Summary
A recent surge in obesity has given impetus to better understand the mechanisms of adipogenesis, particularly brown adipose tissue (BAT) due to its potential utilization for anti-obesity therapy. Postnatal brown adipocytes arise from early muscle-progenitors but how brown fat lineage is determined is not completely understood. Here, we show that a multifunctional protein EWS (Ewing Sarcoma) is essential for determining brown fat lineage during development. BATs from Ews-null embryos and newborns are developmentally arrested. Ews mutant brown preadipocytes fail to differentiate due to loss of Bmp7 expression, a critical early brown adipogenic factor. We demonstrate that EWS, along with its binding partner YBX1 (Y-box binding protein 1), activates Bmp7 transcription. Depletion of either Ews or Ybx1 leads to loss of Bmp7 expression and brown adipogenesis. Remarkably, Ews-null BATs and brown preadipocytes ectopically express myogenic genes. These results demonstrate that EWS is essential for early brown fat lineage determination.
Introduction
Chronic obesity driven by excessive food intake, lack of exercise and/or defects in metabolism will ultimately result in metabolic disorders, such as coronary failure, type 2 diabetes, hypertension, stroke and cancer (Li et al., 2005). White adipose tissue, WAT, comprises the majority of adipose tissues in humans and is the depot of unutilized fat in the form of triglycerides (Cristancho and Lazar, 2011). During starvation, stored triglycerides in WAT are utilized to provide energy. In contrast, BAT oxidizes fat to provide heat to maintain core body temperature and is rich in mitochondria (Frontini and Cinti, 2010; Tseng et al., 2010). Conversion of triglycerides to heat is accomplished in mitochondria via actions of Uncoupling Protein 1 (UCP1), a BAT-specific mitochondrial protein. BAT is highly vascularized and densely innervated by the sympathetic nervous system. In response to cold environment or diet, BAT is activated by β-adrenergic stimulation to expand energy and to maintain thermal or energy balance, a process termed adaptive thermogenesis. With recent findings of active BAT in adult humans (Nedergaard et al., 2007) and an inverse correlation between high BAT activity and obesity (Cypess et al., 2009; van Marken Lichtenbelt et al., 2009), utilization of BAT as an anti-obesity therapy and possibly for other metabolic disorders holds great promise.
BAT exists in two distinct types that differ in their cell of origin. During embryogenesis, brown preadipocytes arise from early muscle progenitors (Atit et al., 2006; Timmons et al., 2007), which express myogenic transcription factor, Myf5 (Seale et al., 2008), and form mature classical BAT in newborns. A second type of brown fat cells is generated during adaptive thermogenesis in response to β-adrenergic stimulation. These brown fat-like cells are generated from Myf5-negative cells (termed brite or beige cells) residing in perivascular regions of various WAT depots (Gupta et al., 2012; Tran et al., 2012). Interestingly, a recent study showed an existence of bipotent adipocyte progenitors in WAT depots, which express PDGFRα and give rise to either brown or white adipocytes in response to β-adrenergic stimulation or high-fat diet, respectively (Lee et al., 2012).
The development of classical BAT has been intensely studied and many transcription factors such as peroxisome proliferator-activated receptor γ (PPARγ), PPARγ coactivator 1 alpha (PGC-1α, CCAAT-enhancer-binding protein-α (CEBP-α, CEBP-β, CEBP-δ, and PRD1-BF1-RIZ1 homologous domain-containing 16 (PRDM16) (Farmer, 2008; Kajimura et al., 2010) and even microRNAs, MiR-193b-365 (Sun et al., 2011), have been identified as important regulators of classical BAT development and function. However, not much is known about the upstream factors that determine the brown fat lineage. It has been demonstrated that BMP7 (Bone-Morphogenic Protein 7) is critical for mesenchymal progenitor cells to commit to brown fat lineage (Tseng et al., 2008); however, the factors that regulate BMP7 expression during the early commitment period are unknown.
Results
Classical BAT development is disrupted in the absence of Ews
We have previously shown that loss of Ews in a mixed 129SvEv/Black Swiss strain results in defects in B-cell development and meiosis, and causes premature cellular senescence in fibroblasts and hematopoietic stem cells, as well as high postnatal mortality (90%) (Cho et al., 2011; Li et al., 2007). Subsequently, we backcrossed Ews+/- to C57BL6/N strain for 12 generations and generated Ews-/- congenic mice, which were born near Mendelian ratio but all mutant mice died within 24h after birth (data not shown). Gross visual examination revealed no major organ defects, except drastically reduced interscapular BATs in all Ews mutants compared to littermates (Figure 1A). As shown by Western blot analysis of wildtype mouse tissues, EWS is abundantly expressed in BAT as well as in WAT and most other tissues (Supplementary Figure 1A). H&E staining confirmed the diminished size of interscapular and subscapular BATs in Ews-null embryos (E18.5) and newborns (P0.5) (Figure 1B and Supplementary Figure 1B). Further examination with Oil Red O revealed a striking absence of multiloculated lipid droplets in Ews mutant BATs compared to wildtype (Supplementary Figure 1C). Importantly, expression of UCP1, a marker of mature and functional BAT, as well as PGC1α and PRDM16, was nearly absent or greatly diminished in all newborn mutant BATs (Figure 1C). Western blot analysis further revealed markedly reduced levels of PPARγ, CEBP-α, -β and -δ, and confirmed near complete absence of UCP1 expression (Figure 1D). Expression of EWS homolog, FUS (Fused in Sarcoma, also called TLS), was not altered. Ews-null newborns (P0.5) in 129SvEv/Black Swiss strain also displayed developmentally arrested BATs with lack of lipids and UCP1 expression (Supplementary Figure 1D). These results reveal an essential and unexpected role for Ews in BAT development. Of note, breeding these mutant mice at thermoneutrality (32°C) did not rescue the early postnatal lethality of Ews-null pups (n=4).
Figure 1. Ews is essential for brown adipose tissue development.
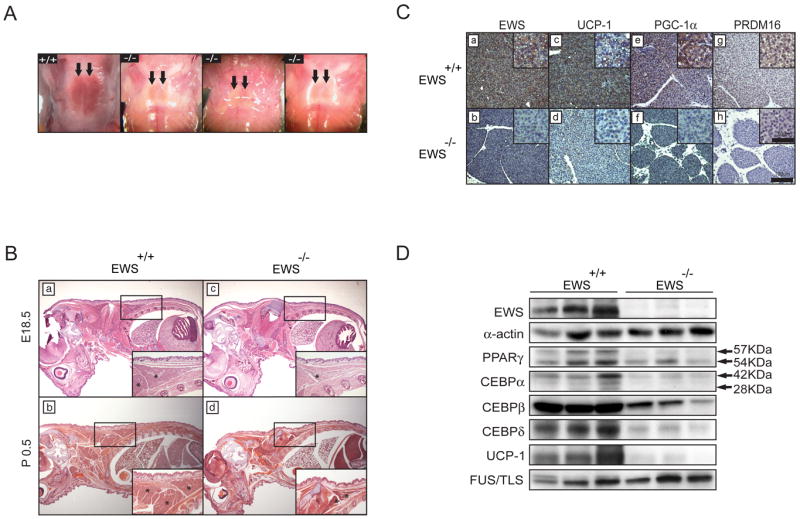
A. Photographs of interscapular BAT in newborns (P0.5). Arrows point to BAT. B. Sagittal section and H&E staining of E18.5 embryos and newborns. Boxed area is shown at higher magnification in the insets. Asterisks indicate interscapular BAT. C. Immunostaining of P0.5 interscapular BAT with anti-EWS, anti-UCP1, anti-PGC1α or anti-PRDM16. Insets show higher magnification (400X). scale bars are indicated. D. Western blot analysis of interscapular BATs from Ews+/+ and Ews-/- newborns (P0.5) (n=3). (See also Figure S1).
Ews is essential for brown adipocyte differentiation
To investigate the mechanism by which Ews regulates BAT development, we derived immortalized brown preadipocytes from Ews wildtype and mutant newborns (Figure 2A). Following the addition of adipogenic cocktail, wildtype preadipocytes differentiated robustly into brown adipocytes (Figure 2B). However, Ews-null preadipocytes failed to differentiate into mature brown adipocytes, while Ews+/- cells showed a slight reduction in brown fat differentiation (Figure 2B). Depletion of Ews with two independent shRNAs also blocked brown fat differentiation (Supplementary Figure 2A and 2B). Upon adipogenic stimulation, wildtype brown preadipocytes expressed high levels of adipogenic transcription factors, Cebpα and Pparγ, and Ews (Figure 2C). In contrast, Ews mutant preadipocytes failed to induce expression of critical adipogenic factors, Pparγ and Prdm16, and expressed significantly reduced levels of Cebpα. Expression of Pgc1α and other brown fat markers, Ucp1, Elovl3 (Elongation of very long chain fatty acids protein3) and Cidea (cell death-inducing DFFA-like effect A), was also significantly dampened in the mutant cells, but expression of a non-adipogenic gene, Lkb1 (Liver kinase B1), was not suppressed.
Figure 2. Ews is required for expression of Bmp7 and brown fat differentiation in brown preadipocytes.
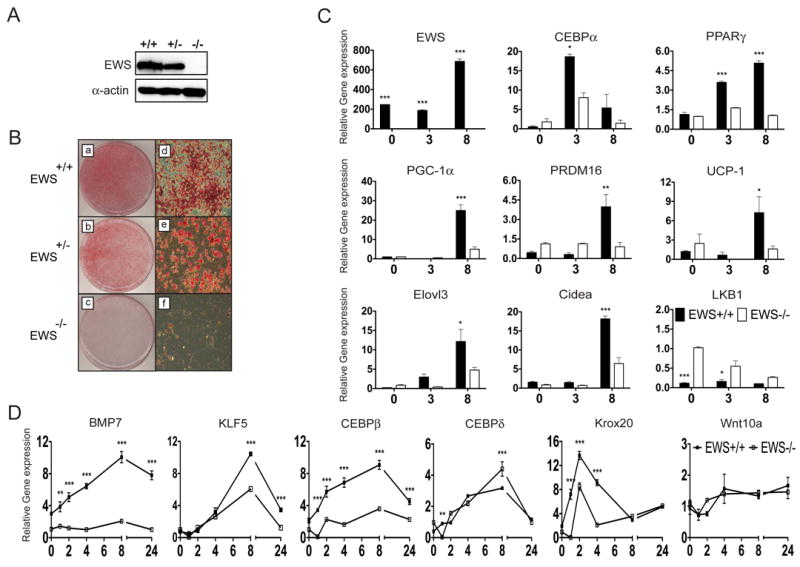
A. Western blot analysis of immortalized brown preadipocytes with anti-EWS and anti-Actin. B. Representative images of Oil Red O staining of brown adipocytes 8 days after addition of adipogenic cocktail. (D-F images are higher magnification, 40X). C. qRT-PCR analysis of indicated transcripts was performed (in triplicate) in brown adipocytes at 0, 3 or 8 days after addition of adipogenic cocktail. Black bar (Ews+/+), White bar (Ews-/-). D. qRT-PCR analysis of brown preadipocytes at indicated hours (0-24) after addition of adipogenic cocktail was performed in triplicates. For C and D, Three independent experiments were performed and data are represented as means ±s.e.m. Two-way ANOVA, *p<0.05, **p<0.01, ***p<0.001. (See also Figure S2).
Loss of Ews leads to a block in early brown fat differentiation and absence of Bmp7 expression
Since PPARγ and CEBPα are critical transcriptional regulators of adipogenesis, we sought to determine whether EWS interacts with PPARγ or CEBPα. However, we did not observe any physical interactions between EWS and these two factors, nor did we observe any effects of EWS on the PPARγ- or CEBPα-mediated transcriptional reporter assays (data not shown). EWS also did not interact with upstream transcription factors, CEBPβ or CEBPδ (data not shown).
Subsequently, we reasoned that EWS might function upstream of these adipogenic factors and examined the expression of very early adipogenic regulators. In wildtype brown preadipocytes, addition of adipogenic cocktail induced a rapid and continuous expression of Bmp7 and other upstream regulators (Figure 2D). Strikingly, Ews-deficient preadipocytes failed to express Bmp7 following the adipogenic stimulation. Expression of upstream transcription factor Cebpβ was also significantly diminished in the absence of Ews, while Cebpδ expression was not affected. Expression of other early transcriptional regulators Krox20 (also known as Egr2) and Klf5 (Kruppel-like factor 5) was modestly reduced, while expression of an upstream inhibitor of adipogenesis, Wnt10a (Kang et al., 2005; Tseng et al., 2005), was not altered.
Ews forms a complex with Ybx1 and the complex activates Bmp7 transcription
BMP7 is the earliest known brown cell-fate determination factor and Bmp7-null newborns display smaller BAT size and markedly reduced UCP1 expression (Tseng et al., 2008), similar to Ews mutants. Recently, YBX1 (also called YB1), a multifunctional protein with roles in transcription and translation (Kohno et al., 2003), was shown to bind and activate the mouse Bmp7 promoter (Wang and Hirschberg, 2011). This was intriguing given that EWS and YBX1 physically and functionally interact (Chansky et al., 2001; Dutertre et al., 2010). We first confirmed the EWS-YBX1 interaction by ectopic expression of EWS and YBX1 in U2OS cells followed by immunoprecipitation (IP) and Western blot analysis (Figure 3A, top panels). To further demonstrate the physiological interaction, we examined endogenous EWS-YBX1 interaction in brown preadipocytes with or without adipogenic stimulation. In the basal state, endogenous EWS interacted weakly with YBX1 (Figure 3A, middle panels), but following short adipogenic stimulation (4h), the EWS-YBX1 interaction was greatly enhanced (Figure 3A, lower panels).
Figure 3. EWS and YBX1 form a complex upon adipogenic stimulation and activate Bmp7 transcription.
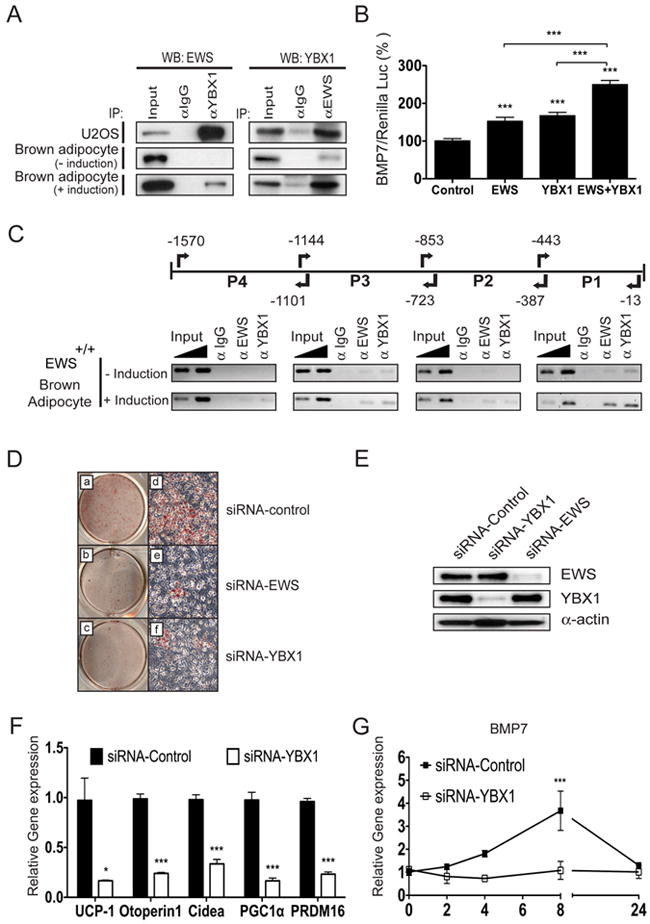
A. Immunoprecipitation (IP)-Western blot analysis of U2OS cells co-transfected with CMV-EWS and CMV-YBX1 plasmids (top panel). Lower panels show IP-Western analysis of endogenous EWS and YBX1 interaction without (middle) or with (bottom) adipogenic stimulation (4h). B. U2OS cells stably transfected with murine Bmp7 promoter-Luc (1395bp) were transfected with CMV-EWS, CMV-YBX1, or both along with Renilla luciferase and luciferase activities were calculated after normalization with Renilla luciferase. Three independent experiments were performed in triplicate. Data are represented as means ±s.e.m. One-way ANOVA, ***p<0.001. C. Chromatin-IP (ChIP) analysis of murine Bmp7 promoter region with anti-EWS and anti-YBX1 with or without adipogenic stimulation (4h). 1 and 2% of total chromatin were used as input. Primers are indicated with arrows. Numbers are relative to the upstream of the translation start site (+1). D. Wildtype brown preadipocytes transfected with siRNAs against Ews, Ybx1 or scrambled control were cultured for 8 days in media containing adipogenic cocktail and stained with Oil Red O. E. Western blot analysis of brown preadipocytes transfected with Ews, Ybx1 or scrambled control siRNAs. F. qRT-PCR analysis of differentiated brown adipocytes transfected with Ybx1 or scrambled control siRNA. G. Bmp7 transcript was analyzed by qRT-PCR at indicated times (0 to 24h) following adipogenic stimulation in brown preadipocytes transfected with Ybx1 or scrambled control siRNA. Three independent experiments were performed and data are represented as means ±s.e.m. *p<0.05, ***p<0.001. (See also Figure S3).
EWS encodes a potent transactivation domain in the amino-terminus (May et al., 1993) and is frequently involved in chromosomal translocations that generate aberrant transcription factors (Sankar and Lessnick, 2011). Thus, we tested the effects of EWS on the transcriptional regulation of Bmp7 using a 1395-bp (-1392 to +3) mouse Bmp7 promoter-reporter construct that contains an YBX1-binding site in the proximal region (-192 to +3) (Wang and Hirschberg, 2011). Expression of EWS or YBX1 alone resulted in a modest activation of the Bmp7 promoter (~1.6-fold) compared to empty vector control (Figure 3B). However, co-expression of EWS and YBX1 resulted in an additive activation (2.6-fold) of the Bmp7 promoter, which was statistically significant than either one alone. The cooperative transcriptional nature of the EWS-YBX1 complex was further examined by Chromatin-IP (ChIP) analysis. In unstimulated brown preadipocytes, a small amount of YBX1 was present on the proximal P1 region (-443 to -13) of Bmp7 promoter, along with barely detectable levels of EWS (Figure 3C), but following short adipogenic stimulation (4h), EWS and YBX1 were rapidly recruited to the P1 region. Less abundant but detectable levels of EWS and YBX1 were also found in the upstream regions. Interestingly, in Ews-null brown preadipocytes, a small amount of YBX1 present on the P1 region of Bmp7 promoter at basal condition was not further enriched upon adipogenic stimulation (Supplementary Figure 3A). YBX1 has been shown to shuttle between nuclear and cytoplasm (Sutherland et al., 2005). However, inactivation of Ews did not alter the nuclear/cytoplasmic ratio or the expression of YBX1 (Supplementary Figure 3B). These results strongly suggest that the recruitment of both factors to the Bmp7 promoter is dependent on the formation of EWS-YBX1 complex.
YBX1 is essential for Bmp7 expression during brown adipocyte differentiation in vitro
The above findings suggest an unexpected role for YBX1 in brown fat development. To test this, we examined the effects of siRNA-mediated depletion of Ybx1 or Ews on brown fat differentiation. Silencing Ybx1 or Ews led to severe inhibition of brown adipocyte differentiation with significantly reduced expression of critical BAT genes such as Ucp1, Pgc1α Prdm16 and others (Figures 3D-F). Importantly, depletion of Ybx1 resulted in a complete loss of Bmp7 expression following adipogenic stimulation (Figure 3G). These results demonstrate that both EWS and YBX1 are critical for Bmp7 expression during the early steps of brown adipocyte differentiation.
Complementation of Ews or Bmp7 restores brown fat differentiation
To determine whether restoring Ews expression in the mutant brown preadipocytes can rescue adipogenesis, we transduced Ews-null preadipocytes with a lentivirus expressing Ews or GFP as a control. Complementation of Ews in the mutant preadipocytes resulted in a partial restoration of adipogenesis whereas the GFP-expressing mutant cells failed to undergo differentiation (Supplementary Figure 4A). Expression of downstream adipogenic factors, Pparγ, Prdm16 and Ucp1, was significantly increased upon Ews complementation (Figure 4A). Ectopic expression of CEBPβ or PPARγ also rescued adipogenesis in the Ews-null cells (Supplementary Figure 4B and 4C), suggesting that these factors function downstream of Ews. Remarkably, re-expression of Ews in the mutant cells restored Bmp7 expression following adipogenic stimulation (Figure 4B) and the expression of other early adipogenic factors (Supplementary Figure 5A). To further demonstrate that BMP7 is the critical missing factor, Ews-null brown preadipocytes were grown in the absence or presence of BMP7. Addition of recombinant BMP7 along with adipogenic cocktail in Ews-null cells resulted in a robust brown fat differentiation (Figure 4C) and restoration of critical adipogenic genes (Figures 4D and Supplementary Figure 5B), while the mutant cells grown in the adipogenic cocktail without BMP7 failed to differentiate nor express the critical adipogenic genes.
Figure 4. Complementation of Ews or BMP7 rescues brown adipogenesis in Ews-/- preadipocytes.
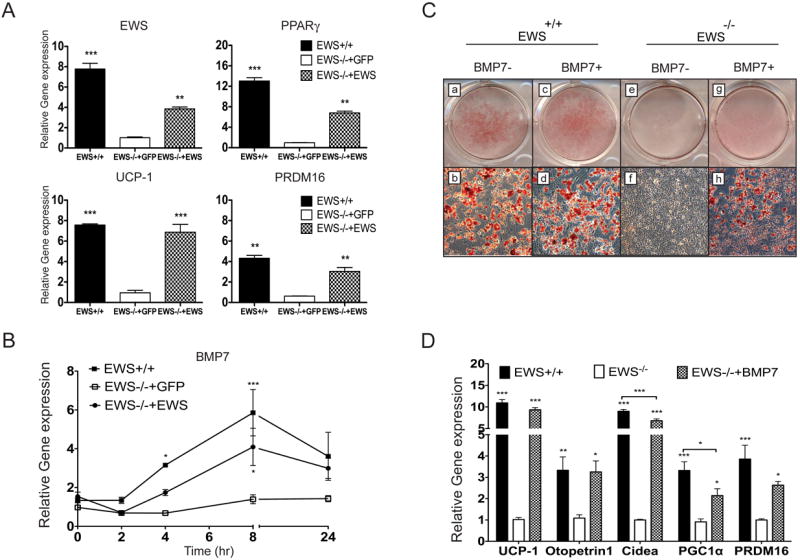
A-B. Ews+/+ or Ews-/- preadipocytes transduced with a lentivirus expressing either Ews or GFP (control) were stimulated with adipogenic cocktail for 8 days (A) or for indicated hours (0 to 24h) (B) and indicated transcripts were analyzed by qRT-PCR. Three independent experiments were performed and data are represented as means ±s.e.m. Two-way ANOVA, *p<0.05, **p<0.01, ***p<0.001. (See also Figure S4). C-D. Ews+/+ or Ews-/- brown preadipocytes were cultured in media containing adipogenic cocktail with or without recombinant BMP7 (100 ng/ml) for 2 days, and continuously cultured in adipogenic cocktail-only containing media for 6 more days. Subsequently, cells were either stained with Oil Red O (C) or analyzed for BAT makers by qRT-PCR analysis (D). Three independent experiments were performed. Data in (D) are represented as means ±s.e.m. One-way ANOVA, *p<0.05, **p<0.01, ***p<0.001. (See also Figure S4 and S5).
Ews-null embryos display markedly reduced Bmp7 expression in early brown fat progenitors
Next, we examined early embryonic BAT development. In contrast to WAT, which is generated postnatally, the development of classical BAT starts between embryonic day 14.5 and 15.5 post-coitus (E14.5-15.5) (Hirning et al., 1989) and completes differentiation prior to birth (Cristancho and Lazar, 2011; Gesta et al., 2007). In wildtype E15.5 embryos, interscapular BAT comprised of densely packed patches of preadipocytes was readily detectable (Figure 5A-a) and expanded rapidly through E17.5 (Figure 5A-b) with clearly visible multiloculated lipid droplets at E18.5 (Figure 5A-d). In contrast, E15.5 Ews-null embryos displayed drastically reduced interscapular BAT comprised of markedly smaller and fewer patches of brown preadipocytes that were loosely organized (Figure 5A-i). There was a limited expansion of brown adipocytes at E17.5 and E18.5 (Figure 5A-j-k), but mutant BATs still contained considerably fewer adipocytes and lacked multiloculated lipid droplets (Figure 5A-l). Immunostaining with Ki-67 antibodies revealed a significantly fewer proliferating preadipocytes in the mutant embryonic BATs compared to wildtype (Figure 5B and 5C). In contrast, immunostaining with activated CASP3 antibodies revealed very few, if any, apoptotic cells in embryonic BATs of either genotype (data not shown). Transmission electron microscope analysis of newborn BATs (P0.5) further confirmed the absence of lipid droplets in mutant adipocytes (compare Figures 5A-h and 5A-p). Expression of critical BAT genes, Pparγ, Cox7a, Ucp1 and Otopetrin1 was significantly reduced in E18.5 Ews-null BATs (Supplementary Figures 6A and 6B).
Figure 5. Loss of Bmp7 expression in E15.5 BATs of Ews-null embryos.
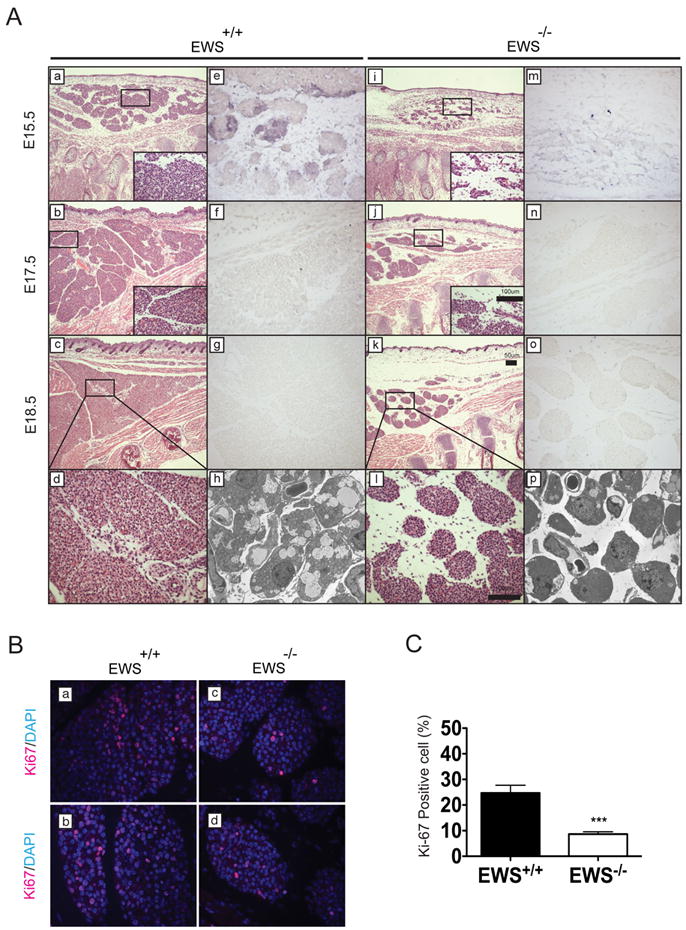
A. Ews+/+ and Ews-/- littermate embryos were harvested at E15.5 (a, e, i and m), E17.5 (b, f, j and n) and E18.5 (c, g, k and o) and paraffin-embedded for H&E (a-d and i-l) or RNA in-situ analysis (e-g and m-o) with antisense mouse Bmp7 probe. Insets and panels (d) and (l) show higher magnification images of the boxed areas. Scale bars are indicated. Transmission electron micrographs of P0.5 newborn BAT from Ews+/+ (h) and Ews-/- (p). B. Interscapular BAT from E18.5 Ews+/+ and Ews-/- littermate embryos were immunostained with Ki-67 and DAPI. Representative images from two independent embryos are shown. C. Quantification of Ki-67 positive brown fat cells. Data are represented as means ±s.e.m. Student t-test, two-tailed, ***p<0.001. (See also Figure S6).
Next, we examined Bmp7 expression in E15.5-18.5 embryos by RNA in-situ analysis. In wildtype E15.5 BATs, Bmp7 was not ubiquitously expressed in all cells but was expressed in intermittently dispersed aggregates of preadipocytes (Figure 5A-e and Supplementary Figure 6C). By E17.5, Bmp7 expression was greatly reduced (Figure 5A-f) and became undetectable at E18.5 (Figure 5A-g). In contrast, Bmp7 expression was dramatically diminished in early brown preadipocytes of E15.5 Ews-null embryos (Figure 5A-m) and became undetectable at E17.5 (Figure 5A-n). There was no detectable signal with a sense Bmp7 probe (Supplementary Figure 6D). As revealed by immunostaining analysis, YBX1 was highly expressed in the early brown fat precursors of E15.5 Ews+/+ and Ews-/- embryos (Supplementary Figure 6E).
Elevated myogenic gene expression in Ews-null brown preadipocytes
As brown preadipocytes share a common lineage with myogenic progenitors (Atit et al., 2006; Seale et al., 2008), we examined the expression of myogenic genes in Ews mutant BATs. As expected, quantitative expression analysis (qRT-PCR) showed a significant reduction in the expression of BAT markers (Ucp1 and Otopetrin1) (Seale et al., 2007) in the mutant BAT compared to wildtype control (Figure 6A). Surprisingly, BATs from Ews-null newborns expressed significantly elevated levels of myogenic genes, embryonic skeletal muscle Myosin heavy chain 3 (Myh3), Tnnt2, Myf5, Myf6 and Myogenin (Myg) (Figure 6A). Immunostaining analysis further confirmed the dramatic increase in the expression of TNNT2 and MYH3 in Ews-null newborns BATs compared to wildtype BATs (Figure 6B). MyoD and Myg transcripts were also moderately elevated in E18.5 BATs of Ews-null embryos (Supplementary Figure 6B). Furthermore, following adipogenic stimulation, we observed increased expression of muscle-specific Tnnt2 and Myg genes in Ews-null brown preadipocytes (Supplementary Figure 7A). Intriguingly, when cultured in myogenic differentiation media (2% horse serum), Ews-null brown preadipocytes, but not wildtype cells, expressed elevated levels of myogenic genes (Figures 6C and 6D). However, unlike C2C12 myoblasts, mutant cells did not form fused muscle fibers. Similarly, depletion of Ews, Ybx1 or Bmp7 by lentivirus-mediated shRNA in wildtype brown preadipocytes also resulted in elevated myogenic gene expression following myogenic stimulation (Supplementary Figure 7B). These results suggest that Ews, along with Ybx1, is critical for brown fat lineage determination in early myogenic progenitors. Nevertheless, the development of muscle, bone and kidney appeared grossly normal in Ews-null newborns (Supplementary Figure 7C) and expression of Bmp7 in kidneys of Ews-null animals was unaltered compared to controls (Supplementary Figure 7D).
Figure 6. Ews deficiency leads to ectopic expression of myogenic genes in BATs.
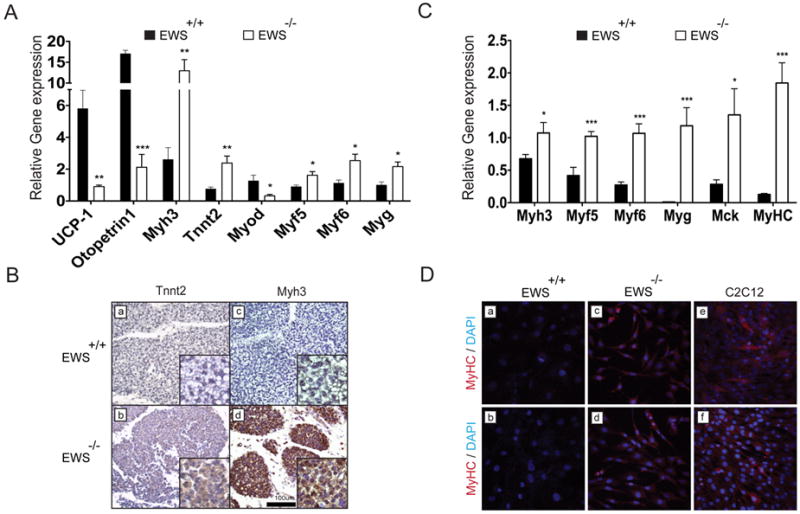
A. SYBR Green qRT-PCR analysis of brown fat (Ucp1 and Otopetrin1) and myogenic (Mhy3, Tnnt2, MyoD, Myf5, Myf6 and Myg) genes in Ews+/+ and Ews-/- newborn BATs (P0.5) (n=5 per genotype). Relative transcript levels were calculated after normalization with β-Actin. Data are represented as means ±s.e.m. *p<0.05, **p<0.01, ***p<0.001. B. Immunohistochemistry of Ews+/+ and Ews-/-newborn BATs (P0.5) with antibodies against TNNT2 and MYH3. Scale bar is indicated. C. qRT-PCR analysis of Ews+/+ or Ews-/- brown preadipocytes cultured for 6 days in 2% horse serum media (myogenic stimulation). Three independent experiments were performed and data are represented as means ±s.e.m. *p<0.05, ***p<0.001. D. Immunostaining of Ews+/+, Ews-/-, or C2C12 cells grown for 6 days in 2% horse serum with anti-MYHC antibody and DAPI (blue). Two representative fields are shown. (See also Figure S7).
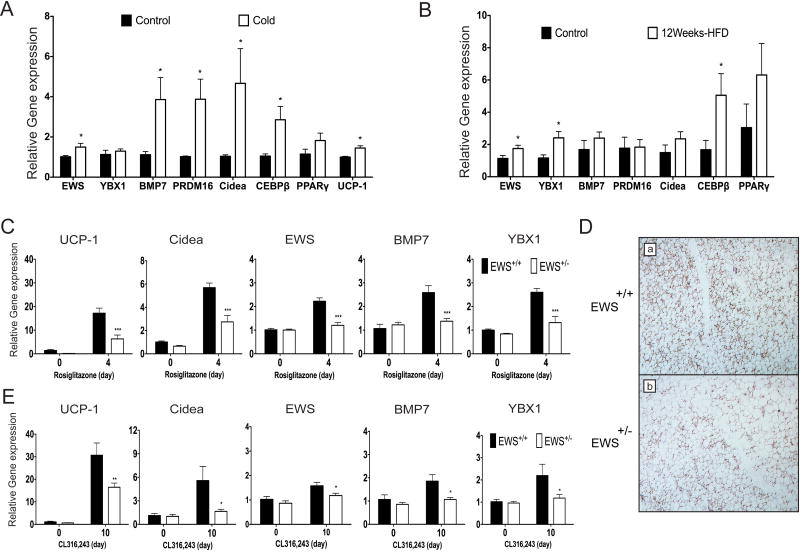
Expression of Ews, Ybx1 and Bmp7 is induced in mature BAT in response to cold or high-fat diet
To examine whether EWS might also play a role in the expansion of mature BAT in response to thermogenic stress, we analyzed the expression of EWS, YBX1 and BMP7 along with other brown fat markers in interscapular BAT of mice under two different thermogenic stimuli: cold or high-fat diet-induced thermogenesis. In either thermogenic stimulation, expression of Ews was modestly induced compared to controls (Figures 7A-B). Expression of Bmp7 in BAT was markedly increased in response to cold along with other brown fat markers (Figure 7A). In response to 12-week high-fat diet intake, a two-fold increase in Ybx1 expression was observed while Bmp7 expression was not significantly altered (Figure 7B). These results indicate that EWS, YBX1 and their transcriptional target BMP7 might play a role in the expansion of mature BAT during adaptive thermogenesis.
Figure 7. Expression of Ews, Ybx1, and Bmp7 is induced in response to thermogenic stimulation and Ews haploinsufficiency leads to reduced brown fat gene expression following rosiglitazone or β3-agonist stimulation.
A-B. qRT-PCR analysis of interscapular BAT from C57BL6 mice after 8h exposure to ambient or cold (4°C) temperature (n=3) (A) or fed with normal or high fat diet for 12 weeks (n=3) (B). C. qRT-PCR analysis of inguinal fat fads from Ews+/+ or Ews+/- mice following 4 daily i.p. injections with PBS or rosiglitazone (10mg/kg) (n=3). D. Immunostaining of UCP1 in inguinal fat from (C). Representative images are shown. 100X magnification. E. qRT-PCR analysis of inguinal fat fads from Ews+/+ or Ews+/- mice following 10 daily i.p. injections with PBS or CL316,243 (1mg/kg) (n=3). Data are represented as means ±s.e.m. Two-way ANOVA, *p<0.05, **p<0.01, ***p<0.001. (See also Figure S7).
A role for EWS in generating thermogenically-induced brown fat
In response to β-adrenergic agonist or PPARγ agonist stimulation, a distinctive type of brown fat cells is generated in WAT depots (Frontini and Cinti, 2010; Wu et al., 2013). Thermogenically-induced brown fat cells (brite or beige cells) are generated more abundantly in subcutaneous WAT depots (e.g. inguinal fat) than in visceral WAT. A recent study has shown that BMP7 is also important for the generation of brown adipocytes from progenitor cells isolated from subcutaneous WAT and skeletal muscle (Schulz et al., 2011). To examine whether EWS might play a role in the formation of brite/beige cells in vivo, we administered rosiglitazone for 4 days or β3-agonist CL316,243 for 10 days to Ews+/+ and Ews+/- mice and analyzed the expression of brown fat markers in inguinal WAT. In wildtype mice, expression of Ucp1 and Cidea was significantly increased following rosiglitazone or CL316,243 treatment (Figure 7C-E). Intriguingly, rosiglitazone or β3-adrenergic stimulation also induced expression of Ews, Ybx1 and Bmp7 in inguinal fat. In contrast, rosiglitazone- or β3-agonist-induced expression of Ews, Ybx1, Bmp7 Ucp1, and Cidea was significantly reduced in inguinal fat of Ews+/- mice compared to Ews+/+ littermates (Figure 7C-E). Expression of brite/beige cell-specific gene, Tmem26 (Wu et al., 2012), was also significantly reduced (Supplementary Fig. 7E). These results suggest that EWS may play an important role in the generation of thermogenically-induced brown fat cells in response to rosiglitazone or β-adrenergic stimulation.
Discussion
Obesity epidemic, driven by excessive energy intake and inactivity leading to accumulation of fat in mostly visceral WAT, has become a major global health threat. Recent discoveries of active BATs in adults (Cypess et al., 2009; Nedergaard et al., 2007; van Marken Lichtenbelt et al., 2009) coupled with the energy burning capacity of BAT in humans (Ouellet et al., 2012) have spurred excitement over the possibility of utilizing BAT expansion and/or activation as a new therapeutic approach. Thus, precise understanding of the upstream regulators of brown adipogenesis will facilitate the development of novel approaches to counteract obesity and related metabolic disorders. To date, BMP7 is the earliest known brown adipogenic determination factor (Tseng et al., 2008), but the upstream genes that regulate BMP7 expression have not been identified. Our study fills this void by demonstrating that EWS, along with YBX1, directly activates BMP7 expression during the early brown adipocyte development. The transcriptional regulation of BMP7 by EWS appears to be specific to brown fat tissue as BMP7 expression in kidneys of Ews-null mice was unaltered. Interestingly, adipogenic stimulation rapidly induces or stabilizes EWS-YBX1 interaction, but the mechanism by which the EWS-YBX1 complex is formed or stabilized is not clear. Identifying the upstream signaling pathways that lead to formation of the EWS-YBX1 complex to induce brown adipogenesis will provide further mechanistic insights into the early BAT development. Additionally, our findings revealed a new role for YBX1 in brown fat development. Mice deficient in Ybx1 die perinatally and the BAT development was not reported (Lu et al., 2005). Hence, in vivo confirmation of YBX1 in BAT development awaits further studies.
Loss of Ews in early brown preadipocytes not only leads to a block in BAT development but also results in elevated myogenic gene expression. Interestingly, shRNA-mediated depletion of either Ybx1 or Bmp7 in brown preadipocytes also led to increased expression of myogenic genes. These findings provide further evidence that EWS, along with YBX1 and its transcriptional target BMP7, is critical for the brown fat specification in the early Myf5+ progenitors. Although Ews-null brown preadipocytes expressed myogenic genes in response to myogenic stimulation, these cells were incapable of forming fused muscle fibers in culture. Moreover, BATs from Ews-null mice did not show any features of muscle differentiation other than the elevated myogenic gene expression. A similar observation was reported for Prdm16-null mice (Seale et al., 2008), in which ectopic myogenic expression was detected in mutant BAT tissues with no signs of muscle differentiation. These findings indicate that in the absence of Ews or Prdm16, early Myf5+ brown fat progenitors will ectopically express myogenic genes, perhaps by default as a result of loss of brown fat specification, but are incapable of generating fully differentiated muscle. This might be due to a lack of external myogenic signals required to generate myofibers or could indicate that Ews-null or Prdm16-null preadipocytes have partially committed toward non-myogenic lineage. It is also possible that EWS might suppress myogenic differentiation in the Myf5+ progenitors. However, ectopic expression of EWS in C2C12 myoblast cell line did not inhibit myogenic differentiation (data not shown).
A role for the EWS-YBX1-BMP7 axis in thermogenesis is suggested by their increased expression under different thermogenic stimuli and by reduced expression of brown fat specific genes in inguinal WAT of Ews heterozygous mice stimulated with β3-agonist or rosiglitazone. However, it will require a conditional Ews-null mouse to unequivocally demonstrate a role for EWS in adaptive thermogenesis. It also remains to be determined whether EWS requires YBX1 and subsequent activation of BMP7 for the formation of brite/beige cells in response to rosiglitazone or β-adrenergic stimulation.
It is evident that EWS has additional functions outside of BAT development as our previous studies demonstrated its roles in B-cell development, meiosis, and premature senescence (Cho et al., 2011; Li et al., 2007). EWS encodes a multifunctional RNA binding protein that, despite lacking a sequence-specific double-stranded DNA binding domain, functions in transcription (Araya et al., 2003) as well as in splicing (Dutertre et al., 2010; Paronetto et al., 2011). This is similar to other RNA binding proteins such as PGC1α (Girnun, 2012) and FUS/TLS (Tan et al., 2012). Given that FUS and EWS are paralogs with sequence and functional similarities, it will be interesting to examine whether FUS also has a role in adipogenesis. However, our findings clearly showed that FUS does not compensate for the loss of EWS during classical BAT development. Interestingly, in majority of myxoid liposarcoma, a subtype of liposarcoma, a balanced chromosomal translocation t(12;16)(q13;p11) is found which results in a fusion of FUS to DDIT3 (FUS-DDIT3, also called FUS/TLS-CHOP) (Crozat et al., 1993; Rabbitts et al., 1993) and in minor cases, a t(12;22)(q13;q12) translocation is found, leading to a fusion of EWS-DDIT3 (Panagopoulos et al., 1996). These, along with another study (Zinszner et al., 1994), suggest that N-terminal domains of EWS and FUS are functionally interchangeable. DDIT3 (CHOP) encodes a member of CEBP-family of transcription factor and by virtue of forming a heterodimer with other CEBP-family proteins, it functions in a dominant-negative manner (Ron and Habener, 1992). Notably, it has been shown that FUS-DDIT3 forms a complex with CEBPβ and inhibits adipogenesis by blocking CEBPβ transcriptional activity (Adelmant et al., 1998; Kuroda et al., 1997). The findings in this study suggest an additional mechanism by which EWS-DDIT3 (or FUS-DDIT3) might further interfere with adipogenesis by inhibiting the functions of remaining EWS (e.g. regulation of BMP7).
Experimental Procedures
Cell lines and siRNA transfection
U2OS, HKE293 and C2C12 cells were cultured in DMEM supplemented with 10% FBS, 100U/ml penicillin and 100ug/ml streptomycin (Invitrogen) at 5% CO2. For siRNA transfection, cells were transfected with 50nM of pooled siRNAs against Ews (3 independent), Ybx1 (2 independent) or scrambled control using Lipofectamine RNAiMAX (Invitrogen). The following siRNAs were purchased and mixed before transfection.
mEws-1: 5’-GCA GUU ACU CUC AGC AGA AdTdT-3’ (Sigma)
mEws-2: 5’-GAG ACU AGU CAA CCU CAA UdTdT-3’ (Sigma)
mEws-3: 5’-CUG ACA ACA GUG CAA UUU AdTdT-3’ (Sigma)
mYbx1-1: 5’-AGA AGG UCA UCG CAA CGA AdTdT-3’ (Ambion)
mYbx1-2: 5’-CCA CGC AAU UAC CAG CAA AdTdT-3’ (Ambion)
Scrambled Silencer Negative Control#1 (Ambion)
Antibodies
Rabbit polyclonal EWS antibody has been described (Li et al., 2007) or purchased from Bethyl Laboratiories. Following antibodies were used: mouse anti-GAPDH, goat anti-PGC1α, rabbit anti-PRDM16 and mouse anti-PPARγ (Santa Cruz Biotechnology), rabbit anti-YBX1 (Bethyl Laboratories), mouse anti-TLS (BD Biosciences), rabbit anti-CEBP/α, rabbit anti-CEBP/β, rabbit anti-CEBP/δ (Cell Signaling), mouse anti-α-Actin (Sigma), rabbit anti-UCP1 (Abcam), mouse anti-Ki67 clone TEC-3 (DAKO) and rabbit anti-TNNT2 (Proteintech). Anti-MYHC and anti-MYH3 antibodies were provided by Alexandra McPherron (NIDDK).
Lentivirus vectors
Lentivirus shRNAs against mouse Ews, Ybx1 and Bmp7 were purchased from Sigma. Full-length mouse Ews cDNA was cloned into the EcoRI/NotI sites of pCDH-GFP-puro lentivirus vector (SBI). Lentivirus vector harboring PPARγ was provided by Kai Ge (NIDDK) and lentivirus vector containing CEBPβ was purchased from Addgene (#15712).
Animal handling
Ews+/- mice in mixed background (129SvEv/Black Swiss) were crossed with C57BL6 for 12 generations. Ews+/- mice (C57BL6N12) were intercrossed to generate Ews-null mice. A multiplex PCR was used for genotyping (see below for primers). All animal procedures were approved and handled according to the guidelines provided by the NIH Animal Research Advisory Committee and by the Tulane University Institutional Animal Care and Use Committee.
Expression analysis of BAT and inguinal fat under different thermogenic challenges
Interscapular BATs from C57BL6 mice housed individually at ambient temperature or at 4°C for 8h, or fed with normal or high-fat diet for 12-weeks were isolated and the expression of the indicated genes was analyzed by qRT-PCR (n=3 for each group). Ews+/+ or Ews-/- mice were injected daily (i.p.) with rosiglitazone (10 mg/kg) or PBS for 4 days (n=3 per genotype) and inguinal fat pads were isolated and analyzed by qRT-PCR for the expression of indicated genes. Similar experiments were performed with ten daily injections (i.p.) of β3-agonist, CL316,243 (1mg/kg).
Generation of brown preadipocyte and differentiation
Brown preadipocytes were isolated from postnatal 0.5 day Ews+/+, Ews+/-, or Ews-/- mouse interscapular brown fat pads and immortalized using a retrovirus expressing SV40 large T antigen as described (Fasshauer et al., 2000). Immortalized brown preadipocytes were cultured in DMEM supplemented with 10% FBS. To induce differentiation into mature brown adipocytes, preadipocytes were cultured for four days in differentiation media supplemented with insulin and triiodothyronine (T3, Sigma), followed by two-day incubation with induction media (beginning of this induction period was considered as differentiation day 0) supplemented with insulin, triiodothyronine, indomethacin, dexamethasone, troglitazone and IBMX (this is referred to as the adipogenic cocktail). After induction, cells were cultured in differentiation media (exchanged every 2 days) for 6 days (Fasshauer et al., 2001). For BMP7-induced adipogenesis, Ews+/+ or Ews-/- preadipocytes were treated with induction media containing recombinant mouse BMP7 (R&D systems) for 2 days (100ng/ml), and cultured in differentiation media for 6 days as described. Cells were fixed with 4% formalin and stained for 2h with 0.5% Oil Red O solution (in 70% isopropyl alcohol).
Histology, immunohistochemistry and transmission electron microscopy
Wildtype and mutant embryos (E15.5, E17.5 and E18.5) and newborns (P0.5) were euthanized, fixed in 10% neutral formalin (Sigma), embedded in paraffin and sectioned for H&E staining. Oil Red O staining was performed on freshly frozen newborn brown fat tissues. Immunohistochemistry was performed using ABC elite kit and ImmPACT DAB peroxidase substrate (Vector labs). Samples were counterstained by hematoxylin GS (Vector labs), dehydrated and mounted for light microscopic analysis (Leica Microsystems). Ki-67 immunostaining was performed with mouse anti-Ki67 and Alexa594-conjugated secondary antibodies and mounted with DAPI containing mounting media (Vector labs). Randomly chosen fields were photographed (at least 4 fields per embryo) and Ki-67 positive cells were counted and quantified against the total number of nuclei. For transmission electron microscopy, freshly isolated BAT (P0.5) was immediately fixed for 4h in 2.5% glutaraldehyde and processed for TEM analysis.
Immunoprecipitation-Western blot analysis
Ews+/+ and Ews-/- preadipocytes were grown in the absence or presence of adipogenic cocktail for 4h and nuclear extracts were prepared. EWS or YBX1 was immunoprecipitated with either anti-EWS or anti-YBX1 antibody, followed by SDS-PAGE and Western blotting was performed with anti-EWS or anti-YBX1. The same procedure was used for U2OS cells transfected with CMV-Ews and CMV-Ybx1.
Chromatin immunoprecipitation (ChIP) assay
ChIP was performed as described (Kim et al., 2007) using rabbit anti-EWS (Li et al., 2007) or anti-YBX1 antibody, Magna chip G kit (Millipore) and Taq PCR Master kit (Qiagen). Primers used for ChIP are listed below.
Gene expression analysis by qRT-PCR
All tissues were harvested, immediately stored in RNAlater (Ambion) and kept in -80°C. Total RNA was isolated using RNeasy mini kit (Qiagen) and quantified using NanoDrop (NanaDrop Technology). cDNAs were synthesized using SuperScript cDNA synthesis kit (Invitrogen), and analyzed by real-time PCR with either TaqMan probes or SYBR Green method (Applied Biosystems). The relative quantity of each transcript was calculated by comparative Ct method normalized against either Gapdh or β-Actin.
Mouse Bmp7 promoter-reporter assay
A 1.4-kb mouse Bmp7 promoter-reporter plasmid (pGL3-Bmp7-luc-1395) (Wang and Hirschberg, 2011) was kindly provided by Dr. Raimund Hirschberg (Los Angeles Biomedical Research Institute, CA). Stable U2OS cells harboring pGL3-Bmp7-luc-1395 plasmid was generated using puromycin selection. Stable U2OS cells were transfected with empty pCMVSport6 (control), pCMVSport6-EWS or pCMVSport6-YBX1, or both, along with Renilla luciferase plasmid (Promega). After 48h, cells were harvested and measured for luciferase activity using Dual-Luciferase Reporter assay system (Promega).
RNA in situ hybridization
A 270-bp mouse Bmp7 cDNA was amplified (forward; 5’-CGA GAC CTT CCA GAT CAC AGT-3’, reverse; 5’-ATG AAG GGT TGC TTG TTC TG-3’), cloned into pCRII plasmid (invitrogen) and verified by sequencing. The antisense or sense probes was synthesized using DIG RNA labeling kit (Roche). Embryos were harvested at different times (E15.5, E17.5 and E18.5), fixed immediately in 10% formalin and prospectively genotyped (tail biopsy). Paraffin-embedded embryos were sectioned (10μm), hybridized with either antisense or sense Bmp7 RNA at 55°C overnight and visualized using alkaline phosphatase-coupled anti-digoxigenin antibody and BCIP-NBT substrate (Roche).
Myogenic differentiation
Ews+/+ and Ews-/- preadipocytes were grown in DMEM with 2% horse serum for 6 days, and total RNA was isolated and analyzed by qRT-PCR for the indicated myogenic genes. For immunofluorescence with anti-MYHC, cells grown in DMEM containing 2% horse serum were fixed, blocked with 5% goat serum, incubated overnight with anti-MYHC and visualized with Alexa Fluor 594 goat anti-mouse secondary antibody (Molecular Probe). DAPI was used to visualize nuclei.
Statistical analysis
Statistical analysis was performed by analysis of variance (ANOVA) test or student’s t-test using GraphPad Prism 5 software (GraphPad Software, CA). Data are represented as means ±s.e.m. and significance was set at p<0.05.
Primers
All the primers used for genotyping, ChIP and realtime PCR as well as the TaqMan probe information are in the Supplementary information.
Supplementary Material
Highlights.
Embryonic BAT development is arrested in Ews KO animals
Ews KO cells fail to undergo BAT differentiation due to loss of Bmp7 expression
EWS and YBX1 activate Bmp7 transcription and promote brown fat differentiation
Ews haploinsufficiency attenuates the formation of thermogenic brown fat cells
Acknowledgments
We thank Raimund Hirschberg (Los Angeles Biomedical Research Institute, CA) for providing the murine Bmp7 promoter-Luc plasmids, Huiyan Lu (NIDDK) for in vitro fertilization and generation of Ews mutant 129SvEv/Black Swiss strain, Oksana Gavrilova (NIDDK) for the temperature-controlled chamber, Sandra E. Dunn (University of British Columbia), Alexandra McPherron and Yun Kyung Lee (NIDDK) and Hyeog Kang (NHLBI) for helpful discussions, and Jan Endlich (JFE Enterprises, MD) for transmission electron microscopy. This research was supported in part by the Intramural Research Program of the NIH, NIDDK and by the startup funds from Tulane University (S.B.L.). J.H.P., H.J.K., S.K., J.H., H.L. performed experiments, J.E.L., K.G., E.M., and B.C.L. contributed critical reagents and suggestions, J.H.P. and S.B.L. designed experiments, analyzed data and wrote the manuscript.
Footnotes
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adelmant G, Gilbert JD, Freytag SO. Human translocation liposarcoma-CCAAT/enhancer binding protein (C/EBP) homologous protein (TLS-CHOP) oncoprotein prevents adipocyte differentiation by directly interfering with C/EBPbeta function. J Biol Chem. 1998;273:15574–15581. doi: 10.1074/jbc.273.25.15574. [DOI] [PubMed] [Google Scholar]
- Araya N, Hirota K, Shimamoto Y, Miyagishi M, Yoshida E, Ishida J, Kaneko S, Kaneko M, Nakajima T, Fukamizu A. Cooperative interaction of EWS with CREB-binding protein selectively activates hepatocyte nuclear factor 4-mediated transcription. J Biol Chem. 2003;278:5427–5432. doi: 10.1074/jbc.M210234200. [DOI] [PubMed] [Google Scholar]
- Atit R, Sgaier SK, Mohamed OA, Taketo MM, Dufort D, Joyner AL, Niswander L, Conlon RA. Beta-catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Dev Biol. 2006;296:164–176. doi: 10.1016/j.ydbio.2006.04.449. [DOI] [PubMed] [Google Scholar]
- Chansky HA, Hu M, Hickstein DD, Yang L. Oncogenic TLS/ERG and EWS/Fli-1 fusion proteins inhibit RNA splicing mediated by YB-1 protein. Cancer Res. 2001;61:3586–3590. [PubMed] [Google Scholar]
- Cho J, Shen H, Yu H, Li H, Cheng T, Lee SB, Lee BC. Ewing sarcoma gene Ews regulates hematopoietic stem cell senescence. Blood. 2011;117:1156–1166. doi: 10.1182/blood-2010-04-279349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristancho AG, Lazar MA. Forming functional fat: a growing understanding of adipocyte differentiation. Nat Rev Mol Cell Biol. 2011;12:722–734. doi: 10.1038/nrm3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozat A, Aman P, Mandahl N, Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363:640–644. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre M, Sanchez G, De Cian MC, Barbier J, Dardenne E, Gratadou L, Dujardin G, Le Jossic-Corcos C, Corcos L, Auboeuf D. Cotranscriptional exon skipping in the genotoxic stress response. Nat Struct Mol Biol. 2010;17:1358–1366. doi: 10.1038/nsmb.1912. [DOI] [PubMed] [Google Scholar]
- Farmer SR. Molecular determinants of brown adipocyte formation and function. Genes Dev. 2008;22:1269–1275. doi: 10.1101/gad.1681308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasshauer M, Klein J, Kriauciunas KM, Ueki K, Benito M, Kahn CR. Essential role of insulin receptor substrate 1 in differentiation of brown adipocytes. Mol Cell Biol. 2001;21:319–329. doi: 10.1128/MCB.21.1.319-329.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasshauer M, Klein J, Ueki K, Kriauciunas KM, Benito M, White MF, Kahn CR. Essential role of insulin receptor substrate-2 in insulin stimulation of Glut4 translocation and glucose uptake in brown adipocytes. J Biol Chem. 2000;275:25494–25501. doi: 10.1074/jbc.M004046200. [DOI] [PubMed] [Google Scholar]
- Frontini A, Cinti S. Distribution and development of brown adipocytes in the murine and human adipose organ. Cell Metab. 2010;11:253–256. doi: 10.1016/j.cmet.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell. 2007;131:242–256. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Girnun GD. The diverse role of the PPARgamma coactivator 1 family of transcriptional coactivators in cancer. Semin Cell Dev Biol. 2012;23:381–388. doi: 10.1016/j.semcdb.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RK, Mepani RJ, Kleiner S, Lo JC, Khandekar MJ, Cohen P, Frontini A, Bhowmick DC, Ye L, Cinti S, et al. Zfp423 expression identifies committed preadipocytes and localizes to adipose endothelial and perivascular cells. Cell Metab. 2012;15:230–239. doi: 10.1016/j.cmet.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirning U, Schmid P, Schulz WA, Kozak LP, Hameister H. In developing brown adipose tissue c-myc protooncogene expression is restricted to early differentiation stages. Cell Differ Dev. 1989;27:243–248. doi: 10.1016/0922-3371(89)90704-1. [DOI] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Spiegelman BM. Transcriptional control of brown fat development. Cell Metab. 2010;11:257–262. doi: 10.1016/j.cmet.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Bajnok L, Longo KA, Petersen RK, Hansen JB, Kristiansen K, MacDougald OA. Effects of Wnt signaling on brown adipocyte differentiation and metabolism mediated by PGC-1alpha. Mol Cell Biol. 2005;25:1272–1282. doi: 10.1128/MCB.25.4.1272-1282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Kim MS, Hancock AL, Harper JC, Park JY, Poy G, Perantoni AO, Cam M, Malik K, Lee SB. Identification of novel Wilms’ tumor suppressor gene target genes implicated in kidney development. J Biol Chem. 2007;282:16278–16287. doi: 10.1074/jbc.M700215200. [DOI] [PubMed] [Google Scholar]
- Kohno K, Izumi H, Uchiumi T, Ashizuka M, Kuwano M. The pleiotropic functions of the Y-box-binding protein, YB-1. Bioessays. 2003;25:691–698. doi: 10.1002/bies.10300. [DOI] [PubMed] [Google Scholar]
- Kuroda M, Ishida T, Takanashi M, Satoh M, Machinami R, Watanabe T. Oncogenic transformation and inhibition of adipocytic conversion of preadipocytes by TLS/FUS-CHOP type II chimeric protein. Am J Pathol. 1997;151:735–744. [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Petkova AP, Mottillo EP, Granneman JG. In vivo identification of bipotential adipocyte progenitors recruited by beta3-adrenoceptor activation and high-fat feeding. Cell Metab. 2012;15:480–491. doi: 10.1016/j.cmet.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Watford W, Li C, Parmelee A, Bryant MA, Deng C, O’Shea J, Lee SB. Ewing sarcoma gene EWS is essential for meiosis and B lymphocyte development. J Clin Invest. 2007;117:1314–1323. doi: 10.1172/JCI31222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Bowerman S, Heber D. Health ramifications of the obesity epidemic. Surg Clin North Am. 2005;85:681–701. v. doi: 10.1016/j.suc.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Lu ZH, Books JT, Ley TJ. YB-1 is important for late-stage embryonic development, optimal cellular stress responses, and the prevention of premature senescence. Mol Cell Biol. 2005;25:4625–4637. doi: 10.1128/MCB.25.11.4625-4637.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May WA, Lessnick SL, Braun BS, Klemsz M, Lewis BC, Lunsford LB, Hromas R, Denny CT. The Ewing’s sarcoma EWS/FLI-1 fusion gene encodes a more potent transcriptional activator and is a more powerful transforming gene than FLI-1. Mol Cell Biol. 1993;13:7393–7398. doi: 10.1128/mcb.13.12.7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedergaard J, Bengtsson T, Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. Am J Physiol Endocrinol Metab. 2007;293:E444–452. doi: 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]
- Ouellet V, Labbe SM, Blondin DP, Phoenix S, Guerin B, Haman F, Turcotte EE, Richard D, Carpentier AC. Brown adipose tissue oxidative metabolism contributes to energy expenditure during acute cold exposure in humans. J Clin Invest. 2012;122:545–552. doi: 10.1172/JCI60433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagopoulos I, Hoglund M, Mertens F, Mandahl N, Mitelman F, Aman P. Fusion of the EWS and CHOP genes in myxoid liposarcoma. Oncogene. 1996;12:489–494. [PubMed] [Google Scholar]
- Paronetto MP, Minana B, Valcarcel J. The Ewing sarcoma protein regulates DNA damage-induced alternative splicing. Mol Cell. 2011;43:353–368. doi: 10.1016/j.molcel.2011.05.035. [DOI] [PubMed] [Google Scholar]
- Rabbitts TH, Forster A, Larson R, Nathan P. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat Genet. 1993;4:175–180. doi: 10.1038/ng0693-175. [DOI] [PubMed] [Google Scholar]
- Ron D, Habener JF. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 1992;6:439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- Sankar S, Lessnick SL. Promiscuous partnerships in Ewing’s sarcoma. Cancer Genet. 2011;204:351–365. doi: 10.1016/j.cancergen.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TJ, Huang TL, Tran TT, Zhang H, Townsend KL, Shadrach JL, Cerletti M, McDougall LE, Giorgadze N, Tchkonia T, et al. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proc Natl Acad Sci U S A. 2011;108:143–148. doi: 10.1073/pnas.1010929108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Xie H, Mori MA, Alexander R, Yuan B, Hattangadi SM, Liu Q, Kahn CR, Lodish HF. Mir193b-365 is essential for brown fat differentiation. Nat Cell Biol. 2011;13:958–965. doi: 10.1038/ncb2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland BW, Kucab J, Wu J, Lee C, Cheang MC, Yorida E, Turbin D, Dedhar S, Nelson C, Pollak M, et al. Akt phosphorylates the Y-box binding protein 1 at Ser102 located in the cold shock domain and affects the anchorage-independent growth of breast cancer cells. Oncogene. 2005;24:4281–4292. doi: 10.1038/sj.onc.1208590. [DOI] [PubMed] [Google Scholar]
- Tan AY, Riley TR, Coady T, Bussemaker HJ, Manley JL. TLS/FUS (translocated in liposarcoma/fused in sarcoma) regulates target gene transcription via single-stranded DNA response elements. Proc Natl Acad Sci U S A. 2012;109:6030–6035. doi: 10.1073/pnas.1203028109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons JA, Wennmalm K, Larsson O, Walden TB, Lassmann T, Petrovic N, Hamilton DL, Gimeno RE, Wahlestedt C, Baar K, et al. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc Natl Acad Sci U S A. 2007;104:4401–4406. doi: 10.1073/pnas.0610615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran KV, Gealekman O, Frontini A, Zingaretti MC, Morroni M, Giordano A, Smorlesi A, Perugini J, De Matteis R, Sbarbati A, et al. The vascular endothelium of the adipose tissue gives rise to both white and brown fat cells. Cell Metab. 2012;15:222–229. doi: 10.1016/j.cmet.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng YH, Butte AJ, Kokkotou E, Yechoor VK, Taniguchi CM, Kriauciunas KM, Cypess AM, Niinobe M, Yoshikawa K, Patti ME, et al. Prediction of preadipocyte differentiation by gene expression reveals role of insulin receptor substrates and necdin. Nat Cell Biol. 2005;7:601–611. doi: 10.1038/ncb1259. [DOI] [PubMed] [Google Scholar]
- Tseng YH, Cypess AM, Kahn CR. Cellular bioenergetics as a target for obesity therapy. Nat Rev Drug Discov. 2010;9:465–482. doi: 10.1038/nrd3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng YH, Kokkotou E, Schulz TJ, Huang TL, Winnay JN, Taniguchi CM, Tran TT, Suzuki R, Espinoza DO, Yamamoto Y, et al. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature. 2008;454:1000–1004. doi: 10.1038/nature07221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, Schrauwen P, Teule GJ. Cold-activated brown adipose tissue in healthy men. N Engl J Med. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- Wang S, Hirschberg R. Y-box protein-1 is a transcriptional regulator of BMP7. J Cell Biochem. 2011;112:1130–1137. doi: 10.1002/jcb.23027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang AH, Khandekar M, Virtanen KA, Nuutila P, Schaart G, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Cohen P, Spiegelman BM. Adaptive thermogenesis in adipocytes: is beige the new brown? Genes Dev. 2013;27:234–250. doi: 10.1101/gad.211649.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinszner H, Albalat R, Ron D. A novel effector domain from the RNA-binding protein TLS or EWS is required for oncogenic transformation by CHOP. Genes Dev. 1994;8:2513–2526. doi: 10.1101/gad.8.21.2513. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.