Abstract
Background and Objective: Wound infections are often difficult to treat due to various bacterial pathogens. Pseudomonas aeruginosa is one of the common invaders of open wounds. Precise diagnosis of this etiological agent in wound infections is of critical importance particularly in treatment of problematic cases. The existing diagnostic methods have certain limitations particularly related to specificity. Our objective was to to establish a comprehensive and reliable multiplex PCR to confirm diagnosis of P. aeruginosa.
Methods: A multiplex PCR test was developed for rapid and comprehensive identification of P. aeruginosa. Four highly specific genes were targeted simultaneously for detection of genus, species and exotoxin production (16S rDNA, gyrB, oprL and ETA) in P. aeruginosa; additionally one internal control gene (invA) of Salmonella was used. The specificity of the multiplex PCR was confirmed using internal and negative controls. Amplified fragments were confirmed by restriction analysis and DNA sequencing.
Results: The developed method was applied on 40 morphologically suspected P. aeruginosa isolates (from 200 pus samples) and 18 isolates were confirmed as P. aeruginosa. In comparison, only 12 could be identified biochemically.
Conclusions: Combination of the four reported genes in multiplex PCR provided more confident and comprehensive detection of P. aeruginosa which is applicable for screening of wound infections and assisting treatment strategy.
Key Words: Multiplex PCR, wound infections, P. aeruginosa
INTRODUCTION
Wound infections are complications caused by bacteria which result in belated healing and can sometimes be even life-threatening.1 These infections also considerably contribute to increased health care costs.2 Pseudomonas aeruginosa (P. aeruginosa) has been recognized as a frequent inhabitant of chronic non-healing wounds3 and is one of the foremost opportunistic bacteria isolated from wounds which cause high morbidity and mortality despite antimicrobial therapy.4 P. aeruginosa infections are generally detected by standard microbiological techniques such as phenotypic and biochemical profiles,5 however these commercial tests tend to be lengthy and unreliable.5,6 Molecular techniques, such as polymerase chain reaction (PCR) are rapid and reliable for the identification of microbial pathogens,6 many PCR based diagnostic methods have been developed for P. aeruginosa.7-9
However, most of the protocols target only a single gene fragment which is inadequate for comprehensive and reliable diagnosis.9,10 The reason is that P. aeruginosa strains from patients demonstrate high genotypic diversity11 and several studies have confirmed the absence of one or more of the virulence genes in some P. aeruginosa strains.12 To overcome these problems, several multiplex PCR protocols have been reported,13 but due to genetic exchanges among P. aeruginosa and closely related bacteria, most multiplex PCRs have low specificity.14 Therefore, there is need to establish a comprehensive and reliable multiplex PCR to confirm diagnosis of P. aeruginosa.
This study deals with optimization of a multiplex PCR targeting four different gene fragments specific for P. aeruginosa (16S rDNA, gyrB, oprL and ETA) simultaneously for comprehensive and confirmatory identification. Application of this method on clinical samples demonstrated improved efficiency and reproducibility.
METHODS
Bacterial isolation: P. aeruginosa strain # MS6 (previously confirmed as P. aeruginosa) was taken from National Institute for Biotechnology and Genetic Engineering (NIBGE) stock cultures. Two hundred wound (pus) samples were collected from non-hospitalized outdoor patients from Allied Hospital, Faisalabad, Pakistan in the year 2011. Samples were collected on sterile cotton wool swabs which were transported to laboratory immediately. In case of a delay, the samples were kept at 4°C in tryptic soy broth (TSB) till transportation. Salmonella enterica serovars Typhi (as internal control) and isolates of Staphylococcus aureus, Escherichia coli, Klebsiella aerogenes, Proteus vulgaris and Proteus mirabilis were also taken from NIBGE stock cultures and used as negative controls. The swabs were streaked on MacConkey agar plates and kept overnight at 37°C to observe colony morphology. Five different colonies from each of the MacConkey agar plates, suspected as P. aeruginosa, were processed further for biochemical identification using RapidONE Remel kit (Thermo Fisher Scientific, Kansas, USA) according to manufacturer’s instructions.
DNA extraction: Morphologically identified P. aeruginosa from wound samples, P. aeruginosa MS6, internal control (S. Typhi) and negative controls strains were cultured in TSB. Genomic DNA of the overnight cultures was extracted by the conventional phenol-chloroform method.15 Integrity of the DNA samples was checked by electrophoresis on 1% agarose gel and purity was determined by ratio of A260/A280 using a spectrophotometer (Spectro22 Labomed 22. Inc, USA).
PCR amplification: First set of primers (Pa16S-F and Pa16S-R) was specific for 16s rDNA gene of the genus Pseudomonas.14 Second (gyrB-F and gyrB-R) and third (oprL-F and oprL-R) primer sets targeted species gene sequences of gyrB and oprL genes respectively.9,7 Fourth set (ETA-F and ETA-R) was used for amplification of exotoxin production related gene fragment (ETA).16 For internal control, the invA gene of S. Typhi was targeted using invA-F and invA-R primers.17 All sets of oligonucleotide primers were synthesized by Gene link (New York, USA). Sequences and lengths of targeted gene fragments are given in Table-I. Preliminarily, the PCR amplification conditions were optimized with P. aeruginosa MS6 for each of the four genes separately and then for the multiplex PCR.
Table-I.
Primers used in multiplex PCR
| Primers | Sequences (5 / - 3 / ) | Genes | Amplicon size (bp) |
|---|---|---|---|
| gyrB-F | CCTGACCATCCGTCGCCACAAC | gyrB | 2229 |
| gyrB-R | CGCAGCAGGATGCCGACGCC | ||
| ETA-F | GACAACGCCCTCAGCATCACCA | ETA | 39716 |
| ETA-R | CGCTGGCCCATTCGCTCCAGCG | ||
| oprL-F | ATG GAAATGCTGAAATTCGGC | oprL | 5047 |
| oprL-R | CTTCTTCAGCTCGACGCGACG | ||
| Pa16S-F | GGGGGATCTTCGGACCTCA | 16SrDNA | 61814 |
| Pa16S-R | TCCTTAGAGTGCCCACCCG | ||
| invA-F | GTGAAATTATCGCCACGTTCGGGCAA | invA | 28417 |
| InvA-R | TCATCGCACCGTCAAAGGAACC |
Each 50μl of the multiplex PCR mixture, in addition to the template DNA, contained 10 x buffer 5 μl, 1.5 mM MgCl2, 0.4mM of each dNTP, 5U of Taq DNA polymerase (Fermentas, USA), 0.25 μM of the primers targeting oprL gene and 0.5μM of each of the primers targeting invA, gyrB, ETA and Pa16S gene fragments. The thermal cycler (PTC 06 ICCC, Pakistan) conditions for the multiplex PCR were: 94°C for 5 minutes followed by 35 cycles of 94°C for 1 min, 58°C for 1 min, 72°C for 1.5 minutes; and a final extension step at 72°C for 7 minutes. Similar multiplex PCR conditions were applied to the DNA templates of negative control isolates. On completion of PCR cycles, the amplified products were electrophoresed on 2% agarose gel, stained with ethidium bromide (5 μg /100 ml) and visualized under UV illumination and documentation system (Viopro platinum, Uvitech, Cambridge, UK). The optimized multiplex PCR conditions were applied on morphologically identified clinical wound samples.
Restriction analysis: The amplified products of the targeted genes, gyrB, ETA, oprL and 16S rDNA, were subjected to restriction analysis with site specific restriction endonucleases. BsuRI was used to restrict amplified products of gyrB and 16S rDNA while CfrI and NcoI restriction endonucleases were used for ETA and oprL respectively. Each of the restriction mixtures contained 5µl (10 U) of enzyme, 3 µl of enzyme buffer, 8 µl of PCR-amplified product and 18µl of deionized water followed by overnight incubation at 37°C. Restricted fragments were electrophoresed on 2.5% agarose gel and visualized under UV illumination and documentation system (Viopro platinum, Uvitech, Cambridge, UK).
DNA sequencing: Sequencing of the amplified gene fragments of P. aeruginosa MS6 (gyrB, ETA, oprL and 16S rDNA) was done by a commercial vendor, Macrogen Inc. (Seoul, Korea) and the sequencing chromatographs were analyzed with Seqman software (DNASTAR, Inc. Wisconsin, USA) followed by submission to GenBank with accession numbers [GenBank: JN717229, GenBank: JN717230, GenBank: JN717231, GenBank: JN717232] respectively.
RESULTS
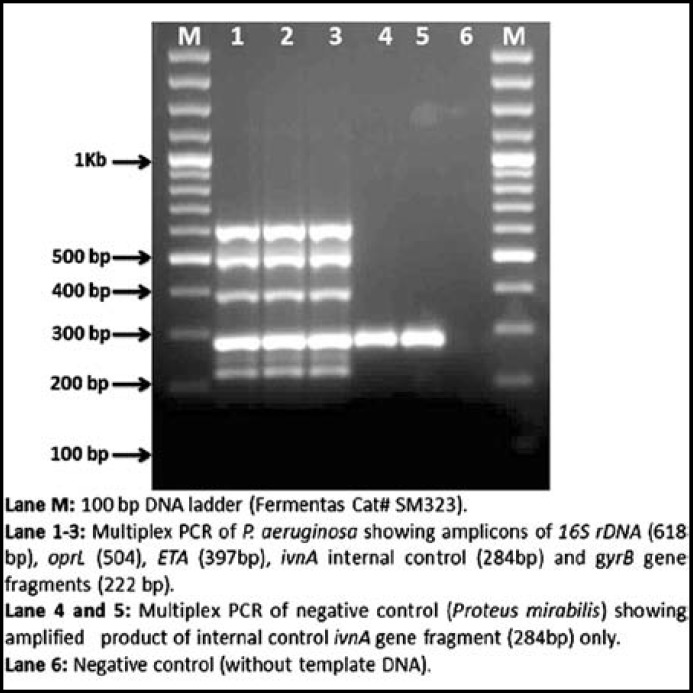
Identification of P. aeruginosa: From 200 clinical pus samples, 40 isolates were suspected as P. aeruginosa on the basis of formation of large, translucent, pale, mucoid colonies on MacConkey agar plates. On nutrient agar, typical greenish blue color (due to production of pyocanin and fluorescin pigments) was observed spreading throughout the medium. Biochemically, 12 isolates were identified as P. aeruginosa by RapidONE Remel kit (Thermo Fisher Scientific, Kansas, USA). However, the number of positive isolates increased from 12 to 18 by multiplex PCR developed for this study. In all cases, amplification products of internal control (284 bp), specific P. aeruginosa gene fragments gyrB (222 bp), ETA (397 bp), oprL (504 bp) and 16S rDNA (618 bp) were obtained (Fig.1). There was no amplification in case of negative control bacteria.
Fig.1.
Multiplex PCR of P. aeruginosa isolates. Lane M: 100 bp DNA ladder (Fermentas Cat# SM323). Lane 1-3: Multiplex PCR of P. aeruginosa showing amplicons of 16S rDNA (618 bp), oprL (504), ETA (397bp), ivnA internal control (284bp) and gyrB gene fragments (222 bp). Lane 4 and 5: Multiplex PCR of negative control (Proteus mirabilis) showing amplified product of internal control ivnA gene fragment (284bp) only. Lane 6: Negative control (without template DNA).
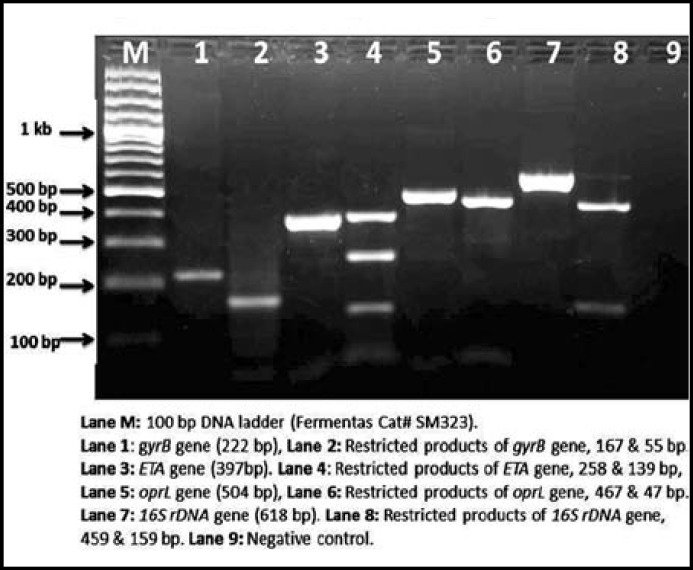
Restriction analysis and DNA sequencing: Restriction of all the four amplified gene fragments resulted in their cleavage into smaller fragments confirming band sizes as retrieved from website www.nebcutter.com. The 16S rDNA gene product (618 bp) produced 459 and 159 bp fragments, the oprL gene product (504 bp) was cleaved into 467 and 47 bp fragments, the ETA gene product (397 bp) yielded 258 and 139 bp fragments and the gyrB gene product (222 bp) was cleaved into fragments of 167 and 55 bp (Fig.2). The sequences of the amplified gene fragments (gyrB, ETA, oprL and 16S rDNA) of P. aeruginosa strain # MS6 were compared with already reported gene sequences of P. aeruginosa on NCBI database using BLAST search and top two similarity results were noted. The gyrB gene fragment sequence was found 99% identical with PAO1 [GenBank: AE004091.2] and 98% identical with NCGM2 [GenBank: AP012280.1]. The ETA gene was found 99% identical with PAO1 [GenBank: AE004091.2] and 98% identical with NCGM2 [GenBank: AP012280.1]. The oprL gene fragment sequence was found 99% identical with each of PAO1 [GenBank: AE004091.2] and M18 [GenBank: CP002496.1]. The 16S rDNA gene fragment sequence was found 97% identical with each of ZDC-2 [GenBank: JQ249910.1] and CW512 [GenBank: FM207514.1].
Fig.2.
Restriction analysis of P. aeruginosa MS6 strain. Lane M: 100 bp DNA ladder (Fermentas Cat# SM323). Lane 1: gyrB gene (222 bp), Lane 2: Restricted products of gyrB gene, 167 & 55 bp. Lane 3:ETA gene (397bp). Lane 4: Restricted products of ETA gene, 258 & 139 bp. Lane 5: oprL gene (504 bp), Lane 6: Restricted products of oprL gene, 467 & 47 bp. Lane 7:16S rDNA gene (618 bp). Lane 8: Restricted products of 16S rDNA gene, 459 & 159 bp. Lane 9: Negative control
DISCUSSION
Wound infections often become complicated and problematic due to invasion of multiple organisms and most of them are multi drug resistant. One of the major causes of complications in the wound infections is P. aeruginosa and its early and precise diagnosis is of substantial importance.8 The delay in accurate diagnosis may prolong the hospitalization and effective treatment.5 In routine, the microbiological culture is the mainstay for detection of P. aeruginosa. Although some other detection methods promise better sensitivity but these methods still need evaluation and validation,14,16 because Pseudomonas species are sometime indistinguishable from other closely related microbes.18 The biochemical tests lack specificity as in one study, 52 non-typical P. aeruginosa isolates were not identified by API 20 E kit.19 We had similar observations with Rapid ONE Remel kit during this study.
Many researchers have made attempts to develop molecular methods especially PCR for the detection of P. aeruginosa,20 but due to various limitations such as genetic diversity and the fact that genome sequences of closest species are not available, a comprehensive and definitive methodology is still lacking.
A multiplex PCR on respiratory samples targeting 3 genes (algD, chit A and 16S rDNA) detected P. aeruginosa in 78.7% samples whereas culture was positive in 56% cases only.21 A real time PCR on multiple targets for identification of P. aeruginosa reported the lesser specificity of ETA and algD genes as compared to oprl.5 Similar results about lesser specificity of ETA and algD genes have also been reported by other researchers.21 A study using quantitative PCR (qPCR) to target the oprL gene showed 85% specificity and concluded that qPCR may have a predictive value for impending P. aeruginosa infection for only a limited number of patients.22 The specificity of another developed multiplex PCR was reported as only 45.5%.23 P. aeruginosa from various sources were studied and it was found that the combination of oprl, oprL, 16S rDNA, ETA and fliC genes leads to false positive result because of genetic conservation and it showed difficulties in building up a reliable screening with these targets, however the gyrB and 16S-23S rDNA ITS genes were highly specific.7
We selected four highly specific gene fragments of P. aeruginosa (16S rDNA, gyrB, oprL and ETA) and optimized a multiplex PCR for its comprehensive and reliable identification with 100% specificity as no amplification was found in case of negative controls. The specificity of each of the four targeted genes has been reported earlier in different studies separately. The 16S rDNA gene showed 96.5% specificity using real time PCR.24 The gyrB gene coding a type II topoisomerase has also been noted to be a better candidate for the identification of bacterial species. 25 The specificity of the oprL gene based PCRs for P. aeruginosa has also been reported as 80%.7
After multiplex PCR, amplicons were confirmed by restriction analysis which provided the relevant fragments of exact sizes and the nucleotide sequencing of the four targeted gene fragments showed more than 97% identity with P. aeruginosa strain PAO1. Main benefit of this developed multiplex PCR is the detection of gyrB, ETA, OprL, and 16S rDNA genes simultaneously that, in presence of internal control, eliminates the chances of false positive results which are due to phenotypic and monogenic resemblance of P. aeruginosa with closely related species.
CONCLUSIONS
We conclude that the unique combination of these four genes in multiplex PCR provides more confident and reliable detection of P. aeruginosa for screening of wound infections that can be helpful to the clinicians for effective antimicrobial therapy.
Authors contribution
MS: Main lab work and preparing draft of paper. AA: Practical guidance in molecular and microbiology work. AH: Concept and finalization of manuscript.
ACKNOWLEDGEMENTS
The research facilities and funds for this work were provided by National Institute for Biotechnology and Genetic Engineering (NIBGE), Faisalabad and Higher Education Commission (HEC), Islamabad, Pakistan.
Conflict of interest: Authors have no conflict of interest.
References
- 1.Bryan CS, Dew CE, Reynolds KL. Bacteremia associated with decubitus ulcers. Arch Intern Med. 1983;143:2093–2095. [PubMed] [Google Scholar]
- 2.Bennett G, Dealey C, Posnett J. The cost of pressure ulcers in the UK. Age Ageing. 2004;33:230–235. doi: 10.1093/ageing/afh086. [DOI] [PubMed] [Google Scholar]
- 3.James GA, Swogger E, Wolcott R, Pulcini E, Secor P, Sestrich J, et al. Biofilms in chronic wounds. Wound Repair Regen. 2008;16:37–44. doi: 10.1111/j.1524-475X.2007.00321.x. [DOI] [PubMed] [Google Scholar]
- 4.Lau GW, Hassett DJ, Britigan BE. Modulation of lung epithelial functions by Pseudomonas aeruginosa. Trends Microbiol. 2005;13:389–397. doi: 10.1016/j.tim.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 5.Qin X, Emerson J, Stapp J, Stapp L, Abe P, Burns JL. Use of real-time PCR with multiple targets to identify Pseudomonas aeruginosa and other nonfermenting gram-negative bacilli from patients with cystic fibrosis. J Clin Microbiol. 2003;41:4312–4317. doi: 10.1128/JCM.41.9.4312-4317.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Procop GW. Molecular diagnostics for the detection and characterization of microbial pathogens. Clin Infect Dis. 2007;1:99–111. doi: 10.1086/519259. [DOI] [PubMed] [Google Scholar]
- 7.de Vos D, Lim A, Jr , Pirnay JP, Struelens M, Vandenvelde C, Duinslaeger L, et al. detection and identification of Pseudomonas aeruginosa in clinical samples such as skin biopsy specimens and expectorations by multiplex PCR based on two outer membrane lipoprotein genes, oprI and oprL. J Clin Microbiol. 1997;35:1295–1299. doi: 10.1128/jcm.35.6.1295-1299.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lavenir R, Jocktane D, Laurent F, Nazaret S, Cournoyer B. Improved reliability of Pseudomonas aeruginosa PCR detection by the use of the species-specific ecfX gene target. J Microbiol Methods. 2007;70:20–29. doi: 10.1016/j.mimet.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Motoshima M, Yanagihara K, Fukushima K, Matsuda J, Sugahara K, Hirakata Y, et al. Rapid and accurate detection of Pseudomonas aeruginosa by real-time polymerase chain reaction with melting curve analysis targeting gyrB gene. Diagn Microbiol Infect Dis. 2007:53–58. doi: 10.1016/j.diagmicrobio.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 10.McCulloch E, Lucas C, Ramage G, Williams C. Improved early diagnosis of Pseudomonas aeruginosa by real-time PCR to prevent chronic colonisation in a paediatric cystic fibrosis population. J Cyst Fibros. 2011:21–24. doi: 10.1016/j.jcf.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Vosahlikova S, Drevinek P, Cinek O, Pohunek P, Maixnerova M, Urbaskova P, et al. High genotypic diversity of Pseudomonas aeruginosa strains isolated from patients with cystic fibrosis in the Czech Republic. Res Microbiol. 2007;158:324–329. doi: 10.1016/j.resmic.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Bradbury RS, Roddam LF, Merritt A, Reid DW, Champion AC. Virulence gene distribution in clinical, nosocomial and environmental isolates of Pseudomonas aeruginosa. J Med Microbiol. 2010;59:881–890. doi: 10.1099/jmm.0.018283-0. [DOI] [PubMed] [Google Scholar]
- 13.Xu W, Zhai Z, Huang K, Zhang N, Yuan Y, Shang Y, et al. A novel universal primer-multiplex-PCR method with sequencing gel electrophoresis analysis. PLoS One. 2012;7:17. doi: 10.1371/journal.pone.0022900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spilker T, Coenye T, Vandamme P, LiPuma JJ. PCR-Based Assay for Differentiation of Pseudomonas aeruginosa from Other Pseudomonas Species Recovered from Cystic Fibrosis Patients. J Clin Microbiol. 2004;42:2074–2079. doi: 10.1128/JCM.42.5.2074-2079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haque A, Ahmed N, Peerzada A, Raza A, Bashir S, Abbas G. Utility of PCR in diagnosis of problematic cases of typhoid. Jpn J Infect Dis. 2001;54:237–239. [PubMed] [Google Scholar]
- 16.Khan AA, Cerniglia CE. Detection of Pseudomonas aeruginosa from clinical and environmental samples by amplification of the exotoxin A gene using PCR. Appl Environ Microbiol. 1994;60:3739–3745. doi: 10.1128/aem.60.10.3739-3745.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rahn K, De Grandis SA, Clarke RC, McEwen SA, Galan JE, Ginocchio C, et al. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol Cell Probes. 1992;6:271–279. doi: 10.1016/0890-8508(92)90002-f. [DOI] [PubMed] [Google Scholar]
- 18.Doring G, Pier GB. Vaccines and immunotherapy against Pseudomonas aeruginosa. Vaccine. 2008;26:1011–1024. doi: 10.1016/j.vaccine.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 19.Warren AE, Boulianne-Larsen CM, Chandler CB, Chiotti K, Kroll E, Miller SR, et al. Genotypic and phenotypic variation in Pseudomonas aeruginosa reveals signatures of secondary infection and mutator activity in certain cystic fibrosis patients with chronic lung infections. Infect Immun. 2011;79:4802–4818. doi: 10.1128/IAI.05282-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laine L, Perry JD, Lee J, Oliver M, James AL, De La Foata C, et al. A novel chromogenic medium for isolation of Pseudomonas aeruginosa from the sputa of cystic fibrosis patients. J Cyst Fibros. 2009;8:143–149. doi: 10.1016/j.jcf.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 21.da Silva Filho LV, Tateno AF, Velloso Lde F, Levi JE, Fernandes S, Bento CN, et al. Identification of Pseudomonas aeruginosa, Burkholderia cepacia complex, and Stenotrophomonas maltophilia in respiratory samples from cystic fibrosis patients using multiplex PCR. Pediatr Pulmonol. 2004;37:537–547. doi: 10.1002/ppul.20016. [DOI] [PubMed] [Google Scholar]
- 22.da Silva Filho LV, Levi JE, Oda Bento CN, da Silva Ramos SR, Rozov T. PCR identification of Pseudomonas aeruginosa and direct detection in clinical samples from cystic fibrosis patients. J Med Microbiol. 1999;48:357–361. doi: 10.1099/00222615-48-4-357. [DOI] [PubMed] [Google Scholar]
- 23.Hoiby N, Frederiksen B, Pressler T. Eradication of early Pseudomonas aeruginosa infection. J Cyst Fibros. 2005;4(Suppl 2):49–54. doi: 10.1016/j.jcf.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 24.Deschaght P, Schelstraete P, Lopes dos Santos Santiago G, Van Simaey L, Haerynck F, Van Daele S, et al. Comparison of culture and qPCR for the detection of Pseudomonas aeruginosa in not chronically infected cystic fibrosis patients. BMC Microbiol. 2010;10:245. doi: 10.1186/1471-2180-10-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melendez JH, Frankel YM, An AT, Williams L, Price LB, Wang NY, et al. Real-time PCR assays compared to culture-based approaches for identification of aerobic bacteria in chronic wounds. Clin Microbiol Infect. 2010;16:1762–1769. doi: 10.1111/j.1469-0691.2010.03158.x. [DOI] [PubMed] [Google Scholar]