtuins, SIRT4 and SIRT5, which lack robust deacetylase activity. SIRT5 has recently been described as a protein lysine desuccinylase and demalonylase [10, 11] and regulates the urea cycle enzyme CPS1 [12]. Less is known about the function of SIRT4. Interestingly SIRT4 has weak ADP-ribosylase activity [13] and has a potent role in regulating lipid oxidation [14]. Thus, more work is required to obtain a better understanding of how the mitochondrial sirtuins function to regulate metabolism in the mitochondria.
In this chapter, we describe how the Seahorse extracellular flux analyzer can be applied to the study of mitochondrial sirtuins. The described methods focus on setting up the seahorse assays to measure the oxidation of specific substrates in a human hepatoma cell line, Huh7. We have optimized and validated the assay conditions. We also generated a stable SIRT4 knocked-down (KD) Huh7 cell line and compared the usage of substrates such as glucose, gluta-mine, and fatty acids in the SIRT4 KD and control cells.
Additionally, we optimized the conditions for the use of well-established mitochondrial drugs (oligomycin, FCCP, antimycin) in these cell lines and evaluated the mitochondrial function in SIRT4 KD, control, and uninfected Huh7 cells.
2 Materials
All media and solutions are prepared using cell culture grade distilled water.
2.1 Huh7 and SIRT4 KD Cells in Culture
Growth Media: add 10 % FBS to high-glucose DMEM, store it at 4 °C. Warm up the media to 37 °C every time before using in cells.
0.25 % Trypsin-EDTA (1×).
Puromycin dihydrochloride.
Lipofectamine 2000.
Optimem.
The pSicoRMS2 lentiviral constructs encoding scramble and SIRT4 shRNA were kindly provided by Dr. Eric Verdin at the Gladstone Institutes, San Francisco, CA, USA.
2.2 Seahorse Assay
XF24 Extracellular Flux Analyzer (SeahorseBioscience, MA, USA).
Prep Station (SeahorseBioscience).
XF24 FluxPak (SeahorseBioscience) (see Note 1).
XF Cell Mito Stress Test Kit (SeahorseBioscience) contains four pre-weighed mitochondrial drugs : Oligomycin A, carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP), Antimycin A and Rotenone (see Note 2). Dissolve each of the compounds in DMSO to make 2.5 mM stock solution. Aliquot and store at −20 °C.
Unbuffered base Dulbecco's Modified Eagle's Media (DMEM) (see Note 3): DMEM; 100× GlutaMax; NaCl; Phenol Red sodium salt. Dissolve 2 g of NaCl, 8.3 g of DMEM and 15 mg of Phenol Red in 900 mL of H2O; and add 10 mL of 100× GlutaMax (see Note 4). Mix well and adjust pH with 1 N NaOH to pH 7.3. Make up to 1 L with water. Filter the media to sterilize and store at 4 °C.
2.3 Basal OCR and ECAR Measurements and Mitochondrial Function Tests in Huh7 Cells
Assay media used for basal OCR and ECAR measurements and mitochondrial function tests in Huh7 cells: take 148.5 mL unbuffered base DMEM (see Note 5), and add 1.5 mL of 45 % glucose solution to make a final concentration of 25 mM (see Note 6). Warm in 37 °C water bath and adjust pH to 7.4 with 1 N NaOH before use (see Note 7).
Remove one aliquot of Oligomycin, FCCP, and Antimycin stock solution (2.5 mM) from −20 °C freezer and thaw at room temperature for 30 min on the seahorse assay day. Dilute each of the compounds in above prepared assay media to a concentration at 10 times of the final working concentration.
2.4 Glutamine Oxidation in Huh7 and SIRT4 KD Cells
Take 148.5 mL unbuffered base DMEM, and mix with 1.5 mL 100× l-Glutamine (see Note 8).
2.5 Glucose Oxidation in Huh7 and SIRT4 KD Cells
Prepare new unbuffered base DMEM without adding glutamax, mix 148.5 mL of this media with only 1.5 mL of 45 % glucose solution.
2.6 Fatty Acid Oxidation (FAO) Measurement by Seahorse Assay
10× KHB buffer stock: mix appropriate amount of buffer components to the following concentration: 1.1 M NaCl, 47 mM KCl, 20 mM MgSO4, 12 mM Na2HPO4. Add 900 mL of H2O and stir to allow each component to fully dissolve. Adjust pH to 7.4 and then adjust volume to 1,000 mL with water. Sterilize by filtering and store at 4 °C.
50 mM carnitine stock: dissolve 81 mg carnitine in 10 mL H2O. Use a syringe filter to sterilize the solution. Prepare 1 mL aliquots and store at −20 °C (see Note 9).
1× KHB buffer: Mix 12 mL of 10× KHB, 1.2 mL of 50 mM carnitine with 106.8 mL of H2O to make 120 mL of 1× KHB containing 0.5 mM carnitine (see Note 10).
Preparing 20 mL stock of BSA-conjugated palmitate solution (2 mM sodium palmitate/0.34 mM BSA) and control BSA solution (0.34 mM BSA) (see Note 11): weigh out 906.8 mg of ultra fatty acid-free bovine serum albumin (BSA) (see Note 12) and add to the glass beaker containing the pre-warmed (37 °C) 20 mL of 150 mM NaCl solution while maintaining the solution warmed at 37 °C (see Note 13). When BSA is fully dissolved, use the syringe filter to filter the entire 20 mL BSA solution. Take 10 mL of the BSA solution and quickly mix with 10 mL of 150 mM NaCl solution to make 0.34 mM stock, which will be used as control substrate in FAO measurement. Make 1 and 2 mL aliquots in glass vials and store in −20 °C. Keep the remaining 10 mL of non-diluted BSA solution in 37 °C water bath while preparing the palmitate solution. Weigh out 12.24 mg of the sodium palmitate, and add it to 8.8 mL of 150 mM NaCl solution in a small glass beaker with a stir bar. Place it into a beaker/water bath and heat to 70 °C while stirring. When the palmitate solution becomes clear, quickly transfer the 70 °C palmitate solution to the 10 mL of non-diluted BSA solution while stirring at 37 °C. Stir the mixture at 37 °C for 1 h (see Note 13). Adjust the pH to 7.4 with 1 N NaOH, and then adjust the final volume to 20 mL with 150 mM NaCl. Prepare 1 and 2 mL aliquot in glass vials and store in −20 °C.
3 Methods
3.1 Generation of SIRT4 Knocked-Down Huh7 Cells
The lentivirus was generated by standard molecular biology protocols using pSicoRMS2 constructs containing scramble or SIRT4 shRNA, as well as puromycin and mCherry selection markers [15]. Briefly, a four plasmid transfection mix was made containing 5 μg pSicoRMS2, 2.5 μg pMDLg/pRRE, 1.25 μg pRSV-Rev, and 1.5 μg pVSVg and 20 μL Lipofectamine 2000 in Optimem. The mixture was used to transfect a T75 plate of 25 % confluent HEK293T cells.
Change culture media 24 h after infection.
Collect media from cells 36 h after infection.
To infect Huh7 cells, mix viral supernatant 1:1 with fresh media and apply directly to cells.
Change culture media 24 h after infection. Use puromycin in fresh culture media and select for infected cells by culturing in the presence of puromycin for 72 h (see Note 14). The optimal concentration of puromycin was found to be 1 g/μL for Huh7 cells, as this caused complete cell death of uninfected cells after 24 h.
Visualize selected cells on a fluorescent microscope to confirm all cells express mCherry.
Isolate RNA using the Qiagen RNeasy isolation kit and prepare cDNA from 1 μg of RNA using the Bio-Rad iScript cDNA synthesis kit. Determine the efficiency of knockdown using real-time qPCR with primers to SIRT4 and an endogenous control. Primers designed to mCherry can also be used to determine infection efficiency between scramble and SIRT4 knockdown cell lines.
3.2 Huh7 and SIRT4 KD Cells Maintenance
When the cells are confluent, remove the growth media and wash with 10 mL of PBS once. Aspirate PBS off and add 1 mL of trypsin to cover the whole surface of the cells. Incubate at 37 °C for 3 min before aspirating off the trypsin (see Note 15). Keep the cells for an additional 1–2 min in the incubator, and then tap the flask to detach the cells from the flask. Add 10 mL fresh growth media to flush all the cells off and collect them in a 50 mL conical tube. Mix 1.25 mL of these cells (1/8) with 14 mL of fresh growth media and plate them in a new T75 flask.
Maintain the Huh7 and KD cells in T75 flask at 37 °C with 5 % CO2. The KD cells need to be cultured for one or two more passages in puromycin-free media before being used for seahorse measurement (see Note 14).
3.3 Basal OCR and ECAR Measurement and Mitochondrial Function Tests in Huh7 Cells (See Note 16)
Prepare materials as in below on day 1 (1 day before the seahorse assay):
Pre-incubate one sensor cartridge with 1 mL/well XF24 calibrant solution on one utility plate in a 37 °C non-CO2 incubator overnight.
Turn on the XF24 analyzer with XF24 software running to allow instrument to stabilize at 37 °C before the assay.
Prepare cells as following. Collect the cells as described in Subheading 3.2 and dilute cells so that every 100 μL of the cell/media mixture should contain the desired number of cells for one well of the XF24 cell culture plate. Based on the results from the optimization experiments (see Note 16), plate Huh7 cells at 60,000 cells/well in the XF24 culture plate. Load 100 μL of the cell/media mixture into each well of the XF24 culture plate, and allow the cells to adhere for 2 h in the incubator before adding additional 150 μL of growth media for a total volume of 250 μL per well (see Note 17). Plate the cells in all wells except for A1, B4, C3, and D6, which will be filled with growth media only and used as background correction wells during the seahorse measurement. Place the same cell line at least in three replicates on each plate.
Leave the cells in the incubator at 37 °C with 5 % CO2 overnight and proceed with the seahorse assay the next day.
Seahorse measurement takes place on day 2 following the steps described in below:
Inspect cells in the XF24 culture plate under microscope to assure confluence and even seeding.
Make fresh assay media as described in Subheading 2. Warm it up to 37 °C and adjust pH to 7.4. Transfer the assay media to a 500 mL square reagent bottle that can fit into the seahorse prep station. Leave the bottle in the prep station, which can maintain the temperature at 37 °C. The media change can be started any time from now.
At this point, load the designed seahorse assay program into XF24 software and have the program ready to go. The mitochondrial drugs also need to be taken out for thawing.
Perform media change (see Note 18) and incubate the cells with the assay media in a 37 °C non-CO2 incubator for 60 min (±5 min in all of our assays) for equilibration. Start the seahorse assay program no more than 30 min after media change in the cells (see Note 19).
Dilute thawed mitochondrial drugs in assay media to 10 times of the final working concentration (see Note 20). Load 50–100 μL through each port as needed (see Note 21). Keep the cartridge loaded with compound in the 37 °C non-CO2 incubator for 10 min or more to allow it to warm up before starting the calibration (see Note 19).
At 30 min post media change in the cells, load the sensor cartridge from step 10 (containing compounds in ports) into XF24 flux analyzer instrument tray, and start calibration.
After calibration is complete, replace the utility plate with the pre-incubated (60 min) cell plate and continue the program. The detailed program is as seen in Table 1.
At the end of the assay, take the cell plate out and examine the cells under the microscope to ensure the cells are still attached and look healthy after the measurement. It is optional to lyze the cells in the plate for protein quantitation. Download and analyze the seahorse measurement results (Fig. 1).
Table 1.
Program used for basal OCR, ECAR measurements and mitochondrial function test
| Command | Time (min) |
|---|---|
| Calibrate | Fixed |
| Equilibrate | Fixed |
| Loop Start | 3 |
| Mix | 3 |
| Wait | 2 |
| Measure | 3 |
| Loop End | |
| Inject from port A | |
| Loop Start | 3 |
| Mix | 3 |
| Wait | 2 |
| Measure | 3 |
| Loop End | |
| Inject from port B | |
| Loop Start | 3 |
| Mix | 3 |
| Wait | 2 |
| Measure | 3 |
| Loop End | |
| Inject from port C | |
| Loop Start | 3 |
| Mix | 3 |
| Wait | 2 |
| Measure | 3 |
| Loop End | |
| Program End | |
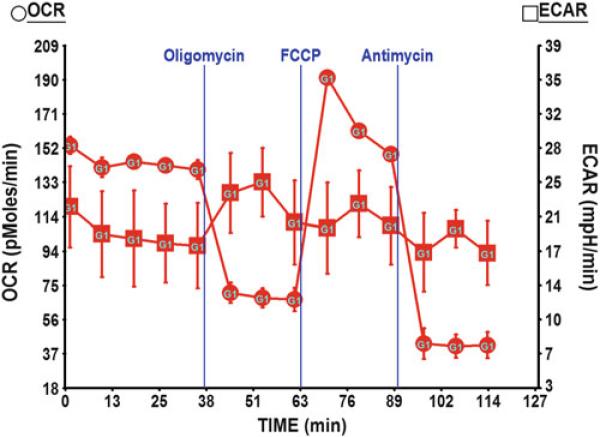
Fig. 1.
OCR and ECAR measurement in Huh7 cells. In the assay media containing 25 mM glucose, Huh7 showed moderate level of basal OCR and relatively high ECAR indicating its preference using glucose for glycolysis. With the optimized concentration of mitochondrial drugs, we saw a ~60 % reduction in OCR following the injection of oligomycin. OCR jumped up by 40 % upon the injection of FCCP indicating the release of maximal respiration capacity. Finally, injection of antimycin blocked all the mitochondrial respiration and revealed the remaining OCR in Huh7 that was from the non-mitochondrial respiration
3.4 Compare the Substrate Utilization and Mitochondrial Function in Uninfected, Scramble (SCR), and SIRT4 KD Huh7 Cells
60,000/well of Huh7, Huh7-SIR4KD, and Huh7-SCR were placed in multiple replicates in one XF24 cell culture plate on day 1.
The rest of the procedures follow what has been described in Subheading 3.3.
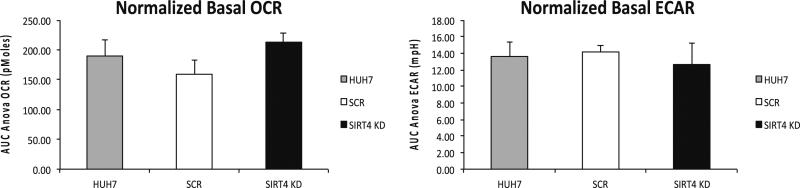
Basal levels of OCR and ECAR, proton lead associated oxygen consumption, maximal mitochondrial respiration and nonmitochondrial respiration were analyzed in these cells (Fig. 2; see Note 22).
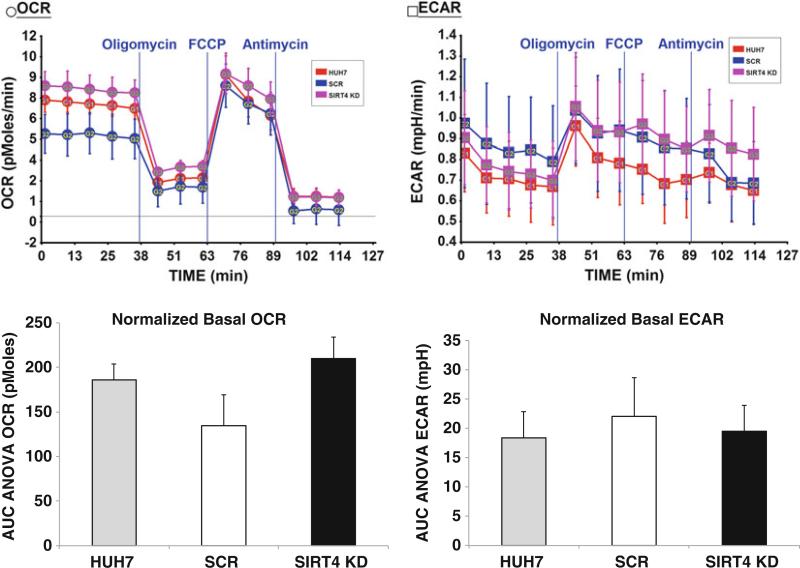
Fig. 2.
OCR and ECAR comparison in uninfected, SCR, and SIRT4 KD Huh7 cells. Seahorse assay was run with the assay media containing 2 mM Glutamax and 25 mM Glucose. Data for each cell line are the average of six or seven replicates and have been normalized to the total protein amount. Basal OCR was higher in SIRT4 KD cells, while basal ECAR remained at similar level in all cell lines. The difference in basal OCR was more significant when comparing the SIRT4 KD cells line to the SCR control (p = 0.014) than to the uninfected Huh7 cells (p = 0.071). ECAR and the disturbed bioenergetics profile post-mitochondrial compounds injection did not show significant difference among the three analyzed cell lines
3.5 Study Glutamine Oxidation in Uninfected, SCR, and SIRT4 KD Huh7 Cells
Use the assay media that contains only glutamine in the base DMEM for this experiment. All the rest of the procedures are the same as described in Subheadings 3.3 and 3.4 (Fig. 3; see Note 23).
Fig. 3.
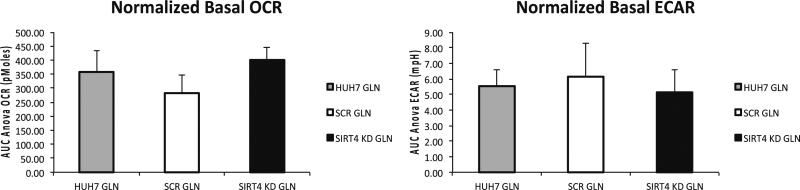
Glutamine oxidation in uninfected, SCR, and SIRT4 KD Huh7 cells. Seahorse assay was run with the assay media containing 4 mM Glutamine. Data represents the average of five replicates for each cell line and has been normalized to the total protein amount. Basal OCR were at the maximal level, while basal ECRA were at minimal level in this media condition for all three analyzed cell lines. The glutamine oxidation in SIRT4 KD cells was the highest comparing to SCR and uninfected Huh7 cells. Again, the OCR difference was more significant when comparing to SCR (p = 0.0089) than comparing to the uninfected Huh7 cells (p = 0.32)
3.6 Study Glucose Oxidation in Uninfected, SCR, and SIRT4 KD Huh7 Cells
Use the assay media that contains only glucose in the base DMEM without glutamax for this experiment. All the rest of the procedures are the same as described in Subheadings 3.3 and 3.4 (Fig. 4; see Note 24).
Fig. 4.
Glucose oxidation in uninfected, SCR, and SIRT4 KD Huh7 cells. Seahorse assay was run with the assay media containing only 25 mM glucose. Data represents the average of six or seven replicates for each cell line and has been normalized to the total protein amount. With the presence of glucose in assay media, ECAR remained at regular level and no significant difference was observed in the three analyzed cell lines. Basal OCR was again increased in SIRT4 KD cells. The increase in basal OCR in SIRT4 KD cells was more significant when comparing to the SCR control (p = 0.00037) than to the uninfected Huh7 cells (p = 0.08)
3.7 FAO Measurement in Uninfected, SCR, and SIRT4 KD Huh7 Cells
On day 1, prepare the seahorse assay and the cells in the same way as described in Subheadings 3.3 and 3.4.
On day 2, follow the same workflow as described in Subheading 3.3. However, for FAO measurement, use 1× KHB buffer as the assay media (see Note 10). In addition, instead of using mitochondrial drugs, the FAO substrate palmitate-BSA and BSA control are used for FAO measurement. Thaw 1 mL aliquot of the palmitate-BSA and BSA control in 37 °C water bath for 5 min when both are used to test FAO procedure. When comparing the FAO between cell lines (Huh7, Huh7-SCR, and Huh7-SIRT4KD, etc.), thaw only one 2 mL aliquot of palmitate-BSA. The thawed palmitate-BSA or BSA control is loaded directly into port A at 75 μL/well without pre-dilution.
Follow the rest of the procedure as described in Subheading 3.3 to run the seahorse assay program. The program used for FAO measurement is as seen in Table 2.
Table 2.
Sample program for fatty acid oxidation measurements
| Command | Time (min) |
|---|---|
| Calibrate | Fixed |
| Equilibrate | Fixed |
| Loop Start | 5 |
| Mix | 3 |
| Wait | 2 |
| Measure | 3 |
| Loop End | |
| Inject from port A | |
| Loop Start | 3 |
| Mix | 3 |
| Wait | 2 |
| Measure | 3 |
| Loop End | |
| Program End | |
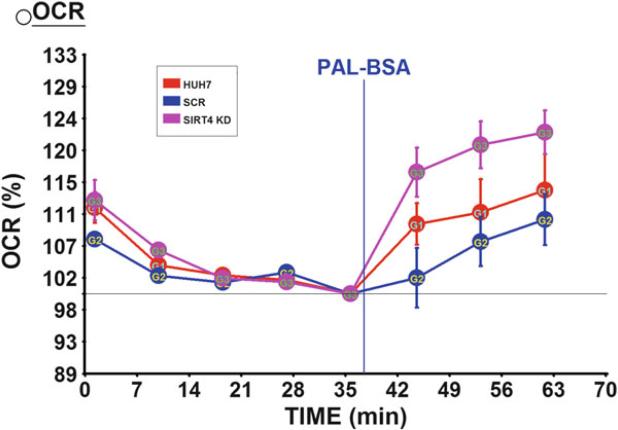
Fig. 5.
Disruption of SIRT4 resulted in increased FAO in Huh7 cells. FAO was measured in a seahorse assay by looking at the percentage increase of OCR in response to the injection of palmitate. Data represents the average of five replicates for each cell line
Acknowledgment
We would like to acknowledge the American Heart Association grants 12SDG8840004 and 12IRG9010008 for funding support (MDH).
Footnotes
The XF24 FluxPak contains 20 XF24 cell culture microplates, 18 XF24 extracellular flux assay kit (each package of the assay kit includes one sensor cartridge, one utility plate, and one lid), and one bottle of 500 mL of XF24 calibrant solution, pH 7.4. The cell culture microplates and calibrant solution (SeahorseBioscience) can be purchased separately. However, the sensor cartridges can only be purchased with the FluxPak.
XF Cell Mito Stress Test Kit contains four mitochondrial drugs (Oligomycin, FCCP, Antimycin, and Rotenone) that can be subsequently added to the cells during a seahorse assay to examine the mitochondrial function. Normally, intracellular substrate oxidation produces ATP and causes oxygen consumption, which is predominantly controlled by the parallel re-entry pathways through the ATP synthase and proton leak. Addition of Oligomycin blocks the ATP synthase and the residual respiration is due to the proton leak. The decrease upon adding Oligomycin approximates to the proton current flowing through the ATP synthase before the inhibitor was added. The decrease compared to basal provides the coupling efficiency. Addition of a carefully calibrated concentration of the protonophore FCCP introduces a high artificial proton conductance into the membrane. This maximal respiration is now controlled by electron transport chain activity and/or substrate delivery. The increased respiratory capacity above basal respiration provides the spare respiratory capacity. Finally, electron transport chain inhibitors (Antimycin or Rotenone) are added; any residual respiration is non-mitochondrial. We have found in our seahorse assays, Antimycin and Rotenone can be equally effective as inhibitors of mitochondrial respiration. Only Antimycin was used in the seahorse assays described in this method. Antimycin A, a complex III inhibitor, binds to the Qi site of cytochrome C reductase, thereby inhibiting the oxidation of ubiquinol in the electron transport chain of oxidative phosphorylation. The inhibition of this reaction disrupts the formation of the proton gradient across the inner membrane. Therefore, the production of ATP is subsequently inhibited, as protons are unable to flow through the ATP synthase complex in the absence of a proton gradient.
Unbuffered Base DMEM can be purchased directly from SeahorseBioscience, which contains 2 mM GlutaMax. The powdered DMEM from sigma contains only the essential components of DME and gives the flexibility to remove l-glutamine from the media, which will potentially facilitate focusing the tests on the oxidation of substrates other than l-glutamine. Using the self-prepared media will also dramatically reduce the cost.
When preparing self-made, unbuffered base DMEM, it is helpful to add the Phenol Red after all other components are well dissolved in water because it can be seen better this way.
It is optional to add or not to add GlutaMax in the base DMEM medium depending on the assay needs. We have used both media as described in this method.
The volume of the assay media is based on using the prep station to change the media in one full (24 wells) XF24 culture plate, because additional volume is required to prime the prep station. If the prep station is not available, prepare 50 mL of the assay media for each full XF24 culture plate and change media manually.
25 mM glucose are used in the assay media to study the basal OCR and ECAR, as well as the mitochondrial function tests because Huh7 cells are normally cultured in media containing high glucose (25 mM). For additional consideration of the assay media, also see Notes 10 and 16.
The pH is temperature-dependent. Because the seahorse measurement is conducted at 37 °C, the assay media needs to be warmed up to 37 °C before adjusting the pH. Filtration of the assay media after the pH adjustment is usually not necessary, as the cells would not typically be put back in culture after the seahorse measurement.
Since we are going to test glutamine oxidation, glucose is completely removed from the assay media so that we can force the cells to use glutamine as their substrate. We add l-glutamine in addition to the glutamax in the media to ensure that the cells have enough fuel.
Frozen carnitine stock is good for up to 1 month and may be used within 3 days of thawing when kept at 4 °C [16]. We always use freshly made carnitine that has been kept frozen for no more than 2 weeks.
In our assays with Huh7 cells, we have found FAO worked the best when no glucose was added in the 1× KHB media. FAO was still measurable when 2.5 mM glucose was present in the assay media, but did not work so well when higher concentration of glucose was used. We think because Huh7 cells have the tendency to use glucose for glycolysis, the presence of glucose will prevent the cells from using other available substrates. Thus the glucose can be completely removed for Huh7 cells when measuring FAO or other substrate utilization to avoid the cells using glucose as the fuel. However, some other cell lines may require the presence of glucose for survival. Therefore, optimization experiments are required for each cell line to determine the ideal glucose concentration that can be used in the assay media.
When the molar ratio of palmitate and BSA is kept at 6:1, we can easily get the clear well-dissolved palmitate/BSA solution. This ratio might be increased a little; however, plamitate will not be fully dissolved in the solution at the ratio of 12:1.
For FAO measurement, make sure to use the fatty acid-free BSA here.
This can be achieved by placing a larger beaker with water on a heated stir plate and maintaining the “water bath” temperature at 37 °C. The glass beaker containing BSA solution can be placed inside in the larger beaker water bath.
In our studies we found that continuous culture of Huh7 cells in puromycin caused a stress on the cells which lead to a decrease in oxidative rate. We recommend using puromycin for selection for 3–5 days, and then remove puromycin during continuous culture of the cells. Since cells are not constantly selected we advise monitoring the levels of infection by mCherry fluorescence, as well as SIRT4 and mCherry mRNA levels.
Removing trypsin may not be necessary, but we prefer to do so, especially when the cells are going to be used for the seahorse assay. Because Huh7 cells are strongly adherent cells, the initial incubation with trypsin usually will not disassociate them from the flask. Therefore, trypsin can be easily aspirated off without losing the cells.
Cells differ in their growth rate, energy demand, preferred nutrients, and tolerance to stress conditions. Optimization of ideal assay conditions for each cell line is the key ensuring the successful seahorse measurement and obtaining meaningful and reproducible results downstream. Typically, one would optimize the following parameters: assay media components; cell seeding density; the concentration of injected compounds; and mix/wait/measure cycle timing [16]. For seahorse assay media, we recommend starting with the media that has the same components as the growth media for that cell line, except that no FBS is added. The components in FBS can be complex and vary quite a bit from lot to lot. Additionally, FBS has buffering capacity and will interfere with the ECAR reading in the seahorse assay. For cells that require FBS for survival during the seahorse measurement, a small amount (<1 %) of FBS can be considered to use in the assay media. We did not use any FBS in our assays as Huh7 cells remain healthy in the media without it.
Besides testing the assay media, a couple of optimization experiments were also done with the focus on determining the ideal seeding density using Huh7 cells in seahorse assays and the working concentration of the mitochondrial drugs that is not toxic to the cells but efficient to stimulate cellular response and allows for looking at the mitochondrial function. In the meantime, we made sure that DMSO (used as solvent for mitochondrial drugs) had no effect on our cells at the concentration we aimed to use. We did not include many replicates in the early optimization experiments. Instead, we set up those optimization experiments in a way that allow us to look for trends and gather information and indication for the later real experiment. Limited to the length of this article, we do not describe details of the setup of the optimization experiments.
The cells were prepared this way for all described seahorse experiments in this article. Always follow the two-step seeding process as recommended in the seahorse assay manual to ensure a momogenous cell layer. Since Huh7 cells are strongly adherent, they become well attached after 1 h. For cells that are not strongly adherent, plan 3–5 h of time to allow the cells fully attached before adding additional media.
Use the same program in the prep station to change media in each experiment for consistency. Basically, growth media were aspirated off first. 675 μL/well of assay media were then dispensed to wash the cells once. After the assay media was aspirated, a final of 675 μL of assay media were added in each well. The media change can be done manually by following the same steps. When manually aspirating the media off, make sure to leave a small amount (50 μL) of media behind in each well to avoid detaching the cells when media is added back. The volume of assay media per well can vary from 500 μL to 1 mL. We use 675 μL/well in our assays described here.
Calibration is the first step in the seahorse assay program checking on the sensors in calibrant buffer. This step takes about 30 min to complete. The measurement on the cells plate can only be started after the calibration is finished. Therefore, the seahorse assay program should be started no more than 30 min after media change in the cells to avoid extended incubation of the cells in the assay media. As mentioned earlier, seahorse assay media is unbuffered media usually containing a limited amount of nutrients and no FBS, therefore extended incubation of the cells in the assay media may result in depletion of nutrients or a dramatic change in media pH, which are unfavorable conditions for the cells to function normally. Therefore, the subsequent seahorse measurements will not be reliable, reproducible, and informative.
On the other hand, the calibration should not be done too early either because the data accuracy may be compromised. Ideally, each experiment should follow the same time frame (±5 min), which will generate consistent results and allow for data comparison across the experiments. Good planning prior to a seahorse assay can usually ensure a smooth workflow. Also, having the seahorse assay program preloaded onto the computer before starting the media change in the cells is usually helpful to keep up with the time.
From the optimization experiments, we learned Oligomycin, FCCP, and Antimycin worked at 1, 0.4, and 1 μM, respectively when seeding Huh7 cells at 60,000/well. We therefore diluted Oligomycin, FCCP, and Antimycin in the assay media to 10, 4, and 10 μM, respectively, which were ready to be loaded in the ports of the sensor cartridge. Because the stock concentration of these drugs is 2.5 mM, dilution of each drug in the assay media is more than 2,500 fold in our assay. At this dilution, DMSO has been shown no effect on the performance of Huh7 cell. Additionally, when the dilution fold is this high, we do not have to worry about the pH change in the assay media that is possibly introduced by the drug solution. Therefore, it is important to make a stock compound or test agent at a high concentration when it is possible.
The volume load from each port should be 1/10 of the final volume in each well in the XF culture plate. That is why we have our compounds diluted to 10 times of their final working concentration. In our assay, we load 675 μL/well assay media in the XF culture plate, we therefore load 75 μL of 10 μM Oligomycin in port A, 83 μL of 4 μM FCCP in port B, and 93 μL of 10 μM Antimycin in port C, respectively. All wells are loaded with prepared compound, including the background correction wells.
At the end of the assay, assay media were aspirated off and 200 μL of protein lysis buffer were added into each well to lyse the cells. Collect the supernatant of the cell lysis and perform BCA assay to quantitate protein. The results shown in Fig. 2 through Fig. 4 were normalized to the protein amount. Figure 2 indicated that the disruption of SIRT4 increased the substrate oxidation (OCR) but not glycolysis (ECAR) at basal level in Huh7 cells. The perturbed OCR and ECAR in response to mitochondrial compounds did not vary in SIRT4 KD cells and the control cell lines. In our experiments, we have also noticed that the increased basal OCR in SIRT4 KD cells was consistently significant when comparing to the scramble control, but was not always significant when comparing to the uninfected Huh7 cells. We attributed the discrepancy in basal OCR between the scramble control and uninfected cell control to the viral infection and puromycin selection in scramble cells. The OCR difference between the SCR and uninfected cells was never significant. We think the scramble cells are more reasonable than the uninfected cells to serve as a control in our experiments. However, we included uninfected Huh7 cells in all our assays for complete information.
Since we saw elevated substrate oxidation in SIRT4 KD cells from Fig. 2, we designed seahorse assays to study specific substrate oxidation. Figure 3 showed results from an experiment that only glutamine was used in the base DMEM assay media. The results indicated that glutamine oxidation was slightly increased in SIRT4 KD cells. Additionally, in the media condition without presence of glucose, glycolysis (ECAR) level is at minimal level in all cell lines.
Figure 4 showed that in the assay media containing only glucose, glycolysis remained at similar level between the SIRT4 KD cells and the control cells; however, basal OCR was again higher in SIRT4 KD cells. Together with the results in Figs. 2 and 3, SIRT4 KD Huh7 cells showed increased mitochondrial respiration. SIRT4 gene knockdown did not seem to have any impact on glycolysis level in Huh7 cells.
Because we inject the substrate of FAO through the port after the basal measurement, we look at the OCR increase percentage above the basal to compare the FAO level in each cell line. This can be achieved by normalizing the data to the last measurement at basal level as shown in Fig. 5. Previously, a report showed that shRNA-SIRT4 adenovirus-mediated Sirt4 KD in primary mouse myotubes and hepatocytes increased FAO, which was measured with [3H] palmitate [14]. Our data reproduced this finding in human Huh7 cells with the label-free seahorse assay.
References
- 1.Clark LC, Jr, Kaplan S, Matthews EC, Edwards FK, Helmsworth JA. Monitor and control of blood oxygen tension and pH during total body perfusion. J Thorac Surg. 1958;36(4):488–496. [PubMed] [Google Scholar]
- 2.Ferrick DA, Neilson A, Beeson C. Advances in measuring cellular bioenergetics using extracellular flux. Drug Discov Today. 2008;13(5–6):268–274. doi: 10.1016/j.drudis.2007.12.008. doi: 10.1016/j.drudis. 2007.12.008. [DOI] [PubMed] [Google Scholar]
- 3.Hirschey Matthew D. Old enzymes, new tricks: sirtuins are NAD+-dependent de- acylases. Cell Metab. 2011;14(6):718–719. doi: 10.1016/j.cmet.2011.10.006. doi: 10.1016/j.cmet.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson KA, Hirschey MD. Mitochondrial protein acetylation regulates metabolism. Essays Biochem. 2012;52:23–35. doi: 10.1042/bse0520023. doi: 10.1042/bse0520023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Jr, Alt FW, Kahn CR, Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464(7285):121–125. doi: 10.1038/nature08778. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirschey MD, Shimazu T, Jing E, Grueter CA, Collins AM, Aouizerat B, Stancakova A, Goetzman E, Lam MM, Schwer B, Stevens RD, Muehlbauer MJ, Kakar S, Bass NM, Kuusisto J, Laakso M, Alt FW, Newgard CB, Farese RV, Jr, Kahn CR, Verdin E. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol Cell. 2011;44(2):177–190. doi: 10.1016/j.molcel.2011.07.019. doi: 10.1016/j.molcel.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Picklo MJ., Sr Ethanol intoxication increases hepatic N-lysyl protein acetylation. Biochem Biophys Res Commun. 2008;376(3):615–619. doi: 10.1016/j.bbrc.2008.09.039. doi: 10.1016/j.bbrc.2008.09.039. [DOI] [PubMed] [Google Scholar]
- 8.Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010;12(6):662–667. doi: 10.1016/j.cmet.2010.11.015. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 9.Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM, Jacobson MP, Verdin E. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 2010;12(6):654–661. doi: 10.1016/j.cmet.2010.11.003. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, He B, Chen W, Zhang S, Cerione RA, Auwerx J, Hao Q, Lin H. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334(6057):806–809. doi: 10.1126/science.1207861. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, Luo H, Zhang Y, He W, Yang K, Zwaans BMM, Tishkoff D, Ho L, Lombard D, He T-C, Dai J, Verdin E, Ye Y, Zhao Y. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics. 2011;10(12) doi: 10.1074/mcp.M111.012658. doi:10.1074/mcp.M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137(3):560–570. doi: 10.1016/j.cell.2009.02.026. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits gluta-mate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126(5):941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 14.Nasrin N, Wu X, Fortier E, Feng Y, Bare OC, Chen S, Ren X, Wu Z, Streeper RS, Bordone L. SIRT4 regulates fatty acid oxidation and mitochondrial gene expression in liver and muscle cells. J Biol Chem. 2010;285(42):31995–32002. doi: 10.1074/jbc.M110.124164. doi: 10.1074/jbc.M110.124164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, Van Parijs L, Jaenisch R, Jacks T. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci U S A. 2004;101(28):10380–10385. doi: 10.1073/pnas.0403954101. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.SeahorseBioscience XF24 extracellular flux analyzer and prep station installation and operation manual. Seahorse Bioscience Inc. 2009:1–159. [Google Scholar]