SUMMARY
Despite extensive study, few therapeutic targets have been identified for glioblastoma (GBM). Here we show that patient derived glioma sphere cultures (GSCs) that resemble either the proneural (PN) or mesenchymal (MES) transcriptomal subtypes differ significantly in their biological characteristics. Moreover, we found that a subset of the PN GSCs undergo differentiation to a MES state in a TNFα/NF-κB dependent manner with an associated enrichment of CD44 subpopulations and radio-resistant phenotypes. We present data to suggest that the tumor microenvironment cell types such as macrophages/microglia may play an integral role in this process. We further show that the MES signature, CD44 expression, and NF-κB activation correlate with poor radiation response and shorter survival in patients with GBM.
INTRODUCTION
Glioblastoma (GBM) is the most common malignant primary central nervous system (CNS) tumor in adults and remains resistant to current therapies (Furnari et al., 2007) Ample evidence exists to argue that GBM, as defined by histopathologic criteria, actually represents multiple distinct molecular entities (Huse et al., 2011). GBM can be segregated into subtypes based on gene expression signatures. While the precise classifications have varied in the literature (Cooper et al., 2010; Huse et al., 2011; Phillips et al., 2006; Verhaak et al., 2010) two subtypes, termed proneural (PN) and mesenchymal (MES), appear robust and generally consistent among the classification schemes. GBMs in the MES subclass are predominantly primary tumors that arise de novo, and, in some studies, exhibit a worse prognosis compared to PN tumors (Colman et al., 2010; Pelloski et al., 2005; Phillips et al., 2006), which may be related to the fact that a subset of the PN tumors display mutations in the isocitrate dehydrogenase 1 gene (IDH1) as well as the glioma-CpG island methylator phenotype (G-CIMP), both favorable prognostic factors (Noushmehr et al., 2010; Verhaak et al., 2010). Conversely MES tumors are G-CIMP−, exhibit wild type (WT) IDH1, and contain alterations in NF1 (Noushmehr et al., 2010; Verhaak et al., 2010).
While a wealth of data on molecular alterations in GBM continues to accumulate, the availability of relevant models that mirror these alterations is limited. Current evidence points toward the existence of a small fraction of tumor initiating cells in the bulk tumor that also exhibit radio-resistant properties (reviwed in Chen et al., 2012). However, the genetic and epigenetic alterations underlying TICs derived from glioma sphere cultures (GSCs) are less characterized. While initial studies identified CD133 as a tumor initiating marker, CD133− subpopulations that resemble the MES subtype also retain the capacity to form tumors in orthotopic transplantation models (reviewed in Stopschinski et al., 2012). Consequently, additional cell surface antigens have been proposed as tumor initiating markers for GSCs including CD44 (Brescia et al., 2012; Jijiwa et al., 2011), a marker that is enriched in cancer stem cells as well as those that undergo epithelial to MES transition (EMT) (Zoller, 2011). Interestingly, MES transition has also been shown to occur in GBM and can be induced by master transcription factors (TFs), STAT3, C/EBPβ and TAZ (Bhat et al., 2011; Carro et al., 2010). Whether this transition occurs in a cell-intrinsic manner or can be influenced by factors secreted in the tumor microenvironment is not known. Furthermore, whether MES differentiation leads to enrichment of the CD44 subpopulation in a fashion similar to other solid tumors remains unexplored. Finally, PN tumors have been found to give rise to MES recurrences, suggestive of a PN to MES transition (Phillips et al., 2006). Therefore, understanding the mechanistic basis of MES differentiation may have implications for the treatment of GBM.
RESULTS
Patient derived GSCs bear resemblance to PN and MES signatures
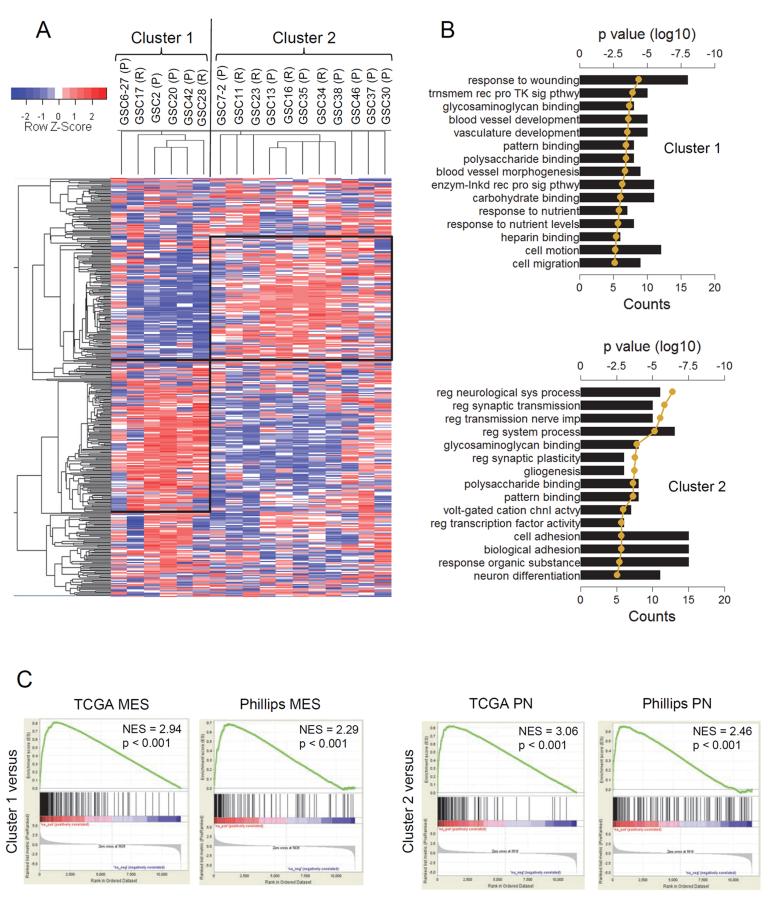
In the context of molecular subtypes reported for GBM, we examined if GSCs isolated from patient derived tumors show similar characteristics. Forty one GBM tumors were subjected to culture conditions according to published protocols and successful expansion as neurospheres was observed in thirty three cases (Table S1). Seventeen GSCs that were expanded earliest were chosen for microarray analysis to identify molecular subtypes using unsupervised algorithms. Using 500 probe-sets with the highest variability in gene expression, two clusters of co-expressed genes were readily apparent by hierarchical clustering (Figure 1A). These two clusters for the most part were well-defined, although some GSCs did not readily fit in this pattern (e.g., GSC6–27, 30 and 46). The primary or recurrent status of the parental tumor of origin had no bearing on the cluster segregation (cluster 1 = 33% versus cluster 2 = 36% recurrent tumors, Figure 1A). To understand the functional significance of these two gene clusters, we performed Gene Ontology (GO) analysis using the Database for Annotation, Visualization and Integrated Discovery (DAVID) webtool (Dennis et al., 2003). Cluster 1 GO terms were enriched for wound response, vasculature formation, and cell motility gene signatures (Figure 1B), whereas cluster 2 showed predominant association with differentiated neural or glial cell functions and homeostatic activities (Figure 1B). Importantly, cluster 1 showed significant similarity only to the MES GBM subclass by Gene Set Enrichment Analysis (GSEA, Subramanian et al., 2005) (Figure 1C), with 89 out of the top 500 enriched genes being MES (Figure S1A and Table S2). Similarly, cluster 2 was predominantly comprised of PN genes (98/500, Figure 1C, S1A and Table S2). Supervised clustering using the TCGA classification (Verhaak et al., 2010) showed a similar grouping of the GSCs at the first branch of the dendrogram compared to the unsupervised clustering (Figure S1B). GSC 6–27 and 30 displayed characteristics of both MES and PN gene signatures. GSC11 and 30 were also enriched for Classical (CL) signatures. EGFR amplification, usually restricted to the CL subtype, was seen in 5 out of the 14 GSCs and appeared distributed between the PN and MES subtypes (3 PN and 2 MES, Figure S1C). NF1 homozygous inactivating mutations were observed in GSC6–27 (exon 39), and GSC28 (exon 50, and exon 38; Figure S1D), both GSCs that had MES characteristics consistent with the TCGA analyses (Verhaak et al., 2010). Quantitative RT-PCR (qRT-PCR) and immunoblotting of basal expression of key PN/MES markers (Table S3) were concordant with microarray results (Figure S1E and F).
Figure 1. Patient Derived GSCs Bear Resemblance to PN and MES Signatures.
(A) Unsupervised hierarchical analysis of the top 500 highest median absolute deviation genes from expression microarray of seventeen GSCs. Expression data was Z-score corrected for display; relatively lower expression is shown in blue and higher expression is shown in red (see color key). Two large clusters, cluster 1 (128 genes) and cluster 2 (102 genes) were identified (shown as black boxes). The vertical black line identifies the first dendrogram splitting of the GSCs. Primary (P) or recurrent (R) status of the GSCs is indicated. (B) The top twenty gene ontology (GO) terms associated with cluster 1 (left) and cluster 2 (right) from the unsupervised GSC hierarchical cluster analysis. GO terms are ranked by p value. The black bars show the number of genes that is common between the GO term's gene set and the respective cluster gene set. The golden line is the log10 of the p value as determined by DAVID functional analysis. (C) GSEA enrichment plots of GSC cluster 1 high (top row) and cluster 2 high (bottom row) gene lists versus queried gene lists (see Supplemental Experimental Procedures for data source). The normalized enrichment scores (NES) and the p values are shown for each plot. See also Figure S1 and Table S1–S5.
To rule out the possibility of non-neoplastic cells being enriched in the GSC isolation procedure, we tested for loss of heterozygosity (LOH) on Chromosome 10q, a frequently deleted region in adult GBM (Pietsch and Wiestler, 1997), and found LOH in 12 out of the 13 GSCs confirming their neoplastic origin (Figure S1G). Implantation of 5 × 105 or fewer unsorted GSCs caused formation of high grade gliomas (HGGs) in a majority of the cases (13/17) with predominant histologic features of this disease (Table S4, S5 and Figure S1H). A subset of the tumors exhibited microvascular proliferation, and/or pseudo-palisading necrosis, both hallmarks of GBM (Figure S1H). Thus, despite differential gene expression signatures, GSCs formed tumors that were histologically similar.
GSCs differ in the transcriptome and epigenetic profiles when compared to the originating tumor
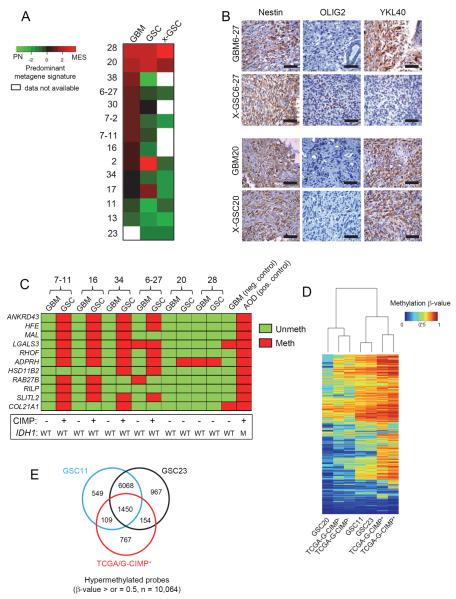
Next, we examined if gene expression patterns observed in the GSCs and xenografts matched with the respective parental GBMs from which they were derived. In order to determine the association of a sample with either a PN or MES gene expression signature, we calculated a metagene score for each sample using a set of four PN (DLL3, OLIG2, ASCL1, and NCAM1), and four MES (YKL40, SERPINE1, TIMP1, and TGFBI) genes, seven out of the eight of which were subset defining in published datasets (Phillips et al., 2006; Verhaak et al., 2010). Using this approach, each sample could be ranked according to its metagene score as being predominantly PN or MES. Surprisingly, we found that most GSCs that arose from MES tumors lost MES characteristics and exhibited a higher PN metagene (Figure 2A). GSC20 and GSC28, both of which originated from highly MES tumors with histological hallmarks of gliosarcoma (Figure S2A), maintained their MES state in culture and as xenografts. With the exception of GSC2 and GSC17 (which lost MES features gained in culture), all PN GSCs maintained their PN status when examined in xenografts. Alterations in growth factor supplements to culture media did not influence gene expression signatures in established GSCs, although neurosphere formation was dependent on EGF (Figure S2B and S2C). Immunohistochemical (IHC) analysis on xenografts showed absence of the MES marker YKL40 in GSCs derived from MES tumors which instead acquired the PN marker OLIG2 (Figure 2B and S2D). This contrast was not observed in the MES GSC20 that retained YKL40 expression in the xenograft, similar to the parental tumor, but lacked OLIG2 expression in both (Figure 2B). These findings imply that the GSC isolation protocol generally favors a loss of MES and gain of PN features from patient to xenograft.
Figure 2. GSCs Differ in the Transcriptome and Epigenetic Profiles When Compared to the Originating Tumor.
(A) Heatmap of the predominant signature of initiating GBM, derived GSC, and xenograft for fourteen samples is shown. A PN and MES qRT-PCR based metagene was calculated for each sample and then compared to each other after Z-score correction. Green shades represent a predominantly PN signature, red a MES one, and black a relatively balanced expression of both, as indicated in the figure. (B) IHC analysis of Nestin, OLIG2 and YKL40 expression in patient matched GBM and xenografts of GSCs. Scale Bar: 50 μm. (C) Methylight profiling of GBMs and their derivative GSCs for G-CIMP status. Eleven markers were tested for presence of methylation on their promoters and coded as red if methylated and green if unmethylated. Samples were deduced as G-CIMP if >50% of the loci showed methylation. A GBM and an anaplastic oligodendroglioma (AOD) sample were used as negative and positive controls respectively. The IDH1 and G-CIMP status of all samples is shown below. (D) Heatmap shows the unsupervised clustering of 2612 G-CIMP signature methylation probes based on p values for the indicated cell types. (E) Venn diagram showing the number of hypermethylated probes (β-values of >0.5) in GSCs 11, 23 and TCGA G-CIMP+ tumors. See also Figure S2.
Given the tight association of the G-CIMP signature with the PN subtype (Noushmehr et al., 2010), we asked if PN GSCs are G-CIMP+. To test this, we used a previously reported G-CIMP signature panel that shows correlation with array based methylation platforms (Noushmehr et al., 2010). Eleven hypermethylated gene regions were chosen based on feasibility and reproducibility in archival tumor derived DNA and deemed G-CIMP+ based on percentage loci that were hypermethylated (> 50%). Consistent with a drift toward a PN signature from tumor to GSC, we observed that PN GSCs were G-CIMP+ compared to their parental tumors that were G-CIMP− (Figure 2C and S2E). To the contrary, MES GSCs 20 and 28 remained G-CIMP− similar to their parental tumors (Figure 2C). To test for the extent of similarity to the previously described G-CIMP signature on a genome wide scale, we profiled three GSCs using the Illumina Infinium methylation array (Figure 2D). Upon cross examination with TCGA tumors, PN GSCs 11 and 23 segregated with G-CIMP+ tumors, while GSC20 clustered with the G-CIMP− cases. Overall, GSCs 11 and 23 showed greater hypermethylation compared to GSC20 (Figure S2F) and showed a ~70% enrichment of G-CIMP signature genes, although numerous distinct non-G-CIMP loci also appeared hypermethylated in these GSCs (Figure 2E). Taken together, our data suggests that PN GSCs can exhibit hypermethylation patterns (henceforth named CIMP) with similarities to G-CIMP even in the absence of IDH1 mutations.
Molecular signatures differ between GBM and their derivative GSCs even in early passages
Based on our initial findings, two possibilities were considered; 1) culturing of freshly resected GBMs in serum free media supplemented with growth factors preferentially induces a PN/CIMP+ signature in culture, or 2) most undifferentiated GSCs are innately PN/CIMP+, but that the microenvironment in human tumors induces a reversible MES/CIMP− differentiation, which is not entirely recapitulated in vitro or in xenografts of immunocompromised mice. To discern which one of these can be attributed to a general MES/CIMP− to PN/CIMP+ drift, and to reduce the potential of artifacts from long-term culture, we examined freshly resected tissues and their derivative serial passage GSCs for gene expression and methylation signatures as soon as sufficient starting material was available for analyses. Strikingly, even in early passages (less than five), we observed that GSCs showed PN characteristics despite having a MES origin (Figure S2G). Moreover, these early passage GSCs were CIMP+ in contrast to their parental tumor which were CIMP− (Figure S2H). These observations, taken together with previous studies showing requirement of extended passages for the induction of the G-CIMP phenotype by IDH1 mutation (Lu et al., 2012; Turcan et al., 2012), favors a model in which, a majority of undifferentiated GSCs already exist in a PN/CIMP+ state and are selectively enriched under proliferating conditions.
CD44 is enriched in the MES subtype and is inversely correlated with OLIG2 expression
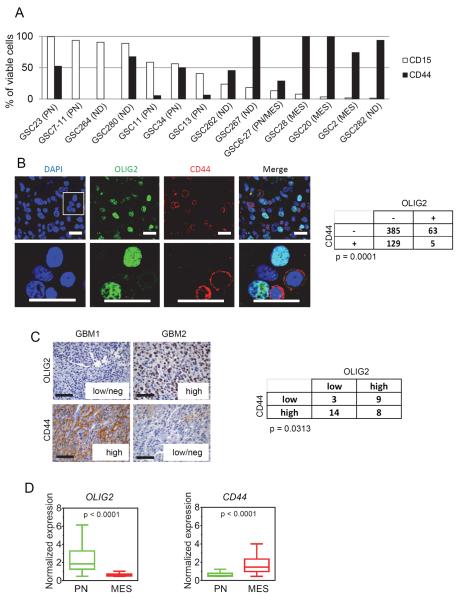
Next, to test if the differential molecular signatures have a bearing on their biological properties, we expanded our repertoire of GSCs. We first examined the expression of cell surface markers that have been used to define tumor initiating potential. We observed enrichment of CD15 specifically in the PN/CIMP+ subclass of GSCs (e.g., GSC11, GSC23, GSC34) that also expressed equal or smaller percentages of CD44 (Figure 3A), although the ratio of CD15 to CD44 varied with passage or confluence of spheres. MES/CIMP− GSCs (e.g., GSC20, GSC28, GSC2) did not express appreciable levels of CD15, but predominantly expressed CD44 (Figure 3A). Comparison of CD15 and CD44 expression among GSCs with a range of passage times showed no correlation (Figure S3A and B). Using OLIG2 as a surrogate for CD15 (Figure S3C) as previously shown (Son et al., 2009), we found a mutually exclusive pattern of inter- and intratumoral staining with CD44 (Figure S3D, 3B and 3C) implying that these were indeed distinct tumor populations. Furthermore, PN tumors expressed higher levels of OLIG2 whereas MES tumors predominantly expressed CD44 and the expression of OLIG2 and CD44 was inversely correlated (Figure 3D and S3E). Additionally, CD44high subpopulations within PN/CIMP+ GSCs showed enrichment of MES markers (Figure S3F). Thus, while unsorted GSCs formed tumors upon transplantation irrespective of exclusive expression of tumor initiation markers, CD15 (e.g., GSC7–11), or CD44 (e.g., GSC20) the proportion of the cell surface expression of CD44 appeared to correlate with a MES state.
Figure 3. CD44 is Enriched in the MES Subtype and is Inversely Correlated with OLIG2 Expression.
(A) CD15 and CD44 of various GSCs were determined by flow cytometry. Bar graph indicates percentage of viable cells that express these markers at the earliest passage tested. ND = not determined. (B) Immunofluorescent staining of OLIG2 (green) and CD44 (red) in human GBM tumors showing a mutually exclusive pattern of staining. Scale bar: 20 μm. The merged image of CD44/OLIG2 is shown on the right against 4',6-diamidino-2-phenylindole (DAPI) stained nuclei (blue). Enlarged inset is shown in the lower panel (scale bar: 20 μm). Quantification of staining in three random fields of three independent tumors and the p value from Chi Square test is shown on the right. (C) Representative IHC images of OLIG2 and CD44 expression in human GBM samples. Scale bar: 50 μm. The table to the right shows the number of tumors expressing OLIG2/CD44. Tumors were classified as low/negative, intermediate, or high depending on the extent of expression in the overall tumors. p value was calculated using Chi Square test. (D) Box plots showing the normalized median expression of OLIG2 and CD44 in TCGA tumors based on Phillips and TCGA classification. Boxes show median 25th and 75th percentile, while whiskers show the 5th and the 95th percentile. p value was determined using a non-parametric Wilcoxon test. See also Figure S3.
PN/CIMP+ and MES/CIMP− GSCs display differential sensitivity to radiation
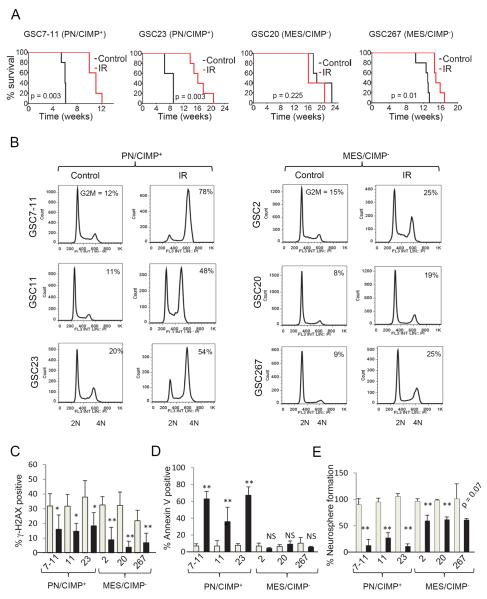
To test if GSCs with varied transcriptome, methylation and cell surface antigen expression patterns also exhibit differential treatment responses, we examined the consequence of clinically relevant fractionated ionizing radiation (IR) (2.5 Gy × 4) on mice 2–3 weeks after orthotopic implantation of GSCs. PN/CIMP+ GSCs (7–11 and 23) showed significantly improved median survival (5–9 weeks) upon IR treatment whereas GSC20 showed no statistical difference and GSC267 showed modest survival (~2 weeks) improvement compared to untreated controls (Figure 4A). Since glioma cells predominantly arrest in the G2/M phase of cell cycle in response to IR (Mir et al., 2010), we asked if the two subtypes of GSCs showed fundamental differences in this mode of arrest. Indeed PN/CIMP+ GSCs showed dramatic accumulation of cells in G2/M, whereas MES/CIMP− GSCs showed only a modest arrest (Figure 4B). In addition, while both subtypes of GSCs showed comparable γ–H2AX foci formation at early time points, MES/CIMP− GSCs (2, 20 and 267) showed enhanced repair ability as evidenced by the reduced number of foci at 24 h compared to PN/CIMP+ GSCs (Figure 4C) consistent with the lack of G2/M arrest. Consequently, the PN/CIMP+ GSCs underwent profound apoptosis (Figure 4D) with reduced neurosphere formation compared to MES/CIMP− GSCs (Figure 4E). We further observed a similar radio-resistant CD44high population within the PN/CIMP+ GSCs (Figure S4A–C).
Figure 4. PN/CIMP+ and MES/CIMP− GSCs Display Differential Sensitivity to Radiation.
(A) Kaplan Meier curves showing survival of mice implanted with PN/CIMP+ (7–11 and 23) or MES/CIMP− (20 and 267) GSCs at 5 × 105 cells per mice with or without fractioned intracranial radiation (2.5 Gy × 4). t test was used to assess statistical significance. (B) Cell cycle analysis of GSCs treated with 6 Gy IR. The percentage of cells in the G2/M phase is indicated within each cell cycle plot. (C) γ-H2AX foci formation assay. Gray bars indicate number of foci after 6 h irradiation whereas black bars show foci after 24 h. At least 25 nuclei were counted. Error bar indicates +/− SEM. t test was used to assess statistical significant differences. *p < 0.05, **p < 0.005 (D) Percentage of cells that were positive for annexin V staining 96 hours post irradiation is shown as bar graphs. Gray bars indicate percentage of cells in untreated population whereas black bars show percentage of annexin V positive cells exposed to 6 Gy IR. Error bar indicates +/− SD. t test was used for statistical significance. **p < 0.005, NS = not significant (E) Neurosphere formation efficiency was determined by setting the number of spheres formed in control groups at 100% (gray bars) and compared to those exposed to 3 Gy IR (black bars). Error bar indicates +/− SD. t test was used for statistical significance. **p < 0.005. See also Figure S4.
TNFα mediates MES differentiation in an NF-κB dependent fashion
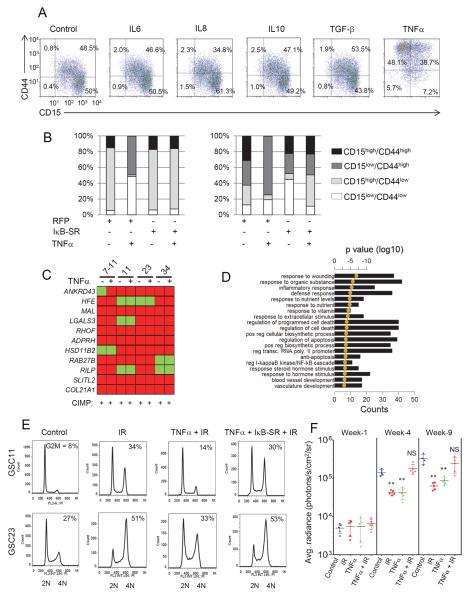
Despite originating from MES tumors, the lack of MES signature in xenografts led us to hypothesize that specific factors in the human tumor microenvironment could alter the transcriptome and epigenetic signatures of GSCs, but these features are not entirely recapitulated in immunocompromised mice. In search of such signaling molecules, we noted that the TCGA analyses showed specific enrichment of genes in the TNFα receptor super-family and the NF-κB pathway in the MES subclass of tumors that also expressed high levels YKL40 and CD44 (Riddick and Fine, 2011; Verhaak et al., 2010). Additionally, prior studies have shown the association of a hypoxic signature and the NF-κB pathway to HGGs (Murat et al., 2009). We hypothesized that cytokines that can trigger NF-κB or alternative TF signaling pathways could result in MES differentiation and influence the proportion of CD44high subpopulations. To test this, we treated PN/CIMP+ GSCs 11, 23 and 34 with similar concentrations of IL6, IL8, IL10, TGF-β or TNFα, all of which are constituents of the GBM microenvironment (Charles et al., 2011). TNFα treatment resulted in a dramatic gain of CD44 expression, an effect not seen with any of the other cytokines tested (GSC34, Figure 5A; GSC11 and 23, data not shown), and this effect was blunted by transduction with a non-degradable mutant form of IκB (IκB super-repressor [IκB-SR]), indicating an NF-κB dependent effect (Figure 5B). Interestingly, the CIMP positivity of the GSCs remained unaltered in response to TNFα treatment (Figure 5C) implying that the regulation of CIMP can be uncoupled from NF-κB mediated MES differentiation in GSCs. However, long term effects of TNFα and/or other potential modifiers of CIMP status are worthy of further exploration.
Figure 5. TNFα Mediates MES Differentiation and Radio-Resistance in an NF-κB-Dependent Fashion.
(A) FACS analysis of expression of CD15 and CD44 in GSC34 after 96 h treatment with 10 ng/ml of indicated cytokines. Percentage of cells in each quadrant is shown. (B) Expression of CD15 and CD44 after TNFα (96 h, 10 ng/ml) with or without pre-treatment with IκB-SR adenovirus of control RFP adenovirus 24 h prior to TNFα treatment by flow cytometry. Stacked bar shows percentage of CD15/CD44 expressing cells after various treatments. (C) Methylight profiling of GSCs after two weeks of TNFα treatment is shown. (D) The top twenty GO terms associated with 1.5 fold or greater TNFα induced genes in GSC11 ranked by lowest p value. Bar graphs show the number of genes overlapping between the GO category and the query gene list. The golden line is the DAVID functional analysis determined log10 of p values. (E) Cell cycle analysis of GSCs after treatments as indicated. The percentage of cells in the G2/M phase is indicated within each cell cycle plot. (F) Tumor volume measurement of GSC23-pCignal lenti-CMV-luc cells injected intracranially into Foxn1nu mice. Mice were imaged 2–3 weeks after implantation as the first timepoint (denoted as week 1) after which the radiation group received four cycles of 2.5 Gy IR on consecutive days. Mice were subject to intracranial administration of TNFα (2 ng/mice) 72 h prior to irrradiation and once every two weeks thereafter. Horizontal black bar shows average radiance (photons/s/cm2/sr) with various treatments and time points. Error bar indicates +/− SEM. t test was used to assess statistical significance. **p < 0.005. NS = not significant. See also Figure S5.
To further characterize the extent of MES differentiation, we performed microarray analysis of GSCs treated with TNFα and found significant enrichment of genes involved in wound healing and vasculature development, as well as, the NF-κB cascade and regulation of cell death related genes (Figure 5D), suggesting that in addition to the canonical NF-κB pathway, TNFα induces a parallel MES differentiation in GSCs, which was further confirmed by qRT-PCR (Figure S5A). Moreover, genes induced by TNFα were significantly similar to the GBM MES subclass (Figure S5B). Interestingly, although a global reduction of the PN signature was not observed, a significant downregulation of OLIG2, PDGFRA, and DLL3 transcripts were seen with TNFα treatment (data not shown). Physiological concentrations (100 pg/ml) of TNFα were sufficient to cause induction of YKL40 and CD44 (Figure S5C and S5D), which was temporally preceded by activation of NF-κB, as judged by serine 536 (ser 536) phosphorylation (Figure S5D). Furthermore, we identified macrophages/microglia as the stromal cell type that can potentially induce MES differentiation (Figure S5E–I) and that MES/CIMP− GSCs show selective susceptibility to minocycline, an inhibitor of microglial activation and NF-κB signaling (Figure S5J–N) (Daginakatte and Gutmann, 2007; Markovic et al., 2011).
Pre-treatment of PN/CIMP+ GSCs with TNFα strongly reduced the G2/M accumulation in response to IR (Figure 5E) as well as the number of γ-H2AX foci (Figure S5O), and these effects were inhibited by pre-treatment with IκB-SR, indicating that TNFα promotes MES differentiation coupled with increased radio-resistance in an NF-κB-dependent manner. We noted that while long term treatment of GSCs with TNFα (5 ng/ml) reduced the neurosphere formation (Figure S5P), exposure of GSCs to IR caused significantly higher neurosphere efficiency in the presence of TNFα compared to untreated controls suggesting a radio-protective effect of TNFα (Figure S5P). To examine this in vivo, we expressed firefly luciferase in GSC23 to monitor tumor kinetics using bio-luminescent imaging. As expected, IR caused a strong decrease in tumor volume (Figure 5F) and a similar growth inhibition was seen with TNFα treatment alone, consistent with our in vitro observations. However, a combination of TNFα and IR caused significant expansion of the tumor and the cells appeared similar in volume to the control group. Overall, our findings indicate that the induction of MES differentiation and enrichment of CD44 by NF-κB activation promotes radio-resistance in PN/CIMP+ GSCs.
NF-κB controls master TFs of MES differentiation in GSCs
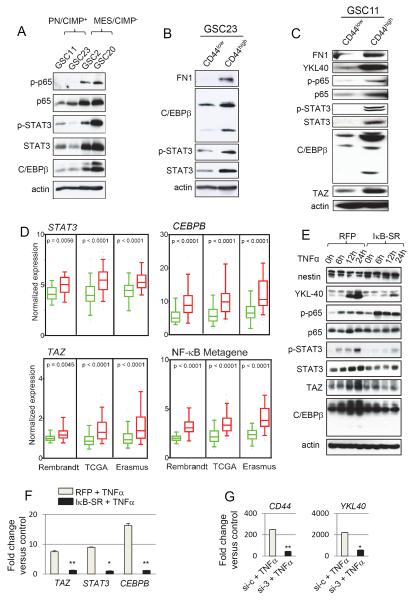
Next, we explored how NF-κB pathway activation integrates into the MES signaling network and its relation to master TFs (STAT3, C/EBPβ, and TAZ) known to induce this signature. We found that both total and phosphorylated forms of p65 (ser 536) were significantly higher in MES/G-CIMP− GSCs 2 and 20 when compared to PN/CIMP+ GSCs 11 and 23 (Figure 6A). The expression of STAT3 and C/EBPβ as well as phosphorylation at tyrosine 705 of STAT3 (which promotes nuclear translocation and DNA binding of STAT3) was also higher in the MES/CIMP− GSCs (Figure 6A). Similar increases in MES proteins and master TFs were seen in the CD44high subpopulation when compared to those that were CD44low (Figure 6B and C). CD44 expression also positively correlated with STAT3, CEBPB and TAZ expression as well as NF-κB pathway activation in human GBMs (Figure 6D). While these TFs showed strong association with the MES signature, classic EMT inducers, SNAIL, SLUG and TWIST1 were not robustly associated (Figure S6A).
Figure 6. NF-κB Controls Master TFs of MES Differentiation in GSCs.
(A) Western blot analysis of phosphorylated p65 (ser 536), total p65, phosphorylated STAT3 (Tyr 705), STAT3 and C/EBPβ in GSCs. (B) and (C) Western blot analysis using indicated antibodies were performed on GSC23 and 11 sorted for CD44high or CD44low subpopulations. (D) Box plots of normalized expression of STAT3, CEBPB, TAZ, and NF-κB metagene in CD44low (green boxes) or CD44high (red boxes) tumors from multiple datasets as indicated. Boxes show median 25th and 75th percentile, while whiskers represent the 5th and the 95th percentile. Outliers are shown as individual points. p value was determined using a non-parametric Wilcoxon test. For the NF-κB metagene, the average expression of 38 NF-κB family members and targets (see Supplemental Experimental Procedures) was condensed into a metagene and plotted. Wilcoxon signed-rank test was used to test statistical significance. (E) Time course western blot analysis of indicated antibodies after TNFα treatment in GSC11 transduced with RFP or IκB-SR adenovirus 24 h prior to TNFα treatment. (F) qRT-PCR analysis of MES signature master TFs STAT3, CEBPB and TAZ in GSC11 treated with TNFα with or without pre-treatment with RFP or IκBSR adenovirus. Error bar indicates +/− SD. t test was used for statistical significance. *p < 0.05 and **p < 0.005. (G) qRT-PCR analysis of YKL40, and CD44 after knockdown of all three master TFs (STAT3, C/EBPβ and TAZ) in GSC11 is shown. Cells were treated with siRNA 72 h prior to treatment with TNFα for an additional 24 h. Error bar indicates +/− SD. t test was used for statistical significance. *p < 0.05 and **p < 0.005. See also Figure S6.
To test whether NF-κB mediates MES reprogramming via master TFs, we treated GSC11 with TNFα at various time points and analyzed the temporal expression of these proteins by western blotting. TNFα induced phosphorylation of p65 preceded the induction of YKL40, STAT3, C/EBPβ and TAZ, an effect that was negated by pre-treatment with IκB-SR, indicating that the master TFs act downstream of the NF-κB pathway (Figure 6E and S6B). Furthermore, up-regulation of STAT3, CEBPB and TAZ mRNA was significantly inhibited by IκB-SR pre-treatment (Figure 6F and S6B). Finally, concomitant silencing of STAT3, C/EBPβ and TAZ caused strong reduction of CD44 and YKL40 induced by TNFα (Figure 6G and S6B). Taken together, these data indicate that NF-κB promotes MES differentiation in GSCs via induction of master TFs. Interestingly, GSC13, which originated from a PN tumor (Figure 1C), did not exhibit MES differentiation even upon long term culture in TNFα suggesting that some PN GSCs are resistant to NF-κB-mediated MES differentiation (Figure S2C).
MES differentiation, CD44 levels and NF-κB activation are predictive of radiation response in GBM
Since activation of NF-κB caused both MES differentiation and enrichment of CD44 populations in GSCs, we performed in-depth analyses of NF-κB activation in GBM. Among previously annotated direct targets of NF-κB, MES GSCs as well as tumors (Table S6) showed a 36% enrichment (induced to a 1.5 fold or greater expression) of these targets when compared to PN counterparts which showed only a 6% enrichment (Figure S7A). Seventeen NF-κB target genes appeared commonly induced in both MES GSCs and GBMs which included CD44, pro-inflammatory cytokines IL1B and IL8, chemokines CCL2 and CXCL5, prostaglandin enzyme cyclooxygenase 2 (COX2), and the downstream target of TNFα stimulation, TNFAIP3 that has been previously shown to be associated with GBM (Hjelmeland et al., 2010; Murat et al., 2009). Thus, the MES phenotype in GSCs and GBM was accompanied by activated NF-κB signaling and CD44 is an integral component of this signature.
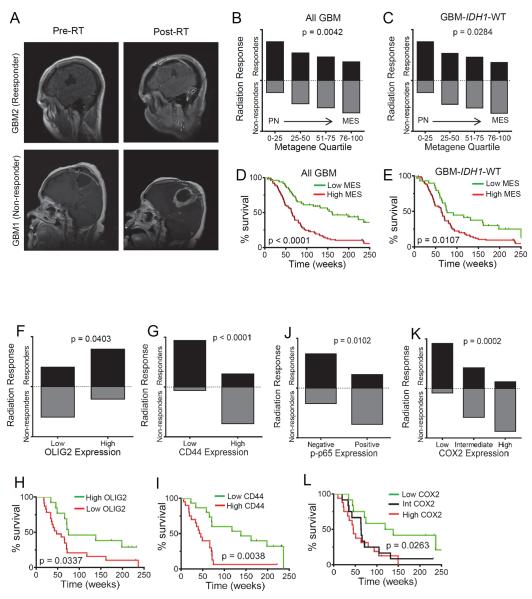
We next examined the association of these variables with radiation response and treatment outcome in a cohort of newly diagnosed GBM patients (Table S7). We used a previously defined radiation response scoring criteria (Pelloski et al., 2005) by comparing the maximal area of enhancement between the pre- (i.e. within 1 month of the start of radiation) and the post-RT magnetic resonance images (MRI) (Figure 7A). Examination of the PN/MES status showed correlation between MES composite metagene (see Supplemental Experimental Procedures for details) and progression following RT (non-responders) whereas a PN composite metagene correlated to patients with stable or reduced disease following RT, even in patients with WT IDH1 tumors (Figure 7B and C). After adjusting for patient age (≤50 vs >50), IDH1 status (mutant vs. WT), and Karnofsky Performance Status (KPS, ≤70 vs >70), only the MES metagene remained a significant predictor of RT response (Table S8). Patients with a higher MES metagene (upper 2/3rd of the metagene quartile) also showed reduced survival irrespective of IDH1 status (Figure 7D and E). Next, we evaluated if CD44 and OLIG2 expression could be used as serviceable markers for MES and PN states respectively. Patients with higher expression of CD44 showed a striking association with poor response to radiation and lower survival compared to those with lower CD44 and conversely patients with high OLIG2 were more likely to be responders to radiation and better survivors (Figure 7F–I; S7B–E). Additionally, using an antibody specific for phosphorylated p65 (ser 276), a transcriptionally active form of NF-κB, as well as its target COX2 (Figure S7F) we found significant association of the expression of these proteins to non-responders compared to those with a favorable response to radiation (Figure 7J and K; S7G and H). While intermediate and high COX2 expression was associated with poor survival (Figure 7L and S7I), p-p65 expression showed a similar trend, but did not reach statistical significance (data not shown).
Figure 7. MES Differentiation, CD44 Levels and NF-κB Activation are Predictive of Radiation Response in GBM.
(A) Representative MRI scans of postoperative/pre-RT and post RT responses of patients typically classified as responder or non-responder. (B) Box plots showing the proportion of patients classified as responders or non-responders against the PN/MES metagene quartiles in all newly diagnosed GBM cases (n=149). Chi square test was used to assess statistical significance. (C) Box plot shown for IDH1 WT cases (n=121). (D) Kaplan Meier curves showing survival of newly diagnosed patients based on PN/MES metagene scores. Low MES represents the bottom one third of the cases whereas high MES were the top two thirds. Log rank test was used to assess statistical significance. (E) Kaplan Meier curves showing survival of newly diagnosed GBM-IDH1 WT patients based on PN/MES metagene scores. Bar graph shows the proportion of OLIG2 (F), CD44 (G), p-p65 (J), and COX2 (K) expression in newly diagnosed GBM-IDH1 WT patients classified as radiation responders or non-responders. Proportion of patients that responded/non-responded were compared using Chi Square test. Kaplan Meier curves showing survival of newly diagnosed GBM-IDH1 WT patients based on OLIG2 (H), CD44 (I) and COX2 (L) are shown below. See also Figure S7, Table S6–S8.
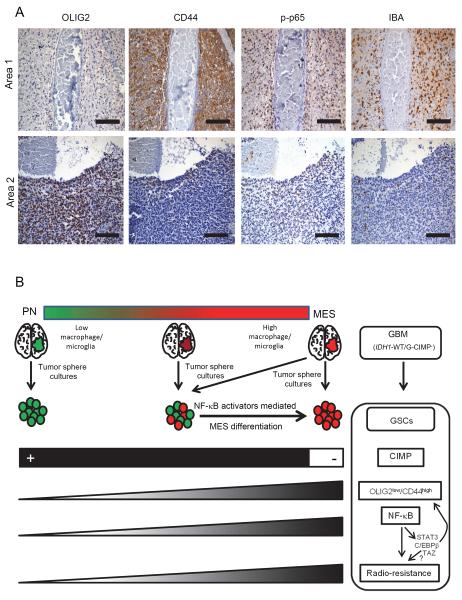
Finally, to characterize intratumoral PN to MES transition in human GBMs, we closely examined the temporal distribution of multiple markers by IHC on serial paraffin embedded sections (Table S9). The expression of these markers ranged from pockets of negative expression to those with strong positivity. Importantly, OLIG2low and CD44high areas (MES signature) coincided with p-p65 positivity (Figure 8A and Table S9). The extent of macrophages/microglia infiltration (as judged by IBA staining) also correlated with the MES regions. This finding highlights intratumoral PN/MES heterogeneity that correlates with activation of NF-κB and macrophages/microglial involvement in GBM.
Figure 8. Features Associated with MES Differentiation Induced by NF-κB in GBM.
(A) Consecutive five micron sections were stained for various markers by IHC. Two independent areas within a same tumor are shown for mutual exclusive expression of OLIG2 from CD44, p-p65 and IBA. Scale bar: 100 μm. (B) Cartoon showing a summary of our findings. We propose that GSCs when isolated from the microenvironment may differ in their molecular signatures from the parental tumor. While GBMs in the extreme ends of PN/MES axis will likely contain (and enrich for) GSCs with similar signatures to the parental tumor, GBMs with intermediate to high MES signatures enrich for PN GSCs that are maintained in a MES state in the human tumor microenvironment (by cell types such as macrophages/microglia). These PN GSCs also tend to be CIMP+ although derived from G-CIMP− tumors which lack the IDH1 mutation. MES differentiation, CD44 enrichment, and radio-resistance can be induced in PN/CIMP− GSCs by activation of NF-κB and downstream master TFs (STAT3, C/EBPβ, and TAZ). In contrast, MES GSCs are CIMP−, predominantly express CD44, are radio-resistant, and exhibit constitutive activation of NF-κB and downstream master TFs. See also Table S9.
DISCUSSION
Differential molecular signatures in GSCs and GBMs
While gene expression profiling of GBM has consistently shown the PN and MES subtypes, parallel efforts on GSCs have been limited, and mouse models representative of transcriptome subtypes of GBM are only beginning to emerge (Chow et al., 2011; Friedmann-Morvinski et al., 2012; Koso et al., 2012; Liu et al., 2011). In this report, we demonstrate that with the exception of a few, GSCs in general show gene expression and epigenetic profile differences from their parental tumors, and that the majority of the GSCs exhibit an overall PN/CIMP+ signature despite originating from predominantly MES/CIMP− GBMs. Placement of these PN/CIMP+ GSCs in intracranial xenografts did not restore the MES phenotype of the parental tumor, indicating a potential shortcoming of immunocompromised xenograft models to fully recapitulate the human tumor microenvironment (Magee et al., 2012). We could not ascertain the CIMP status of the xenografts due to technical difficulties given the small size of these tumors. While TNFα/NF-κB activation induced MES differentiation, it did not alter CIMP methylation patterns, suggesting that other tumor microenvironment derived cytokines could play a role in this process. Coupled with recent reports that resetting the epigenome of GSCs can cause remarkable changes in their malignant behavior (Stricker et al., 2013), future studies utilizing GSCs as a model system should take these factors into consideration.
MES differentiation is mediated by NF-κB induction of master TFs
In addition to the identification of a role for NF-κB in inducing the MES signature, we show that this occurs via the induction of STAT3, C/EBPβ and TAZ, although it remains to be seen if the radio-resistance mediated by NF-κB is also dependent on these master TFs. Interestingly, these same TFs (with the exception of TAZ) play prominent roles in inflammatory response, and past studies have shown considerable crosstalk between these TFs. For example NF-κB promotes an inflammatory response through secretion of TNFα, IL1β, and IL6, of which IL6 triggers STAT3 activation (Ben-Neriah and Karin, 2011; Hayden and Ghosh, 2012). Conversely, studies have shown that nuclear translocation of NF-κB is dependent on acetylation of NF-κB by p300, which requires STAT3 activation (Gravendeel et al., 2009). The inter-dependency of NF-κB and C/EBPβ has also been previously reported in other studies (Acosta et al., 2008; Kuilman et al., 2008). While we show that TNFα can be derived from macrophages/microglia, it is noteworthy that these cell types themselves exhibit plasticity and can be polarized to a pro-inflammatory (M1) or anti-inflammatory (M2) phenotype in the brain microenvironment (Li and Graeber, 2012; Wu et al., 2010). Because most GBMs arise de novo without prior clinical history of a lower grade tumor, it is difficult to study tumor evolution in GBM, i.e., if a MES tumor evolved from an early stage PN tumor is difficult to ascertain and the characteristics and the influence of the microenvironment in the early stages of a GBM is virtually unknown. Here we show evidence for the transcriptomic plasticity of the PN and MES states by IHC analysis of tumors that had regions of both PN and MES markers and their correlation to NF-κB activation and infiltration of macrophages/microglia. Consistent with our findings, recent studies have shown that the MES subclass of GBMs exhibit a high degree of necrosis (Cooper et al., 2012), and macrophages/microglial infiltration (Engler et al., 2012; Li et al., 2012). In addition, tumor evolutionary dynamics has been shown in a recent report wherein multiple transcriptome signatures were found within the same tumor (Sottoriva et al., 2013). We found that in some cases, MES/CIMP− GSCs showed constitutive MES signatures even in instances where NF1 mutations were not seen (e.g., GSCs 2 and 20), and when removed from the microenvironment suggesting that cell intrinsic mechanisms that sustain the MES network also exist.
Association of the MES signature, CD44 and NF-κB signaling with radiation resistance
Patients with GBM currently undergo standard treatment consisting of maximal surgical resection, combined radiation and chemotherapy, and adjuvant chemotherapy with temozolomide (Furnari et al., 2007; Hegi et al., 2005). Radiation has been a mainstay of GBM treatment for decades and the exact molecular mechanisms driving resistance in GBMs is unknown. Our previous studies have shown that patients with a MES signature belong to the poor prognosis subclass and are resistant to standard treatments (Colman et al., 2010), and that PN tumors can recur in a MES state (Phillips et al., 2006). Here we show that PN/CIMP+ GSCs under specific conditions can undergo MES differentiation, with associated radio-resistance. Importantly, we show that in newly diagnosed GBM, an increased MES metagene, CD44 expression, or NF-κB activation are associated with poor radiation response and shorter survival even in the absence of IDH1 mutation. Although NF-κB has been previously implicated in GBM (Bredel et al., 2006; Bredel et al., 2011; Park et al., 2009), our studies identify a role for NF-κB in mediating radiation resistance. We speculate that global MES differentiation induced by NF-κB parallels activation of checkpoint pathways, leading to enhanced DNA damage repair, and unperturbed cell cycle progression in response to IR. Moreover, given that NF-κB has been shown to mediate anti-apoptotic effects and DNA damage repair (Magne et al., 2006), it is conceivable that this pathway acts as a potential link between MES differentiation and radio-resistance. In the context of previous studies showing that the CD133+ GSCs are resistant to radiation, and even more efficiently under the influence of the brain microenvironment (Jamal et al., 2012), it remains to be seen if CD133+ subpopulation within the CD44high GSCs represent a refinement of the radio-resistant cell types.
In summary, (Figure 8B), we show that although GBM patient tumors appear predominantly MES/CIMP− at presentation or progression, the GSCs derived from these tumors using a standard isolation procedure tend to be PN/CIMP+ (despite absence of the IDH1 mutation), suggesting that tumor microenvironmental factors in humans may induce a MES/CIMP− signature. We further show that in a subset of the PN/CIMP− GSCs, MES differentiation with associated enrichment of CD44 expressing subpopulations, and radio-resistance can be induced in an NF-κB dependent fashion. Our data suggest that inhibition of NF-κB activation can directly impact radio-resistance and presents an attractive therapeutic target for GBM.
EXPERIMENTAL PROCEDURES
GSCs isolation and cell culture
Freshly resected tumor tissues were enzymatically and mechanically dissociated into single cells and grown in DMEM/F12 media supplemented with B27 (Invitrogen), EGF (20 ng/ml), bFGF (20 ng/ml). After two to four weeks, free floating neurospheres were collected and thereafter routinely cultured in the above mentioned neurosphere media, with dissociation to single cells every 5–6 days. For growth factor comparison, PDGF (R&D Systems) was used at a final concentration of 10 ng/ml. For cytokine treatment, GSCs were dissociated into single cells with Accutase (Sigma Aldrich) and treated with various concentrations and durations as indicated in figure legends. IL6, IL8, IL10, and TGF-β were obtained from R&D Systems, and TNFα was from Sigma Aldrich. RFP and IκB-SR adenovirus were obtained from Vector Biolabs. The use of human tumor tissue samples and all other tumor related studies were conducted in accordance with the protocols approved by the Institutional Review Board at the UT, M.D. Anderson Cancer Center or by the Medical Ethical Committee at the University Medical Center Groningen. The use of the tissues for the experiments involving isolation of GSCs, DNA and RNA isolation, and/or IHC on human tumors was exempt from requiring consent as per the MDACC Institutional Review Board. Patient materials at UMCG were obtained after routine diagnostics, coded according to the National Code for the Good Use of Patient Material, were exempt from informed consent.
Microarray and bioinformatic analyses
RNA labeling and hybridization to Affymetrix HGU133 version 2.0 genechips was performed by Expression Analysis (Durham, NC). Raw .cel files were processed using R and Bioconductor (Gentleman et al., 2004), using a custom CDF (Sandberg and Larsson, 2007), with background correction, log transformation, and quantile normalization performed using the RMA algorithm. Detailed description of all other bio-informatic analyses is described in Supplemental Experimental Procedures.
Xenograft models and treatments
GSCs were implanted intracranially using the guide screw system in 4–5 week old NOD/SCID or Foxn1nu mice. 5 × 105 cells or fewer (as indicated) were injected intracranially in each mouse after one week of guide screw implantation and randomly distributed between groups. A minimum of five mice were used in each group. For in vivo bio-luminescent imaging, GSCs were engineered to express luciferase by transducing with pCignal lenti-CMV-luc viral particles (SABiosciences). Kinetics of tumor growth was monitored using IVIS 200 system bio-luminescent imaging and tumor volume measured using Living Image 4.1 software. IR was delivered using fractionated doses (2.5Gy × 4) using a 60Co teletherapy unit and a custom gig with validated dosimetry. Mice that presented neurological symptoms (i.e. hydrocephalus, seizures, inactivity, and/or ataxia) or moribund were sacrificed, and brains were fixed in formalin and stained with H&E to confirm the presence of tumor and subject to IHC. All animal procedures were reviewed and approved by the Institutional Animal Care and Use Committee at the M.D. Anderson Cancer Center.
Supplementary Material
SIGNIFICANCE.
In this study, we characterize plasticity between the proneural (PN) and mesenchymal (MES) transcriptome signatures observed in glioblastoma (GBM). Specifically, we show that PN glioma sphere cultures (GSCs) can be induced to a MES state with an associated enrichment of CD44 expressing cells and a gain of radio-resistance, in an NF-κB dependent fashion. Newly diagnosed GBM samples show a direct correlation between radiation response, higher MES metagene, CD44 expression, and NF-κB activation, and we propose macrophages/microglia as a potential microenvironmental component that can regulate this transition. Our results uncover a previously unknown link between subtype plasticity that is regulated by NF-κB. Inhibition of NF-κB activation can directly impact radio-resistance and presents an attractive therapeutic target for GBM.
ACKNOWLEDGEMENTS
We would like to acknowledge the Caroline Ross Endowment Fellowship, American Brain Tumor Association Basic Research Fellowship, Odyssey Special Fellowship and the MDACC Brain Tumor SPORE Career Development grant (to K.P.B); the Brain Tumor Funders' Collaborative, the Dr. Marnie Rose Foundation, and the National Brain Tumor Society (to K.A.); the V Foundation and the SPORE grant P50CA127001 from NIH/NCI (to K.A. and H.C.); Huntsman Cancer Foundation (to H.C.); Ben and Cathy Ivy Foundation Research Award (to F.F.L., K.A., and E.P.S); SPORE Animal Core grant (to F.F.L); RO1-CA1208113 from NIH/NCI (to A.H); and the Dutch Cancer Society grant RUG 2011–5150 (to V.B, H.W.B and W.F.D) for their generous support. We thank Alicia Ledoux, Bhavna Singh, and Susan Cweren for histology, Verlene Henry for animal injections, the Flow Cytometry & Cellular Imaging Core Facility, Small Animal Imaging Facility, and the Division of Surgery Pathology Core Facility (all at MDACC) for technical support. We apologize for not citing all original references due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS The Gene Expression Omnibus accession numbers for the microarray experiments are GSE49009.
REFERENCE
- Acosta JC, O'Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, Fumagalli M, Da Costa M, Brown C, Popov N, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–1018. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- Ben-Neriah Y, Karin M. Inflammation meets cancer, with NF-kappaB as the matchmaker. Nat Immunol. 2011;12:715–723. doi: 10.1038/ni.2060. [DOI] [PubMed] [Google Scholar]
- Bhat KP, Salazar KL, Balasubramaniyan V, Wani K, Heathcock L, Hollingsworth F, James JD, Gumin J, Diefes KL, Kim SH, et al. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 2011;25:2594–2609. doi: 10.1101/gad.176800.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredel M, Bredel C, Juric D, Duran GE, Yu RX, Harsh GR, Vogel H, Recht LD, Scheck AC, Sikic BI. Tumor necrosis factor-alpha-induced protein 3 as a putative regulator of nuclear factor-kappaB-mediated resistance to O6-alkylating agents in human glioblastomas. J Clin Oncol. 2006;24:274–287. doi: 10.1200/JCO.2005.02.9405. [DOI] [PubMed] [Google Scholar]
- Bredel M, Scholtens DM, Yadav AK, Alvarez AA, Renfrow JJ, Chandler JP, Yu IL, Carro MS, Dai F, Tagge MJ, et al. NFKBIA deletion in glioblastomas. N Engl J Med. 2011;364:627–637. doi: 10.1056/NEJMoa1006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brescia P, Richichi C, Pelicci G. Current strategies for identification of glioma stem cells: adequate or unsatisfactory? J Oncol. 2012;2012:376894. doi: 10.1155/2012/376894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles NA, Holland EC, Gilbertson R, Glass R, Kettenmann H. The brain tumor microenvironment. Glia. 2011;59:1169–1180. doi: 10.1002/glia.21136. [DOI] [PubMed] [Google Scholar]
- Chen J, McKay RM, Parada LF. Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell. 2012;149:36–47. doi: 10.1016/j.cell.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow LM, Endersby R, Zhu X, Rankin S, Qu C, Zhang J, Broniscer A, Ellison DW, Baker SJ. Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain. Cancer Cell. 2011;19:305–316. doi: 10.1016/j.ccr.2011.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman H, Zhang L, Sulman EP, McDonald JM, Shooshtari NL, Rivera A, Popoff S, Nutt CL, Louis DN, Cairncross JG, et al. A multigene predictor of outcome in glioblastoma. Neuro Oncol. 2010;12:49–57. doi: 10.1093/neuonc/nop007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper LA, Gutman DA, Chisolm C, Appin C, Kong J, Rong Y, Kurc T, Van Meir EG, Saltz JH, Moreno CS, Brat DJ. The tumor microenvironment strongly impacts master transcriptional regulators and gene expression class of glioblastoma. Am J Pathol. 2012;180:2108–2119. doi: 10.1016/j.ajpath.2012.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper LA, Gutman DA, Long Q, Johnson BA, Cholleti SR, Kurc T, Saltz JH, Brat DJ, Moreno CS. The proneural molecular signature is enriched in oligodendrogliomas and predicts improved survival among diffuse gliomas. PLoS One. 2010;5:e12548. doi: 10.1371/journal.pone.0012548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daginakatte GC, Gutmann DH. Neurofibromatosis-1 (Nf1) heterozygous brain microglia elaborate paracrine factors that promote Nf1-deficient astrocyte and glioma growth. Hum Mol Genet. 2007;16:1098–1112. doi: 10.1093/hmg/ddm059. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Engler JR, Robinson AE, Smirnov I, Hodgson JG, Berger MS, Gupta N, James CD, Molinaro A, Phillips JJ. Increased microglia/macrophages gene expression in a subset of adult and pediatric astrocytomas. PLoS One. 2012;7:e43339. doi: 10.1371/journal.pone.0043339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann-Morvinski D, Bushong EA, Ke E, Soda Y, Marumoto T, Singer O, Ellisman MH, Verma IM. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science. 2012;338:1080–1084. doi: 10.1126/science.1226929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravendeel LA, Kouwenhoven MC, Gevaert O, de Rooi JJ, Stubbs AP, Duijm JE, Daemen A, Bleeker FE, Bralten LB, Kloosterhof NK, et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69:9065–9072. doi: 10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. NF-kappaB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 2012;26:203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- Hjelmeland AB, Wu Q, Wickman S, Eyler C, Heddleston J, Shi Q, Lathia JD, Macswords J, Lee J, McLendon RE, Rich JN. Targeting A20 decreases glioma stem cell survival and tumor growth. PLoS Biol. 2010;8:e1000319. doi: 10.1371/journal.pbio.1000319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse JT, Phillips HS, Brennan CW. Molecular subclassification of diffuse gliomas: seeing order in the chaos. Glia. 2011;59:1190–1199. doi: 10.1002/glia.21165. [DOI] [PubMed] [Google Scholar]
- Jamal M, Rath BH, Tsang PS, Camphausen K, Tofilon PJ. The brain microenvironment preferentially enhances the radioresistance of CD133(+) glioblastoma stem-like cells. Neoplasia. 2012;14:150–158. doi: 10.1593/neo.111794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jijiwa M, Demir H, Gupta S, Leung C, Joshi K, Orozco N, Huang T, Yildiz VO, Shibahara I, de Jesus JA, et al. CD44v6 regulates growth of brain tumor stem cells partially through the AKT-mediated pathway. PLoS One. 2011;6:e24217. doi: 10.1371/journal.pone.0024217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koso H, Takeda H, Yew CC, Ward JM, Nariai N, Ueno K, Nagasaki M, Watanabe S, Rust AG, Adams DJ, et al. Transposon mutagenesis identifies genes that transform neural stem cells into glioma-initiating cells. Proc Natl Acad Sci U S A. 2012;109:E2998–3007. doi: 10.1073/pnas.1215899109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, Aarden LA, Mooi WJ, Peeper DS. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- Li B, Senbabaoglu Y, Peng W, Yang ML, Xu J, Li JZ. Genomic estimates of aneuploid content in glioblastoma multiforme and improved classification. Clin Cancer Res. 2012;18:5595–5605. doi: 10.1158/1078-0432.CCR-12-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Graeber MB. The molecular profile of microglia under the influence of glioma. Neuro Oncol. 2012;14:958–978. doi: 10.1093/neuonc/nos116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Sage JC, Miller MR, Verhaak RG, Hippenmeyer S, Vogel H, Foreman O, Bronson RT, Nishiyama A, Luo L, Zong H. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146:209–221. doi: 10.1016/j.cell.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell. 2012;21:283–296. doi: 10.1016/j.ccr.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magne N, Toillon RA, Bottero V, Didelot C, Houtte PV, Gerard JP, Peyron JF. NF-kappaB modulation and ionizing radiation: mechanisms and future directions for cancer treatment. Cancer Lett. 2006;231:158–168. doi: 10.1016/j.canlet.2005.01.022. [DOI] [PubMed] [Google Scholar]
- Markovic DS, Vinnakota K, van Rooijen N, Kiwit J, Synowitz M, Glass R, Kettenmann H. Minocycline reduces glioma expansion and invasion by attenuating microglial MT1-MMP expression. Brain Behav Immun. 2011;25:624–628. doi: 10.1016/j.bbi.2011.01.015. [DOI] [PubMed] [Google Scholar]
- Mir SE, De Witt Hamer PC, Krawczyk PM, Balaj L, Claes A, Niers JM, Van Tilborg AA, Zwinderman AH, Geerts D, Kaspers GJ, et al. In silico analysis of kinase expression identifies WEE1 as a gatekeeper against mitotic catastrophe in glioblastoma. Cancer Cell. 2010;18:244–257. doi: 10.1016/j.ccr.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat A, Migliavacca E, Hussain SF, Heimberger AB, Desbaillets I, Hamou MF, Ruegg C, Stupp R, Delorenzi M, Hegi ME. Modulation of angiogenic and inflammatory response in glioblastoma by hypoxia. PLoS One. 2009;4:e5947. doi: 10.1371/journal.pone.0005947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Hatanpaa KJ, Xie Y, Mickey BE, Madden CJ, Raisanen JM, Ramnarain DB, Xiao G, Saha D, Boothman DA, et al. The receptor interacting protein 1 inhibits p53 induction through NF-kappaB activation and confers a worse prognosis in glioblastoma. Cancer Res. 2009;69:2809–2816. doi: 10.1158/0008-5472.CAN-08-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelloski CE, Mahajan A, Maor M, Chang EL, Woo S, Gilbert M, Colman H, Yang H, Ledoux A, Blair H, et al. YKL-40 expression is associated with poorer response to radiation and shorter overall survival in glioblastoma. Clin Cancer Res. 2005;11:3326–3334. doi: 10.1158/1078-0432.CCR-04-1765. [DOI] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Pietsch T, Wiestler OD. Molecular neuropathology of astrocytic brain tumors. J Neurooncol. 1997;35:211–222. doi: 10.1023/a:1005843913095. [DOI] [PubMed] [Google Scholar]
- Riddick G, Fine HA. Integration and analysis of genome-scale data from gliomas. Nat Rev Neurol. 2011;7:439–450. doi: 10.1038/nrneurol.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Larsson O. Improved precision and accuracy for microarrays using updated probe set definitions. BMC Bioinformatics. 2007;8:48. doi: 10.1186/1471-2105-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son MJ, Woolard K, Nam DH, Lee J, Fine HA. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4:440–452. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, Tavare S. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110:4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein B, Baldwin AS., Jr. Distinct mechanisms for regulation of the interleukin-8 gene involve synergism and cooperativity between C/EBP and NF-kappa B. Mol Cell Biol. 1993;13:7191–7198. doi: 10.1128/mcb.13.11.7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopschinski BE, Beier CP, Beier D. Glioblastoma cancer stem cells - From concept to clinical application. Cancer Lett. 2012 doi: 10.1016/j.canlet.2012.05.033. [DOI] [PubMed] [Google Scholar]
- Stricker SH, Feber A, Engstrom PG, Caren H, Kurian KM, Takashima Y, Watts C, Way M, Dirks P, Bertone P, et al. Widespread resetting of DNA methylation in glioblastoma-initiating cells suppresses malignant cellular behavior in a lineage-dependent manner. Genes Dev. 2013;27:654–669. doi: 10.1101/gad.212662.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A, Wei J, Kong LY, Wang Y, Priebe W, Qiao W, Sawaya R, Heimberger AB. Glioma cancer stem cells induce immunosuppressive macrophagess/microglia. Neuro Oncol. 2010;12:1113–1125. doi: 10.1093/neuonc/noq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer. 2011;11:254–267. doi: 10.1038/nrc3023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.