Abstract
Multi-drug resistant (MDR) cancer is a significant clinical obstacle and is often implicated in cases of recurrent, non-responsive disease. Targeted nanoparticles were made by synthesizing a poly(D,L-lactide-co-glycolide)/poly(ethylene glycol)/epidermal growth factor receptor targeting peptide (PLGA/PEG/EGFR-peptide) construct for incorporation in poly(epsilon-caprolactone) (PCL) nanoparticles. MDR was induced in a panel of nine human breast and ovarian cancer cell lines using hypoxia. EGFR-targeted polymer blend nanoparticles were shown to actively target EGFR over-expressing cell lines, especially upon induction of hypoxia. The nanoparticles were capable of sustained drug release. Combination therapy with lonidamine and paclitaxel significantly improved the therapeutic index of both drugs. Treatment with a nanoparticle dose of 1 μM paclitaxel/10 μM lonidamine resulted in less than 10% cell viability for all hypoxic/MDR cell lines and less than 5% cell viability for all normoxic cell lines. Comparatively, treatment with 1 μM paclitaxel alone was the approximate IC50 value of the MDR cells while treatment with lonidamine alone had very little effect. PLGA/PEG/EGFR-peptide delivery system actively targets a MDR cell by exploiting the expression of EGFR. This system treats MDR by inhibiting the Warburg effect and promoting mitochondrial binding of pro-apoptotic Bcl-2 proteins (lonidamine), while hyperstabilizing microtubules (paclitaxel). This nanocarrier system actively targets a MDR associated phenotype (EGFR receptor over-expression), further enhancing the therapeutic index of both drugs and potentiating the use of lonidamine/paclitaxel combination therapy in the treatment of MDR cancer.
Keywords: Multi-drug resistant cancer, hypoxia, Warburg effect, nanoparticle, drug delivery, lonidamine, paclitaxel, polymeric nanocarriers
Introduction
The development of multidrug resistant (MDR) cancer is a significant clinical obstacle that often results in non-responsive, recurrent disease and eventual metastasis 1-6. MDR refers to a state of resilience against structurally and/or functionally unrelated drugs 1. Multidrug resistant cancer is the biological result of microenvironmental selection pressures that contribute to tumor progression. These selection pressures include hypoxia and changes in the regulation/expression of oncogenes, tumor suppressors, and apoptotic factors. Cellular responses to concurrent selection pressures and chemotherapeutic agents determine if the cell will become quiescent (G0 phase), engage in apoptosis, sustain its current phase, or develop MDR. MDR can be intrinsic (innate) or acquired through exposure to chemotherapeutic agents 1.
Hypoxia
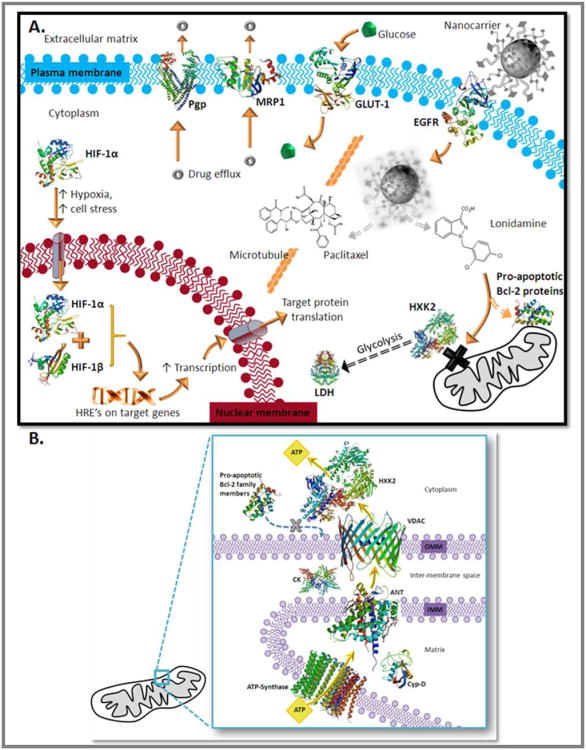
Due to the haphazard and ever-evolving vasculature of a tumor, fluctuating states of chronic and transient hypoxia can occur within the same tumor mass 7. Under conditions of hypoxia and cell stress, cancer cells undergo complex phenotype changes that can result in MDR and resistance to radiation therapy 7-13. The hypoxic transformation begins when Hypoxia Inducible Factor alpha (HIF-1α) translocates from the cytoplasm to the nucleus where it complexes with HIF-1β, forming an active transcription factor that binds to Hypoxia Responsive Elements (HRE's) on target genes, inducing transcription 12, 14. This is demonstrated in Figure 1.A..
Figure 1. A. - MDR Characteristics and the Treatment Strategy.
Under normoxic conditions HIF-1α is located in the cytoplasm associated with a complex of regulatory proteins including the von Hippel–Lindau (VHL) complex. Under hypoxic conditions and cell stress HIF-1α translocates to the nucleus where it complexes with HIF-1β, forming an active transcription factor. The HIF complex binds to hypoxia responsive elements on target genes, increasing transcription and subsequent translation. Target genes include EGFR, GLUT-1, Pgp, and glycolytic enzymes. EGFR overexpression increases the sensitivity of the cell to growth factors; the current treatment strategy capitalizes on this overexpression by using a nanocarrier that is surface modified with EGFR-specific peptides. This active targeting allows facilitated uptake of the formulation, as opposed to non-specific endocytos is which may aid in endosomal escape. Once the nanoparticle is digested by the cell the active agents, paclitaxel and lonidamine are released. Paclitaxel is a microtubule stabilizing agent that hyperstabilizes microtubules preventing disassembly and subsequent cell division while lonidamine is a hexokinase 2 inhibitor. Active targeting of this nanocarrier system to the EGFR receptor should decrease residual toxicity associated with traditional chemotherapy while the combination of paclitaxel with lonidamine offers a unique strategy for terminating the energy supply of MDR cancer and inducing apoptosis. Protein structures were obtained from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB) and are referenced accordingly 62-72.
B. Hexokinase 2 and Lonidamine This figure depicts the association of hexokinase 2 (HXK2) with components of the mitochondrial permeability transition pore complex (mtPTP) and coupling of the components to mitochondrial ATP synthase. ATP synthase is the protein constituent of complex V of oxidative phosphorylation that drives ATP synthesis coupled to the electron transport chain (complex's I-IV; not shown). ATP exits mitochondria by passing from ATP-synthase to the adenine nucleotide translocator (ANT) (also located in the inner mitochondrial membrane; IMM), to the voltage dependent anion channel (VDAC) in the outer mitochondrial membrane (OMM). Creatine kinase (CK), cyclophilin D (Cyp-D), and the peripheral benzodiazepine receptor (not shown) are also associated with the mtPTP. Hexokinase 2 associates with VDAC for the immediate capture of ATP and utilization in glycolysis. Association of hexokinase 2 with VDAC prevents binding of pro-apoptotic Bcl-2 family member proteins to the mtPTP. Protein structures were obtained from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB) and are referenced accordingly 63-64, 67-68, 73-75.
These target genes include P-glycoprotein (P-gp) and multi-drug resistance protein (MRP-1); two drug efflux pumps that are central to the MDR phenotype. One of the most predominate mechanisms of MDR is increased drug efflux through transmembrane pumps known as ATP-Binding Cassette (ABC) transporters 15-17. P-glycoprotein (Pgp) is an ABC transporter associated with malignant transformation, poor prognosis, and the development of MDR cancer 6,15-16, 18. Multi-drug resistance protein 1 (MRP-1) and breast cancer resistance protein (BCRP) are also ABC transporters that contribute to MDR cancer 16, 18-20.
EGFR
Another target of HIF is epidermal growth factor receptor (EGFR) which is correlated with aggressive tumor behavior 21. Many disease cells over-express certain growth factors and receptors to increase survival and maintain homeostasis 22. Over-expression of these receptors leads to membrane clustering. Over-expression of EGFR leads to a higher number EGFR receptors and increased receptor density (clustering) in the cell membrane which potentiates the effects of EGF 22-23. Up-regulated EGFR confers a survival advantage for many types of MDR cancer 21,22-23. As demonstrated in Figure 1.A., this targeted nanocarrier system exploits the upregulation of EGFR through the use of a targeting peptide on the surface of the nanocarriers that binds to the EGFR receptor.
The Warburg Effect
Cancer cells often resort to aerobic glycolysis for energy acquisition, this phenomenon was discovered by Otto Warburg in the 1930's and has since been coined the Warburg Effect 24-28. HIF has been demonstrated to target many glycolytic proteins such as the glucose transporters GLUT-1 and GLUT-3, hexokinase 1 and 2, phosphofructokinase, aldolase, glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase, enolase, pyruvate kinase, and lactate dehydrogenase 7-8, 11-14, 29-33. Reverse activation; glycolytic proteins activating HIF-1α, has also been demonstrated which implies a possible feedback loop between the glycolytic and HIF-1α pathways that may be critical in the transformation of a normal cell into a cancer cell 30.
Hexokinase 2, perhaps the most fundamental enzyme involved in the Warburg effect, is transcriptionally activated by HIF-1α and is overexpressed in many forms of cancer 12, 34-37. Hexokinase catalyzes the first step of glycolysis, conversion of glucose and ATP to glucose-6-phosphate and ADP. The hexokinase 2 isoform is directly associated with the VDAC (voltage dependent anion channel) on the mitochondrial outer membrane and the ANT (adenine nucleotide translocator) on the inner mitochondrial membrane – this association allows hexokinase 2 to directly couple the ATP produced by ATP synthase (transported out of mitochondria by ANT) to aerobic glycolysis (Figure 1.B.) 35-37. The Warburg effect is dependent on both the over-expression of hexokinase 2 and the direct coupling of hexokinase 2 to the VDAC/ANT/ATPsynthase complex 35-37. Also of biological significance, by virtue of its association with mtPTP components, hexokinase 2 has spatial anti-apoptotic activity 35. Hexokinase 2 is associated with the VDAC and the ANT; two proteins that compose the mtPTP complex. Hexokinase 2 association prevents association of pro-apoptotic BcL-2 family member proteins (Bad, Bak, Bax) with the mtPTP complex; pro-apoptotic factor association is necessary for mitochondrial permeability transition and cytochrome c release (the apoptotic cascade) (Figure 1.B.) 35.
Lonidamine (1-[(2,4-Dichlorophenyl)methyl]-1H-indazole-3-carboxylic acid) is an experimental drug originally designed as an antispermatogenic agent that has been discovered to have pro-apoptotic and anti-glycolytic activity 38-40. In the 1980's lonidamine was demonstrated to inhibit aerobic glycolysis via direct inhibition of hexokinase 2 38. Lonidamine has also been shown to induce apoptosis via induction of the mitochondrial permeability transition pore complex 40. This is consistent with the role of hexokinase 2 as a spatial apoptotic inhibitor. In cell based studies, lonidamine has been successful in treating adriamycin resistant breast cancer cells (MCF-7), nitrosourea resistant glioblastoma cells (LB9), and doxorubicin resistant hepatocarcinoma (HepG2) cells 41-43. An anti-angiogenic effect of lonidamine has also been suggested 44. Lonidamine's potential has been explored as a treatment for benign prostatic hyperplasia; clinical trials of lonidamine progressed to phase II in the USA but were halted in 2006 due to the occurrence of elevated liver enzymes in several patients 45-46. This residual toxicity could be circumvented by improving and refining the drug delivery of lonidamine.
Active Targeting is achieved by this system by using nanocarriers that have been engineered to bind to the EGFR receptor by incorporation of a PLGA-PEG-EGFR peptide construct. Active targeting can help a system to overcome biological barriers (active transport verses non-specific endocytosis), decrease the residual toxicity of a system, and increase the therapeutic effect 47.
This study describes the design, development, and characterization of EGFR-targeted nanocarriers for combination paclitaxel/lonidamine delivery to treat MDR cancer. EGFR targeting is achieved by first grafting a PEG-maleimide residue onto poly(DL-lactide-co-glycolide) (PLGA), and subsequently grafting an EGFR-specific peptide to the PEG to create a PLGA-PEG-Peptide construct. This construct is then incorporated into the nanoparticle formulation along with PLGA-PEG and poly(epsilon-caprolactone (PCL) so that the PLGA and PCL form the core of the particles while the PEG and peptide residues modify the surface of the nanoparticles. SEM, dynamic light scattering, and ESCA were used to characterize the nanocarriers. Drug loading and release were also measured. Fluorescent microscopy and fluorescent quantification were used to assess the targeting ability of these nanocarriers in a panel of nine cell lines with various levels of EGFR expression.
The two therapeutic agents selected for combination therapy are paclitaxel and lonidamine. Paclitaxel is a common chemotherapeutic agent that hyper-stabilizes microtubules, preventing cell division. Although the clinical use of paclitaxel is common, its application is often limited by the residual toxicity associated with the drug. Lonidamine is a therapeutic agent with great clinical potential in the treatment of MDR cancer as it acts to inhibit aerobic glycolysis and induce apoptosis. Yet, the clinical application of lonidamine is impeded by the occurrence of residual liver toxicity. The low bioavailability and liver toxicity demonstrated by lonidamine in clinical trials make it an ideal candidate for nanoparticle delivery. The MTS assay was used to assess the efficacy of combination therapy.
Materials and Methods
Polymer and Peptide Conjugation
An established EGFR specific peptide with the following sequence was used to achieve active targeting with the nanoparticle formulation: YHWYGYTPQNVI-GGGGC 48-49. This peptide, termed GE11, was originally synthesized and screened as an EGFR specific peptide by Zonghai Li and colleagues (Shanghai, PR China) 48-49. The “GGGG” sequence functions as an adequate spacer while the carboxyl terminal cysteine of the peptide reacts with the maleimide of the PLGA-PEG construct. The peptide was synthesized by Tufts University Core Facility, Boston, MA.
The peptide-PEG-PLGA construct was synthesized, with slight modifications, according to established methods 50-52. One gram 50:50 poly(DL-lactide-co-glycolide) (PLGA) with an inherent viscosity of 0.15-0.25 and molecular weight of 10,000 (Durect Lactel® Adsorbable Polymers; Pelham, AL) was dissolved in 2 mL chloroform (Fisher Scientific;Pittsburgh, PA) while stirring in a tightly sealed vial. Once dissolved, 45 mg EDC (Fisher Scientific) and 27 mg NHS (Fisher Scientific) was added and the mixture stirred overnight in a tightly sealed vial. The product was precipitated with 8 mL diethyl ether (Fisher Scientific) and centrifuged at 4,000g for 10 minutes. Following centrifugation, the supernatant was discarded and the polymer re-dissolved in 2 mL chloroform. This washing cycle of precipitation/dissolution was repeated three times before drying the activated PLGA-NHS ester under vacuum. Then 0.5 g PLGA-NHS was dissolved in 2 mL chloroform and once fully dissolved, 0.125 g amine- poly(ethylene glycol) PEG-maleimide (MW 2000; JenKem Technology; Allen, TX) and 20ul N,N-diisopropylethylamine (Fisher Scientific) was added, the vial tightly sealed, and the mixture stirred overnight. The product was precipitated with 8 mL of an ice-cold 80/20 mix of diethyl ether/methanol and centrifuged at 4,000g for 10 minutes. After discarding the supernatant the product was re-dissolved in 2 mL chloroform and the washing cycle repeated two more times before drying the PLGA-PEG under vacuum. To prepare a PLGA-PEG conjugate for non-targeted particles, m-PEG-amine (MW 2,000; LaysanBio; Arab, AL) was used in replacement of the amine-PEG-maleimide.
For peptide conjugation to PLGA-PEG, 20 mg of the PLGA-PEG and 2 mg of peptide was dissolved in 800 uL of 50/50 acetonitrile/DMF, covered tightly, and stirred overnight. The product was precipitated with 3 mL of an ice-cold 80/20 mix of diethyl ether/methanol, centrifuged at 4,000g for 10 minutes, the supernatant discarded, and the product re-dissolved in 800 uL 50/50 acetonitrile/DMF. This cycle was repeated two more times and then the PLGA-PEG-peptide was dried under vacuum. NMR was used to assess grafting of PEG to PLGA and conjugation of the peptide to PEG.
Nanoparticle Preparation and Characterization
EGFR targeted and non-targeted polymer blend nanoparticles were synthesized using a solvent displacement method 53-54. Briefly, the PLGA-PEG-peptide conjugate (or the PLGA-PEG conjugate for non-targeted particles), PCL (MW 10,000), and therapeutic agents were dissolved in 2 mL 50/50 acetonitrile/DMF, and placed in a 37°C water bath for 10 minutes to facilitate dissolution. This polymer/drug solution was added drop-wise to 20 mL distilled, deionized water while stirring. The preparation was covered with aerated parafilm, allowed to stir overnight, centrifuged at 10,000g for 30 minutes, and then resuspened in di water. To synthesize targeted nanoparticles the PLGA-PEG-peptide conjugate was added to the nanoparticle formulation at 20% w/w total polymer, with an additional 10% w/w of PLGA-PEG conjugate. Likewise, the PLGA-PEG conjugate was added at a concentration of 20% w/w total polymer. Control nanoparticles synthesized to determine the optimal loading efficiency were composed of either PCL or PLGA, surface modified with PEO (Pluronic® F-108 NF). To assess peptide modification of the nanoparticles samples were sent to the National ESCA and Surface Analysis Center for Biomedical Problems (Seattle, WA). XPS spectra were taken on a Surface Science Instruments S-probe spectrometer. Three spot analysis was done on each batch of nanoparticles.
After determining the optimal dose combination, dual agent loaded nanoparticles were synthesized with a 10:1 molar ratio of lonidamine to paclitaxel. Loading efficiency of the nanoparticles was determined by lyophilizing the nanoparticles to obtain a dry powder, dissolving the particles in acetone, and measuring the absorbance using a Bio-Tek Synergy HT plate reader at 200 nm for paclitaxel and 300 nm for lonidamine (Winooski, RI). Nanoparticle size and zeta potential were characterized using a Brookhaven ZetaPlus Particle Analyzer (Brookhaven Instruments; Holtsville, NY). SEM images of the nanoparticles were obtained using a Hitachi S-4800 microscope.
To measure drug release from nanoparticles, nanoparticles were lyophilized, weighed, and resuspended in two different PBS/0.1% Tween-80 buffers; one at pH 7.4 and one at pH 6.5 – then incubated in a 37°C water bath. At various time points between 15 minutes and ten days an aliquot of eluted drug medium was removed for quantification; this volume was replaced with fresh buffer. Drug release was quantified by measuring the absorbance of the release media using a Bio-Tek Synergy HT plate reader (Winooski, RI).
For initial dose response studies, single agent loaded nanoparticles were synthesized; paclitaxel was loaded at 10% w/w and after determining maximal loading, lonidamine was also loaded at 10% w/w. After determining the optimal dose combination, dual agent loaded nanoparticles were synthesized with a 10:1 molar ratio of lonidamine to paclitaxel.
Cell Culture and Treatment
SKOV3 cells, MDA-MB-231 cells, and OVCAR5 cells were obtained from ATCC (Manassas, VA). The SKOV3-TR cells and the MDA-MB-435 cells were a kind gift from Dr. Duan (Massachusetts General Hospital, Sarcoma Molecular Biology Laboratory). Cells were incubated at 37°C and maintained in RPMI-1640 media (Mediatech, Inc; Manassas, VA) supplemented with 10% fetal bovine serum (Gemini Bio-products; West Sacramento, CA) and 1% penicillin/streptomycin/amphotericin B mixture (Lonza; Walkersville, MD). Hypoxia was used to induce MDR. To create hypoxic conditions using low-oxygen gas; cell culture flasks were placed in a modular incubation chamber (Billups-Rothenberg, Inc.; Del Mar, CA), flushed with a 0.5% O2, 5% CO2, nitrogen balanced gas for five minutes, and incubated at 37°C for various time points.
For cell viability, cells were plated in 96-well plates at 2,000 cells per well and treated with various concentrations of nanoparticle formulations and drug solutions suspended in FBS supplemented media. Control treatments with supplemented media, blank nanoparticles and poly(ethyleneimine) were also conducted. Treatment continued undisturbed for 5 days under normoxic and hypoxic conditions. Cell viability was then measured using the MTS assay according to the manufacturer's protocol (Promega; Madison, WI).
Protein Extraction and Western Blot Analysis
Basal protein was extracted from cells grown to 90% confluency in 75 cm2 tissue culture flasks under normoxic and hypoxic conditions. Basal protein was extracted using a high salt lysis buffer at 4°C. Protein concentrations were quantified using the BCA Protein Assay (Pierce Biotechnology). Protein was separated on 4-20% gradient SDS-PAGE gels (PAGEgel, Inc.; San Diego, CA) and transferred onto PVDF membranes (0.45 μm pore; Millipore, Billerica, MA). Membranes were blocked for 30 minutes with StartingBlock™ buffer (Pierce Biotechnology) before a 1 hour incubation with the primary antibody. Membranes were then washed with TBST for 10 minutes (three times) and subsequently incubated with a horseradish peroxidase conjugated secondary antibody for 1 hour. Membranes were again washed with TBST, flash rinsed with deionized distilled water, incubated for 2-10 minutes in an enhanced chemiluminescence substrate (Pierce Biotechnology), and imaged using a Kodak FX Imaging Station (Rochester, NY). All blocking, probing, and washing steps were conducted at room temperature. P-glycoprotein antibody was purchased from Calbiochem while the EGFR and β-actin antibodies were purchased from Cell Signaling Technology (Danvers, MA). The secondary antibodies were purchased from Abcam (Cambridge, MA).
Nanoparticle Trafficking and Cellular Uptake
To assess and visualize particle uptake kinetics and quantity, nanoparticles were prepared as previously mentioned and loaded with 1% w/w rhodamine 123 (Invitrogen; Carlsbad, CA). Cells were seeded at 10,000 cells per well and treated with rhodamine 123 loaded nanoparticles suspended in supplemented media (5 μM with respect to rhodamine). For competitive binding studies 100-fold molar excess of EGFR antibody (Cell Signaling Technology, Danvers, MA) was added to the media (relative to the amount of peptide on the nanoparticles). At various time points (from 15 minutes to 6 hours), the treatment solution was removed and cells were washed three times with supplemented media. For quantification of uptake, plates were read on a Bio-Tek Synergy HT plate reader (Winooski, RI) at 485ex/528em. For microscopy, wells were also treated with Hoechst 33342 nucleic acid stain (Invitrogen) 15 minutes before washing. Cells were visualized using an Olympus IX51 Microscope.
Statistical Data Analysis
All statistical analysis was done using GraphPad Prism® software. Statistical significance of uptake was determined using one-way ANOVA and Bonferroni's Multiple Comparison Test. For cell viability studies, statistical significance of treatments were determined using a two-tailed, Student's t test at a 95% confidence interval (P< 0.05). For cell viability studies n = 8 for each treatment group. IC50 values for paclitaxel treatment were calculated using variable slope curve fitting of transformed data.
Results and Discussion
Nanoparticle Development and Characterization
The primary objective in designing this drug delivery system was to design a system capable of delivering a therapeutic dose of lonidamine and paclitaxel to the site of a tumor. To accomplish this, the system must: (1) achieve high encapsulation of both drugs and (2) be surface modified with PEG to avoid immediate RES clearance, increasing resident circulation and ability to reach the tumor. A secondary design objective of this formulation was to synthesize a polymer-PEG conjugate and a polymer-PEG-EGFR peptide conjugate and incorporate these constructs to achieve PEG surface modification of the nanoparticles and to achieve active targeting of MDR cells (via EGFR binding). To accomplish this objective, m-PEG-amine (MW 2000) was conjugated to NHS-activated PLGA (to use for PEG modification of the formulation). For the active targeting construct, amine-PEG-maleimide was conjugated to NHS-activated PLGA and an EGFR specific peptide (YHWYGYTPQNVI-GGGGC) was conjugated to the PEG via cysteine/maleimide linkage. NMR spectra of the conjugate and raw materials were acquired to evaluate this reaction (data not shown).
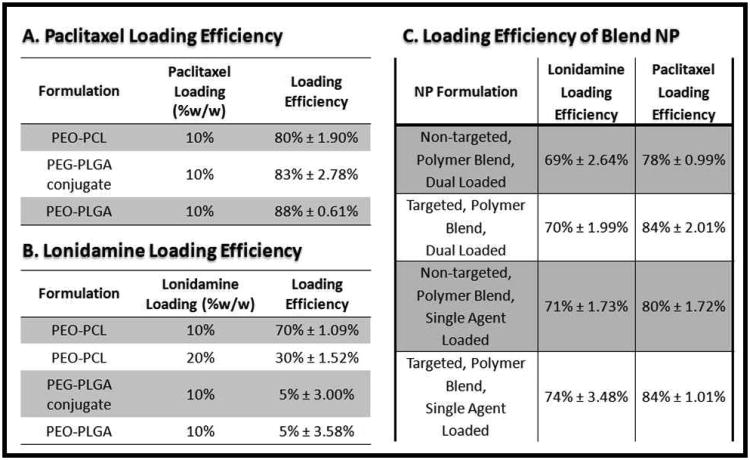
To select the optimal polymer for this drug delivery system, a variety of polymeric nanoparticles were synthesized and drug loading efficiency was evaluated (Figure 2.A. and 2.B.). The loading efficiency of PEO (Pluronic® F-108 NF) modified PCL particles, PEO (Pluronic® F-108 NF) modified PLGA particles, and PLGA particles with the PLGA-PEG construct were compared. All formulations were evaluated for 10% w/w loading efficiency of paclitaxel and of lonidamine; 20% w/w loading of lonidamine was also examined for the PEO-PCL formulation. The highest loading efficiency of paclitaxel was achieved with the PEO-PLGA formulation (88%), yet the PEO-PCL and the PLGA/PEG-PLGA conjugate nanoparticles also achieved high loading efficiencies; 80% and 83% respectively. The loading efficiency of PEO-PCL nanoparticles loaded with 10% w/w lonidamine was 70%; this decreased to 30% loading efficiency when the dose was increased to 20% w/w. For both the PEO-PLGA nanoparticles and for the PLGA/PEG-PLGA construct nanoparticles, lonidamine loading was only 5%. The decrease in lonidamine loading with a higher w/w loading in PCL and the substantially lower loading in PLGA based nanoparticles is most likely due to the hydrophobicity of lonidamine. As PCL is more hydrophobic than PLGA, higher encapsulation is possible with PCL. Yet there seems to be a maximal threshold above which lonidamine encapsulation decreases, most likely due to excessive hydrophobicity of the polymer/drug solution which is not conducive to nano-precipitation via the solvent displacement method.
Figure 2. Nanoparticle Loading Efficiency.
The loading efficiency of different polymer nanoparticle formulations was used as a parameter to determine the optimal composition of the polymer blend nanocarrier system. Paclitaxel (A) and Lonidamine (B) loading were examined in single polymer systems and in polymer blend system (C).
Based on these findings, it was determined that the predominate polymer constituent of the nanoparticle formulation needed to be PCL in order to achieve any appreciable encapsulation of lonidamine. To achieve the design goals of the system (high encapsulation and surface modification with a malleable construct), a polymer blend nanoparticle formulation of PCL, PLGA-PEG construct, and the PLGA-PEG-peptide construct was developed. The PCL and the PLGA of the conjugate should form the core of the nanoparticle with the PEG and peptide residues extending out from this core. To assess the capability of these polymer blend nanoparticles, nanoparticles were synthesized with single agent loading (paclitaxel alone and lonidamine alone) and with dual agent loading and the loading efficiency was determined (Figure 2.C.). Maintaining a predominately PCL based system allowed for encapsulation comparable to PEO-PCL nanoparticles (70% for lonidamine, 80% for paclitaxel). Combining the two drugs into one formulation did not have a great impact on the loading efficiency relative to formulations loaded with single agents.
To further characterize the polymer blend nanoparticles, the size and zeta potential of the combination drug loaded and unloaded polymer blend nanoparticles and targeted polymer blend nanoparticles were measured (Table 1). PEO-PCL nanoparticles were the largest particles which also had the most negative zeta potential (174.3 nm; -33.1 mV) while the PLGA/PEG-PLGA conjugate nanoparticles were the smallest, also bearing the most positive zeta potential (70.1 nm; -26.13 mV). The unloaded polymer blend nanoparticles were smaller than the PEO-PCL nanoparticles with a slight reduction in zeta potential (129.2 nm; -30.12 mV). This size reduction relative to the PEO-PCL nanoparticles may be attributed to the structural difference between the PEO and the PEG-PLGA graft. The PEO (Pluronic® F108 NF) is of an ABA structure where the more hydrophobic (B) block would interact with the core of the nanoparticle while the A-blocks branch out, forming a V-shape with the central V-apex interacting with the PCL core. The PEG-PLGA conjugate on the other hand, does not have this branching structure; the PLGA of the construct interacts with the PCL core while the PEG end protrudes on the surface of the particle. This more linear structure may be conducive to smaller particle sizes as the core becomes more compact (no branching V-type components). This would also explain the smaller size of the PLGA/PEG-PLGA conjugate nanoparticles compared to PEO-PLGA nanoparticles (70.1 nm verses 122.2 nm). Drug loading did not drastically alter the size or zeta potential of the polymer blend nanoparticles (123.4 nm; -31.4 mV for drug loaded and 129.2 nm; -30.12 mV for unloaded). Adding the targeting construct increased the particle size; from 123.4 nm to 139.6 nm for drug loaded nanoparticles and from 129.2 nm to 151.5 nm for unloaded nanoparticles. There was only a slight reduction in zeta potential; this is most likely due to the neutrality of the peptide (isoelectric point near 7.4).
Table 1. Nanoparticle Size and Zeta Potential.
| Nanoparticle Formulations | Size (nm) | Zeta Potential (mV) |
|---|---|---|
| Non-targeted, Polymer Blend, Dual Loaded | 123.4 ± 4.4 | -31.4 ± 2.6 |
| Non-targeted, Polymer Blend, Unloaded | 129.2 ± 6.8 | -30.12 ± 2.6 |
| Targeted, Polymer Blend,Dual Loaded | 139.6 ± 4.1 | -29.6 ± 3.8 |
| Targeted, Polymer Blend,Unloaded | 151.5 ± 3.2 | -28.9 ± 4.3 |
| PEO-PCL,Unloaded | 174.3 ± 10.6 | -33.1 ± 5.9 |
| PEO-PLGA,Unloaded | 122.2 ± 7.5 | -26.8 ± 8.2 |
| PEG-grafted-PLGA,Unloaded | 70.1 ± 2.8 | -26.13 ± 6.4 |
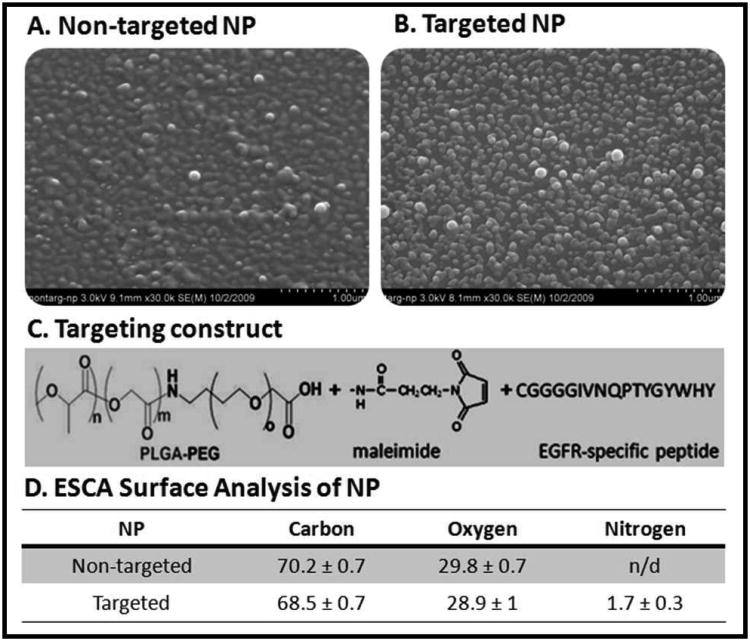
SEM images of the non-targeted and targeted nanoparticles confirmed the nanometer scale of the particles (Figure 3.A. and Figure 3.B.). The targeting construct is depicted in Figure 3.C. To confirm that the EGFR peptide was on the surface of the nanoparticles electron spectroscopy for chemical analysis (ESCA) was used to examine the surface of the particles at an approximate depth of 50 Å (∼ 5 nm) (Figure 3.D). The percent composition of carbon and oxygen was similar for the targeted and non-targeted nanoparticles, suggesting similar PEG modifications and polymer compositions. However, there was no detectable level of nitrogen on the surface of the non-targeted nanoparticles whereas the targeted nanoparticles had approximately 2% nitrogen composition. This correlates to the total w/w ratio of the peptide in the targeted nanoparticles formulation.
Figure 3. Nanoparticle Characterization.
Scanning electron micrographs of Non-targeted polymer blend nanoparticles (A) and targeted polymer blend nanoparticles (B). Scale bar is 1 μM. The targeting construct (C) is incorporated in the targeted nanocarriers so that the PLGA portion interacts with the PCL core while the PEG and EGFR-peptide protrude from the surface of the particle. ESCA analysis of the surface of the nanocarriers (D) confirmed the presence of the EGFR peptide (nitrogen) on the surface of the targeted nanoparticles while there was no detectable (n/d) nitrogen on the non-targeted nanoparticles.
Drug Release Kinetics
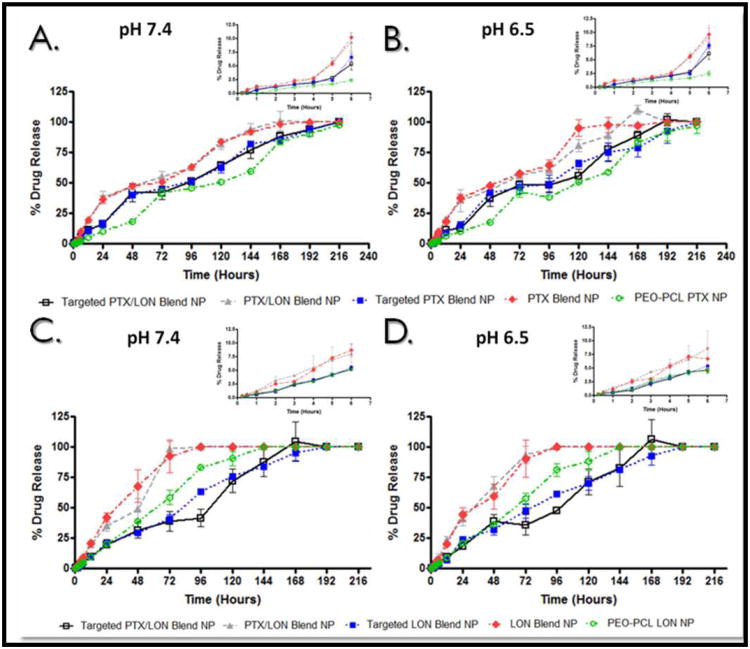
An important characteristic of a nanoparticle formulation is its drug release profile. Just as drug release from common dosage forms can determine clinical application, the release of drugs from a nanocarrier system must be sustained to justify the system and ensure the drug is released at the target site, not during systemic circulation. The release of lonidamine and paclitaxel from combination polymer blend nanoparticles (dual loaded), singly loaded polymer blend nanoparticles (either paclitaxel alone or lonidamine alone), targeted combination polymer blend nanoparticles (dual loaded), targeted singly loaded polymer blend nanoparticles (either paclitaxel alone or lonidamine alone), and from PEO-PCL nanoparticles was measured over the course of ten days (Figure 4). Paclitaxel release is presented in the top panel of Figure 4 (Figure 4.A. and Figure 4.B.) while lonidamine release is depicted in the bottom panel (Figure 4.C. and Figure 4.D.) Release was measured both at physiological pH 7.4 (Figure 4.A. and Figure 4.C.) and at a pH common to the microenvironment of a tumor (6.5) (Figure 4.B. and Figure 4.D.). The figure insets represent the early time points (from 0 to 6 hours).
Figure 4. Drug Release Kinetics from Nanocarriers.
Drug release kinetics from the nanocarriers were measured up to 10 days. Panels (A) and (B) represent paclitaxel drug release at pH 7.4 and pH 6.5 respectively. Panels (C) and (D) represent release of lonidamine at pH 7.4 and pH 6.5 respectively. Figure insets represent release from 15 minutes until 6 hours.
As demonstrated by the early time points (Figure 4, insets), none of the formulations exhibited burst release of lonidamine at pH 7.4 (Figure 4.C.) and at pH 6.5 (Figure 4.D.). The release kinetics of lonidamine from all three formulations did not appear to be greatly affected by pH as the profiles were similar for both pH 7.4 and pH 6.5. By 72 hours, 100% of the dose is released from both dual loaded polymer blend nanoparticles and the singly loaded polymer blend nanoparticles at pH 7.4 (the percentage of release after 72 hours at pH 6.5 is slightly reduced to 93%). After 72 hours only 58% of the lonidamine dose has been released from the PEO-PCL nanoparticles at pH 7.4 (57% at pH 6.5); complete release from the PEO-PCL occurs after 144 hours (both pH values). After 72 hours only 39% of the total lonidamine dose was released from the targeted, dual loaded polymer blend particles and 41% of total lonidamine was released by the targeted, singly loaded polymer blend nanoparticles (neither of these values changed in response to pH). The targeted nanoparticles exhibited the most sustained release for lonidamine; 100% of the total loaded lonidamine was released after 168 hours for the dual loaded formulation and after 192 hours for the singly loaded formulation (there was no change with a change in pH). This prolonged release may be attributable to the spatial interactions of the polymer/drug mixture in the nanoparticle core, the density of the particle, and the surface protection of the formulation. The PEO-PCL particles may be more rigid and stable (owing to the B-block interaction of PEO with PCL), this rigidity may enhance the association of lonidamine with the polymers and as PCL degradation is very slow, this association may extend the release of the drug. The targeted nanoparticles, on the other hand, have extensive surface modification relative to the other formulations; this may prolong their degradation. The targeted nanoparticles may also have a very dense core of drug/PCL/PLGA tightly packed by the peptide and PEG protrusions.
Paclitaxel release kinetics were also sustained over the course of several days for the five formulations (Figure 4.A. and Figure 4.B.). None of the formulations exhibited burst release at early time points at both pH 7.4 (Figure 4.A., inset) and pH 6.5 (Figure 4.B., inset). The dual loaded polymer blend nanoparticles have a much greater retention of paclitaxel than lonidamine; after 72 hours at both pH 7.4 and pH 6.5, 100% of lonidamine is released where as at 72 hours only 52% of total loaded paclitaxel is released (57% at pH 6.5). Singly loaded lonidamine and paclitaxel polymer blend nanoparticles also exhibit this retention difference. This could be due to the bulkier structure of paclitaxel; it may be that paclitaxel is more tightly associated with the core polymers. One hundred percent of the loaded paclitaxel dose is not released until 168 hours for both the dual loaded and singly loaded polymer blend nanoparticles at pH 7.4 (this value is the same for the dual loaded polymer blend nanoparticles at pH 6.5 but is reduced to 144 hours for the singly loaded polymer blend nanoparticles at pH 6.5). As with lonidamine, the PEO-PCL nanoparticles demonstrate more prolonged release of paclitaxel; it is not until 216 hours (at both pH values) that this formulation releases 100% of the paclitaxel dose. Similar to lonidamine release kinetics, the targeted formulations exhibit sustained release for paclitaxel; it is not until 216 hours that 100% of the paclitaxel dose is released by the targeted formulations (this is released to 192 hours for the dual loaded targeted particles at pH 6.5). The sustained release kinetics of lonidamine and paclitaxel from the targeted combination polymer blend nanoparticles ensures that the therapeutics will not completely leach out of the particles during circulation, but will be released once the particles are internalized and digested by the target cells.
MDR and EGFR Expression in the Cell Panel
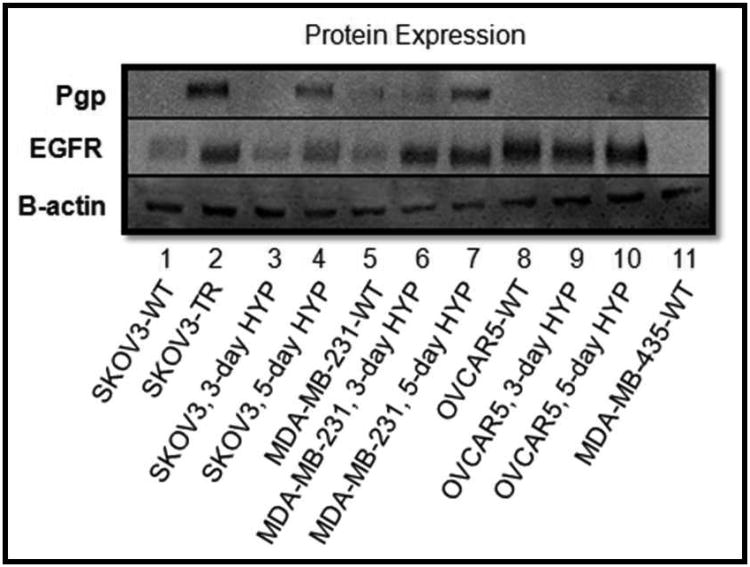
In certain cancer cells, hypoxia has been shown to contribute to MDR 7-13. We have conducted extensive analysis of the relationship between hypoxia and MDR in this panel of cell lines (work submitted for publication). In the current study, we used hypoxia to induce MDR in a panel of cell lines. Pgp expression was used as a positive marker for MDR (Figure 5). EGFR expression was also analyzed. As demonstrated in Figure 5, hypoxia induced the expression of Pgp in SKOV3 cells after 5 days of hypoxia and in MDA-MB-231 cells after 3 days of hypoxic exposure. The OVCAR5 cells appeared resistant to hypoxic induction of Pgp. The established MDR cell line, SKOV3-TR cells were a positive control for Pgp expression. Hypoxia also increased the expression of EGFR in the SKOV3 cell line and in the MDA-MB-231 cell line. The OVCAR5 cells have a high basal level of EGFR. The MDA-MB-435 cells were used as a negative control for EGFR expression.
Figure 5. Protein Expression Analysis.
Basal protein was extracted from the panel of cell lines grown under normoxic and hypoxic conditions (three and five days of hypoxia). Protein was probed for expression of the MDR marker, Pgp as well as for EGFR. β-actin was used as a loading control.
Cell Uptake of EGFR-Targeted Nanoparticles
To confirm that the nanoparticles were being taken up by the cells, nanoparticles were loaded with rhodamine 123 and incubated with the panel of cells (SKOV3-TR, SKOV3, MDA-MB-231, OVCAR5, and MDA-MB-435 cells) under normoxic and hypoxic conditions. At time intervals spanning from 15 minutes to 6 hours images of the cells were taken (Figures 6-10, Panel A; only the 15 and 30 minute time points are shown) and the fluorescent intensity was quantified as a percentage of the total administered dose (Figures 6-10, Panel B). The panel of cell lines were treated with targeted nanoparticles (T), targeted nanoparticles in the presence of competitive EGFR antibody (TC), non-targeted nanoparticles (N), and non-targeted nanoparticles in the presence of competitive EGFR antibody (NC). Cells were treated with a 5 μM dose of the nanoparticles (relative to rhodamine loading) and examined after 15 minutes of treatment, 30 minutes, 1 hour, 2 hours, 3 hours, and 6 hours. Nuclei were stained with Hoechst 33342 (blue). For competitive studies, a 100-fold molar excess of the EGFR antibody (relative to the peptide content of the nanoparticle dose) was added to the cells 10 minutes before treatment with the particles.
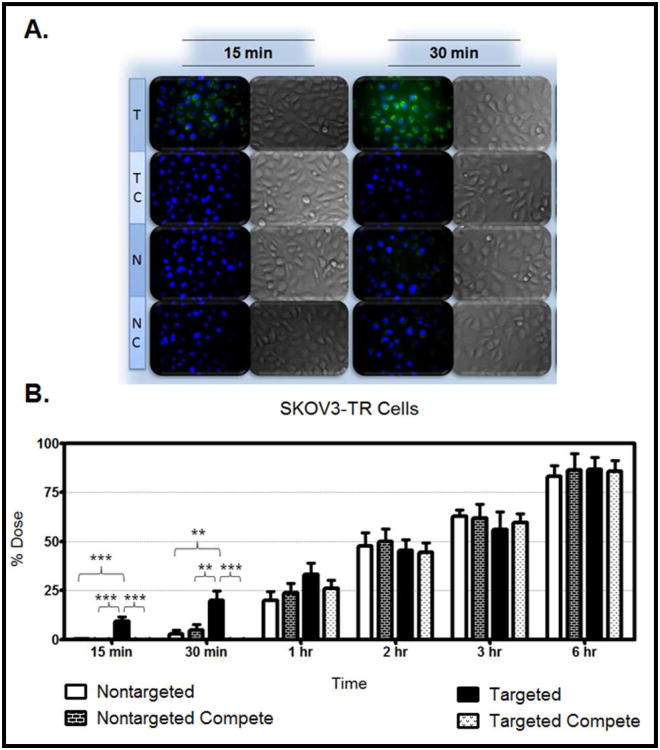
Figure 6. Nanoparticle Uptake in SKOV3TR Cells.
Panel A: Nanoparticle formulations were loaded with rhodamine 123 (green). SKOV3TR cells were treated with targeted nanoparticles (T), targeted nanoparticles in the presence of excess competitive EGFR antibody (TC), non-targeted nanoparticles (N), and non-targeted nanoparticles in the presence of excess competitive EGFR antibody (NC). Cells were imaged after 15 minutes and 30 minutes of treatment (A). Cell nuclei were stained with Hoechst 33342 (blue). The fluorescent images represent a merge of the blue and green fluorescence; corresponding DIC images are also shown. Targeted nanoparticle uptake is apparent at the early time points and is blocked by the competitive antibody. Panel B: Cells were treated with the rhodamine 123 loaded formulations, and the amount of rhodamine 123 taken up by the cells was quantified and converted to a percentage of the administered dose. The SKOV3TR cells were treated with non-targeted nanoparticles (white bar), non-targeted nanoparticles in the presence of excess competitive EGFR antibody (black/white brick bar), targeted nanoparticles (black bar), and targeted nanoparticles in the presence of excess competitive EGFR antibody (black/white speckled bar). Cells were treated for 15 minutes, 30 minutes, 1 hour, 2 hours, 3 hours, and 6 hours. Fluorescence was quantified using a Bio-Tek Synergy HT plate reader. Each treatment represents an n=8.
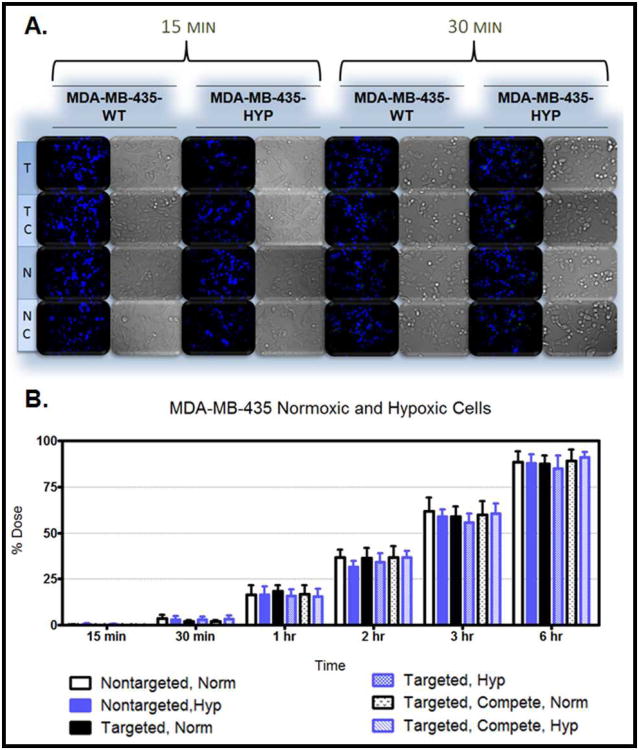
Figure 10. Nanoparticle Uptake in MDA-MB-435-WT and Hypoxic Cells.
Panel A: Nanoparticle formulations were loaded with rhodamine 123 (green). MDA-MB-435 wild type (WT) and hypoxic (HYP) cells were treated with targeted nanoparticles (T), targeted nanoparticles in the presence of excess competitive EGFR antibody (TC), non-targeted nanoparticles (N), and non-targeted nanoparticles in the presence of excess competitive EGFR antibody (NC). Cells were imaged after 15 minutes and 30 minutes of treatment (A). Cell nuclei were stained with Hoechst 33342 (blue). The fluorescent images represent a merge of the blue and green fluorescence; corresponding DIC images are also shown. There is no apparent difference between the targeted and non-targeted nanoparticle uptake in this EGFR-negative cell line. Antibody/nanoparticle competition was also not apparent. Panel B: Cells were treated with the rhodamine 123 loaded formulations, and the amount of rhodamine 123 taken up by the cells was quantified and converted to a percentage of the administered dose. Normoxic treatments are shown in black while the hypoxic treatments are shown in blue. Cells were treated with the following nanoparticle formulations; non-targeted nanoparticles (solid white bar for normoxic cells; solid blue bar for hypoxic cells), targeted nanoparticles (solid black bar for normoxic cells; blue checkered bar for hypoxic cells), and targeted nanoparticles in the presence of excess competitive EGFR antibody (black/white speckled bar for normoxic cells; blue diagonal line bar for hypoxic cells). Cells were treated for 15 minutes, 30 minutes, 1 hour, 2 hours, 3 hours, and 6 hours. Fluorescence was quantified using a Bio-Tek Synergy HT plate reader. Each treatment represents an n=8.
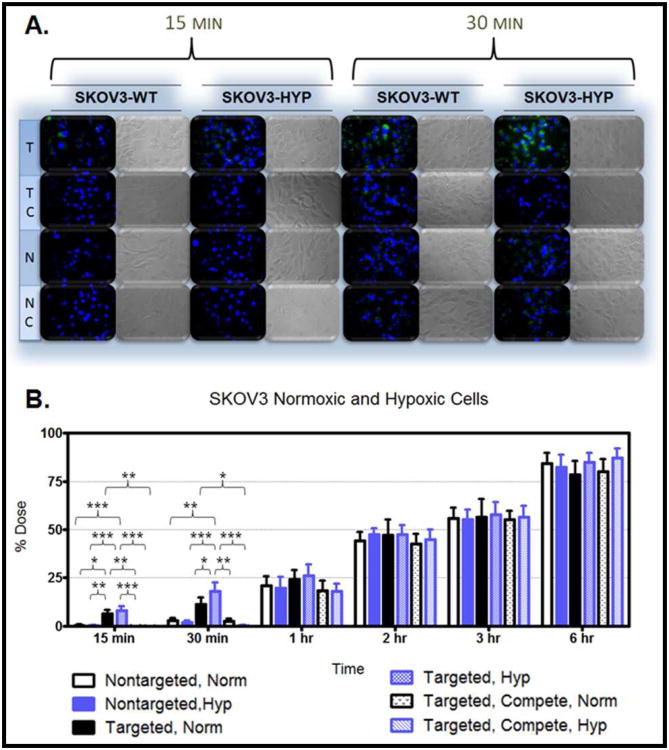
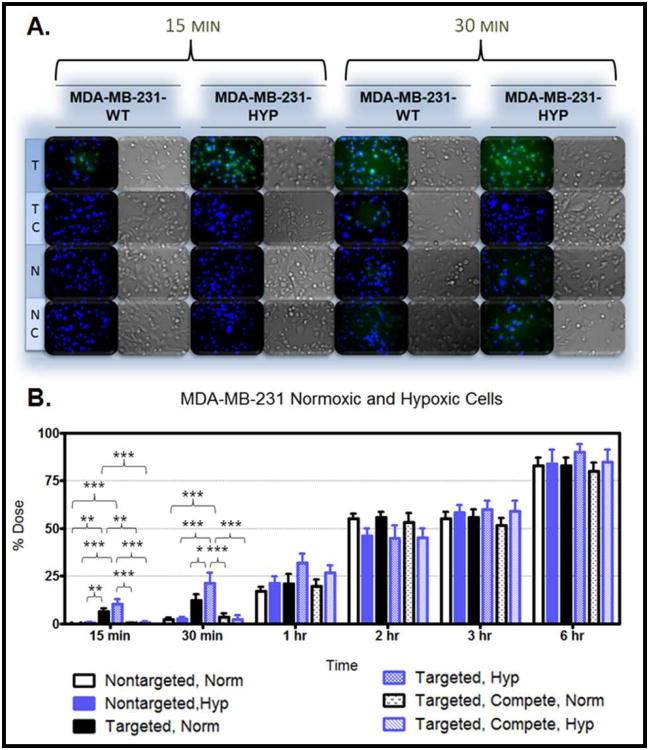
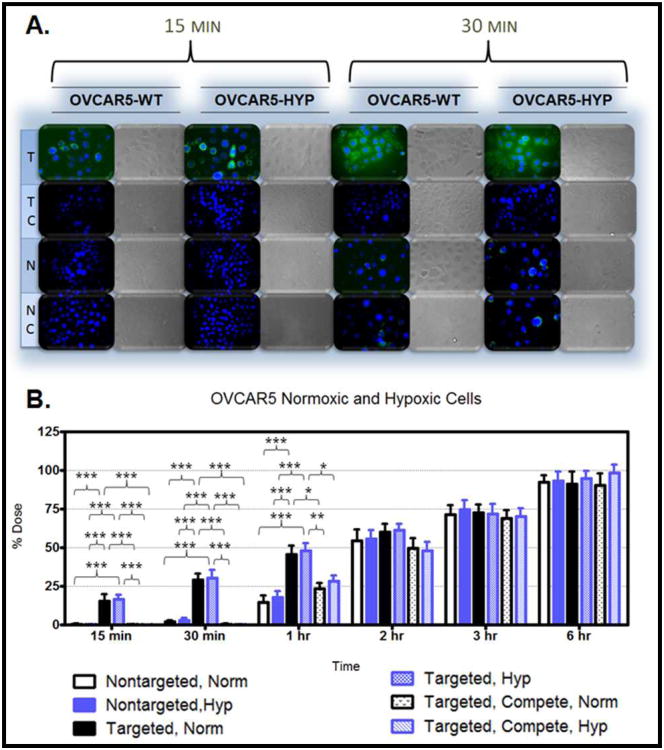
Figure 6.A. demonstrates the uptake in the MDR cells after 15 minutes and 30 minutes of treatment. The cellular uptake after 15 minutes and 30 minutes of treatment was also monitored in hypoxic and normoxic derivatives of SKOV3 cells (Figure 7.A.), MDA-MB-231 cells (Figure 8.A.), OVCAR5 cells (Figure 9.A.), and MDA-MB-435 cells (Figure 10.A.). Although the microscopy demonstrates the differences in uptake kinetics between the different formulations and the between the time points; the data is qualitative and does not demonstrate clear differences between the hypoxic and normoxic derivatives. As such, quantitative nanoparticle uptake was measured on a Bio-Tek Synergy HT plate reader and the results are presented in Panel B of Figures 6-10.
Figure 7. Nanoparticle Uptake in SKOV3-WT and Hypoxic Cells.
Panel A: Nanoparticle formulations were loaded with rhodamine 123 (green). SKOV3 wild type (WT) and hypoxic (HYP) cells were treated with targeted nanoparticles (T), targeted nanoparticles in the presence of excess competitive EGFR antibody (TC), non-targeted nanoparticles (N), and non-targeted nanoparticles in the presence of excess competitive EGFR antibody (NC). Cells were imaged after 15 minutes and 30 minutes of treatment (A). Cell nuclei were stained with Hoechst 33342 (blue). The fluorescent images represent a merge of the blue and green fluorescence; corresponding DIC images are also shown. Targeted nanoparticle uptake is apparent at the early time points and is blocked by the competitive antibody. Panel B: Cells were treated with the rhodamine 123 loaded formulations, and the amount of rhodamine 123 taken up by the cells was quantified and converted to a percentage of the administered dose. Normoxic treatments are shown in black while the hypoxic treatments are shown in blue. Cells were treated with the following nanoparticle formulations; non-targeted nanoparticles (solid white bar for normoxic cells; solid blue bar for hypoxic cells), targeted nanoparticles (solid black bar for normoxic cells; blue checkered bar for hypoxic cells), and targeted nanoparticles in the presence of excess competitive EGFR antibody (black/white speckled bar for normoxic cells; blue diagonal line bar for hypoxic cells). Cells were treated for 15 minutes, 30 minutes, 1 hour, 2 hours, 3 hours, and 6 hours. Fluorescence was quantified using a Bio-Tek Synergy HT plate reader. Each treatment represents an n=8.
Figure 8. Nanoparticle Uptake in MDA-MB-231-WT and Hypoxic Cells.
Panel A: Nanoparticle formulations were loaded with rhodamine 123 (green). MDA-MB-231 wild type (WT) and hypoxic (HYP) cells were treated with targeted nanoparticles (T), targeted nanoparticles in the presence of excess competitive EGFR antibody (TC), non-targeted nanoparticles (N), and non-targeted nanoparticles in the presence of excess competitive EGFR antibody (NC). Cells were imaged after 15 minutes and 30 minutes of treatment (A). Cell nuclei were stained with Hoechst 33342 (blue). The fluorescent images represent a merge of the blue and green fluorescence; corresponding DIC images are also shown. Targeted nanoparticle uptake is apparent at the early time points and is blocked by the competitive antibody. Panel B: Cells were treated with the rhodamine 123 loaded formulations, and the amount of rhodamine 123 taken up by the cells was quantified and converted to a percentage of the administered dose. Normoxic treatments are shown in black while the hypoxic treatments are shown in blue. Cells were treated with the following nanoparticle formulations; non-targeted nanoparticles (solid white bar for normoxic cells; solid blue bar for hypoxic cells), targeted nanoparticles (solid black bar for normoxic cells; blue checkered bar for hypoxic cells), and targeted nanoparticles in the presence of excess competitive EGFR antibody (black/white speckled bar for normoxic cells; blue diagonal line bar for hypoxic cells). Cells were treated for 15 minutes, 30 minutes, 1 hour, 2 hours, 3 hours, and 6 hours. Fluorescence was quantified using a Bio-Tek Synergy HT plate reader. Each treatment represents an n=8.
Figure 9. Nanoparticle Uptake in OVCAR5-WT and Hypoxic Cells.
Panel A: Nanoparticle formulations were loaded with rhodamine 123 (green). OVCAR5 wild type (WT) and hypoxic (HYP) cells were treated with targeted nanoparticles (T), targeted nanoparticles in the presence of excess competitive EGFR antibody (TC), non-targeted nanoparticles (N), and non-targeted nanoparticles in the presence of excess competitive EGFR antibody (NC). Cells were imaged after 15 minutes and 30 minutes of treatment (A). Cell nuclei were stained with Hoechst 33342 (blue). The fluorescent images represent a merge of the blue and green fluorescence; corresponding DIC images are also shown. Targeted nanoparticle uptake is apparent at the early time points and is blocked by the competitive antibody. Panel B: Cells were treated with the rhodamine 123 loaded formulations, and the amount of rhodamine 123 taken up by the cells was quantified and converted to a percentage of the administered dose. Normoxic treatments are shown in black while the hypoxic treatments are shown in blue. Cells were treated with the following nanoparticle formulations; non-targeted nanoparticles (solid white bar for normoxic cells; solid blue bar for hypoxic cells), targeted nanoparticles (solid black bar for normoxic cells; blue checkered bar for hypoxic cells), and targeted nanoparticles in the presence of excess competitive EGFR antibody (black/white speckled bar for normoxic cells; blue diagonal line bar for hypoxic cells). Cells were treated for 15 minutes, 30 minutes, 1 hour, 2 hours, 3 hours, and 6 hours. Fluorescence was quantified using a Bio-Tek Synergy HT plate reader. Each treatment represents an n=8.
The degree of targeted nanoparticle uptake at the early time points (15 minutes and 30 minutes) paralleled the expression level of EGFR in the panel of cell lines. The highest expression of EGFR was in the OVCAR5 normoxic and hypoxic derivatives, followed by the SKOV3TR cells and the hypoxic MDA-MB- 231 cells, then the SKOV3 hypoxic cells, the SKOV3 normoxic and MDA-MB231 normoxic cells, and lastly the MDA-MB-435 cells which had no EGFR expression. Inhibition of targeted nanoparticle uptake in the presence of excess EGFR antibody is indicative of competitive binding between the nanoparticles and the free antibody. This inhibition was significant in all EGFR-expressing cell lines after 15 and 30 minutes of treatment and also after 1 hour of treatment in the OVCAR5 derivatives (Figures 6-10). No competition was observed in the EGFR-negative MDA-MB-435 cell line (Figure 10). This competitive binding verifies that the targeted nanoparticles are engaging the EGFR receptor.
The degree of targeted nanoparticle uptake relative to non-targeted nanoparticle uptake was also significant at the early time points for all EGFR expressing cell lines. There was no appreciable uptake of the non-targeted nanoparticles in any cell line after 15 minutes of treatment. After 30 minutes of treatment, non-targeted nanoparticle uptake ranged from 1.92% of the total administered dose (in the SKOV3 derivatives) to 2.20% (in the normoxic MDA-MB-435 cells). There was no significance between the non-targeted nanoparticles in the presence and absence of free EGFR antibody (Figures 6-10).
As indicated by the quantitative data, there was significance between targeted nanoparticle uptake in normoxic and hypoxic derivatives of the SKOV3 and MDA-MD-231 cells after 30 minutes of treatment. The lack of significance at the 15 minute time point may be because 15 minutes is not adequate time to demonstrate a difference between the hypoxic and normoxic uptake by saturating the receptors (although it is adequate time to demonstrate a difference between the non-targeted and targeted nanoparticles). At the 30 minute time point in the the SKOV3 and MDA-MD-231 cell lines there was no difference between the accumulation of targeted nanoparticles in normoxic cells and between targeted nanoparticles in the presence of competitive EGFR antibody under normoxic conditions, and between non-targeted nanoparticles under normoxic and hypoxic conditions. For the MDA-MB-231 cell line there was also no difference between the uptake of targeted nanoparticles under normoxic conditions and targeted nanoparticles in the presence of competitive EGFR antibody under hypoxic conditions. At this time point in these cell lines, targeting enhances uptake proportionate to EGFR expression.
The lack of significance between the uptake of targeted nanoparticles in normoxic and hypoxic derivative of the OVCAR5 cells may be due to the excessively high basal expression of EGFR in the normoxic derivatives. Targeting does show a clear engagement of the EGFR receptor at 15 minutes (demonstrated by competitive binding) and enhanced uptake relative to non-targeted nanoparticles at 15 minutes for the SKOV3 cells and the MDA-MD-231 cells and upto the 1 hour time point for the OVCAR5 cells.
The difference between non-targeted nanoparticle uptake (no appreciable uptake) and targeted nanoparticle uptake at the early time points indicate that the two different formulations may engage in distinct mechanisms of uptake. It has been established that non-targeted nanoparticles are internalized via non-specific endocytosis 47, 55-59. Most likely, the EGFR targeted nanoparticles are internalized by mechanisms similar to endogenous EGFR substrates (EGF). Studies have shown that after endogenous substrate binding to EGFR and by environmental activation (radiation), EGFR and the bound substrate are internalized via a process characteristic to this protein tyrosine kinase 60-61. This characterized receptor/substrate internalization is a rapid process, occurring within 10 to 20 minutes of EGFR stimulation 61. This process is more similar to a flip-flop mechanism than to a membrane budding process 60-61. As the EGFR-targeted nanoparticles are an extension of an EGFR substrate (the peptide), the mechanism of targeted nanoparticle uptake is most likely via the receptor/substrate internalization process.
After one hour of treatment for every cell line (except the highly EGFR expressing OVCAR5 derivatives), there is no significance between the targeted and non-targeted nanoparticle uptake (Figure 6-10, Panel B). This suggests that the kinetic differences between non-specific endocytosis and receptor/substrate internalization are normalizing. Also at this time point, there is no observed competition between the targeted nanoparticles and the free antibody in all cell lines (again, except for the OVCAR5 derivatives). It is possible that after 1 hour, the free antibody has already engaged EGFR receptors and been internalized, restoring the receptors for targeted nanoparticle binding. It is also likely that after 1 hour of treatment uptake of all formulations by non-specific endocytosis masks any kinetic differences due to competitive receptor/substrate internalization.
The uptake studies verify that cellular uptake kinetics are undoubtedly cell-type specific. However, there is a general trend of increasing particle uptake in each cell line from 15 minutes until 6 hours at which point between 80% of the total administered dose (for the MDA-MB-231 normoxic cells treated with targeted nanoparticles in the presence of competitive antibody) and 100% of the administered dose (for the OVCAR5 hypoxic derivatives treated with non-targeted nanoparticles) had been taken up. Interestingly, there does not appear to be any appreciable nanoparticle uptake in the MDA-MB-231 cells between 2 and 3 hours (Figure 8). The MDA-MB-231 cells may reach a saturation threshold (after 2 hours) at which point the cell increases metabolism of the internalized particles and vesicles, restores phospholipids to the cell membrane, apparently ceasing additional uptake, before resuming internalization at a slower rate. This would explain the dramatic increase in particle uptake between 1 hour and 2 hours (an average of 39% of the total dose), the lack of change between 2 and 3 hours, and the subsequent slow uptake between 3 hours and 6 hours (an average rate of 10% total dose per hour). Also of note, the MDA-MB-435 cells appear to have a slower initiation of particle uptake. After two hours of treatment approximately 35% of the administered dose was taken up by the MDA-MB-435 cells which is 10% lower than uptake by the SKOV3 derivatives (with no significance between the nanoparticle formulations). For each cell line, the particle uptake kinetics visualized during microscopy (Figures 6-10, Panel A) were consistent with the kinetics observed in quantifying cumulative nanoparticle uptake (Figures 6-10, Panel B) and confirmed interaction of the targeted nanoparticles with the EGFR receptor.
It is not unusual that the benefit of active targeting was only demonstrated during the early time points. This analysis was done in a stationary cell culture system where saturation (even by non-targeted nanoparticles) is fairly rapid and inevitable as particle uptake becomes dominated by non-specific endocytosis. However, an in vivo study would be expected to demonstrate the true potential of a targeted nanocarrier system. We have also completed in vivo studies which show improved pharmacokinetic parameters and enhanced efficacy of this targeted system (work to be published).
Efficacy of Combination Therapy
To determine the optimal combination of lonidamine and paclitaxel, a dose response study was conducted in three cell lines (SKOV3 ovarian cancer cells, MDA-MB-231 breast cancer cells, and OVCAR5 ovarian cancer cells) under normoxic and hypoxic conditions as well as in MDR cells (SKOV3-TR ovarian cancer cells). Cells were treated with drug loaded nanoparticle formulations and with solution forms of the drugs. First, single agent treatment was examined (data not shown). Dose response to paclitaxel encapsulated in nanoparticles and paclitaxel solution was measured over the range of 0.001 μM to 10 μM. The cells that were the most resistant to paclitaxel were the MDR cells (SKOV3-TR), the hypoxic SKOV3 cells, and the hypoxic MDA-MB-231 cells. The OVCAR5 cells were the most sensitive to paclitaxel. As paclitaxel is a Pgp substrate, these responses can be correlated with the expression profile of Pgp in the cells (Figure 5). Hypoxia induced the expression of Pgp in SKOV3 and MDA-MB-231 cells (which are the most resistant, second to the MDR cells), yet the OVCAR5 cells did not express basal levels of Pgp and expression was not induced by hypoxia (most sensitive to paclitaxel).
The IC50 values for paclitaxel treatment are listed in Table 2. The IC50 values for MDR SKOV3 cells treated with nanoparticle formulations of paclitaxel are 92 times the IC50 values for the wild-type, normoxic SKOV3 cells with similar treatment; the MDR cells treated with paclitaxel solution had an IC50 value 79 times that of wild-type normoxic cells treated with solution. While the IC50 values for the hypoxic SKOV3 cells are 72 times that of the wild-type, normoxic SKOV3 cells treated with nanoparticle encapsulated paclitaxel and the hypoxic SKOV3 cells treated with solution have an IC50 value 64 times that of wild-type normoxic SKOV3 cells treated with paclitaxel solution. This increased IC50 value in hypoxic cells verifies the dynamic cellular transformation that occurs during hypoxia; shifting the cells toward a MDR character (portrayed in Figure 5). Likewise, the IC50 for hypoxic MDA-MB-231 cells treated with nanoparticle paclitaxel is 70 times that of the wild-type, normoxic cells treated with the formulation and hypoxic MDA-MB-231 cells treated with solution paclitaxel had an IC50 65 times that of wild-type, normoxic breast cancer cells treated with solution. Yet, the IC50 values of the hypoxic OVCAR5 cells treated with paclitaxel nanoparticles was only 13 times that of the wild-type, normoxic, OVCAR5 cells treated with the same formulation and the IC50 of the hypoxic OVCAR5 cells treated with paclitaxel solution was 20 times that of the wild-type, normoxic OVCAR5 cells treated with solution paclitaxel. This minimal increase in IC50 values for the OVCAR5 cells subjected to hypoxia coincides with the lack of hypoxic transformation in the OVCAR5 cells demonstrated by the protein analysis (Figure 5). The IC50 values again clarify that hypoxia seemed effective in inducing MDR character in the SKOV3 cells and in the MDA-MB-231 cells, yet this selection pressure did not reach the threshold for transforming OVCAR5 cells. Lonidamine treatment alone had very little effect at 1 μM and 10 μM doses and for most cells did not even reach an IC50 value with a 100 μM dose (data not shown). The response of the different cell types to lonidamine is also consistent with the protein expression profile and glycolytic character of the cells (previous work submitted for publication). The OVCAR5 cells had extremely low glycolytic character and as lonidamine is a hexokinase-2 inhibitor, lonidamine alone had very little effect even at a 100 μM dose.
Table 2. IC50 Values From Paclitaxel Treatment.
| Cell Line | Condition | Treatment | IC50 (μM) | ||
|---|---|---|---|---|---|
| SKOV3-TR | MDR | NP | 1.011 ± 0.009 | ||
| SKOV3-TR | MDR | SOL | 0.950 ± 0.021 | ||
| SKOV3 | NORMOXIC | NP | 0.011 ± 0.004 | ||
| SKOV3 | NORMOXIC | SOL | 0.012 ± 0.006 | ||
| SKOV3 | HYPOXIC | NP | 0.791 ± 0.040 | ||
| SKOV3 | HYPOXIC | SOL | 0.770 ± 0.067 | ||
| MDA-MB-231 | NORMOXIC | NP | 0.012 ± 0.004 | ||
| MDA-MB-231 | NORMOXIC | SOL | 0.013 ± 0.006 | ||
| MDA-MB-231 | HYPOXIC | NP | 0.845 ± 0.049 | ||
| MDA-MB-231 | HYPOXIC | SOL | 0.840 ± 0.051 | ||
| OVCAR5 | NORMOXIC | NP | 0.003 ± 0.003 | ||
| OVCAR5 | NORMOXIC | SOL | 0.004 ± 0.003 | ||
| OVCAR5 | HYPOXIC | NP | 0.038 ± 0.015 | ||
| OVCAR5 | HYPOXIC | SOL | 0.083 ± 0.038 | ||
Based on the dose response of single agents, doses of 0.1 μM paclitaxel (below the IC50 of MDR and hypoxic cells), 1 μM (the approximate IC50 of MDR and hypoxic cells), and 10 μM paclitaxel (above the IC50 of MDR and hypoxic cells) were selected for a combination study with 1, 10, and 100 μM lonidamine (data not shown). As a result of the dose responses of the cell lines to paclitaxel/lonidamine combination treatment, 1 μM paclitaxel and 10 μM lonidamine were selected as the optimal dose combination. This dose enhanced the efficacy of both drugs in the MDR cell line and in the hypoxic and normoxic SKOV3 and MDA-MB-231 cells.
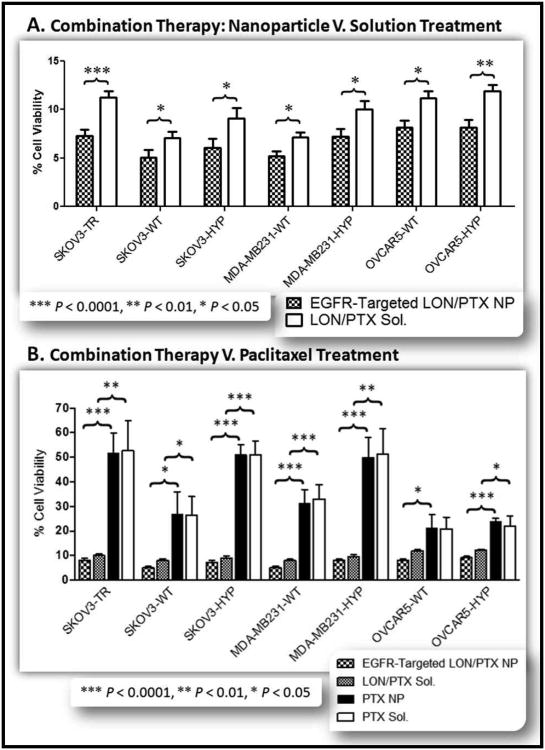
After selection of the optimal dose combination, EGFR-targeted nanoparticles dual loaded with lonidamine and paclitaxel at a dose ratio of 10:1 were synthesized using PCL, the PLGA-PEG conjugate (10% w/w and the PLGA-PEG-EGFR peptide construct (20% w/w). Cells were treated with the nanoparticles at a dose of 1 μM paclitaxel/10 μM lonidamine and viability was assessed after five days of treatment (Figure 11). Treatment with the lonidamine/paclitaxel loaded EGFR targeted nanoparticles resulted in less than 10% cell viability for all cell lines and less than 5% cell viability for the wild-type, normoxic SKOV3 and MDA-MB-231 cells (Figure 11.A.). For all cell lines, there was significance between the EGFR-targeted combination blend nanoparticles and drug solution (Figure 11.A.). This enhanced efficacy is most likely due to the sustained drug release from the nanoparticles.
Figure 11. Lonidamine and Paclitaxel Combination Therapy.
A panel of seven cell lines was used to evaluate the efficacy of combination lonidamine/paclitaxel therapy. The cell lines included SKOV3TR cells, SKOV3 wild type cells (WT), SKOV3 hypoxic cells (HYP), MDA-MB-231 wild type cells, MDA-MB-231 hypoxic cells, OVCAR5 wild type cells, and OVCAR5 hypoxic cells. (A). Each cell line was treated with EGFR-Targeted lonidamine/paclitaxel loaded nanoparticles (EGFR-Targeted LON/PTX NP; black and white checkered bar) and with lonidamine/paclitaxel solution (LON/PTX Sol; solid white bar). All treatment doses were 10 μM lonidamine and 1 μM paclitaxel. Combination therapy dramatically reduced the cell viability for all cell lines to below 10% cell viability for all cell lines treated with the combination nanoparticles and to approximately 5% cell viability for the wild type SKOV3 and MDA-MB-231 cells. Each treatment represents an n=7. (B.) Combination paclitaxel/lonidamine treatment with 10 μM lonidamine and 1 μM paclitaxel was compared to treatment with 1 μM paclitaxel alone (in nanoparticle and solution forms). The panel of cells were treated with EGFR-Targeted lonidamine/paclitaxel loaded nanoparticles (EGFR-Targeted LON/PTX NP; black and white checkered bar), lonidamine/paclitaxel solution (LON/PTX Sol; gray checkered bar), paclitaxel nanoparticles (PTX NP; solid black bar), and with paclitaxel solution (PTX Sol; solid white bar). As illustrated, 1 μM paclitaxel is the approximate IC50 for the MDR SKOV3TR cells and for the hypoxic SKOV3 cells and the hypoxic MDA-MB-231 cells. This is reduced by 40% when combined with lonidamine treatment. Each treatment represents an n=7.
Treatment with the dual loaded lonidamine/paclitaxel EGFR-targeted nanoparticles was compared to treatment with nanoparticles only loaded with paclitaxel; also, treatment with a solution combination of lonidamine and paclitaxel was compared to treatment with paclitaxel solution alone (Figure 11.B.). Combination treatment was at a dose of 1 μM paclitaxel and 10 μM lonidamine; this was compared to treatment with 1 μM paclitaxel alone. Combination treatment with lonidamine/paclitaxel nanoparticles enhanced the cell kill efficacy relative to treatment with paclitaxel nanoparticles (extremely significant, p < 0.0001, for the MDR, all hypoxic cell derivatives, and the normoxic MDA-MB-231 cells; and significant, p < 0.05, for the SKOV3 wild-type normoxic cells and the OVCAR5 wild-type normoxic cells). Solution treatment with a combination of lonidamine and paclitaxel enhanced the cell kill efficacy relative to treatment with paclitaxel solution alone for all cell lines except for the wild-type, normoxic OVCAR5 cells (significant at p < 0.0001, for the SKOV3 hypoxic cells and for the MDA-MB-231 normoxic cells; very significant, p < 0.01, for the MDR and MDA-MB-231 hypoxic cells; significant, p < 0.05, for the SKOV3 normoxic cells and for the OVCAR5 hypoxic cells; and not significant for the OVCAR5 normoxic cells). As previously mentioned, the decreased response of the OVCAR5 normoxic cells to lonidamine is most likely due to the lower glycolytic character of these cells. For the hypoxic and normoxic SKOV3 and MDA-MB-231 cells as well as for the MDR cells, combination treatment with lonidamine and paclitaxel at the dose ratio of 10 μM lonidamine: 1 μM paclitaxel greatly enhances the cell kill efficacy relative to treatment with 1 μM paclitaxel alone (Figure 11.B.).
Conclusions
This novel drug delivery system achieves the functional goals of the system; improved efficacy with combination therapy and active EGFR targeting. The nanocarriers are capable of appreciable drug encapsulation and sustained drug release. ESCA analysis confirmed that the surface of the nanocarriers were modified with the targeting construct. This was further confirmed with the demonstrated targeting of the nanocarriers in the panel of cell lines; uptake kinetics emulated the expression of EGFR in the cell lines and competitive inhibition by an EGFR antibody was apparent. Combination therapy with 1 μM paclitaxel and 10 μM lonidamine resulted in 5-10% cell viability whereas treatment with 1 μM paclitaxel alone was the approximate IC50 of the MDR cells. This drug delivery system is a well characterized platform for the delivery of paclitaxel and lonidamine to treat MDR cancer.
Acknowledgments
This study was supported by the National Cancer Institute, National Institutes of Health through grants R01 CA-119617 and R01 CA-119617S1 (ARRA Supplement), and R21 CA-135594. Scanning electron microscopy was performed by William Fowle at the Electron Microscopy Center at Northeastern University.
Contributor Information
Lara Milane, Email: lara.milane@gmail.com.
Zhenfeng Duan, Email: ZDUAN@PARTNERS.ORG.
Mansoor Amiji, Email: m.amiji@neu.edu.
References
- 1.Harris AL, Hochhauser D. Mechanisms of multidrug resistance in cancer treatment. Acta oncologica (Stockholm, Sweden) 1992;31(2):205–213. doi: 10.3109/02841869209088904. [DOI] [PubMed] [Google Scholar]
- 2.Jamroziak K, Robak T. Pharmacogenomics of MDR1/ABCB1 gene: the influence on risk and clinical outcome of haematological malignancies. Hematology (Amsterdam, Netherlands) 2004;9(2):91–105. doi: 10.1080/10245330310001638974. [DOI] [PubMed] [Google Scholar]
- 3.Leighton JC, Jr, Goldstein LJ. P-glycoprotein in adult solid tumors. Expression and prognostic significance. Hematology/oncology clinics of North America. 1995;9(2):251–273. [PubMed] [Google Scholar]
- 4.Tredan O, Galmarini CM, Patel K, Tannock IF. Drug resistance and the solid tumor microenvironment. Journal of the National Cancer Institute. 2007;99(19):1441–1454. doi: 10.1093/jnci/djm135. [DOI] [PubMed] [Google Scholar]
- 5.Tsuruo T, Naito M, Tomida A, Fujita N, Mashima T, Sakamoto H, Haga N. Molecular targeting therapy of cancer: drug resistance, apoptosis and survival signal. Cancer science. 2003;94(1):15–21. doi: 10.1111/j.1349-7006.2003.tb01345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yague E, Arance A, Kubitza L, O'Hare M, Jat P, Ogilvie CM, Hart IR, Higgins CF, Raguz S. Ability to acquire drug resistance arises early during the tumorigenesis process. Cancer research. 2007;67(3):1130–1137. doi: 10.1158/0008-5472.CAN-06-2574. [DOI] [PubMed] [Google Scholar]
- 7.Cosse JP, Michiels C. Tumour hypoxia affects the responsiveness of cancer cells to chemotherapy and promotes cancer progression. Anti-cancer agents in medicinal chemistry. 2008;8(7):790–797. doi: 10.2174/187152008785914798. [DOI] [PubMed] [Google Scholar]
- 8.Brahimi-Horn MC, Chiche J, Pouyssegur J. Hypoxia and cancer. Journal of molecular medicine (Berlin, Germany) 2007;85(12):1301–1307. doi: 10.1007/s00109-007-0281-3. [DOI] [PubMed] [Google Scholar]
- 9.Guppy M. The hypoxic core: a possible answer to the cancer paradox. Biochemical and biophysical research communications. 2002;299(4):676–680. doi: 10.1016/s0006-291x(02)02710-9. [DOI] [PubMed] [Google Scholar]
- 10.Hockel M, Vaupel P. Tumor hypoxia: definitions and current clinical, biologic, and molecular aspects. Journal of the National Cancer Institute. 2001;93(4):266–276. doi: 10.1093/jnci/93.4.266. [DOI] [PubMed] [Google Scholar]
- 11.Kizaka-Kondoh S, Inoue M, Harada H, Hiraoka M. Tumor hypoxia: a target for selective cancer therapy. Cancer science. 2003;94(12):1021–1028. doi: 10.1111/j.1349-7006.2003.tb01395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Semenza GL. Targeting HIF-1 for cancer therapy. Nature reviews. 2003;3(10):721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 13.Shannon AM, Bouchier-Hayes DJ, Condron CM, Toomey D. Tumour hypoxia, chemotherapeutic resistance and hypoxia-related therapies. Cancer treatment reviews. 2003;29(4):297–307. doi: 10.1016/s0305-7372(03)00003-3. [DOI] [PubMed] [Google Scholar]
- 14.Harris AL. Hypoxia--a key regulatory factor in tumour growth. Nature reviews. 2002;2(1):38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 15.Chinn LW, Kroetz DL. ABCB1 pharmacogenetics: progress, pitfalls, and promise. Clinical pharmacology and therapeutics. 2007;81(2):265–269. doi: 10.1038/sj.clpt.6100052. [DOI] [PubMed] [Google Scholar]
- 16.Gillet JP, Efferth T, Remacle J. Chemotherapy-induced resistance by ATP-binding cassette transporter genes. Biochimica et biophysica acta. 2007;1775(2):237–262. doi: 10.1016/j.bbcan.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nature reviews. 2002;2(1):48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 18.Kimura Y, Morita S, Matsuo M, Ueda K. Mechanism of multidrug recognition by MDR1/ABCB1. Cancer science. 2007;98(9):1303–1310. doi: 10.1111/j.1349-7006.2007.00538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buys TP, Chari R, Lee EH, Zhang M, MacAulay C, Lam S, Lam WL, Ling V. Genetic changes in the evolution of multidrug resistance for cultured human ovarian cancer cells. Genes, chromosomes & cancer. 2007;46(12):1069–1079. doi: 10.1002/gcc.20492. [DOI] [PubMed] [Google Scholar]
- 20.Lemos C, Jansen G, Peters GJ. Drug transporters: recent advances concerning BCRP and tyrosine kinase inhibitors. British journal of cancer. 2008;98(5):857–862. doi: 10.1038/sj.bjc.6604213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franovic A, Gunaratnam L, Smith K, Robert I, Patten D, Lee S. Translational up-regulation of the EGFR by tumor hypoxia provides a nonmutational explanation for its overexpression in human cancer. Proceedings of the National Academy of Sciences of the United States of America; 2007; pp. 13092–13097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rojo F, Albanell J, Rovira A, Corominas JM, Manzarbeitia F. Targeted therapies in breast cancer. Seminars in diagnostic pathology. 2008;25(4):245–261. doi: 10.1053/j.semdp.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 23.Hsieh MY, Yang S, Raymond-Stinz MA, Steinberg S, Vlachos DG, Shu W, Wilson B, Edwards JS. Stochastic simulations of ErbB homo and heterodimerisation: potential impacts of receptor conformational state and spatial segregation. IET systems biology. 2008;2(5):256–272. doi: 10.1049/iet-syb:20070073. [DOI] [PubMed] [Google Scholar]
- 24.Altenberg B, Greulich KO. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics. 2004;84(6):1014–1020. doi: 10.1016/j.ygeno.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 25.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134(5):703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 26.Lopez-Lazaro M. The warburg effect: why and how do cancer cells activate glycolysis in the presence of oxygen? Anti-cancer agents in medicinal chemistry. 2008;8(3):305–312. doi: 10.2174/187152008783961932. [DOI] [PubMed] [Google Scholar]
- 27.Warburg O. Science. 3215. Vol. 124. New York, N.Y: 1956. On respiratory impairment in cancer cells; pp. 269–270. [PubMed] [Google Scholar]
- 28.Young CD, Anderson SM. Sugar and fat – that's where it's at: metabolic changes in tumors. Breast Cancer Res. 2008;10(1):202. doi: 10.1186/bcr1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu H, Savaraj N, Priebe W, Lampidis TJ. Hypoxia increases tumor cell sensitivity to glycolytic inhibitors: a strategy for solid tumor therapy (Model C) Biochemical pharmacology. 2002;64(12):1745–1751. doi: 10.1016/s0006-2952(02)01456-9. [DOI] [PubMed] [Google Scholar]
- 30.Lu H, Forbes RA, Verma A. Hypoxia-inducible factor 1 activation by aerobic glycolysis implicates the Warburg effect in carcinogenesis. The Journal of biological chemistry. 2002;277(26):23111–23115. doi: 10.1074/jbc.M202487200. [DOI] [PubMed] [Google Scholar]
- 31.Lum JJ, Bui T, Gruber M, Gordan JD, DeBerardinis RJ, Covello KL, Simon MC, Thompson CB. The transcription factor HIF-1alpha plays a critical role in the growth factor-dependent regulation of both aerobic and anaerobic glycolysis. Genes & development. 2007;21(9):1037–1049. doi: 10.1101/gad.1529107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robey IF, Lien AD, Welsh SJ, Baggett BK, Gillies RJ. Neoplasia. 4. Vol. 7. New York, N.Y: 2005. Hypoxia-inducible factor-1alpha and the glycolytic phenotype in tumors; pp. 324–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Semenza GL. Hypoxia-inducible factor 1 and cancer pathogenesis. IUBMB life. 2008;60(9):591–597. doi: 10.1002/iub.93. [DOI] [PubMed] [Google Scholar]
- 34.Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25(34):4647–4662. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- 35.Mathupala SP, Ko YH, Pedersen PL. Hexokinase II: cancer's double-edged sword acting as both facilitator and gatekeeper of malignancy when bound to mitochondria. Oncogene. 2006;25(34):4777–4786. doi: 10.1038/sj.onc.1209603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pedersen PL. Warburg, me and Hexokinase 2: Multiple discoveries of key molecular events underlying one of cancers' most common phenotypes, the “Warburg Effect”, i.e., elevated glycolysis in the presence of oxygen. Journal of bioenergetics and biomembranes. 2007;39(3):211–222. doi: 10.1007/s10863-007-9094-x. [DOI] [PubMed] [Google Scholar]
- 37.Pedersen PL. Voltage dependent anion channels (VDACs): a brief introduction with a focus on the outer mitochondrial compartment's roles together with hexokinase-2 in the “Warburg effect” in cancer. Journal of bioenergetics and biomembranes. 2008;40(3):123–126. doi: 10.1007/s10863-008-9165-7. [DOI] [PubMed] [Google Scholar]
- 38.Floridi A, Paggi MG, D'Atri S, De Martino C, Marcante ML, Silvestrini B, Caputo A. Effect of lonidamine on the energy metabolism of Ehrlich ascites tumor cells. Cancer research. 1981;41(11 Pt 1):4661–4666. [PubMed] [Google Scholar]
- 39.Giorgioni G, Ruggieri S, Di Stefano A, Sozio P, Cinque B, Di Marzio L, Santoni G, Claudi F. Glycosyl and polyalcoholic prodrugs of lonidamine. Bioorganic & medicinal chemistry letters. 2008;18(7):2445–2450. doi: 10.1016/j.bmcl.2008.02.046. [DOI] [PubMed] [Google Scholar]
- 40.Ravagnan L, Marzo I, Costantini P, Susin SA, Zamzami N, Petit PX, Hirsch F, Goulbern M, Poupon MF, Miccoli L, Xie Z, Reed JC, Kroemer G. Lonidamine triggers apoptosis via a direct, Bcl-2-inhibited effect on the mitochondrial permeability transition pore. Oncogene. 1999;18(16):2537–2546. doi: 10.1038/sj.onc.1202625. [DOI] [PubMed] [Google Scholar]
- 41.Biroccio A, Del Bufalo D, Fanciulli M, Bruno T, Zupi G, Floridi A. bcl-2 inhibits mitochondrial metabolism and lonidamine-induced apoptosis in adriamycin-resistant MCF7 cells. International journal of cancer. 1999;82(1):125–130. doi: 10.1002/(sici)1097-0215(19990702)82:1<125::aid-ijc21>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 42.Del Bufalo D, Biroccio A, Soddu S, Laudonio N, D'Angelo C, Sacchi A, Zupi G. Lonidamine induces apoptosis in drug-resistant cells independently of the p53 gene. The Journal of clinical investigation. 1996;98(5):1165–1173. doi: 10.1172/JCI118900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li YC, Fung KP, Kwok TT, Lee CY, Suen YK, Kong SK. Mitochondrial targeting drug lonidamine triggered apoptosis in doxorubicin-resistant HepG2 cells. Life sciences. 2002;71(23):2729–2740. doi: 10.1016/s0024-3205(02)02103-3. [DOI] [PubMed] [Google Scholar]
- 44.Del Bufalo D, Trisciuoglio D, Scarsella M, D'Amati G, Candiloro A, Iervolino A, Leonetti C, Zupi G. Neoplasia. 5. Vol. 6. New York, N.Y: 2004. Lonidamine causes inhibition of angiogenesis-related endothelial cell functions; pp. 513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brawer MK. Lonidamine: basic science and rationale for treatment of prostatic proliferative disorders. Reviews in urology. 2005;7(7):S21–26. [PMC free article] [PubMed] [Google Scholar]
- 46.Ditonno P, Battaglia M, Selvaggio O, Garofalo L, Lorusso V, Selvaggi FP. Clinical Evidence Supporting the Role of Lonidamine for the Treatment of BPH. Reviews in urology. 2005;7(7):S27–33. [PMC free article] [PubMed] [Google Scholar]
- 47.Sinha R, Kim GJ, Nie S, Shin DM. in cancer therapeutics: bioconjugated nanoparticles for drug delivery. Molecular cancer therapeutics. 2006;5(8):1909–1917. doi: 10.1158/1535-7163.MCT-06-0141. [DOI] [PubMed] [Google Scholar]
- 48.Li Z, Zhao R, Wu X, Sun Y, Yao M, Li J, Xu Y, Gu J. Identification and characterization of a novel peptide ligand of epidermal growth factor receptor for targeted delivery of therapeutics. Faseb J. 2005;19(14):1978–1985. doi: 10.1096/fj.05-4058com. [DOI] [PubMed] [Google Scholar]
- 49.Song S, Liu D, Peng J, Sun Y, Li Z, Gu JR, Xu Y. Peptide ligand-mediated liposome distribution and targeting to EGFR expressing tumor in vivo. International journal of pharmaceutics. 2008;363(1-2):155–161. doi: 10.1016/j.ijpharm.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 50.Cheng J, Teply BA, Sherifi I, Sung J, Luther G, Gu FX, Levy-Nissenbaum E, Radovic-Moreno AF, Langer R, Farokhzad OC. Formulation of functionalized PLGA-PEG nanoparticles for in vivo targeted drug delivery. Biomaterials. 2007;28(5):869–876. doi: 10.1016/j.biomaterials.2006.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gu F, Zhang L, Teply BA, Mann N, Wang A, Radovic-Moreno AF, Langer R, Farokhzad OC. Precise engineering of targeted nanoparticles by using self-assembled biointegrated block copolymers. Proceedings of the National Academy of Sciences of the United States of America; 2008; pp. 2586–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Townsend SA, Evrony GD, Gu FX, Schulz MP, Brown RH, Jr, Langer R. Tetanus toxin C fragment-conjugated nanoparticles for targeted drug delivery to neurons. Biomaterials. 2007;28(34):5176–5184. doi: 10.1016/j.biomaterials.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chawla JS, Amiji MM. Biodegradable poly(epsilon -caprolactone) nanoparticles for tumor-targeted delivery of tamoxifen. International journal of pharmaceutics. 2002;249(1-2):127–138. doi: 10.1016/s0378-5173(02)00483-0. [DOI] [PubMed] [Google Scholar]
- 54.Shenoy DB, Amiji MM. Poly(ethylene oxide)-modified poly(epsilon-caprolactone) nanoparticles for targeted delivery of tamoxifen in breast cancer. International journal of pharmaceutics. 2005;293(1-2):261–270. doi: 10.1016/j.ijpharm.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 55.Akhtar S, Benter IF. Nonviral delivery of synthetic siRNAs in vivo. The Journal of clinical investigation. 2007;117(12):3623–3632. doi: 10.1172/JCI33494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gratton SE, Ropp PA, Pohlhaus PD, Luft JC, Madden VJ, Napier ME, DeSimone JM. The effect of particle design on cellular internalization pathways. Proceedings of the National Academy of Sciences of the United States of America; 2008; pp. 11613–11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kingsley JD, Dou H, Morehead J, Rabinow B, Gendelman HE, Destache CJ. Nanotechnology: a focus on nanoparticles as a drug delivery system. J Neuroimmune Pharmacol. 2006;1(3):340–350. doi: 10.1007/s11481-006-9032-4. [DOI] [PubMed] [Google Scholar]
- 58.Moghimi SM, Hunter AC, Murray JC. Nanomedicine: current status and future prospects. Faseb J. 2005;19(3):311–330. doi: 10.1096/fj.04-2747rev. [DOI] [PubMed] [Google Scholar]
- 59.Xu ZP, Niebert M, Porazik K, Walker TL, Cooper HM, Middelberg AP, Gray PP, Bartlett PF, Lu GQ. Subcellular compartment targeting of layered double hydroxide nanoparticles. J Control Release. 2008;130(1):86–94. doi: 10.1016/j.jconrel.2008.05.021. [DOI] [PubMed] [Google Scholar]
- 60.Dittmann K, Mayer C, Kehlbach R, Rodemann HP. Radiation-induced caveolin-1 associated EGFR internalization is linked with nuclear EGFR transport and activation of DNA-PK. Molecular cancer. 2008;7:69. doi: 10.1186/1476-4598-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Orth JD, Krueger EW, Weller SG, McNiven MA. A novel endocytic mechanism of epidermal growth factor receptor sequestration and internalization. Cancer research. 2006;66(7):3603–3610. doi: 10.1158/0008-5472.CAN-05-2916. [DOI] [PubMed] [Google Scholar]
- 62.Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, Chang G. Science. Vol. 323. RCSB Protein Data Bank; 2009. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding; pp. 1718–1722. PDB ID: 3G61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bayrhuber M, Meins T, Habeck M, Becker S, Giller K, Villinger S, Vonrhein C, Griesinger C, Zweckstetter M, Zeth K. Proc Natl Acad Sci USA. Vol. 105. RCSB Protein Data Bank; 2009. Structure of the Human Voltage-Dependent Anion Channel; pp. 15370–15375. PDB ID: 2JK4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bruncko M, Oost TK, Belli BA, Ding H, Joseph MK, Kunzer A, Martineau D, McClellan WJ, Mitten M, Ng SC, Nimmer PM, Oltersdorf T, Park CM, Petros AM, Shoemaker AR, Song X, Wang X, Wendt MD, Zhang H, Fesik SW, Rosenberg SH, Elmore SW. J Med Chem. Vol. 50. RCSB Protein Data Bank; 2007. Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-xL; pp. 641–662. PDB ID: 2O21. [DOI] [PubMed] [Google Scholar]
- 65.Card PB, Erbel PJ, Gardner KH. J Mol Biol. Vol. 353. RCSB Protein Data Bank; 2005. Structural Basis of ARNT PAS-B Dimerization: Use of a Common Beta-sheet Interface for Hetero- and Homodimerization; pp. 664–677. PDB ID: 1X0O. [DOI] [PubMed] [Google Scholar]
- 66.Hon WC, Wilson MI, Harlos K, Claridge TD, Schofield CJ, Pugh CW, Maxwell PH, Ratcliffe PJ, Stuart DI, Jones EY. Nature. Vol. 417. RCSB Protein Data Bank; 2002. Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL; pp. 975–978. PDB ID: 1LQB. [DOI] [PubMed] [Google Scholar]
- 67.Nury H, Dahout-Gonzalez C, Trezeguet V, Lauquin G, Brandolin G, Pebay-Peyroula E. FEBS Lett. Vol. 579. RCSB Protein Data Bank; 2005. Structural Basis for Lipid-Mediated Interactions between Mitochondrial Adp/ATP Carrier Monomers; p. 6031. PDB ID: 2C3E. [DOI] [PubMed] [Google Scholar]
- 68.Rabeh WM, Zhu H, Nedyalkova L, Tempel W, Wasney G, Landry R, Vedadi M, Arrowsmith CH, Edwards AM, Sundstrom M, Weigelt J, Bochkarev A, Park H. Structural Genomics Consortium (SGC) Crystal structure of human hexokinase II. RCSB Protein Data Bank; 2006. PDB ID: 2NZT. [Google Scholar]
- 69.Ramaen O, Leulliot N, Sizun C, Ulryck N, Pamlard O, Lallemand JY, Van Tilbeurgh H, Jacquet E. J Mol Biol. Vol. 359. RCSB Protein Data Bank; 2006. Structure of the Human Multidrug Resistance Protein 1 Nucleotide Binding Domain 1 Bound to Mg(2+)/ATP Reveals a Non-Productive Catalytic Site; p. 940. PDB ID: 2CBZ. [DOI] [PubMed] [Google Scholar]
- 70.Read JA, Winter VJ, Eszes CM, Sessions RB, Brady RL. Proteins. Vol. 43. RCSB Protein Data Bank; 2001. Structural basis for altered activity of M- and H-isozyme forms of human lactate dehydrogenase; pp. 175–185. PDB ID: 1I0Z. [DOI] [PubMed] [Google Scholar]
- 71.Verdon G, Albers SV, Dijkstra BW, Driessen AJ, Thunnissen AM. J Mol Biol. Vol. 330. RCSB Protein Data Bank; 2003. Crystal structures of the ATPase subunit of the glucose ABC transporter from Sulfolobus solfataricus: nucleotide-free and nucleotide-bound conformations; pp. 343–358. PDB ID: 1OXT. [DOI] [PubMed] [Google Scholar]
- 72.Yun CH, Boggon TJ, Li Y, Woo S, Greulich H, Meyerson M, Eck MJ. Cancer Cell. Vol. 11. RCSB Protein Data Bank; 2007. Structures of Lung Cancer-Derived Egfr Mutants and Inhibitor Complexes: Mechanism of Activation and Insights Into Differential Inhibitor Sensitivity; p. 217. PDB ID: 2ITY. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Eder M, Fritz-Wolf K, Kabsch W, Wallimann T, Schlattner U. Proteins: Struct Funct Genet. Vol. 39. RCSB Protein Data Bank; 2000. Crystal Structure of Human Ubiquitous Mitochondrial Creatine Kinase; p. 216. PDB ID: 1QK1. [DOI] [PubMed] [Google Scholar]
- 74.Rastogi VK, Girvin ME. Nature. Vol. 402. RCSB Protein Data Bank; 1999. Structural changes linked to proton translocation by subunit c of the ATP synthase; pp. 263–268. PDB ID: 1C17. [DOI] [PubMed] [Google Scholar]
- 75.Schlatter D, Thoma R, Kueng E, Stihle M, Mueller F, Boroni E, Hennig M. Acta Crystallogr Sect D. Vol. 61. RCSB Protein Data Bank; 2005. Crystal Engineering Yields Crystals of Cyclophilin D Diffracting to 1.7 A Resolution; p. 513. PDB ID: 2BIT. [DOI] [PubMed] [Google Scholar]