Abstract
Fungal species are continuously being studied to not only understand disease in humans and plants but also to identify novel antibiotics and other metabolites of industrial importance. Genetic manipulations, such as gene deletion, gene complementation, and gene over-expression, are common techniques to investigate fungal gene functions. Although advances in transformation efficiency and promoter usage have improved genetic studies, some basic steps in vector construction are still laborious and time-consuming. Gateway cloning technology solves this problem by increasing the efficiency of vector construction through the use of λ phage integrase proteins and att recombination sites. We developed a series of Gateway-compatible vectors for use in genetic studies in a range of fungal species. They contain nutritional and drug-resistance markers and can be utilized to manipulate different filamentous fungal genomes.
Keywords: Genetic engineering, filamentous fungi, Gateway technology, recombination
1. Introduction
Filamentous fungi comprise a large group of biologically relevant organisms. Many filamentous fungal species produce useful metabolites, such as pigments, vitamins, and antibiotics, whereas some other species are infectious and cause disease in humans, other animals, and plants. For this reason, researchers have studied fungal species extensively not only to understand human and plant disease, but also to identify new antibiotics and other relevant metabolites for industrial purposes. Genetic manipulations, such as the creation of deletion or over-expression strains, are often used when examining virulence factors and/or protein functions.
Plasmids are routinely created for gene deletion and over-expression studies via several methods. One method involves the use of restriction enzymes to isolate a DNA insert and to linearize a destination vector. The two segments of DNA are then ligated, resulting in a final vector whereby the DNA insert has been introduced into the destination vector. This method can be challenging, because the restriction enzymes used must be either identical or compatible for both the insert and vector. Furthermore, if a single restriction enzyme is used to linearize the vector, dephosphorylation of the digested end is required to prevent self-ligation. Even with proper steps and controls, researchers often face low colony yields and false positives.
A second method that can be used for vector construction or direct genetic manipulations involves fusion polymerase chain reaction (f-PCR). In this case, primers are engineered to contain overlaps of different DNA fragments to be joined. For example, the 3' primer for the first DNA fragment would contain a series of nucleotides that are identical to the 5' end of the second DNA fragment, and the 5' primer for the second DNA fragment would contain a series of nucleotides that are identical to the 3' end of the first DNA fragment (Spiliotis, 2012; Szewczyk et al., 2006; Yu et al., 2004). Two separate products are obtained from the first round of PCR and then fused together in the second reaction to create one PCR product. This fused product can either be transformed directly into an organism for genetic studies or can undergo additional cloning methods to create a novel plasmid (Spiliotis, 2012; Szewczyk et al., 2006; Yu et al., 2004). One caveat to using f-PCR to create a deletion construct is the need for multiple fragments to be fused together. Whether gene deletion or placement of epitope tags is the goal, all constructs must have a selective marker plus target DNA fragments. When combining all fragments together, the final product can become quite long, which can result in a decrease in PCR efficiency and thus product yield. Furthermore, to construct a plasmid using an f-PCR product, restriction enzymes and ligases must be employed, which introduces limitations discussed above.
Many of the limitations of the previous methods can be alleviated by the use of recombinatorial cloning systems. Numerous systems exist, including Recombineering, Sequence and Ligation-Independent Cloning (SLIC), Ligation-Independent Cloning (LIC), Mating-assisted genetically integrated cloning (MAGIC), Gateway, DNA assembler, and Cre-loxP. Some, such as Recombineering, MAGIC, and DNA assembler, are carried out in vivo (Li and Elledge, 2005; Liu et al., 2003; Mosberg et al.; Shao and Zhao, 2009; Sharan et al., 2009), while others, such as Cre-loxP, SLIC, LIC, and Gateway can be performed as in vitro reactions (Hartley et al., 2000; Li and Elledge; Li and Elledge, 2007; Magnani et al., 2006; Siegel et al., 2004; Yamamura, 2012). Depending on the intended outcome, all systems have their advantages. For instance, MAGIC, which relies on bacterial mating, facilitates the cloning of an insert into multiple destination plasmids (Li and Elledge, 2005). SLIC, Gateway, and DNA assembler have been shown to be efficient for inserting multiple fragments into a single destination vector (Li and Elledge; Li and Elledge, 2007; Magnani et al., 2006; Shao and Zhao, 2009). Recombineering and Cre-loxP are frequently used for conditional knockouts in mice, as cloning can be controlled through selective Cre recombinase expression (Liu et al., 2003; Yamamura, 2012). Cre-loxP and Gateway recombinatorial cloning are similar in the fact that they both involve site-specific DNA recombination reactions.
Cre recombinase mediates the site-specific recombination of DNA sequences between loxP sites (Deng, 2012; Siegel et al., 2004; Yamamura, 2012). The loxP site consists of two 13-base pair (bp) inverted repeats separated by an 8-bp nonpalindromic core region. Based on the orientation and placement of loxP sites, DNA fragments can be excised, translocated, or inverted. For example, if a DNA fragment is flanked by loxP sites that are oriented in the same direction, the fragment will be excised and circularized. If, however, the DNA fragment is flanked by loxP sites that are oriented in opposite directions, the fragment will be inverted. Translocation of DNA fragments is possible if the loxP sites are located on different chromosomes. This Cre-loxP system has been employed for both rapid vector cloning and in vivo genetic techniques, predominantly in yeast and mice (Deng, 2012; Siegel et al., 2004; Yamamura, 2012).
With Gateway technology, λ phage integrase recombination proteins mediate site-specific recombination reactions. DNA inserts, which are flanked by appropriate recombination sites (e.g., attB1 and attB2), are incubated with λ phage integrase recombination proteins and donor/destination vectors, which also contain appropriate recombination sites (e.g., attP1 and attP2 or attR1 and attR2). Two reactions can be carried out: (BP) attB × attP → attL + attR, involving integrase (Int) and integration host factor (IHF), and (LR) attL × attR → attB + attP, involving Int, IHF, and excisionase. Orientation of the DNA insert can be controlled because only certain recombination sites will recombine with others; for example, attB1 will only recombine with attP1 (Hartley et al., 2000; Magnani et al., 2006).
Gateway technology has been used in numerous laboratories to manipulate fungal genomes. During the past several years, we have created a series of vectors to facilitate genetic engineering in Aspergillus fumigatus and Aspergillus nidulans using Gateway technology. Other laboratories have also created Gateway-enabled vectors, but the vectors published thus far appear to focus more on protein tagging and fluorescent labeling (Mabashi et al., 2006; Toews et al., 2004). The Gateway-enabled plasmids we have created focus more on gene deletion, gene integration, and high-copy gene expression. Although our laboratory has only used these vectors in A. fumigatus and A. nidulans, they can potentially be used in a variety of other fungal organisms, including other Aspergillus sp., Neurospora sp., Fusarium sp., Penicillium sp., and other fungal species of agricultural and industrial importance (Abe et al., 2006; Bugeja et al., 2012; Pahirulzaman et al., 2012; Paz et al., 2011). This is due to the fact that the selectable markers we chose are versatile and can be used across different species. Our aim was to create recombinatorial vectors as efficient tools in gene modification techniques in multiple fungal systems.
2. Materials and Methods
2.1. DNA manipulations
We gel purified all PCR and digestion products using a QIAquick gel purification kit (Qiagen, Hilden, Germany) and following manufacturer's instructions. We isolated all plasmid DNA using a PureLink Miniprep kit (Invitrogen, Grand Island, NY) according to manufacturer's instructions. We further purified plasmid DNA by banding on cesium chloride ethidium bromide gradients (Maniatis et al., 1982).
2.2. Construction of vectors
2.2.1. pDONR 221-AnargB, pDONR 221-AnpyrG, pDONR 221-AfpyroA, pDONR 221-AfpyrG, pDONR 221-HPH and pDONR 221-BAR
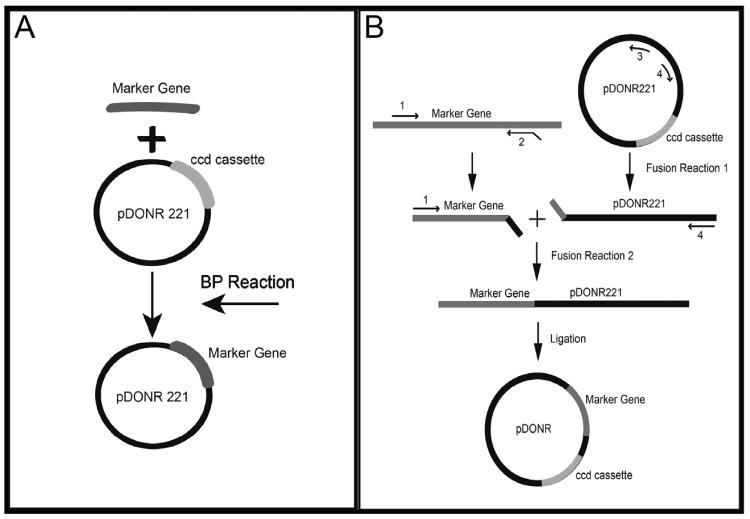
We amplified the coding region plus short non-coding regions (NCRs) flanking each selective marker using Accuprime Pfx DNA polymerase per the manufacturer's instructions. We used genomic DNA from A. nidulans 1155 as a template for AnargB, AnpyrG, and the glufosinate resistance gene (bar). We used genomic DNA from A. fumigatus Af293 as a template for AfpyroA and AfpyrG. We used pCB1003 (http://www.fgsc.net/fgn41/carroll.html) as a template for the hygroR cassette. We constructed primers with attB sites to facilitate the use of the MultiSite Gateway System (Invitrogen, Grand Island, NY). We recombined PCR fragments into pDONR 221 using the BP recombination reaction (Fig. 1A) (Invitrogen, Grand Island, NY). We transformed reaction mixes into TOP10 cells (Invitrogen, Grand Island, NY) via electroporation, as recommended by the manufacturer. We grew transformed cells on lysogeny agar (LA) (1% tryptone, 0.5% yeast extract, 10 mM NaCl, 2.5 mM KCl, 1.5% agar) plus 50 μg/ml kanamycin at 37°C overnight. We picked colonies and transferred them to 2 ml lysogeny broth (LB) (described above for LA, but without agar) plus 50 μg/ml kanamycin to grow overnight in a 37°C shaking incubator. We isolated plasmid DNA and digested it with specific enzymes to verify the correct insertion of each PCR fragment.
Figure 1. Schematic illustration of pDONR221 and pDONR vector construction.
(a) Schematic of the BP reaction used to create the plasmids for gene deletion. (b) Schematic of the f-PCR used to create the plasmids for gene integration. All gene integration plasmids were created by this methods, except pDONR HPH A and pDONR HPH B, which were created using restriction digest and ligation.
2.2.2. pDONR A, pDONR G, pDONR NG, pDONR B, pDONR BAR, pDONR HPH A, and pDONR HPH B
We constructed new Gateway-compatible vectors containing unique nutritional and drug-resistance markers for the complementation of deletion mutants. We used f-PCR to facilitate the integration of the selective markers into pDONR221 (Fig. 1B). For the first reaction, we created two cassettes: a selective marker cassette and a linearized pDONR221 cassette. We obtained both cassettes via PCR using e2TAK DNA polymerase (Takara Bio Inc., Otsu, Shiga, Japan) according to the manufacturer's recommendations. We used genomic DNA from A. nidulans 1155 as a template for AnpyrG and BAR. We used genomic DNA from A. fumigatus Af293 as a template for the AfpyroA, AfpyrG, and AfargB PCR products. Each selective marker cassette contained a unique 5' SpeI site and a 3' extension identical to the first 30 bps of the pDONR221 fragment. The pDONR 221 cassette contained a 5' extension identical to the last 30 bps of the selective marker cassette and a unique 3' SpeI site.
For the second reaction, we fused both cassettes using the selective marker forward primer and the pDONR221 reverse primer. We amplified a 50-μl reaction containing 50 fmol of each fragment, 0.3 μM of each primer, 500 μM of deoxynucleoside triphosphates, buffer 3 at a 1X concentration, and 1 μl of Expand Long DNA Template Mix (Roche Applied Science, Indianapolis, IN) per the manufacturer's instructions. We digested the PCR fragment with SpeI and circularized it by ligation using T4 DNA ligase (Invitrogen, Grand Island, NY) and manufacturer's recommendations. We transformed the ligation mixture into ccd-compatible chemically competent cells (Invitrogen, Grand Island, NY) via heat shock, as recommended by the manufacturer. We grew transformed cells on LA plus 50 μg/ml kanamycin at 37°C overnight. We picked colonies and transferred them to 2 ml LB plus 50 μg/ml kanamycin to grow overnight in a 37°C shaking incubator. We isolated plasmid DNA and digested it with specific enzymes to verify the correct fragment orientation.
We constructed pDONR HPH A and pDONR HPH B via blunt-end ligation, hence plasmid A and plasmid B, representing two orientations of the hygroR cassette. We amplified the hygroR cassette from pCB1003 using AccuPrime Pfx DNA polymerase (Life Technologies Grand Island, NY) per the manufacturer's recommendations. We digested pDONR 221 with EcoRV and the hygroR cassette with HpaI. We then treated pDONR 221 with Antarctic phosphatase (NEB, Ipswich, MA), according to the manufacturer's instructions. We ligated the two fragments using T4 DNA ligase (Invitrogen, Grand Island, NY) per the manufacturer's instructions. We transformed the ligation mixture into TOP10 electrocompetent cells and plated transformed cells on LA plus 50 μg/ml kanamycin at 37°C overnight. We picked colonies and transferred them to 2 ml LB plus 50 μg/ml kanamycin to grow overnight in a 37°C shaking incubator. We isolated plasmid DNA and digested it with specific enzymes to verify the fragment orientation.
2.2.3. pDONR AMA and pDONR AMA/HPH
We created pDONR AMA by ligating the ccdB region (attP1-ccdB-Chloramphenicol resistance gene [CmR]-attP2) from pDONR 221 into pRG3-AMA-NotI. We amplified the ccdB region from pDONR 221 using AccuPrime Pfx DNA polymerase (Life Technologies, Grand Island, NY) per the manufacturer's instructions. We engineered primers so the ccdB PCR product would have SphI at the 5' end and KpnI at the 3' end. We digested both pRG3-AMANotI and the ccdB cassette with SphI and KpnI. We then treated pRG3-AMA-NotI with Antarctic phosphatase (NEB, Ipswich, MA) according to the manufacturer's instructions. We ligated the two fragments using T4 DNA ligase (Invitrogen, Grand Island, NY) according to the manufacturer's recommendations. We transformed the ligation mixture into ccd-compatible chemically competent cells (Invitrogen, Grand Island, NY) by heat shock according to the manufacturer's instructions. We grew transformed cells on LA plus 75 μg/ml ampicillin at 37°C overnight. We picked colonies and transferred them to 2 ml LB plus 75 μg/ml ampicillin to grow overnight in a 37°C shaking incubator. We isolated plasmid DNA and digested it with specific enzymes to verify the correct fragment orientation.
We created pDONR AMA/HPH by ligating an AMA1 cassette into pDONR HPH B. We linearized pDONR HPH B via amplification using e2TAK DNA polymerase (Takara Bio Inc., Otsu, Shiga, Japan) according to the manufacturer's recommendations. We engineered primers so the linearized pDONR HPH B fragment would contain a HindIII site on both ends. We digested pDONR HPH B with HindIII and treated with Antarctic phosphatase (NEB, Ipswich, MA) according to the manufacturer's instructions. We digested pDONR AMA with HindIII, which separated the AMA1 fragment from the rest of the vector. We ligated the two fragments using T4 DNA ligase (Invitrogen, Grand Island, NY) per the manufacturer's recommendations. We transformed the ligation mixture into ccd-compatible chemically competent cells (Invitrogen, Grand Island, NY) by heat shock as recommended by the manufacturer. We grew transformed cells on LA plus 50 μg/ml hygromycin at 37°C overnight. We picked colonies and transferred them to 2 ml LB plus 50 μg/ml hygromycin to grow overnight in a 37°C shaking incubator. We isolated plasmid DNA and digested it with specific enzymes to verify the correct fragment orientation.
2.3. Creation of an mkkA deletion mutant using pDONR 221-AfpyrG
To create deletion constructs, we combined the inserts from three pDONR vectors (Table 1) using an LR recombination reaction and pDEST R4-R3 as the destination vector (Invitrogen, Grand Island, NY). The pDEST vector consisted of a 5' NCR, a selective marker, and a 3' NCR (Magnani et al., 2006). We engineered a NotI site into the forward primer of the 5' NCR to create a unique site for digestion to linearize the vector. We transformed LR reaction mixes into TOP10 cells (Invitrogen, Grand Island, NY) via electroporation according to the manufacturer's instructions. We grew transformed cells on LA plus 75 μg/ml ampicillin at 37°C overnight. We picked colonies and transferred them to 2ml LB plus 75 μg/ml ampicillin to grow overnight in a 37°C shaking incubator. We isolated plasmid DNA and digested it with specific enzymes to verify the correct fragment orientation.
Table 1.
Vectors, selective markers, and primers used in this study.
| Vector | Selective marker | Primers |
|---|---|---|
| pDONR 221-AnargB | argB A. nidulans | ArgB1-ggggacaagtttgtacaaaaaagcaggctaagctttatttcgcggtttt ArgB2-ggggaccactttgtacaagaaagctgggtgtcgacctacagccattggg |
| pDONR 221-AnpyrG | pyrG A. nidulans | attB1-AnpyrG5F-ggggacaagtttgtacaaaaaagcaggctgcctatgccagcattgagcaatg attB2-AnpyrG3R-ggggaccactttgtacaagaaagctgggtcagtggagcgaaccaattctctc |
| pDONR 221-AfpyroA | pyroA A. fumigatus | attBpyroA1-ggggacaagtttgtacaaaaaagcaggctaagttgatcgatgaatgacattc attBpyroA2-ggggaccactttgtacaagaaagctgggtctaaggcacccagcatgagtgc |
| pDONR 221-AfpyrG | pyrG A. fumigatus | attB1-AfpyrGF-ggggacaagtttgtacaaaaaagcaggctgcctcaaacaatgctcttcac attB2-AfpyrGR-ggggaccactttgtacaagaaagctgggtattctgtctgagaggaggcac |
| pDONR 221-HPH | HPH Escherichia coli | attB1-HPH
F-ggggacaagtttgtacaaaaaagcaggctgaattcgtcgacgttaactgg attB2-HPH R-ggggaccactttgtacaagaaagctgggtgaattcgtcgacgttaactgc |
| pDONR 221-BAR | BAR Streptomyces hygroscopicus | attB BAR
1-ggggacaagtttgtacaaaaaagcaggctgcaagcccagatgagaaccgac attB BAR 2-ggggaccactttgtacaagaaagctgggtcggagagagttgaacctggacg |
| pDONR A | pyroA A. fumigatus | pyroA
F-GCGCGCACTAGTgcatcccaaggggaaggtagc pyroA FR-ctagcgttaacgcgagagtagggaactgcctaaggcacccagcatgagtgc pDONR221 FFA-cagttgacggcactcatgctgggtgccttaggcagttccctactctcgcgt pDONR221 R1-GCGCGCACTAGTaattgcgttgcgctcactgcc |
| pDONR G | pyrG A. fumigatus | pyrG
F-GCGCGCACTAGTatgctcttcaccctcttcgcg pyrG FR-ctagcgttaacgcgagagtagggaactgccctgagaggaggcactgatgcg pDONR221 FFG-ttggcatcacgcatcagtgcctcctctcagggcagttccctactctcgcgt pDONR221 R1-GCGCGCACTAGTaattgcgttgcgctcactgcc |
| pDONR NG | pyrG A. nidulans | AnpyrG
F-GCGCGCACTAGTctatgccagcattgagcaatg AnpyrG FR-gagtagggaactgccagtggagcgaaccaattctct pDONR221 FFG-AN-ttggttcgctccactggcagttccctactctcgcgt pDONR221 R1-GCGCGCACTAGTaattgcgttgcgctcactgcc |
| pDONR B | argB A. fumigatus | AfargB
F-GCGCGCACTAGTaacgctgacgatgaggctgat AfargB FR-gagtagggaactgccaaacggttcttggctggaagg pDONR221 FF-AfargB-agccaagaaccgtttggcagttccctactctcgcgt pDONR221 R1-GCGCGCACTAGTaattgcgttgcgctcactgcc |
| pDONR HPH A pDONR HPH B | HPH Escherichia coli | HPH
F-ACTAGTgaattcgtcgacgttaactgc HPH FR-ctagcgttaacgcgagagtagggaactgcccgtcgacgttaactggttccc |
| pDONR BAR | BAR (Bernhard Straubinger 1992) Streptomyces hygroscopicus | BAR
F-GCGCGCACTAGTaagcccagatgagaaccgac BAR FR-gagtagggaactgccggagagagttgaacctggacg pDONR221 FF-BAR-ggttcaactctctccggcagttccctactctcgcgt pDONR221 R1-GCGCGCACTAGTaattgcgttgcgctcactgcc |
| pDONR AMA | pyr4 Neurospora crassa | ccd
F-GGGGCATGCcccaaataatgattttatttt ccd R-GGGGGTACCaaataatgattttattttgactga |
| pDONR AMA/HPH | HPH Escherichia coli | pDONR HindIII
F-GCGCGCAAGCTTaagcggaagagcgcccaatac pDONR 221 R3-GCGCGCAAGCTTggcggtaatacggttatccacagaatcag |
| pDONR P4-P1R-mkkA 5' FR | mpkkA
1-ggggacaactttgtatagaaaagttgaaGCGGCCGCtcaatatatcgtcaactcaga mpkkA 2- ggggactgcttttttgtacaaacttgcggcgagactgttttgcctctc |
|
| pDONR P2R-P3-mkkA 3' FR | mpkkA 3-
ggggacagctttcttgtacaaagtggaactctttggtctttgacgctta mpkkA 4- ggggacaactttgtataataaagttgcacttcccggggtctacttcgt |
|
| pDONR A-mkkA CR | mkkA C1 -
ggggacaagtttgtacaaaaaagcaggctaagcggccgcgagcccagaccctagatatcg mkkA C2- ggggaccactttgtacaagaaagctgggtctacagcctgcaagcttttcag |
|
| pDONR AMA-gliZ | gliZ attB 1-
ggggacaagtttgtacaaaaaagcaggctgcgaccgcagctgattggag gliZ attB 2- ggggaccactttgtacaagaaagctgggtcgattccctttgtgccgcc |
|
attB sites are underlined. All primers are listed 5' to 3'. Bolded sequences are extension regions for f-PCR. Uppercase regions are SpeI site in all primers, except ccd F (SphI), ccd R (KpnI), pDONR HindIII F (HindIII), pDONR R3 (HindIII), and mpkkA 1 (NotI). All nutritional markers are flanked by their respective promoter and terminator regions. HPH is preceded by the A. nidulans trpC promoter region (http://www.fgsc.net/fgn41/carroll.html), and BAR is flanked by the amdS promoter region from A. nidulans (with increased expression from I9 and I66 mutations) and the glucoamylase terminator from A. niger (Bernhard Straubinger 1992).
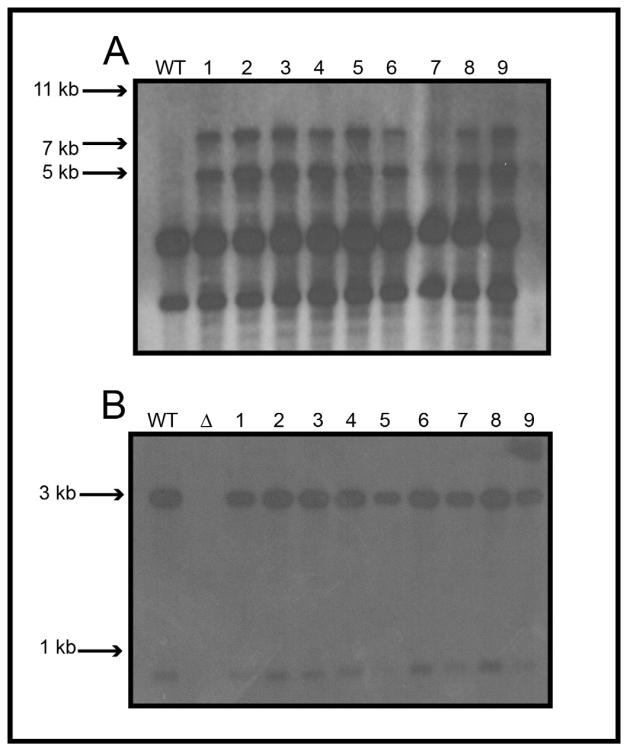
We grew A. nidulans 1155 (obtained from the Fungal Genetics Stock Center) in malt extract glucose (MEG) (2% malt extract, 0.2% peptone, 1% glucose, trace elements and vitamin mix as modified (Reyes et al., 2006)) supplemented with 5 mM uridine and 10 mM uracil. We performed the transformation as previously described (May, 1989), using pDEST R4-R3-mkkA 5' NCR-AfpyrG-mkkA 3' NCR. We grew transformants on minimal medium with vitamins (MMV) (1× MM salts [20 mM ammonium tartrate, 7 mM KCl, 2 mM MgSO4:7H2O], 1% glucose, 12 mM KPO4 [pH 6.8], trace elements, vitamin mix as previously described, and 1.25% agar) supplemented with 0.2 M sucrose at 37°C for 3–5 days. We screened for mutants that were prototrophic for uridine and uracil. We prepared genomic DNA from transformant strains (Jin et al., 2004), and we identified deletion mutants via Southern blot analysis (Fig. 2A). We made a DNA probe using the 5' and 3' NCRs of mkkA.
Figure 2. Southern hybridization analysis for the ΔmkkA and mkkA(R) strains.
(a) Southern hybridization for the ΔmkkA deletion mutant. WT is the wild-type strain. WT should display a band at 11909 bps and the deletion mutants should display bands at 7161 bps and 4965 bps. Note that there are two extra bands in all lanes. We used the deletion construct as the probe and the extra bands are possibly due to hybridization of part of the pDEST R4-R3 plasmid. (b) Southern hybridization for the mkkA(R) strain. WT is the wild-type strain and Δ represents the ΔmkkA mutant used in the study. WT and complemented strains should display two bands at 2999 bps and 865 bps, while the ΔmkkA deletion strain should have no band.
2.4. Complementation of the mkkA deletion mutant using pDONR A
We obtained the coding region of mkkA (flanked by promoter and terminator regions) by PCR using e2TAK DNA polymerase (Takara Bio Inc., Otsu, Shiga, Japan) according to the manufacturer's recommendations. We used A. nidulans 1155 genomic DNA as a template. Primers were constructed with attB1 and attB2 sites to facilitate the use of the Gateway System (Invitrogen, Grand Island, NY) (Table 1). We recombined the PCR fragment into pDONR A using the BP recombination reaction (Invitrogen, Grand Island, NY). We transformed the reaction mix into TOP10 cells (Invitrogen, Grand Island, NY) via electroporation according to the manufacturer's instructions. We grew the transformation mix on LA plus 50 μg/ml kanamycin at 37°C overnight. We picked colonies and transferred them to 2 ml of LB plus 50 μg/ml kanamycin to grow overnight in a 37°C shaking incubator. We isolated plasmid DNA and digested it with specific enzymes to verify the correct insertion.
We grew ΔmkkA in MEG. We performed the transformation using previously described methods (May, 1989). We grew transformants on MM (as previously described above, but without vitamin mix) supplemented with 0.2 M sucrose at 37°C for 3–4 days. We screened for mutants that were prototrophic for pyridoxine. We prepared genomic DNA from transformant strains (Jin et al., 2004) and verified complementation via Southern blot analysis (Fig. 2B). We made a DNA probe using the coding region of mkkA. To verify proper complementation, we grew ΔmkkA, mkkA(R), and controls on MAG (MEG with 2% agar) at 37°C for 2 days. A pyrG+ control strain (TS1) was created by crossing 1155 (obtained from the Fungal Genetics Stock Center) with strain 20.1.8 (Wu and Miller, 1997).
2.5. High-copy expression of gliZ using pDONR AMA
We obtained the coding region of gliZ (flanked by promoter and terminator regions) via PCR using e2TAK DNA polymerase (Takara Bio Inc., Otsu, Shiga, Japan) per the manufacturer's instructions. We used A. fumigatus Af293 genomic DNA as a template for the reaction. Primers were constructed with attB1 and attB2 sites to facilitate the use of the Gateway System (Invitrogen, Grand Island, NY). We recombined the PCR fragment into pDONR AMA using the BP recombination reaction (Invitrogen, Grand Island, NY). We transformed the reaction mix into TOP10 cells (Invitrogen, Grand Island, NY) via electroporation as recommended by the manufacturer. We grew the transformation mix on LA plus 75 μg/ml ampicillin at 37°C overnight. We picked colonies and transferred them to 2 ml of LB plus 75 μg/ml ampicillin to grow overnight in a 37°C shaking incubator. We isolated plasmid DNA and digested it with specific enzymes to verify the correct insertion.
We grew Af293.1 in MEG supplemented with 5 mM uridine and 10 mM uracil. We performed the transformation as previously described (May, 1989), with the following changes. Once protoplasts were formed and incubated for 3 hours, we carried out all the subsequent reactions at half the specified volume. We used 500–750 ng of vector DNA for each reaction. We plated two amounts of protoplasts (20 μl and 50 μl) each in 4 ml of yeast extract glucose (YAG) top agar (0.5% yeast extract, 1% glucose, trace elements and vitamin mix as previously described, 10 mM MgCl2, and 1% agar) supplemented with 1 M sucrose. These changes were necessary owing to the fact that using the AMA1 construct greatly increases transformation efficiency. Using these two amounts of protoplasts ensured that at least one plate would harbor single colonies, as opposed to an overcrowding of colonies. We grew transformants on YAG (as previously described, but with 1.5% agar) supplemented with 0.2 M sucrose at 37°C for 1–2 days. We screened for mutants that were prototrophic for uridine and uracil. To verify proper plasmid presence, we prepared genomic DNA from transformant strains (Jin et al., 2004). We transformed 1 μl of genomic DNA into TOP10 cells (Invitrogen, Grand Island, NY) via electroporation as recommended by the manufacturer. We grew the transformation mix on LA plus 75 μg/ml ampicillin at 37°C overnight. We picked colonies and transferred them to 2 ml of LB plus 75 μg/ml ampicillin to grow overnight in a 37°C shaking incubator. We isolated plasmid DNA and digested it with specific enzymes.
2.6. RNA dot blot analysis
We performed RNA dot blot analysis on our pDONR AMA-gliZ strain to measure RNA levels and gene activity. We grew stationary cultures (5×106 spores/ml) in 25 ml of complete medium (CM) (MMV [as previously described], 0.1% yeast extract, 0.2% peptone, 0.1% tryptone, and 1% CM supplement [27 mM adenine HCl, 33.5 mM methionine, 173 mM arginine, and 1.3 mM riboflavin]) at 37°C for 48 hours. We collected total RNA from freeze-dried mycelia using the TRIzol method (Reyes et al., 2006). We mixed total RNA with denaturing solution (50% formamide, 16% formaldehyde, 1X borate buffer [20X borate buffer: 0.4M boric acid, 4 mM EDTA, pH 8.3, with NaOH], and 0.025% bromophenol blue) and incubated for 10 minutes at 65°C. We quenched samples in ice for 10 minutes and added equal volume 20x sodium chloride-sodium citrate (SSC) (3 M NaCl, 0.3 M sodium citrate [pH 7.0]). We equilibrated a nylon membrane in 10xSSC for 10 minutes and placed it in a 96-well dot blot apparatus (Bio-Rad, Hercules, CA) attached to a vacuum manifold. We collected samples in 100-μl volumes via aspiration. Each dot contained 3 μg of RNA. Immediately before and after the samples were collected, we aspirated 50 μl of 10x SSC through the filter in duplicate. This created clean, reproducible binding of the RNA. Once all the samples were collected, we air dried the filter overnight and then baked it in an oven for 2 hours. For prehybridization and hybridization, the filter was sealed in a bag using a heated sealer. We prehybridized the filter at 42°C for 4–6 hours and hybridized the filter at 42°C overnight. We made DNA probes using only the coding region of each gene of interest (gliA, gliZ, gliP, gliT, and Actin). We washed the filter and exposed it to a Typhoon 8600 phosphorImager (GE Healthcare Life Sciences, Pittsburgh, PA) overnight. We quantified the intensity of hybridization using ImageQuant 5.1 software (GE Healthcare Life Sciences, Pittsburgh, PA). We quantified Actin mRNA levels as a control and standardized data to Actin, before normalizing to the pDONR AMA empty vector control.
All Gateway-enabled plasmids created in this study will be available at the Fungal Genomic Stock Center.
3. Results
The goal of this work was to create a variety of recombinatorial plasmids containing different marker genes to increase efficiency in vector manipulations used for genetic engineering. The markers chosen are, for the most part, versatile and can be used in multiple biologically relevant fungal organisms. All vectors, selective markers, and primers are listed in Table 1.
3.1. Plasmids for use in gene replacement
We designed several plasmids (Fig. 3A) intended for use with the MultiSite Gateway system (Invitrogen) to join two or more DNA segments (Magnani et al., 2006). We use these vectors to create deletion constructs with a 5' NCR and a 3' NCR flanking the selective marker in a destination vector, such as pDEST R4-R3, and give an example of this below. This can be accomplished in two steps: (1) Clone the 5' NCR and the 3' NCR into donor vectors pDONR P4-P1R and pDONR P2R-P3, respectively, using a BP recombination reaction and (2) Recombine the 5' NCR, the selective marker, and the 3' NCR into a destination vector (e.g., pDEST R4-R3) using an LR recombination reaction to form the final deletion construct. Importantly, different pDONR vectors contain different attP sites, and therefore require different attB sites to surround the appropriate DNA insert. For example, pDONR P4-P1R contains attP4 and attP1R, which will only recombine with attB4 and attB1R (Hartley et al., 2000; Magnani et al., 2006). This allows for directionality of the DNA insert in the recombination reactions. When using nutritional markers for gene deletion, we recommend using a selective marker from an organism other than the one being transformed. For example, if we were deleting a gene from an A. fumigatus strain, then we suggest using either argB or pyrG from A. nidulans. Using genes from different species decreases the chances of the deletion vector integrating at the nutritional gene locus, thereby increasing the efficiency of homologous recombination at the target locus.
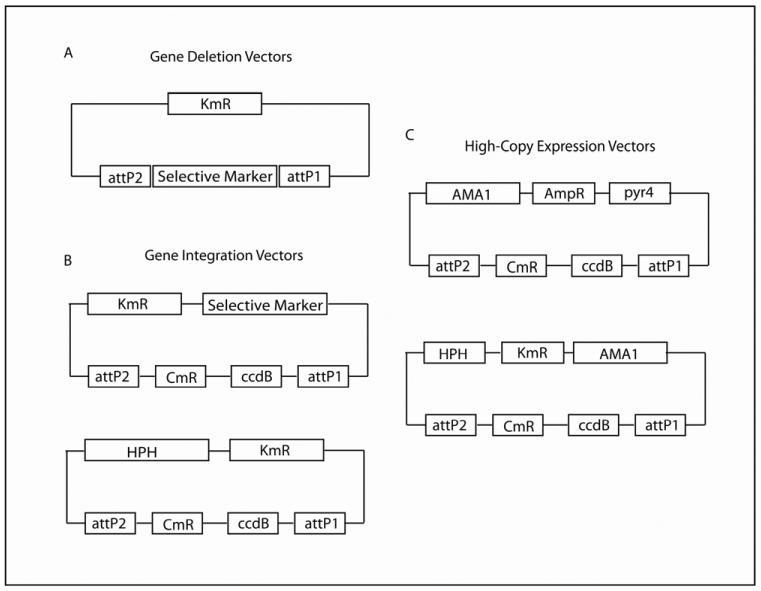
Figure 3. General structure of vectors created.
(a) All gene deletion vectors have a selective marker that was inserted in between the attP sites. The ccdB region has been removed. (b) All gene integration vectors have a selective marker and a ccdB region flanked by attP recombination sites. Of note, in pDONR HPH A and pDONR HPH B, the hph resistance marker is in a different spot in the vector than the selective markers in the other pDONR vectors. The pDONR 221 and pDONR vectors are kanamycin resistant in bacteria. (c) Both high-copy expression vectors contain AMA1 from A. nidulans. pDONR AMA has the ccdB region, flanked by attP sites, which was cloned into the pRG3-AMA-Not1 vector. This vector contains the pyr4 selective marker from N. crassa and is ampicillin resistant in bacteria. pDONR AMA/HPH has an hph resistance marker and the ccdB region flanked by attP recombination sites. This vector is kanamycin resistant in bacteria. Empty pDONR and pDONR AMA plasmids are also chloramphenicol resistant, until they lose the ccdB region through recombination.
3.2. Plasmids for integrative recombination
We created several pDONR vectors (Fig. 3B) to facilitate the complementation of gene deletions. We engineered these vectors using pDONR 221 as the base vector. Because these vectors still harbor the ccdB cassette, they can only be transformed into ccd-compatible bacterial strains until a gene of interest is recombined into the attP sites. With complementation, using a nutritional marker from the same strain you are working with can be advantageous. For example, if you are complementing a gene deletion created in A. nidulans, then it would be beneficial to use a vector that contains a nutritional marker from A. nidulans. Therefore, the complementation vector will have a higher chance of integrating at the nutritional gene locus, rather than inserting in a random position that could cause adverse positional effects or change gene expression. For some auxotrophic mutants, the entire nutritional locus has been deleted. In this case, the complementation construct is more likely to integrate randomly in the genome. All nutritional marker genes are flanked by a portion of the 5' NCR and 3' NCR, which contributes to targeted integration of the construct at the nutritional locus. In addition, when creating complementation plasmids, cloning the gene of interest, flanked by at least 1 kb of 5' NCR and 3' NCR, into the complementation vector is advantageous and can increase the likelihood of the construct integrating at a defined locus and not in a random position in the genome.
3.3. Plasmids for high-copy gene expression
We created two pDONR AMA vectors (Fig. 3C) for use in high-copy gene expression and genetic screens. Because these vectors still harbor the ccdB cassette, they can only be transformed into ccd-compatible bacterial strains until a gene of interest is recombined into the attP sites. These plasmids are autonomously replicating circular plasmids that are maintained as such following transformation. This is especially useful for genetic screens because the plasmids are easily recovered from fungal genomes by transformation of the genomic DNA into bacteria (Efimov, 2003; Osherov et al., 2001; Ukil et al., 2008). These plasmids are also valuable in over-expression studies, because there are, on average, anywhere from 10 to 30 copies present per genome (Aleksenko and Clutterbuck, 1997; Gems et al., 1991). Although the use of these vectors results in increased transformation efficiency in many fungal species, they are mitotically unstable and therefore require the use of a selective medium to remain present in the genome. These features are possible based on the presence of AMA1 (autonomous maintenance in Aspergillus) from A. nidulans (Aleksenko and Clutterbuck, 1997; Gems et al., 1991). The AMA1 cassette consists of a 300-bp central spacer flanked by two inverted copies of a MATE1 (mobile Aspergillus transformation enhancer) element. This MATE1 element is a genomic repeat region that is present throughout the genome of A. nidulans (Aleksenko and Clutterbuck, 1997; Gems et al., 1991).
We created pDONR AMA by cloning the ccdB cassette (attP1-ccdB-chloramphenicol resistance gene [CmR]-attP2) from pDONR 221 into the KpnI and SphI sites of pRG3-AMA-Not1 (Osherov and May, 2000). This vector contains pyr4 from Neurospora crassa and complements pyrG- mutants of A. nidulans and A. fumigatus. We generated pDONR AMA/HPH by cloning the AMA1 region from pDONR AMA into pDONR HPH B. This vector behaves as pDONR AMA, but contains a hygromycin resistance cassette (hygroR) as a selective marker. Due to the instability of the vector, any strain transformed with pDONR AMA/HPH will require growth on media containing hygromycin to prevent loss of the plasmid. attB1 and attB2 are needed for recombination into both of these vectors, since the ccdB cassette is from pDONR 221.
3.4. Deletion and complementation of mkkA using pDONR 221-AfpyrG and pDONR A
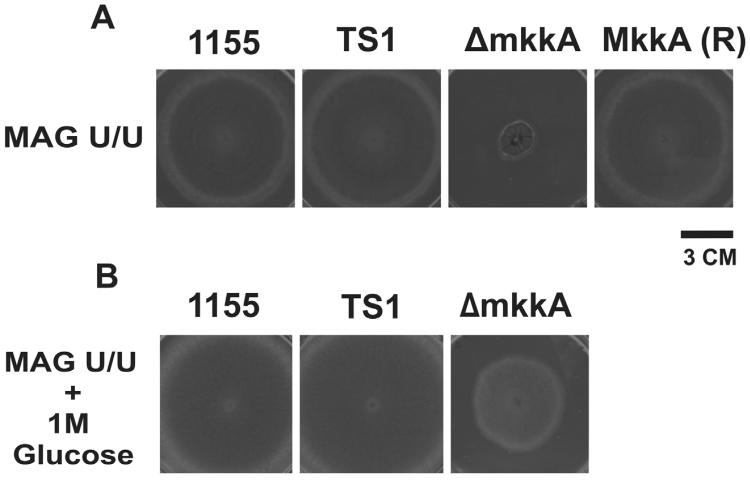
We created the deletion construct in 6 days from PCR of the 5' and 3' NCRs to transformation of the deletion construct into A. nidulans. We generated the deletion mutant by replacing the entire coding region of mkkA (AN4189.3) with AfpyrG. We screened transformants for proper nutritional prototrophy. Of the colonies screened, 100% were positive for homologous recombination of AfpyrG at the mkkA locus. The majority of ΔmkkA transformants exhibited extremely poor growth, with almost no conidiation. ΔmkkA grew best on rich medium supplemented with 1 M glucose as an osmotic stabilizer (Fig. 4A & B). For complementation, we transformed pDONR A-mkkA into ΔmkkA. We created the complementation vector in 3 days from PCR of the mkkA cassette to transformation of the vector into ΔmkkA. We screened transformants for the ability to grown in the absence of pyridoxine. Of the colonies we screened, 100% had obtained the mkkA coding region via a successful integration event. Upon complementation, we were able to restore conidiation of ΔmkkA back to wild-type levels (Fig. 4A).
Figure 4. Plate growth of ΔmkkA and mkkA (R).
(a) The A. nidulans mkkA deletion mutant and complemented strain mkkA(R) grown alongside An1155 and TS1 (controls). The use of pDONR A-mkkA as an integration vector restores ΔmkkA growth to wild-type levels. (b) The mkkA deletion mutant is severely defective in growth, but growth is partially restored in the presence of 1 M glucose as an osmotic stabilizer.
3.5. High-copy expression of gliZ
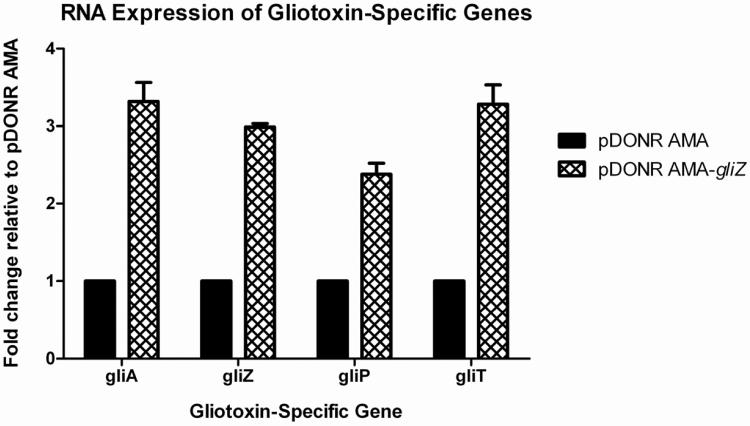
GliZ is the transcriptional activator of the gliotoxin biosynthetic cluster in A. fumigatus. This cluster, like many other secondary metabolite clusters, is under coordinate regulation through GliZ, therefore, induction of gliZ results in the induction of all other genes within the gliotoxin cluster (Bok et al., 2006). We amplified gliZ (Afu6g09630) with flanking promoter and terminator regions. We created a gliZ high-copy vector (pDONR AMA-gliZ) in 3 days and transformed pDONR AMA-gliZ into Af293.1. We verified the presence of the proper plasmid via digestion analysis. We grew strains in conditions conducive to low gliotoxin cluster gene expression and isolated total RNA. We quantified mRNA levels of multiple genes within the gliotoxin cluster via dot blot analysis. As shown in figure 3, gliZ is transcribed at higher than normal levels as a result of the autonomous nature of the pDONR AMA-gliZ plasmid. Consequently, other genes within the gliotoxin cluster are being induced to higher than normal levels as well (Fig. 5).
Figure 5. Transcription of gliotoxin-specific genes in a gliZ high-copy strain.
The presence of extra copies of gliZ causes an increase in RNA levels of multiple genes within the gliotoxin biosynthetic cluster. We collected total RNA and quantified mRNA levels of designated genes by RNA Dot Blot Analysis. Data are expressed as fold change relative to a pDONR AMA empty vector control. All data was standardized to Actin before normalizing to the empty vector control.
4. Conclusions
Numerous recombinatorial methods have been developed to increase efficiency in genetic manipulations. Some of these methods (recombineering, MAGIC, and DNA assembler) are performed in vivo (Li and Elledge, 2005; Liu et al., 2003; Mosberg et al.; Shao and Zhao, 2009; Sharan et al., 2009), while others (Cre-loxP, SLIC, LIC, and Gateway) can be carried out as in vitro reactions (Hartley et al., 2000; Li and Elledge; Li and Elledge, 2007; Magnani et al., 2006; Siegel et al., 2004; Yamamura, 2012). These methods offer different advantages and disadvantages, depending on the intended outcome. Two methods that have been used quite often, Cre-loxP and Gateway, have increase the efficiency of plasmid construction via site-specific recombination events.
The Cre-loxP system has been used in conditional knockout studies in different organisms and is advantageous if one is interested in expressing any gene of interest, fluorescent genes, luciferase genes, or mutated cDNAs under the control of the endogenous promoter of a targeted gene, although the insertion of loxP sites around the targeted gene is first required to be able to perform these techniques (Deng, 2012; Siegel et al., 2004; Yamamura, 2012). When loxP sites are in place, Cre recombinase is all that is required for the recombination event to occur. One caveat of the Cre-loxP system is the lack of directional cloning with wild-type loxP sites. DNA fragments that are flanked by these identical wild-type loxP sites will insert in either orientation, which can be troublesome if the direction of the insert is important. In addition, if a DNA fragment, flanked by wild-type loxP sites, is inserted into a genomic sequence, these loxP sites can be targeted by Cre recombinase at any time, which results in unwanted excision of the fragment. To alleviate this problem, several mutated loxP sites have been created to facilitate directional recombination and prevent re-excision (Deng, 2012; Siegel et al., 2004; Yamamura, 2012). While these mutant sites have been shown to be stable, they are not commercially available in a multitude of vectors.
The Gateway system has been used in numerous eukaryotic organisms for genetic manipulations. Similar to the Cre-loxP system, DNA fragments can be inserted into vectors using recombination sites (att sites). These att sites are available in a variety of “flavors” and have specific att complements. For example, attL2 will only recombine with attR2 and attB1 will only recombine with attP1. This eliminates the need to mutate att sites and facilitates directional cloning of any DNA fragment. One of the other factors that make this system highly efficient is the ccdB cassette, which is flanked by the att sites in donor and destination vectors. This gene encodes a toxin from Escherichia coli, which is responsible for post-segregational killing of daughter cells that do not contain the F plasmid (Bernard and Couturier, 1992). The presence of the ccdB gene in Gateway vectors ensures that any vectors that do not recombine properly will kill the bacterial strain used for transformation, leaving only recombined vectors containing the insert of interest. For more information on the ccdB counter-selection technique, please refer to Bernard and Couturier 1992. Additionally, in contrast to the Cre-loxP system, numerous Gateway-enabled vectors are commercially available for different genetic techniques, making the Gateway system a powerful tool for genetic engineering. One caveat to Gateway technology is the need for multiple recombination proteins to carry out reactions, although these proteins are readily available as easy to use kits, which further enhances the efficiency of the reaction.
Taking advantage of recombinatorial cloning techniques, we have created a series of vectors to facilitate gene deletion, gene complementation, and high-copy gene expression in a variety of fungal species. Our aim was to create a set of versatile recombinatorial vectors to be used as efficient tools for genetic manipulations in biologically relevant fungal species. Gateway-enabled vectors have been constructed in other laboratories, but mostly appear to focus on protein fusions and fluorescence tagging (Mabashi et al., 2006; Toews et al., 2004). The benefit of our plasmids is that they are intended to serve as versatile constructs. The specific selective markers we chose have been shown to be compatible with several other fungal species, such as Aspergillus species, Penicillium species, and Neurospora species (Abe et al., 2006; Bugeja et al., 2012; Pahirulzaman et al., 2012; Paz et al., 2011). Therefore, even though our studies focused solely on A. fumigatus and A. nidulans, we hypothesize that these plasmids will serve as efficient tools in other biologically and agriculturally relevant fungal organisms. We do not claim that the plasmids we have constructed will work ubiquitously, as there are always certain species that are incompatible. For instance, the autonomously replicating plasmids work well with Aspergillus species, as hyphae contain septae to separate nuclei. However, the AMA1-containing plasmids may not function as described in fungal organisms that produce coenocytic hyphae.
Our laboratory has used the pDONR 221 vectors for numerous gene deletions, and the pDONR vectors for gene complementation. In the present study, we demonstrated the competence of the pDONR 221-AfpyrG vector and the pDONR A vector with the mkkA deletion mutant and mkkA complemented strain, respectively, in A. nidulans. Construction of the pDEST R4-R3 deletion vector and pDONR A complementation vector was quick and efficient. We have used most of the Gateway-enabled vectors in our own experiments and have not encountered any predicament with recombination or gene expression within the fungal strain being used. To our knowledge, the presence of the att sites does not cause any setbacks with gene expression or growth. We have also used both pDONR AMA and pDONR AMA/HPH in high-copy expression experiments and have given an example in the present study with the gliZ high-copy strain in A. fumigatus. Recombination experiments have been rapid and successful for both pDONR AMA vectors, usually only taking 1 day for recombination and an additional 2 days for insert verification. They behave as autonomously replicating plasmids and the presence of the att sites do not interfere with gene expression. Although AMA1-containing plasmids have been used successfully in numerous Aspergillus species and even a few Penicillium species, one caveat to using AMA1-based plasmids is their mitotic instability (Efimov, 2003; Fierro et al., 1996; Fierro et al., 2004; Gems et al., 1991; Osherov et al., 2001). This can pose problems, as the AMA1-based plasmid has a higher chance of being lost in progeny than an integrative plasmid. This instability is not constant in all fungal species that have been studied using these autonomously replicating plasmids. For instance, mitotic stability of AMA1-based plasmids is higher in P. nalgiovense than in P. chrysogenum (Fierro et al., 1996; Fierro et al., 2004). In our lab, we have used AMA1-based plasmids frequently with great success and find that as long as strains are grown on selective medium, the incidence of plasmid loss decreases.
As our results demonstrate, using Gateway-enabled vectors expedites the process of vector construction for genetic techniques. Not only is Gateway technology widely available and easy to use, but it also has many advantages over traditional vector construction, such as increased effectiveness and built-in screening. The variety of different att sites is ideal for directional cloning of DNA fragments and can be used to fuse multiple DNA fragments together or simply attach a tag. We have constructed a series of Gateway-enable vectors containing various selective markers to streamline genetic manipulation. These vectors are meant to be used as versatile genetic tools, as they can be used in a variety of fungal species.
Highlights
We created a series of Gateway-enabled plasmids for use in filamentous fungi
These plasmids can be used in gene deletion, gene integration, and high-copy gene expression
The plasmids contain a variety of nutritional and drug-resistance markers that are functional across multiple fungal groups
Acknowledgements
This work was funded by grants AI051144 and AI074673 from the National Institutes of Health, NIAID, to G.S.M. While this work was done, T.J.S was supported by the Schissler Foundation Fellowship. All sequencing was performed by the MD Anderson Core DNA Analysis facility funded by the Core grant CA016672 (SMF).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. Author Contributions C.K.C created and analyzed the pDONR 221-AfpyrG, pDONR 221-AnpyrG, pDONR 221-BAR, pDONR 221-HPH, pDONR HPH A and B plasmids. T.J.S created and analyzed all other plasmids. T.J.S performed the experiments used as examples in this paper. T.J.S wrote the entire paper with editing from G.S.M. G.S.M supervised the project.
References
- Abe A, et al. Construction of pDESTR, a GATEWAY vector for gene disruption in filamentous fungi. Curr Microbiol. 2006;52:210–5. doi: 10.1007/s00284-005-0238-0. [DOI] [PubMed] [Google Scholar]
- Aleksenko A, Clutterbuck AJ. Autonomous plasmid replication in Aspergillus nidulans: AMA1 and MATE elements. Fungal Genet Biol. 1997;21:373–87. doi: 10.1006/fgbi.1997.0980. [DOI] [PubMed] [Google Scholar]
- Bernard P, Couturier M. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J Mol Biol. 1992;226:735–45. doi: 10.1016/0022-2836(92)90629-x. [DOI] [PubMed] [Google Scholar]
- Bok JW, et al. GliZ, a transcriptional regulator of gliotoxin biosynthesis, contributes to Aspergillus fumigatus virulence. Infect Immun. 2006;74:6761–8. doi: 10.1128/IAI.00780-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugeja HE, et al. Tools for high efficiency genetic manipulation of the human pathogen Penicillium marneffei. Fungal Genet Biol. 2012;49:772–8. doi: 10.1016/j.fgb.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Deng C-X. The Use of Cre-loxP Technology and Inducible Systems to Generate Mouse Models of Cancer. In: Ried J. E. G. a. T., editor. Genetically Engineered Mice for Cancer Research: Design, Analysis, Pathways, Validation and Pre-Clinical Testing. Spring Science + Business Media, LLC; 2012. pp. 17–36. [Google Scholar]
- Efimov VP. Roles of NUDE and NUDF proteins of Aspergillus nidulans: insights from intracellular localization and overexpression effects. Mol Biol Cell. 2003;14:871–88. doi: 10.1091/mbc.E02-06-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierro F, et al. Autonomously replicating plasmids carrying the AMA1 region in Penicillium chrysogenum. Curr Genet. 1996;29:482–9. doi: 10.1007/BF02221518. [DOI] [PubMed] [Google Scholar]
- Fierro F, et al. High efficiency transformation of Penicillium nalgiovense with integrative and autonomously replicating plasmids. Int J Food Microbiol. 2004;90:237–48. doi: 10.1016/s0168-1605(03)00306-4. [DOI] [PubMed] [Google Scholar]
- Gems D, et al. An autonomously replicating plasmid transforms Aspergillus nidulans at high frequency. Gene. 1991;98:61–7. doi: 10.1016/0378-1119(91)90104-j. [DOI] [PubMed] [Google Scholar]
- Hartley JL, et al. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–95. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, et al. Simple chemical extraction method for DNA isolation from Aspergillus fumigatus and other Aspergillus species. J Clin Microbiol. 2004;42:4293–6. doi: 10.1128/JCM.42.9.4293-4296.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MZ, Elledge SJ. SLIC: a method for sequence- and ligation-independent cloning. Methods Mol Biol. 852:51–9. doi: 10.1007/978-1-61779-564-0_5. [DOI] [PubMed] [Google Scholar]
- Li MZ, Elledge SJ. MAGIC, an in vivo genetic method for the rapid construction of recombinant DNA molecules. Nat Genet. 2005;37:311–9. doi: 10.1038/ng1505. [DOI] [PubMed] [Google Scholar]
- Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods. 2007;4:251–6. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- Liu P, et al. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–84. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabashi Y, et al. Development of a versatile expression plasmid construction system for Aspergillus oryzae and its application to visualization of mitochondria. Biosci Biotechnol Biochem. 2006;70:1882–9. doi: 10.1271/bbb.60052. [DOI] [PubMed] [Google Scholar]
- Magnani E, et al. From Gateway to MultiSite Gateway in one recombination event. BMC Mol Biol. 2006;7:46. doi: 10.1186/1471-2199-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, et al. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- May GS. The highly divergent beta-tubulins of Aspergillus nidulans are functionally interchangeable. J Cell Biol. 1989;109:2267–74. doi: 10.1083/jcb.109.5.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosberg JA, et al. Lambda red recombineering in Escherichia coli occurs through a fully single-stranded intermediate. Genetics. 186:791–9. doi: 10.1534/genetics.110.120782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osherov N, et al. Resistance to itraconazole in Aspergillus nidulans and Aspergillus fumigatus is conferred by extra copies of the A. nidulans P-450 14alpha-demethylase gene, pdmA. J Antimicrob Chemother. 2001;48:75–81. doi: 10.1093/jac/48.1.75. [DOI] [PubMed] [Google Scholar]
- Osherov N, May G. Conidial germination in Aspergillus nidulans requires RAS signaling and protein synthesis. Genetics. 2000;155:647–56. doi: 10.1093/genetics/155.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahirulzaman KA, et al. A toolkit for heterologous expression of metabolic pathways in Aspergillus oryzae. Methods Enzymol. 2012;517:241–60. doi: 10.1016/B978-0-12-404634-4.00012-7. [DOI] [PubMed] [Google Scholar]
- Paz Z, et al. One step construction of Agrobacterium-Recombination-ready-plasmids (OSCAR), an efficient and robust tool for ATMT based gene deletion construction in fungi. Fungal Genet Biol. 2011;48:677–84. doi: 10.1016/j.fgb.2011.02.003. [DOI] [PubMed] [Google Scholar]
- Reyes G, et al. Novel mitogen-activated protein kinase MpkC of Aspergillus fumigatus is required for utilization of polyalcohol sugars. Eukaryot Cell. 2006;5:1934–40. doi: 10.1128/EC.00178-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Zhao H. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009;37:e16. doi: 10.1093/nar/gkn991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan SK, et al. Recombineering: a homologous recombination-based method of genetic engineering. Nat Protoc. 2009;4:206–23. doi: 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RW, et al. Recombinatorial cloning using heterologous lox sites. Genome Res. 2004;14:1119–29. doi: 10.1101/gr.1821804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiliotis M. Inverse fusion PCR cloning. PLoS One. 2012;7:e35407. doi: 10.1371/journal.pone.0035407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk E, et al. Fusion PCR and gene targeting in Aspergillus nidulans. Nat Protoc. 2006;1:3111–20. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- Toews MW, et al. Establishment of mRFP1 as a fluorescent marker in Aspergillus nidulans and construction of expression vectors for high-throughput protein tagging using recombination in vitro (GATEWAY) Curr Genet. 2004;45:383–9. doi: 10.1007/s00294-004-0495-7. [DOI] [PubMed] [Google Scholar]
- Ukil L, et al. Copy number suppressors of the Aspergillus nidulans nimA1 mitotic kinase display distinctive and highly dynamic cell cycle-regulated locations. Eukaryot Cell. 2008;7:2087–99. doi: 10.1128/EC.00278-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Miller BL. Aspergillus asexual reproduction and sexual reproduction are differentially affected by transcriptional and translational mechanisms regulating stunted gene expression. Mol Cell Biol. 1997;17:6191–201. doi: 10.1128/mcb.17.10.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura K. A. a. K.-i. Genetic Manipulations using Cre and mutant loxP sites. Springer Science+Business Media, LLC; 2012. [Google Scholar]
- Yu JH, et al. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol. 2004;41:973–81. doi: 10.1016/j.fgb.2004.08.001. [DOI] [PubMed] [Google Scholar]