Abstract
Telomerase adds telomeric repeats to chromosome ends using an internal RNA template and specialized telomerase reverse transcriptase (TERT), thereby maintaining genome integrity. Little is known about the physical relationships among protein and RNA subunits within a biologically functional holoenzyme. Here we describe the architecture of Tetrahymena thermophila telomerase holoenzyme determined by electron microscopy. Six of the 7 proteins and the TERT-binding regions of telomerase RNA (TER) have been localized by affinity labeling. Fitting with high-resolution structures reveals the organization of TERT, TER, and p65 in the RNP catalytic core. p50 has an unanticipated role as a hub between the RNP catalytic core, p75-p19-p45 subcomplex, and the DNA-binding Teb1. A complete in vitro holoenzyme reconstitution assigns function to these interactions in processive telomeric repeat synthesis. These studies provide the first view of the extensive network of subunit associations necessary for telomerase holoenzyme assembly and physiological function.
Telomerase is a unique endogenous eukaryotic reverse trancriptase (RT) required for maintenance of linear chromosome ends and is a highly regulated determinant of cellular aging, stem cell renewal, and tumorigenesis1,2. TER contains a region of sequence complementarity to the telomeric repeat that is used as a template for addition of successive repeats to the 3′ end of chromosomes, for example addition of the repeat TTAGGG in humans. The specialized telomerase catalytic cycle of telomeric repeat synthesis has been extensively investigated for the human enzyme and biochemically similar enzymes from model organisms such as the telomere-rich ciliate Tetrahymena thermophila3. While only TERT and TER are required for telomerase catalytic activity in vitro, the physiologically functional holoenzyme is a multisubunit RNP4,5. Affinity purification, mass spectrometry, and subunit tagging assays have established 8 telomerase-specific, reciprocally co-purifying subunits of Tetrahymena telomerase holoenzyme, each essential for telomere length maintenance6,7 (Fig. 1a). Three of these subunits are required for in vivo assembly of a catalytically active RNP: TERT, TER, and the La-family protein p657. The p65 La and RRM1 domains increase p65 affinity for TER, but only the C-terminal xRRM2 domain (Fig. 1a) is critical for the TER folding that enhances TERT RNP assembly in vitro and in vivo8-11. Two TER regions, loop 4 (L4) next to the p65 ×RRM2 interaction site and the TERT high-affinity binding element (TBE) 5′ to the template (Fig. 1a), bind to the TERT high-affinity RNA binding domain (TRBD)12-14. The TERT TRBD, RT domain, and C-terminal extension (CTE) form a ring that encircles an active site cavity, with a fourth TERT N-terminal (TEN) domain (Fig. 1a) positioned in a TER-dependent but otherwise unknown location15-19.
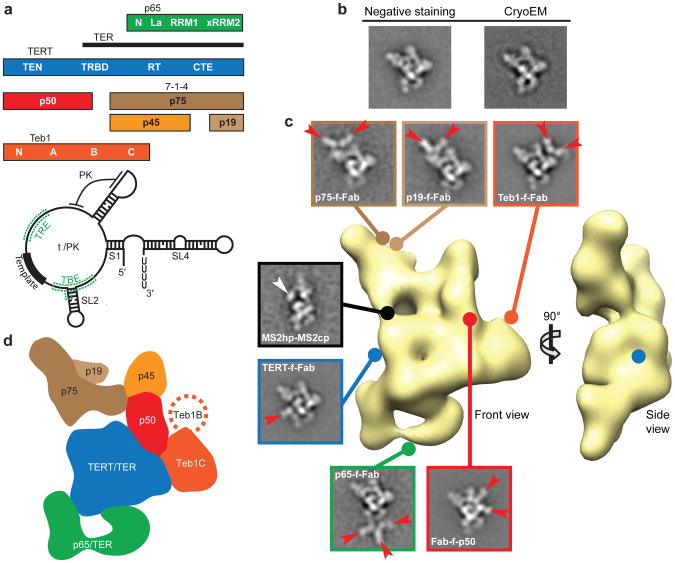
Figure 1. EM reconstruction of Tetrahymena telomerase holoenzyme and subunit localization.
a, Holoenzyme subunits and domains (top) and TER secondary structure (below). b, Representative class averages of negative staining EM and cryoEM images of TERT-f telomerase. c, 3D reconstruction of Teb1-f telomerase (front and side views) and class averages of affinity labeled telomerase particles. Lines with circle heads indicate attachment point of Fab (red arrows) and MS2cp (white arrow). Side-lengths of class averages in this and subsequent figures are 350 Å. d, Subunit schematic (front view).
Little is known about the configuration and roles of the 5 additional Tetrahymena telomerase holoenzyme proteins required for in vivo telomere elongation (p75, p50, p45, p19, and Teb1; Fig. 1a); only Teb1 has a known domain structure. Like the paralogous large subunit of the single-stranded DNA (ssDNA) binding factor Replication Protein A, Teb1 has four OB-fold domains, NABC (Fig. 1a); A and B bind ssDNA with high affinity and C is necessary for holoenzyme association6,20,21. Teb1 is required for the particularly high DNA product interaction stability of an endogenously assembled Tetrahymena telomerase holoenzyme, evident as high repeat addition processivity (RAP) in vitro6,20,21. The holoenzyme subunits p75, p19, and p45, here designated 7-1-4, form a subcomplex that remains assembled upon micrococcal nuclease-induced dissociation of the other holoenzyme subunits in vitro6,21. Little is known about p50, which appears sub-stoichiometric on silver stained gels of affinity purified holoenzyme6.
Here we report the native electron microscopy (EM) structure and a complete in vitro reconstitution of Tetrahymena telomerase holoenzyme. This first physical and functional network architecture of a telomerase holoenzyme provides unprecedented detail about the structure of the RNP catalytic core and reveals the organization of holoenzyme subunits that confer processivity and bridge telomerase to telomeres.
Overall structure and localization of subunits
Tetrahymena telomerase holoenzymes were prepared for EM (Supplementary Fig. 1) by affinity purification6 from 10 different strains bearing N- or C-terminal 3 × FLAG and tandem Protein A tags (zzf or fzz, respectively) on TERT or other holoenzyme subunits (Supplementary Table). Comparison of the class averages from negative staining EM images of telomerase purified using TERT-fzz (TERT-f telomerase) to those from cryoelectron microscopy (cryoEM) images (Fig. 1b and Supplementary Fig. 2a,b) shows that the negative stain did not change the structure within experimental resolution. In addition to the predominant (“stable”) conformation of the complete holoenzyme particle, constituting 31% of the total particles, the class averages show additional particle subpopulations of varying conformation and subunit composition (described below and Supplementary Fig. 2). The class averages from both negative staining EM images and cryoEM images show a strongly preferred orientation with almost all particles sitting on the support carbon film in the same way (Supplementary Fig. 2). To overcome the problems of preferred orientation and structural variability, the EM image acquisition and three-dimensional (3D) reconstructions were carried out using an automated random conical tilt (RCT) method22 (Supplementary Fig. 3). The 3D structures of TERT-f and Teb1-f telomerase holoenzymes are indistinguishable (Supplementary fig. 4a), and Teb1-f gave higher yields of holoenzyme. A 3D structure (Fig. 1c) of the ∼500 kDa Teb1-f telomerase holoenzyme from 3D reconstruction of 2,220 particles with a resolution of ∼25 Å (Supplementary Fig. 4b) demonstrates well-ordered holoenzyme density occupying about 200 × 150 × 80 Å.
To locate each protein subunit within the overall holoenzyme structure, telomerase was purified from strains harboring fzz-tagged TERT, p75, p65, p45, p19, or Teb1 in place of the corresponding untagged protein6. During purification, the zz portion of the tag was removed by proteolytic cleavage, leaving only a short f-tag (3 × FLAG) that was labeled using monoclonal anti-FLAG Fab (see Methods). Each tagged protein can bind up to three Fabs, as illustrated in some of the class averages and 3D maps (Fig. 1c and Supplementary Fig. 5), which pinpoint to a single spot and localize the binding site of Fab unambiguously. Affinity purification of the biologically functional C-terminally tagged p50 (p50-fzz) does not enrich sufficient holoenzyme for silver staining detection in SDS-PAGE6. Instead we used N-terminally zzf-tagged p50 (zzf-p50) expressed in partial replacement of the endogenous locus, which did purify holoenzyme to homogeneity (Supplementary Fig. 6). All holoenzyme protein subunits were localized by Fab labeling except p45, which was identified by process of elimination (see Methods). In addition, we affinity purified telomerase from a strain with biologically functional tagged TER harboring a small hairpin tag (MS2hp), which is recognized by the MS2 coat protein (MS2cp), appended to TER stem 2 (S2)23 (Fig. 1c and Supplementary Fig. 5g). The MS2cp dimer bound to this tag appears as extra density in the class averages and thus provides the approximate location of the TBE 5′ to the template (Fig 1a).
The positions of the TERT and Teb1 C-termini and the p50 N-terminus could be precisely mapped in the 3D reconstructions of the respective Fab-labeled telomerase particles (Supplementary Fig. 5a-c), while a less precise location of the C-termini of p65, p75, and p19 (which due to low purification yield did not give sufficient particles for RCT data collection) was obtained from the class averages (Supplementary Table, Fig. 1c, and Supplementary Fig. 5d-f). Approximate subunit boundaries (schematized in Fig. 1d) could be modeled after fitting the holoenzyme density with known TERT15-18, TER8,24-26, p658 and Teb120 high-resolution domain structures combined with comparisons of class averages and 3D reconstructions across different complexes as described below. These data reveal that, in the “front” view in Fig. 1c (schematized in Fig. 1d) with p65 at the bottom, TERT occupies the center of the particle and 7-1-4 subunits form the top. Teb1 projects from the middle layer to the right. The p50 subunit is also part of the middle layer, networked between TERT, 7-1-4, and Teb1 (Fig. 1c,d). Numerous inter-subunit appositions generate an intricate and highly contoured surface.
The structure of the RNP catalytic core
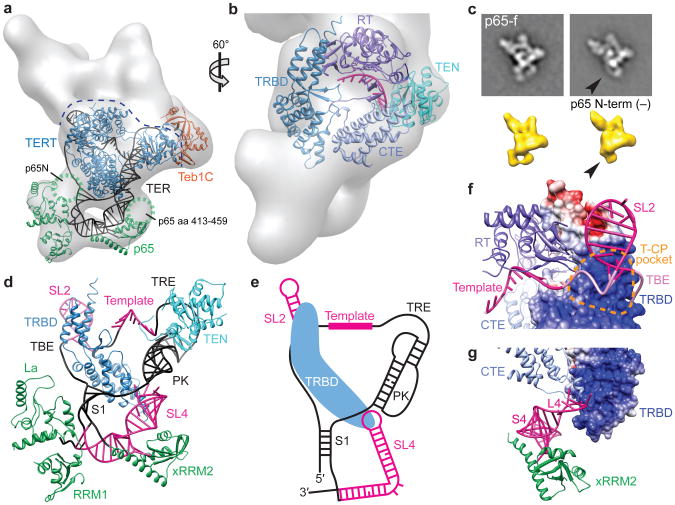
Based on the determined subunit locations we fit the crystal and NMR structures of TER and protein domains in the EM density map of Teb1-f telomerase to generate a model of the entire RNP catalytic core (Fig. 2a and Supplementary Video). The RT and CTE domains of Tetrahymena TERT were homology modeled27 from the Tribolium castaneum TERT crystal structure18 and combined with the crystal structures of Tetrahymena partial TRBD15 and TEN16 domains as described in Methods. The modeled TERT TRBD-RT-CTE fits in only one orientation (Fig. 2a,b and Supplementary Fig. 7a,b). The position of the TERT CTE is consistent with the location of the C-terminus identified in the 3D reconstruction of TERT-f-Fab telomerase (Fig. 1c and Supplementary Fig. 5a)17. The TEN domain was placed into density remaining after determination of subunit boundaries of the adjacent Teb1C and p50, and oriented based on the homology model of human TERT28. The modeled Tetrahymena TERT TRBD interacts with the CTE, which is consistent with the Tribolium TERT crystal structures17,18 and isolated human TERT domain interactions19. There is EM density linking the TEN and CTE regions and the TEN and TRBD regions, which could correspond to the ∼70 amino acids of potentially unstructured TEN-TRBD linker29,30 and adjacent TRBD sequence missing from high-resolution structures15,16, which crosslinks to ssDNA and improves template boundary definition19,31.
Figure 2. Structure of the RNP catalytic core.
a, 3D reconstruction of Teb1-f telomerase with TERT, p65, and TER (black), plus Teb1C modeled into the EM density. The dashed line indicates the top boundary of TERT/TER. b, Zoomed and rotated view of (a) showing TERT domains TEN, CTE, TRBD, and RT, with TER template and essential Mg2+ at the active site in magenta. c, Class averages of p65-f telomerase (top) and 3D reconstructions (bottom) of TERT-f telomerase with p65 (left) and p65 missing N-terminal density (right, black arrows). d, TER model structure (well determined, magenta; remaining, black) and interactions with TERT TRBD and TEN and p65 La, RRM1, and xRRM2 domains. e, Secondary structure schematic of TER with TRBD. f, Modeled interaction between TBE (pink) and TRBD T-CP pocket. g, Modeled interactions of distal SL4 with CTE and bottom of TRBD. In f,g TRBD is shown as GRASP surface.
The C-terminus of p65 is at the bottom of the particle, below TERT (Fig. 1d and Supplementary Fig. 5d). In the class averages and 3D reconstructions we observed particle subpopulations lacking part of the density at the bottom left of the particle (Fig. 2c and Supplementary Figs. 2, 3c, and 5d), which must arise from loss of p65 N-terminal domain(s) due to partial proteolysis of this subunit common during holoenzyme purification6 and/or consequent loss of positional constraints on the remaining La and RRM1 domains. The structure of the p65 C-terminal domain xRRM2:stem 4 (S4) complex8 could therefore be localized and fit to the density present in all the 3D reconstructions (Fig. 2a,d and Supplementary Fig. 7c,d). The remaining parts of S1-SL4 and p65 La and RRM1 could then be modeled in (see Methods). The p65 N-terminal domain (near S1) and the long β2-β3 loop within xRRM2 that was deleted in the crystal structure could occupy the remaining unmodeled density (Fig. 2a)8,26. Together these fittings provide the overall topology of p65-TER interaction (Fig. 2a,d).
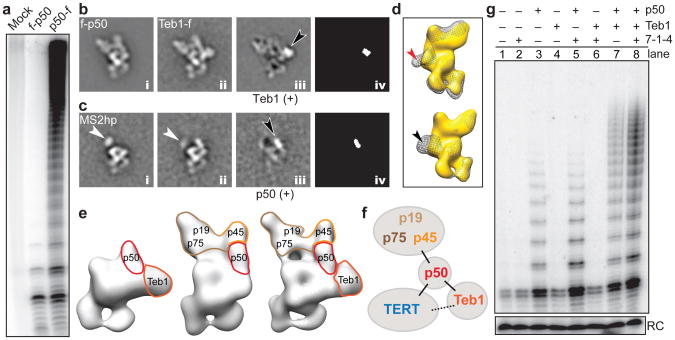
Figure 3. p50 anchors TERT, 7-1-4, and Teb1.
a, Primer extension assay of FLAG antibody purifications from cell extracts lacking a tagged subunit (mock) or with f-p50 or p50-f. b, Class averages of f-p50 telomerase (i), Teb1-f telomerase (ii), difference map by subtracting i from ii (iii), and map of statistically significant (>4 σ) regions in the difference map (iv). Black arrow points to Teb1 density. c, MS2hp telomerase class averages containing MS2cp without p50 (i), with p50 (ii), difference map (iii), and statistically significant regions (iv) as in (b). White and black arrows point to MS2cp and p50 densities, respectively. 7-1-4 is not seen in these class averages. d, Back view of 3D reconstructions of TERT-f telomerase lacking Teb1 (gold) overlaid with (top) Fab-f-p50 (gray mesh) and (bottom) Teb1-f telomerase (gray mesh) showing that Fab (red arrow) occupies the same site as Teb1 (black arrow). e, 3D reconstructions of TERT-f telomerase lacking 7-1-4 (left), lacking Teb1 (middle), and holoenzyme (right). f, Schematic of subunit interactions. g, Primer extension assay of the RNP catalytic core (TERT-TER-p65) reconstituted with additional combinations of 7-1-4, p50, and/or Teb1BC. Reactions were for 5 min. RC is a radiolabeled oligonucleotide added to telomerase products as a precipitation recovery control.
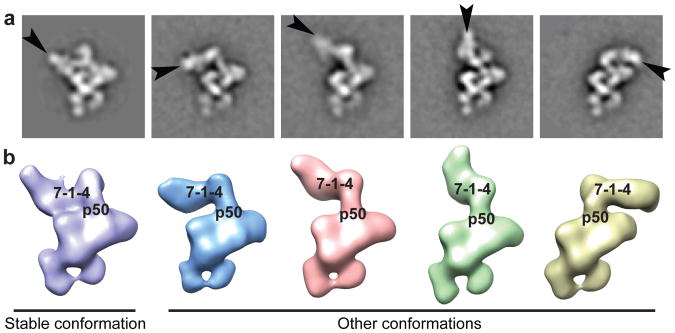
Figure 5. Positional dynamics of 7-1-4.
a, Teb1-f telomerase class averages with p75 indicated (black arrows). b, 3D reconstructions of Teb1-f telomerase showing different positions of 7-1-4.
Tetrahymena TER contains two major domains, the template/pseudoknot (t/PK) and SL4, which are connected by S1 (Fig. 1a). Starting with the defined locations of the template in the active site of TERT18, S2 by MS2cp labeling, and SL4 in complex with p65 ×RRM28 (Fig. 2d, magenta), and considering topological restrictions based on the length of the single-stranded regions of TER, a model structure of the PK, and S1, we traced a potential trajectory of the entire TER (Fig. 2a,d,e) fit into the remaining EM density. The template recognition element (TRE) appears to be close to the TEN domain, as implicated biochemically32, and the bottom of the PK is close to L4 (Fig. 2a,d). The locations of TER elements in the model are consistent with the large body of biochemical data on TER structure and function4,5,33,34. Of particular significance are the well-determined locations of the two TRBD binding elements, the TBE and L49,10,13,32, which we find bind to two distinct regions of the TRBD, near the top left and bottom of the TERT ring, respectively (Fig. 2b,d,e).
Remarkably, only the apical loop of SL4 connects to the density assigned to TERT, resulting in a U-shape at the bottom of the holoenzyme formed by TER S1/SL4 bound by p65. (Fig. 2a,d,e). The interaction of distal SL4 with TERT explains how it can stimulate telomerase activity when added to the t/PK in trans35,36 and how binding of p65 ×RRM2 to S4 mediates hierarchical assembly of p65-TER with TERT8-10. The EM structure and model reveal that the TRBD bridges the TBE and L4, which are ∼40 Å apart. In the model, distal SL4 approaches the 3′ end of the PK, consistent with reported high FRET between L4 and an internal PK position37. Although detailed interactions await a high-resolution structure, in the model the TBE contacts the TRBD in the T-pocket and CP-motif region (T-CP pocket) (Fig. 2f), which is consistent with biochemical data12,14,15, and distal SL4 is flanked by the CTE and TRBD (Fig. 2g), potentially contributing to folding of TERT around TER.
p50 anchors the accessory proteins
The catalytic activity associated with telomerase purified using zzf-p50 has low RAP, in contrast with the high-RAP activity enriched by the much lower yield of holoenzyme purified by p50-fzz (Fig. 3a). The Fab labeling showed that the N-terminus of p50 and the C-terminus of Teb1 are relatively close together (Fig. 1c and Supplementary Fig. 5b,c). Comparison of the class averages of f-p50 and Teb1-f telomerase shows that all f-p50 telomerase particles lack density for Teb1 and reveals the boundary of Teb1 by difference map (Fig. 3b). The boundary of p50 was further defined by comparing class averages from a small subclass of particles containing only the RNP catalytic core, which was observed only in the MS2cp-bound MS2hp telomerase, with class averages containing p50 (Fig. 3c). Superpositions of 3D reconstructions of Fab-f-p50 and Teb1-f telomerase on TERT-f telomerase lacking Teb1 (see below) show that Fab bound to the N-terminus of f-p50 occupies the same space as Teb1 (Fig. 3d). Together, these results identify the locations of Teb1 and p50, show that p50 interacts directly with the RNP catalytic core, and reveal that Teb1 is in close proximity to the N-terminus of p50 and to TERT. The holoenzyme structure therefore explains the low-RAP activity of purified N-terminally tagged p50 (Fig. 3a), since enzyme purification by the N-terminal zzf-p50 tag disrupts Teb1 binding. Beyond density assigned to the RNP catalytic core, Teb1, and p50, the remaining density in the top part of the 3D map is occupied by the 7-1-4 subcomplex. Taken together, 3D reconstructions reveal a central location for p50 in holoenzyme architecture, as an interaction hub between TERT, 7-1-4, and Teb1 (schematized in Fig. 3f).
Assigning function to subunit architecture
In parallel with structural analysis, we developed a method for holoenzyme reconstitution from entirely recombinant subunits. RNP catalytic core with TERT-f was preassembled in rabbit reticulocyte lysate (RRL) and then combined with RRL-expressed p50 and/or 7-1-4 and/or bacterially expressed Teb1BC (see Methods)21 (Fig. 3g). Direct primer-extension assays showed that addition of 7-1-4 or Teb1 alone did not alter the low-RAP product synthesis profile of the RNP catalytic core, but addition of p50 alone stimulated enzyme activity and the synthesis of multi-repeat products (Fig. 3g, lane 3). For p50-containing enzymes, 7-1-4 alone increased the amount of product (Fig. 3g, lane 5), Teb1 alone increased product length (lane 7), and their combination into complete holoenzyme was synergistic for increased activity level and long product synthesis (lane 8). These in vitro biochemical activities of p50, 7-1-4, and Teb1 were evident within a 5 min reaction time that allowed all product DNAs to be resolved by gel electrophoresis (Fig. 3g) or with the additional time and [dGTP] that supported very long product synthesis (Supplementary Fig. 8). The p50-dependent activity of 7-1-4 and Teb1 explains the presence of p50 in virtually all of the EM structures.
Previous reconstitution assays combined RRL-assembled recombinant RNP catalytic core, bacterially expressed Teb1, and endogenously assembled Tetrahymena proteins that remained bound to p45-f after micrococcal nuclease treatment of purified holoenzyme20,21,38. We found that the 7-1-4 complex isolated by this method does not completely dissociate p50, which as a full-length protein co-migrates with p45-f (Supplementary Fig. 6). The presence of residual p50 accounts for the reconstitution of holoenzyme-like catalytic activity by complementation of the RNP catalytic core and Teb1 in previous assays20,21,38. Reconstitutions using entirely recombinant holoenzyme subunits demonstrate a p50-dependent influence of 7-1-4 or Teb1 on RNP catalytic activity (Fig. 3g), consistent with their locations in the EM structures (Fig. 3e,f).
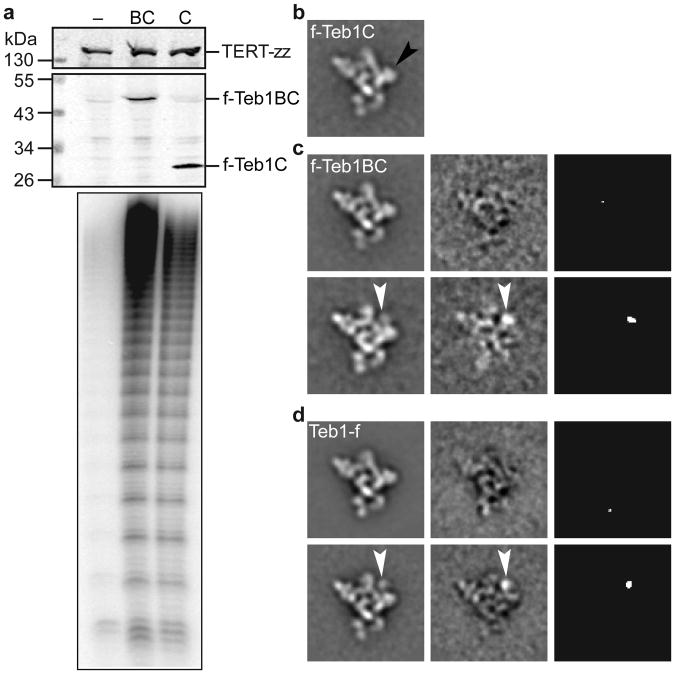
Teb1 domains are positionally flexible
All telomerase preparations except for those with tagged Teb1 have ∼5% of particles missing density for Teb1 in the class averages and 3D reconstructions (Fig. 3e and Supplementary Fig. 2). This is not surprising, because Teb1 is sensitive to proteolysis and dissociation during holoenzyme affinity purification6. To identify specific Teb1 domains in the structures, we constructed strains in which TERT was C-terminally tagged at its endogenous locus with only tandem Protein A domains (TERT-zz) and an N-terminally f-tagged Teb1C or Teb1BC was expressed from an integrated transgene. Holoenzyme particles isolated by tandem affinity purification of TERT-zz/f-Teb1C or TERT-zz/f-Teb1BC had the expected high RAP (Fig. 4a) and virtually identical class averages and 3D reconstructions to each other and to those from purified Teb1-fzz (Fig. 4b-d and Supplementary Fig. 4a). Because Teb1C mediates Teb1 association with other holoenzyme proteins21 and the N-terminal tag on Teb1BC absolutely requires the associated holoenzyme to contain Teb1B, we conclude that the Teb1B domain is positionally flexible and therefore did not appear in the class averages and 3D reconstructions. Similarly, for Teb1-fzz purified particles, which could contain all of the Teb1NABC domains, only Teb1C is visible in the majority of class averages. Teb1C was modeled into the EM density map (Fig. 2a) based on the position of the C-terminus identified by Fab labeling and the best fit with the shape of the EM density. The orientation around the y-axis is not definitive. We observed a small set of class averages (containing <5% of the particles) of f-Teb1BC and Teb1-f telomerases that show a weak extra density above Teb1C (Fig. 4c,d), which we assign to Teb1B. The conformational flexibility of Teb1 NAB domains relative to Teb1C is consistent with structural studies of the paralogous large subunit of Replication Protein A, which has three DNA-binding OB-fold domains that become ordered relative to each other only upon binding to ssDNA39.
Figure 4. Contribution of Teb1 domains to holoenzyme structure and activity.
a, Cell extract western blots and two-step purified enzyme primer extension assays of f-Teb1BC (BC, lane 2) and f-Teb1C (C, lane 3) telomerase. Cell extract with TERT-zz alone (–, lane 1) is a negative control for specificity of f-Teb1BC and f-Teb1C binding to FLAG antibody. b-d, Comparison of class averages (left column) of f-Teb1C (b), f-Teb1BC (c), and Teb1-f (d) telomerases. Density assigned to Teb1B (white arrows) was seen in <5% of particles, while density for Teb1C (black arrow in b) occupies a fixed position in all particles. Class averages without and with Teb1B density are represented by the upper and lower rows of c and d, respectively. Difference maps (middle column) by subtracting b from the respective class averages and maps of statistically significant (>4 σ) regions in the difference maps (right column) in c and d show Teb1B density (white arrows).
7-1-4 subcomplex has multiple orientations
Neither high-resolution structures nor exact biological functions of the 7-1-4 subunits are known. Both the EM structures and previous biochemical data6,21 indicate they constitute a stably assembled subcomplex. Because no class averages lacking an individual 7-1-4 subunit were observed, inter-subunit boundaries could only be inferred from the class averages and Fab labeling of the C-termini of p19 and p75, which showed that these two subunits are close together. In all class averages and 3D reconstructions, the major class of particles had a 7-1-4 conformation with p75 positioned across the top of the RNP catalytic core (Fig. 1c and Supplementary Fig. 2) in the stable conformation (Fig. 5). All telomerase samples show other positions of 7-1-4 in which it hinges away from the RNP catalytic core while maintaining a physical connection to p50 (Fig. 5 and Supplementary Fig. 3c). In these other conformations, the region of the class average where 7-1-4 is located appears “fuzzier” than the rest of the particle, consistent with conformational variability in this part of telomerase holoenzyme. In ∼10% of class averages from every purification, including those with tagged p75 or p45, no 7-1-4 density is visible, indicating that some 7-1-4 positions are not well defined relative to the RNP catalytic core (Fig. 3e and Supplementary Fig. 2). Binding of MS2cp to MS2hp holoenzyme appears to favor p75 displacement from the stable position (Fig. 1c and Supplementary Fig. 5g). Analysis of the various conformations indicates that 7-1-4 is rotating as an intact substructure (Fig. 5). Telomerase product DNA can be crosslinked to p45 as well as the TERT TEN domain31,40, which suggests that at least some orientations of 7-1-4 must bring p45 close to DNA.
Implications for holoenzyme assembly and function
TERT and TER comprise a minimal set of telomerase subunits for repeat synthesis in vitro5,41, but despite extensive efforts the 3D arrangement and molecular interactions between TERT and TER have not been previously defined. Although divergent in size and sequence, TERs contain a t/PK domain and almost without exception a separate activating domain (SL4 in Tetrahymena), each with a TERT binding site33,42. Here we have unambiguously located two elements of Tetrahymena TER that constitute known binding sites for TERT and modeled a path of the entire TER. The functional equivalent of Tetrahymena TER distal SL4 in vertebrate TER is P6/P6.1, which crosslinks to the same region of the TRBD where L4 is located in our model43. Our model structure thus provides insight into human telomerase TERT-TER interactions and explains the separable interactions of the TERT binding elements on the t/PK and activating domains of TER.
Using classification and/or 3D reconstruction of 16 differently labeled telomerase samples (Supplementary Table), we were able to identify the subunit arrangement of the holoenzyme. The EM results together with reconstituted enzyme activity assays suggest a subdivision of holoenzyme functional units: the TERT-TER-p65 catalytic core, p50-Teb1, and 7-1-4. Both structure determination and activity assays of reconstituted holoenzyme subcomplexes establish a central role for p50, a subunit lacking any predicted domain folds. The EM data reveal that 7-1-4 is a structural unit with dynamic orientation relative to the rest of the holoenzyme. We suggest that the positional dynamics of 7-1-4 coordinate telomerase holoenzyme with additional telomere synthesis or processing activities.
In the model for holoenzyme subunit interactions, the TERT TEN domain, the lower region of p50, and Teb1C have a triangulated arrangement that suggests coordination by pairwise direct contacts. The TERT TEN domain is required for RAP and has been implicated in both ssDNA and TER binding5,41,42. Although Teb1 appears to contact TERT, subcomplex structures never have Teb1 in the absence of p50, and reconstituted enzyme activity assays do not show Teb1 function in the absence of p50. The assembly of p50 with the RNP catalytic core dramatically increases processive repeat synthesis, suggesting that p50 stabilizes enzyme association with ssDNA in potential functional analogy to yeast Est3 and vertebrate TPP144-48, favoring a productive TERT TEN domain conformation and/or providing additional ssDNA contact. Subsequent p50-dependent recruitment of Teb1 further enhances activity, which we speculate could occur through interaction of p50 and the Teb1C OB fold. The physical location of p50 determined by EM and its functional significance determined by reconstitution assays suggest that p50 could be a critical determinant of holoenzyme assembly in vivo. This first view of the architecture of a complete telomerase holoenzyme provides unprecedented opportunity to understand the biological mechanisms for coupling of telomerase to its telomere substrates.
Methods Summary
Tetrahymena strain constructions and steps of tag-based affinity purification were done as described6 and in Methods. To label the tagged subunit for EM, telomerase particles were first purified using anti-FLAG M2 antibody resin then bound to rabbit-IgG resin. The telomerase-bound IgG resin was then incubated with Fab derived from anti-FLAG M2 IgG, and elution was effected by protease cleavage. Negatively stained EM specimens were prepared with fresh telomerase samples, stained with 0.8% uranyl formate, and examined with an FEI Tecnai F20 electron microscope operated at 200 kV. Frozen hydrated specimens were prepared using Quantifoil grids and imaged with an FEI Titan Krios electron microscope operated at 120 kV. The image processing tasks, including image classification and RCT reconstruction, were performed as described in Methods.
Telomerase activity assays were performed at room temperature using purified telomerase complexes on FLAG antibody resin with standard Tetrahymena holoenzyme reaction conditions using 0.3 μM 32P-labeled dGTP. Holoenzyme reconstitution used synthetic genes encoding TERT-f, p75, p65, p50, p45, and p19 for expression in RRL; TER purified following in vitro transcription by T7 RNA polymerase; and N-terminally His6-tagged Teb1BC purified following bacterial expression21.
Full Methods and any associated references are available in the online version of the paper.
Methods
Strain construction, growth, and analysis
Tetrahymena strains TERT-fzz, Teb1-fzz, p75-fzz, p65-fzz, p50-fzz, MS2hp, p45-fzz, and p19-fzz were previously described6,23. New strains zzf-p50, TERT-zz/f-Teb1C, and TERT-zz/f-Teb1BC were selected for targeted replacement of the endogenous locus (zzf-p50 or TERT-zz) and/or for MTT1 promoter-driven transgene integration at BTU1 (f-Teb1C, f-Teb1BC) using the neo2 and bsr2 cassettes6,49. Cells were grown to mid-log phase (2-4 × 105 cells/mL) at 30°C in modified Neff medium (0.25% proteose peptone (EMD Milipore Chemicals, Darmstadt, Germany), 0.25% BD Bacto yeast extract (Becton, Dickinson, Franklin Lakes, NJ), 0.2% glucose, 30 μM FeCl3). For large-scale purifications, transgene expression was induced by addition of 6 μM CdCl2 1 h before harvesting. Cell extract preparation and affinity purifications for subunit identification and activity assays were performed as described6. Activity assays were done using standard Tetrahymena holoenzyme reaction conditions (50 mM Tris•acetate, pH 8.0, 10 mM spermidine, 5 mM 2-mercaptoethanol, 2 mM MgCl2, 100 nM d(GT2G3)3, 200 μM dTTP, and 0.3 μM 32P-labeled dGTP) for 5 min or with 3.0 μM 32P-labeled dGTP for 15 min6. Genotypes were verified by Southern blots and western blots using rabbit IgG (Sigma) to detect the zz tag or anti-FLAG M2 mouse monoclonal antibody (Sigma) to detect the f tag.
Cell extract preparation and affinity purification for EM
Cells were harvested by centrifugation (5 min, 2,100g), washed in 20 mM HEPES•NaOH, pH 8.0, and then resuspended in lysis buffer (30 mL/L of growth) containing H2EG50 (20 mM HEPES•NaOH, pH 8.0, 50 mM NaCl, 1 mM EDTA•TS, 1 mM TCEP•HCl, and 10% glycerol) supplemented with 0.1% Triton X-100, 0.2% IGEPAL CA-630, 0.1 mM PMSF, 5 μM proteosome inhibitor MG-132, and 1,000-fold dilution of Sigma Protease Inhibitor Cocktail P8340. To effect cell lysis, resuspensions were rotated end-over-end for 20 min and cleared by ultracentrifugation (1 h, 145,000g).
To minimize proteolysis, all purification steps were carried out at 4 °C and additional washes were used in the early steps of purification as described below. Rabbit-IgG agarose slurry (Sigma) (3 μL/mL of cell extract) was washed 3× in lysis buffer, then added to cell extract for binding with end-over-end rotation overnight. Resin was collected by centrifugation (1 min, 3,200g) and washed 3× with no incubation and then 3× with end-over-end rotation (10 min) in wash buffer (20 mM HEPES•NaOH, pH 8.0, 50 mM NaCl, 1 mM MgCl2, 1 mM TCEP•HCl, 10% glycerol, and 0.1% IGEPAL CA-630). Telomerase was eluted with tobacco etch virus (TEV) protease (30 nM) in wash buffer for 1 h. After elution, supernatant containing holoenzyme was incubated with washed anti-FLAG M2 affinity gel (Sigma) for 1 h (4 μL slurry/g of pelleted cells). After binding, anti-FLAG M2 affinity gel was washed as above. Elution was effected by addition of glycerol-free wash buffer containing 3 × FLAG peptide (Sigma, 200 ng/μL).
Fab labeling for subunit identification
Anti-FLAG Fab was prepared by incubating monoclonal mouse anti-FLAG M2 IgG (Sigma) with resin-immobilized papain (Thermo Scientific Pierce, Rockford, IL) and purified according to the manufacturer's protocol. To prevent masking of the 3 × FLAG epitope present on the target subunit by the 3 × FLAG peptide required for elution, the tandem purification was done in reverse order with anti-FLAG M2 affinity gel purification followed by rabbit-IgG agarose purification. Washing and elution were done as described above. To effect Fab labeling, holoenzyme-enriched rabbit-IgG agarose was incubated (1 h, 4°C) with wash buffer containing anti-FLAG Fab (7.5 μg/mL). Excess Fab was removed by washing 5× with wash buffer. Labeled holoenzyme was eluted with wash buffer containing His6-TEV protease (2 nM). After elution, His6-TEV protease was removed from the elution by incubating with washed Ni-NTA agarose (30 min, 4°C).
Holoenzyme purification by MS2cp
Tetrahymena expressing MS2hp TER were harvested, lysed, and clarified as described above. N-terminal His6-zzf-MS2cp bacterially expressed from pET28 was purified using Ni-NTA agarose. Rabbit-IgG agarose was incubated with His6-zzf-MS2cp in a 1:1 stoichiometry to anticipated holoenzyme yield based on previous preparations. The enriched rabbit-IgG agarose was then washed 3× and added to clarified extract for over-night incubation. Purification then proceeded as described above.
EM specimen preparation and data collection
For negative staining EM, 2 μL of telomerase sample was applied to a glow-discharged grid coated with carbon film. The sample was left on the carbon film for 10 sec, followed by negative staining with 0.8% uranyl formate. Negatively stained specimens were carefully examined by EM, and only the regions with particles fully embedded in stain were selected for imaging. EM micrographs were recorded on a TIETZ F415MP 16-megapixel CCD camera at 68,027× magnification in an FEI Tecnai F20 electron microscope operated at 200 kV. The micrographs were saved by 2× binning and had a pixel size of 4.4 Å. To overcome the problems of preferred orientation and structural variability of the samples, 3D reconstructions were carried out using RCT method, which takes advantage of classification to differentiate different conformations and tilting to get lateral views for 3D reconstruction22. RCT images were collected with the grids tilted at two angles successively (65° and 0°) for each specimen area of interest with the assistance of Leginon automation software50,51. To minimize the potential smearing/stretching on 3D maps due to the missing cone problem in RCT data collection, a high tilt angle (65°) was used. For samples where only 2D image analysis was performed, micrographs were taken without tilting of the grids. Total numbers of micrographs and particles used for analysis are summarized in the Supplementary Table.
For cryoEM, 2.5 μL of sample was applied to a glow-discharged Quantifoil R2/1 grid. The grid was then blotted with filter paper remove excess sample, and flash-frozen in liquid ethane with FEI Vitrobot Mark IV. The grid was loaded into an FEI Titan Krios electron microscope operated at 120 kV. Micrographs were acquired with a Gatan UltraScan4000 16-megapixel CCD camera at 65, 162 × magnification with defocus values ranging from −1.9 to −4.0 μm and an exposure dose of 20 e−/Å2, and without tilting of the grids. The micrographs with a pixel size of 2.3 Å were used for data processing without binning.
2D image reference-free classification
Particles from the untilted micrographs were automatically picked using DoGpicker52. All particles in the micrographs, including a small number of aggregated particles, were initially picked to avoid bias in selecting particles. Particles were boxed out in 96 × 96 pixels (or 144 × 144 pixels for cryoEM images) using batchboxer in EMAN53. The defocus value of each micrograph was determined by CTFFIND54 and the particles were corrected for contrast transfer function (CTF) by phase-flipping with the corresponding defocus and astigmatism values using bsoft55. The phase-corrected particles were then iteratively (normally 9 iterations) classified using the refine2d.py program in EMAN. The classification for each sample was done independently without using any model or reference. The aggregated particles clustered in the few classes showing poor structures were manually eliminated during the iterations of classification. For the samples at very low concentration (Fab-labeled p75-f, p19-f and p65-f telomerase, and MS2cp-labeled MS2hp telomerase), a looser criterion for DoGpicker was used to pick more particles and consequently some “fake” particles from the stain background were also selected. The class averages were visually inspected at several different iterations of classification to remove the “fake” particles that were clustered in a small set of class averages that show no structural features.
For the RCT 3D reconstructions (Supplementary Table), the screened particles from the above steps were re-classified using the Correspondence Analysis method in SPIDER56. The class averages from the above EMAN classification step were used as references for multi-reference alignment in SPIDER. Each new class from this analysis was used for an RCT 3D reconstruction.
RCT 3D reconstruction and refinement
Corresponding particles from tilt pairs (65° and 0°) were picked using ApTiltPicker.py in Appion52 modified to optimize automatic particle picking and verified by visual inspection. The 0° tilt particles were classified as described above. Next, 3D RCT maps were reconstructed from 65° tilt particles. Particles within each class of the 0° tilt data set all have the same conformation and the same orientation except in-plane rotation. Their corresponding 65° tilt particles have different orientations due to in-plane rotation and were combined to reconstruct one 3D map. Each 3D map was iteratively refined with SPIDER followed by FREALIGN57 by refinement of the center of 65° tilt particles in the corresponding class. A representative refinement procedure is shown in Supplementary Fig. 3b. The orientations of 65° tilt particles were calculated from the in-plane rotation of the corresponding 0° tilt particles and the geometric relationship between the 65° and 0° tilt micrographs, and kept fixed during structure refinement. Multiple 3D maps were generated from RCT 3D reconstruction because particles in different conformations were classified into different classes each of which generated an individual 3D map. Representative 3D maps are shown in Supplementary Fig. 3c. Each 3D map was refined using only the 65° tilt particles in its corresponding class. The defocus value of each 65° tilt particle was calculated by CTFTILT program54 and then applied to 3D reconstructions with FREALIGN for proper correction of phase and amplitude in CTF.
To estimate the resolution of the representative 3D map of Teb1-f telomerase in the “stable” conformation, the 2,220 65° tilt particles used to reconstruct that 3D map were split into odd- and even-numbered halves and the halves of data were used to reconstruct two 3D maps with FREALIGN using the determined orientations and image centers, respectively, to calculate Fourier shell correlation (FSC). The resolution is 25 Å at FSC=0.5 (Supplementary Fig. 4b). For comparison, the FSC was also calculated using the recently introduced “gold standard” FSC method58, in which the halves of data (both 65° and 0° tilt) were split at the beginning for independent orientation determination, 3D reconstruction, and refinement to generate two independent 3D maps. The “gold standard” FSC suggests resolutions of 29 Å at FSC=0.5 and 24 Å at FSC=0.143 (Supplementary Fig. 4b). Since a high tilt angle (65°) was used for RCT 3D reconstructions, the missing cone was minimized and did not have notable impact on the structural interpretation of the 3D maps at the present resolution.
Evaluation of sample integrity by size-exclusion chromatography (SEC)
To confirm that the affinity-purified telomerase holoenzymes were intact after purification and that negative staining did not induce significant aggregation, we obtained an SEC profile of a sample of affinity purified TERT-f telomerase and EM images from the peak fractions (Supplementary Fig. 1c,d). The sample (50 μL) was run at 50 μL/min on a Superdex 200 column in the same buffer used for the EM samples except IGEPAL CA-630 was replaced by Tween-80. The SEC profile shows that the telomerase holoenzyme runs as a single peak with a constant A260/A280 ratio indicative of a largely intact RNA-protein complex. The class averages of particles from the peak fraction are identical to those for the affinity-purified sample (data not shown). There are no later eluting peaks from dissociated subunits (which would have different A260/A280 ratios).
Localization of Fab-labeled subunits
Each protein was assigned by identifying the characteristic Fab density in the class averages and 3D reconstructions. The 3 × FLAG tag allows multiple Fabs to bind simultaneously. Fab was observed attached to all tagged telomerase particles except for p45-f. For p45-f telomerase, Fabs were observed bound to free p45-f protein, which is in excess in that strain, while the p45-f telomerase holoenzyme particles appeared unchanged. The location of p45 in the holoenzyme was therefore assigned based on the density remaining after assignment of TERT-f, Teb1-f, p75-f, p65-f, f-p50, and p19-f.
Fitting of atomic models into the 3D EM maps
The homology model of Tetrahymena TERT RT-CTE domains was built using amino acids 520–1117 and the Tribolium TERT crystal structure (PDB ID: 3KYL) as a template with SWISS-MODEL27. The model of Tetrahymena TERT (TRBD, RT, and CTE domains) was built by joining the crystal structure of Tetrahymena TRBD (PDB ID: 2R4G) and the homology model of RT-CTE domains using the Tribolium TERT crystal structure as reference for structural alignment. The model of Tetrahymena TERT was then manually placed in an approximate position in the EM density map, followed by docking into the EM density map accurately using the “Fit in Map” function in UCSF Chimera59. The location of the TEN domain, absent in Tribolium TERT17, was supported by definition of the subunit boundaries of the adjacent Teb1C and p50. The crystal structure of Tetrahymena TERT TEN domain (PDB ID: 2B2A) was manually placed into the EM density map. Its position was iteratively adjusted to fit the empty density adjacent to the boundaries of Teb1 and p50 and the rest of TERT. Since the orientation of the TEN domain could not be determined based on the EM density due to the limit of resolution, we oriented it to be consistent with the homology model of human TERT28. For fitting of p65 ×RRM2:SL4, the putative EM density of p65:TER cut from the full EM density map was used in order to avoid interference from unrelated structures. The model of p65 ×RRM2:SL48 was manually placed in the EM density map based on the location of the C-terminus determined by Fab labeling and then docked using the “Fit in Map” function in UCSF Chimera. Its position was further manually adjusted to avoid clashing with the model of TERT. To confirm the fittings of TERT and p65 ×RRM2:SL4, these two structures were also fit into the EM density map using the colores program in Situs60. The fitting with colores was automated by 6D-space search and contour-based matching, and did not require manual placing of high-resolution structures to approximate positions in EM density maps. The crystal structure of Teb1C (PDB ID: 3U50) was manually placed into the EM density map and its orientation was adjusted so that the position of its C-terminus was consistent with the result from the Fab labeling experiment. The homology model of p65 La-RRM1:AUUUU-3′8,61 was manually placed into the EM density map, and its orientation was adjusted to match the shape of the EM density map. Cross-correlation for the fittings were: TERT TRBD-RT-CTE, 0.91; TERT TEN, 0.84; p65 ×RRM2:SL4, 0.80; p65 La-RRM1:AUUUU-3′, 0.86; and Teb1C, 0.82. Although the position and orientation of p65 ×RRM2:SL4 are well determined, the cross-correlation coefficient reflects the absence of the β2-β3 loop and amino acids linking xRRM2 to RRM1 in the crystal structure8.
Structures of SL2, distal SL4, and the PK
Previously reported NMR structures of SL2 and distal SL425,26 were re-refined using a new NOE restraint list derived from re-analysis of all previously obtained 2D NOESY and 3D NOESY-HMQC spectra and a new set of residual dipolar couplings (RDC) restraints (63 for SL2 and 48 for distal SL4) collected using pf1 bacteriophage for the alignment media. The new PDB accession numbers for SL2 and distal SL4 are 2M22 and 2M21, respectively, and BMRB accession numbers are 18892 and 18891, respectively. A grid search produced optimal values for the magnitude and asymmetry of the RDC alignment tensor of Da = −22.3 Hz, R = 0.38 for SL2, and Da = −17.5 Hz, R = 0.40 for SL4. The structures were calculated from an extended, unfolded RNA conformation using XPLOR-NIH v.2.9.862,63 following standard XPLOR protocols. A model structure of the PK was generated based on the secondary structure and two U-A-U triples predicted by sequence analysis64. XPLOR-NIH was used to calculate the structure from an extended RNA conformation as previously described62,63. A mock NMR restraint list was derived based on the solution structure of the human telomerase PK65. For the two stems, A-form RNA restraints were used for the dihedral angles and NOEs. NOEs for residues in the stems include H6/H8 (Py/Pu) to its own sugar protons and to the sugar protons (H1′, H2′, H3′, H4′) of the n-1 residue, H6/H8 to H6/H8 sequential, and imino sequential NOEs. For the PK loop 1, base-triple hydrogen bond restraints, planarity restraints with PK stem 2 residues, and sequential stacking NOEs were used to form the structured triple helix. PK loop 2 was given H6/H8 sequential NOEs and H/H8-H1′ NOEs within the minor groove of PK stem 1 to place it in the expected conformation.
Modeling of TER
First, the position of the template in TER was determined by matching the crystal structure of Tribolium TERT-nucleic acid complex (PDB ID: 3KYL) with the docked model of Tetrahymena TERT. Second, the position of SL4 was determined by the docking of the atomic model of p65 ×RRM2:SL4 complex into the EM density map. Third, the atomic model of SL2 was manually placed into the EM density map and its position was iteratively adjusted to match the result from MS2cp labeling experiment. S1 (modeled as A-form RNA) and the PK were placed into the EM density map based on physical constraints with the rest of TER. S1 is connected to S4 by 4 nt and to S2 by 10 nt. The model of the 31 nt PK, which is connected to S1 by 3 nt and to the template by the TRE, was fit into the only region of unoccupied density, between the TRBD and TEN domains, remaining after all the other major elements of TERT and TER were localized. Although the modeled positions of the TRE 3′ to the template (Fig. 1a) and the top of the pseudoknot could potentially be swapped with the TEN domain, they would still be on the same side (right side or behind in Fig. 2a or 2b, respectively) of the TERT TRBD-RT-CTE ring that has the active site and bound template. In either case, the TRE appears to be close to the TEN domain. The single-stranded regions of TER (except the template) were initially modeled as ideal A-form single-stranded RNA and their structures were modified using the “Minimize structure” function in UCSF Chimera to connect with their respective folded RNA fragments. The EM density maps and the modeled protein and RNA structures were visualized with UCSF Chimera.
Supplementary Material
Acknowledgments
This work was supported by grants from NSF MCB1022379 and NIH GM48123 to J.F., NIH GM54198 to K.C., GM071940 and AI069015 to Z.H.Z, Ruth L. Kirschstein NRSA postdoctoral fellowship GM101874 to E.J.M., and Ruth L. Kirschstein NRSA pre-doctoral training grant GM007185 fellowship for H.C. and D.D.C. We acknowledge the use of instruments at the Electron Imaging Center for NanoMachines supported by NIH (1S10RR23057 to ZHZ) and CNSI at UCLA.
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions: J.J. and E.J.M. purified and characterized EM samples, collected and analyzed EM data, and wrote the paper; K.H., B.E., and B.M. designed and made strains, expression plasmids, initial purifications, and reconstituted holoenzyme; H.C. purified telomerase; D.D.C. refined and modeled elements of TER; Z.H.Z., K.C., and J.F. supervised the research, analyzed data, and wrote the paper.
Author Information: The authors declare no competing financial interests.
References
- 1.Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31:9–18. doi: 10.1093/carcin/bgp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armanios M, Blackburn EH. The telomere syndromes. Nat Rev Genet. 2012;13:693–704. doi: 10.1038/nrg3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackburn EH, Greider CW, Szostak JW. Telomeres and telomerase: The path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med. 2006;12:1133–1138. doi: 10.1038/nm1006-1133. [DOI] [PubMed] [Google Scholar]
- 4.Egan ED, Collins K. Biogenesis of telomerase ribonucleoproteins. RNA. 2012;18:1747–1759. doi: 10.1261/rna.034629.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Podlevsky JD, Chen JJL. It all comes together at the ends: Telomerase structure, function, and biogenesis. Mutat Res Fundam Mol Mech Mugag. 2012;730:3–11. doi: 10.1016/j.mrfmmm.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Min B, Collins K. An RPA-related sequence-specific DNA-binding subunit of telomerase holoenzyme is required for elongation processivity and telomere maintenance. Mol Cell. 2009;36:609–619. doi: 10.1016/j.molcel.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Witkin KL, Collins K. Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes Dev. 2004;18:1107–1118. doi: 10.1101/gad.1201704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh M, et al. Structural basis for telomerase RNA recognition and RNP assembly by the holoenzyme La family protein p65. Mol Cell. 2012;47:16–26. doi: 10.1016/j.molcel.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Connor CM, Collins K. A novel RNA binding domain in Tetrahymena telomerase p65 initiates hierarchical assembly of telomerase holoenzyme. Mol Cell Biol. 2006;26:2029–2036. doi: 10.1128/MCB.26.6.2029-2036.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stone MD, et al. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature. 2007;446:458–461. doi: 10.1038/nature05600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akiyama BM, Loper J, Najarro K, Stone MD. The C-terminal domain of Tetrahymena thermophila telomerase holoenzyme protein p65 induces multiple structural changes in telomerase RNA. RNA. 2012;18:653–660. doi: 10.1261/rna.031377.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bryan TM, Goodrich KJ, Cech TR. Telomerase RNA bound by protein motifs specific to telomerase reverse transcriptase. Mol Cell. 2000;6:493–499. doi: 10.1016/s1097-2765(00)00048-4. [DOI] [PubMed] [Google Scholar]
- 13.Lai CK, Mitchell JR, Collins K. RNA binding domain of telomerase reverse transcriptase. Mol Cell Biol. 2001;21:990–1000. doi: 10.1128/MCB.21.4.990-1000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai CK, Miller MC, Collins K. Template boundary definition in Tetrahymena telomerase. Genes Dev. 2002;16:415–420. doi: 10.1101/gad.962602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rouda S, Skordalakes E. Structure of the RNA-binding domain of telomerase: Implications for RNA recognition and binding. Structure. 2007;15:1403–1412. doi: 10.1016/j.str.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs SA, Podell ER, Cech TR. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat Struct Mol Biol. 2006;13:218–225. doi: 10.1038/nsmb1054. [DOI] [PubMed] [Google Scholar]
- 17.Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–637. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- 18.Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol. 2010;17:513–518. doi: 10.1038/nsmb.1777. [DOI] [PubMed] [Google Scholar]
- 19.Robart AR, Collins K. Human telomerase domain interactions capture DNA for TEN domain-dependent processive elongation. Mol Cell. 2011;42:308–318. doi: 10.1016/j.molcel.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng Z, et al. Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA. Proc Natl Acad Sci U S A. 2011;108:20357–20361. doi: 10.1073/pnas.1113624108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Min B, Collins K. Multiple mechanisms for elongation processivity within the reconstituted Tetrahymena telomerase holoenzyme. J Biol Chem. 2010;285:16434–16443. doi: 10.1074/jbc.M110.119172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radermacher M, Wagenknecht T, Verschoor A, Frank J. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J Microsc. 1987;146:113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- 23.Cunningham DD, Collins K. Biological and biochemical functions of RNA in the Tetrahymena telomerase holoenzyme. Mol Cell Biol. 2005;25:4442–4454. doi: 10.1128/MCB.25.11.4442-4454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y, et al. Structure of stem-loop IV of Tetrahymena telomerase RNA. EMBO J. 2006;25:3156–3166. doi: 10.1038/sj.emboj.7601195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richards RJ, Theimer CA, Finger LD, Feigon J. Structure of the Tetrahymena thermophila telomerase RNA helix II template boundary element. Nucleic Acids Res. 2006;34:816–825. doi: 10.1093/nar/gkj481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richards RJ, et al. Structural study of elements of Tetrahymena telomerase RNA stem-loop IV domain important for function. RNA. 2006;12:1475–1485. doi: 10.1261/rna.112306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 28.Steczkiewicz K, et al. Human telomerase model shows the role of the TEN domain in advancing the double helix for the next polymerization step. Proc Natl Acad Sci U S A. 2011;108:9443–9448. doi: 10.1073/pnas.1015399108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedman KL, Cech TR. Essential functions of amino-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes Dev. 1999;13:2863–2874. doi: 10.1101/gad.13.21.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Armbruster BN, Banik SSR, Guo C, Smith AC, Counter CM. N-terminal domains of the human telomerase catalytic subunit required for enzyme activity in vivo. Mol Cell Biol. 2001;21:7775–7786. doi: 10.1128/MCB.21.22.7775-7786.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romi E, et al. High-resolution physical and functional mapping of the template adjacent DNA binding site in catalytically active telomerase. Proc Natl Acad Sci U S A. 2007;104:8791–8796. doi: 10.1073/pnas.0703157104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Connor CM, Lai CK, Collins K. Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. J Biol Chem. 2005;280:17533–17539. doi: 10.1074/jbc.M501211200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Theimer CA, Feigon J. Structure and function of telomerase RNA. Curr Opin Struct Biol. 2006;16:307–318. doi: 10.1016/j.sbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 34.Cole DI, et al. New models of Tetrahymena telomerase RNA from experimentally derived constraints and modeling. J Am Chem Soc. 2012 doi: 10.1021/ja305636u. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai CK, Miller MC, Collins K. Roles for RNA in telomerase nucleotide and repeat addition processivity. Mol Cell. 2003;11:1673–1683. doi: 10.1016/s1097-2765(03)00232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mason DX, Goneska E, Greider CW. Stem-loop IV of Tetrahymena telomerase RNA stimulates processivity in trans. Mol Cell Biol. 2003;23:5606–5613. doi: 10.1128/MCB.23.16.5606-5613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu JY, Stone MD, Zhuang X. A single-molecule assay for telomerase structure-function analysis. Nucleic Acids Res. 2010;38:e16. doi: 10.1093/nar/gkp1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eckert B, Collins K. Roles of telomerase reverse transcriptase N-terminal domain in assembly and activity of Tetrahymena telomerase holoenzyme. J Biol Chem. 2012;287:12805–12814. doi: 10.1074/jbc.M112.339853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan J, Pavletich NP. Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 2012;26:2337–2347. doi: 10.1101/gad.194787.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenfeld KK, Ziv T, Goldin S, Glaser F, Manor H. Mapping of DNA binding sites in the Tetrahymena telomerase holoenzyme proteins by UV cross-linking and mass spectrometry. J Mol Biol. 2011;410:77–92. doi: 10.1016/j.jmb.2011.04.040. [DOI] [PubMed] [Google Scholar]
- 41.Collins K. Single-stranded DNA repeat synthesis by telomerase. Curr Opin Chem Biol. 2011;15:643–648. doi: 10.1016/j.cbpa.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blackburn EH, Collins K. Telomerase: An RNP enzyme synthesizes DNA. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bley CJ, et al. RNA–protein binding interface in the telomerase ribonucleoprotein. Proc Natl Acad Sci U S A. 2011;108:20333–20338. doi: 10.1073/pnas.1100270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee J, Mandell EK, Rao T, Wuttke DS, Lundblad V. Investigating the role of the Est3 protein in yeast telomere replication. Nucleic Acids Res. 2010;38:2279–2290. doi: 10.1093/nar/gkp1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yen WF, Chico L, Lei M, Lue NF. Telomerase regulatory subunit Est3 in two Candida species physically interacts with the TEN domain of TERT and telomeric DNA. Proc Natl Acad Sci U S A. 2011;108:20370–20375. doi: 10.1073/pnas.1017855108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Talley JM, DeZwaan DC, Maness LD, Freeman BC, Friedman KL. Stimulation of yeast telomerase activity by the ever shorter telomere 3 (Est3) subunit is dependent on direct interaction with the catalytic protein Est2. J Biol Chem. 2011;286:26431–26439. doi: 10.1074/jbc.M111.228635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zaug AJ, Podell ER, Nandakumar J, Cech TR. Functional interaction between telomere protein TPP1 and telomerase. Genes Dev. 2010;24:613–622. doi: 10.1101/gad.1881810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sexton AN, Youmans DT, Collins K. Specificity requirements for human telomere protein interaction with telomerase holoenzyme. J Biol Chem. 2012;287:34455–34464. doi: 10.1074/jbc.M112.394767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Couvillion MT, Collins K. Biochemical approaches including the design and use of strains expressing epitope-tagged proteins. Methods Cell Biol. 2012;109:347–355. doi: 10.1016/B978-0-12-385967-9.00012-8. [DOI] [PubMed] [Google Scholar]
- 50.Suloway C, et al. Automated molecular microscopy: The new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 51.Suloway C, et al. Fully automated, sequential tilt-series acquisition with Leginon. J Struct Biol. 2009;167:11–18. doi: 10.1016/j.jsb.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: Software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol. 2009;166:205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 54.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 55.Heymann JB. Bsoft: Image and molecular processing in electron microscopy. J Struct Biol. 2001;133:156–169. doi: 10.1006/jsbi.2001.4339. [DOI] [PubMed] [Google Scholar]
- 56.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 57.Grigorieff N. FREALIGN: High-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 58.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 60.Chacón P, Wriggers W. Multi-resolution contour-based fitting of macromolecular structures. J Mol Biol. 2002;317:375–384. doi: 10.1006/jmbi.2002.5438. [DOI] [PubMed] [Google Scholar]
- 61.Jacks A, et al. Structure of the C-terminal domain of human La protein reveals a novel RNA recognition motif coupled to a helical nuclear retention element. Structure. 2003;11:833–843. doi: 10.1016/s0969-2126(03)00121-7. [DOI] [PubMed] [Google Scholar]
- 62.Schwieters CD, Kuszewski JJ, Marius Clore G. Using Xplor–NIH for NMR molecular structure determination. Prog Nucl Magn Reson Spectrosc. 2006;48:47–62. [Google Scholar]
- 63.Schwieters CD, Kuszewski JJ, Tjandra N, Marius Clore G. The Xplor–NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 64.Ulyanov NB, Shefer K, James TL, Tzfati Y. Pseudoknot structures with conserved base triples in telomerase RNAs of ciliates. Nucleic Acids Res. 2007;35:6150–6160. doi: 10.1093/nar/gkm660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Theimer CA, Blois CA, Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol Cell. 2005;17:671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.