Summary
Background and objectives
Relapsing and recurrent peritonitis episodes are important causes of treatment failure in patients undergoing peritoneal dialysis (PD). This study explored whether the level of bacteria-derived DNA fragment in PD effluent predicts the development of relapsing or recurrent peritonitis.
Design, setting, participants, & measurements
The study included 143 patients with PD peritonitis in a dialysis unit between September 2010 and December 2011. Every 5 days until antibiotic treatment ended, PD effluent was collected to determine bacteria-derived DNA fragment level, which is represented by the number of PCR cycles at which bacterial DNA could be detected. Patients were followed for the development of relapsing or recurrent peritonitis.
Results
Thirty-nine patients were excluded because of immediate treatment failure or incorrect diagnosis. Of the other 104 patients, 15 (14.4%) developed relapsing peritonitis, 3 (2.9%) had recurrent peritonitis, and 5 (4.8%) had repeat episodes. Patients with relapsing or recurrent peritonitis episodes had significantly higher levels of bacterial DNA fragment in PD effluent than those without relapsing or recurrence, both 5 days before (31.9±3.4 versus 34.3±3.0 cycles; P=0.002) and on the day of (32.3±2.6 versus 34.1±1.7 cycles; P<0.001) completion of antibiotics. When bacterial DNA fragment detectable by 34 PCR cycles 5 days before the completion of antibiotics is used as the cutoff, it has a sensitivity of 88.9% and specificity of 60.5% for the prediction of relapsing or recurrent peritonitis.
Conclusions
Bacterial DNA fragment levels in PD effluent are significantly higher, both 5 days before and on the date of completion of antibiotics, among patients who subsequently develop relapsing or recurrent peritonitis than among those cured by antibiotics. Further studies are needed to validate these results and confirm the clinical utility of dialysate bacterial DNA fragment level.
Introduction
Peritoneal dialysis (PD) is the first-line treatment of ESRD in Hong Kong (1). Despite the advances in antibiotic therapy and connecting system, peritonitis remains a major complication of PD. Although <4% of the peritonitis episodes resulted in death, peritonitis is a “contributing factor” to death in 16% of deaths in PD patients in Hong Kong (2). Furthermore, peritonitis is the most common cause of treatment failure in PD (1,3,4). In this regard, relapsing and recurrent peritonitis episodes are a major cause of peritoneal failure. Our recent study shows that around 15% of all PD-related peritonitis episodes are followed by relapsing or recurrent peritonitis, often resulting in prolonged hospitalization, expensive treatment, need of catheter removal, and conversion to hemodialysis (5).
Our previous study showed that patients with relapsing peritonitis were younger and had a significantly lower Charlson comorbidity score than the other patients (5). It is also known that peritonitis episodes caused by Staphylococcus aureus and Pseudomonas species have a high risk of relapse (6,7). Timely implementation of a change in therapy for high-risk patients would, at least in theory, be valuable for the prevention of peritonitis recurrence. However, there is no accurate laboratory test to tell which patient is going to develop relapsing or recurrent peritonitis after completion of antibiotic treatment.
Recently, Schindler et al. (8) reported that short bacteria-derived DNA fragments are present in clinically used fluids, such as dialysis fluid. DNA fragments are thought to be derived from microorganisms inhabiting body fluid (9). Most of these microorganisms, including potential pathogens, might subsist in a viable but not culturable state or may need specific culture media (10). Because bacteria-derived DNA fragments could be detected by real-time quantitative PCR (RT-QPCR) with excellent sensitivity and universal primers could be used to detect most of the pathogenic bacteria without a priori knowledge of the bacteriologic cause, it represents an attractive means to test for the presence of viable bacteria in biologic specimens. For example, Bruns et al. (11) and Rogers et al. (12) both showed that fragments of bacterial nucleic acid could be identified in the ascitic fluid of cirrhotic patients. However, there are no published studies on the level of bacteria-derived DNA fragment in the dialysis effluent of PD patients.
The aim of this study is to compare the level of bacteria-derived DNA fragments in PD effluent between patients with and without relapsing peritonitis and to establish the cutoff level of bacteria-derived DNA fragments in PD effluent for the prediction of relapsing or recurrent peritonitis.
Materials and Methods
Case Selection
We recruited 143 patients with PD peritonitis in our dialysis unit between September 2010 and December 2011. The diagnosis of peritonitis was based on at least two of the following (13): (1) abdominal pain or cloudy PD effluent; (2) leukocytosis in PD effluent (white blood cell count >100 cells/ml); and (3) positive Gram stain or culture from PD effluent. Patients with fungal peritonitis or obvious surgical problems who required laparotomy were excluded. All patients were treated according to the guideline of the International Society for Peritoneal Dialysis (13). We did not recruit patients with repeat and recurrent episodes or with peritonitis caused by Mycobacterium or fungal species. Patients were excluded from final analysis if they had early treatment failure or if the diagnosis of peritonitis was not confirmed. In general, all patients received 2 weeks of effective antibiotics, except for episodes caused by S. aureus or Pseudomonas species, which were treated for 3 weeks.
After written inform consent, PD effluent samples were collected every 5 days until the antibiotic treatment was completed. The samples were used to test bacteria-derived DNA fragments as well as to perform routine cell counts and ordinary bacterial cultures. This study was approved by the Joint Chinese University of Hong Kong–New Territories East Cluster Clinical Research Ethics Committee. All experimentation procedures are in compliance with the Declaration of Helsinki.
DNA Extraction from PD Effluent
The method of extraction of bacterial DNA from biologic fluid has been described previously (14). DNA from 2 ml PD effluent was extracted using the EZ1 DNA tissue kit and BioRobot EZ1 with the EZ1 bacteria card (Qiagen), according to the manufacturer’s instructions. Purified DNA was eluted in 50 µl of elution buffer before amplification.
Bacterial DNA Amplification
The method of bacterial DNA amplification has been described previously (14,15). Briefly, bacterial DNA was quantified by RT-QPCR using the ABI Prism 7900 Sequence Detection System (Applied Biosystems, Foster City, CA). Commercially available Taqman primers and probes, including two unlabeled primers and one FAM dye-labeled TaqMan MGB probe were used (all from Applied Biosystems). Unlabeled primers used for RT-QPCR amplification of the bacterial 16S rRNA gene were p16SrRNA+ and p16SrRNA−, which are able to amplify DNA from gram-positive or gram-negative bacteria. Previous studies have demonstrated that amplification of fragments of the 16S rRNA gene is both sensitive and specific for detection of DNA from all of the well known species of bacteria (15–19). Each sample was run in triplicate. RT-QPCR were performed at 50°C for 2 minutes and 95°C for 10 minutes, followed by 40 cycles at 95°C for 45 seconds, 53°C for 1 minute, and 72°C for 1 minute. Because PD effluent was directly used as the template and there is no intrinsic housekeeping gene for comparison, the number of RT-QPCR cycles at which bacterial DNA could be detected is reported.
Outcome Measures
All patients were followed for 1 year after completion of antibiotic therapy. The study did not affect their clinical management. The primary end point of this study was relapsing and recurrent peritonitis. Relapsing peritonitis was defined as an episode that occurs within 4 weeks of completion of therapy of a prior episode with the same organism (or culture negative in the second episode) (13). Recurrent peritonitis was defined as an episode that occurs within 4 weeks of completion of therapy of a prior episode but with a different organism (13). Secondary outcomes include repeat peritonitis, catheter removal, conversion to long-term hemodialysis, death due to peritonitis, and all-cause mortality. Repeat peritonitis was defined as an episode that occurs more than 4 weeks after completion of therapy of a prior episode with the same organism (13).
Statistical Analyses
Statistical analyses were performed by SPSS for Windows software, version 15.0 (SPSS Inc., Chicago, IL). Descriptive data are represented as mean ± SD. Demographic data were compared by one-way ANOVA or unpaired t test as appropriate. To increase the number of events and improve the statistical power of the study, relapsing and recurrent episodes were decided a priori to be considered as one group for analysis. Because the result of bacterial DNA fragment level is skewed, data were compared by Kruskal-Wallis test or Mann-Whitney U test as appropriate. Receiver-operating characteristic (ROC) curves were constructed by standard methods. A P value <0.05 was considered to represent a statistically significant difference. All probabilities are two tailed.
Results
We included 143 peritonitis episodes. Early treatment failure was noted in 31 patients (11 died during antibiotic treatment; 20 had catheter removal because of refractory peritonitis). Another 8 patients were excluded because the diagnosis of peritonitis was not confirmed. The baseline clinical information of the remaining 104 patients is summarized in Table 1. Five patients were treated with machine-assisted PD, and the other 99 patients had continuous ambulatory PD. All patients undergoing continuous ambulatory PD were using double-bag disconnect systems; 13 were using PD solutions with low glucose degradation product. Of the 104 patients, 50 (48.1%) had no history of peritonitis; the median number of previous peritonitis episode was 1 (interquartile range, 0–2).
Table 1.
Baseline demographic and clinical data
| Variable | Relapsing/ Recurrent Peritonitis | P Value | |
|---|---|---|---|
| Yes | No | ||
| Patients (n) | 18 | 86 | |
| Men/women (n/n) | 13/5 | 51/35 | 0.31 |
| Age (yr) | 59.7±14.8 | 61.5±12.9 | 0.60 |
| Duration of dialysis (mo)a | 37.8 (19.3–70.2) | 40.3 (15.5–78.2) | 0.54 |
| Body height (cm) | 160.3±7.9 | 161.4±8.1 | 0.60 |
| Body weight (kg) | 61.7±17.3 | 61.9±12.6 | 0.96 |
| Cases of renal diagnosis, n (%) | 0.06 | ||
| GN | 4 (22.2) | 23 (26.7%) | |
| Diabetic nephropathy | 3 (16.7) | 33 (38.4%) | |
| Hypertensive nephrosclerosis | 3 (16.7) | 9 (10.5%) | |
| Polycystic kidney | 0 | 6 (7.0%) | |
| Obstructive uropathy | 3 (16.7) | 3 (3.5%) | |
| Others/unknown | 3 (16.7) | 11 (12.8%) | |
| Cases of comorbid conditions, n (%) | |||
| Diabetes | 6 (33.3) | 40 (46.5%) | 0.31 |
| Coronary heart disease | 1 (5.6) | 19 (22.1%) | 0.11 |
| Cerebrovascular disease | 5 (27.8) | 24 (27.9%) | 0.99 |
| Peripheral vascular disease | 1 (5.6) | 10 (11.6%) | 0.44 |
| Charlson comorbidity index | 5.6±2.6 | 5.9±2.9 | 0.60 |
Values expressed with a plus/minus sign are the mean ± SD.
Data presented as median (interquartile range).
The microbiological causes of the peritonitis episode are summarized in Table 2. Five patients had concomitant exit site infection. Of the 104 episodes, 95 received cefazolin and ceftazidime as the initial empirical antibiotics cover, while the other 9 received vancomycin and gentamicin (mostly because of suspected penicillin allergy). After initial response to antibiotics, 15 patients (14.4%) developed relapsing episodes, 3 (2.9%) had recurrent episodes, and 5 (4.8%) had repeat episodes. Their microbiological causes are further summarized in Table 2.
Table 2.
Microbiological causes of peritonitis episode
| Variable | All, n (%) | Relapsing, n | Recurrent, n | Repeat, n |
|---|---|---|---|---|
| Total | 104 | 15 | 3 | 5 |
| Gram-positive organisms | ||||
| Staphylococcus aureus | 13 (12.5) | 2 | 0 | 2 |
| CNSS | 14 (13.5) | 4 | 2 | 1 |
| Streptococcus species | 11 (10.6) | 0 | 0 | 0 |
| Gram-negative organisms | ||||
| Pseudomonas species | 6 (5.8) | 2 | 0 | 1 |
| Enterobacteriaceae species | 25 (24.0) | 2 | 1 | 1 |
| Polymicrobial | 13 (12.5) | 0 | 0 | 0 |
| Culture negative | 22 (21.2) | 5 | 0 | 0 |
CNSS, coagulase-negative staphylococcal species.
Bacterial DNA Fragments in PD Effluent
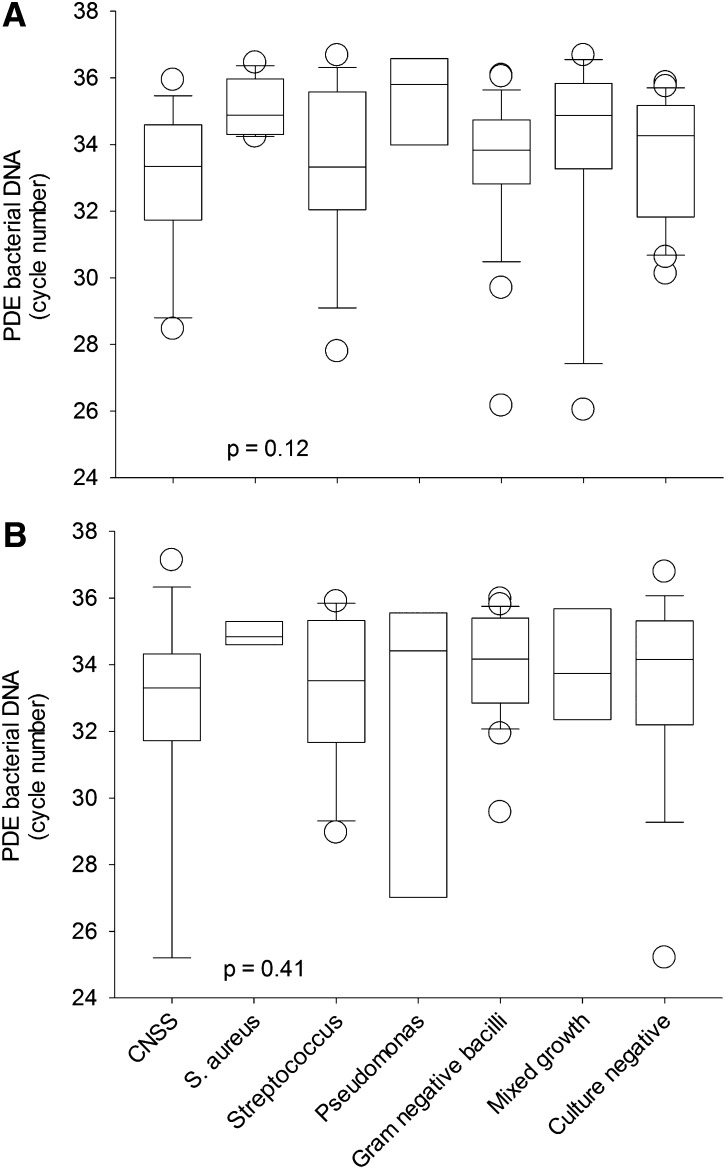
The PD effluent levels of bacterial DNA fragments among different causative organisms are compared and summarized in Figure 1. In short, there was no relation between PD effluent bacterial DNA fragment levels and the causative organisms, both 5 days before and on the day of completion of antibiotic treatment. On the day of completion of antibiotic treatment, PD effluent levels of bacterial DNA fragments in patients without relapsing or recurrence and those with relapsing, recurrent, and repeat peritonitis episodes were 34.1±1.7, 32.5±2.6, 31.3±2.7, and 33.7±1.0 cycles, respectively (Kruskal-Wallis test, P=0.006). Similarly, 5 days before the completion of antibiotic treatment, PD effluent levels of bacterial DNA fragment of these groups were 34.3±3.0, 31.6±3.6, 33.3±1.9, and 33.0±1.8 cycles, respectively (P=0.01). In contrast, PD effluent levels of bacterial DNA fragment at earlier time points were similar between patients without relapsing or recurrence, those with relapsing, recurrent, and repeat peritonitis episodes (details not shown). All patients in this study had a PD effluent nucleated cell count <20 cells/mm3, both 5 days before and on the day of completion of antibiotic treatment. PD effluent nucleated cell count at earlier time points did not predict subsequent development of relapsing or recurrent peritonitis (details not shown).
Figure 1.
Comparison of bacterial DNA fragment levels in peritoneal dialysis effluent (PDE). (A) Five days before and (B) on the day of completion of antibiotic therapy, among different causative organisms. Bacterial DNA fragment levels are expressed as the number of polymerase chain reaction (PCR) cycles; a lower cycle number indicates a higher level of bacterial DNA level. In whisker-box plots, boxes indicate median, 25th and 75th percentiles, and whiskers indicate 5th and 95th percentiles. Data were compared by Kruskal-Wallis test. CNSS, coagulase-negative staphylococcal species.
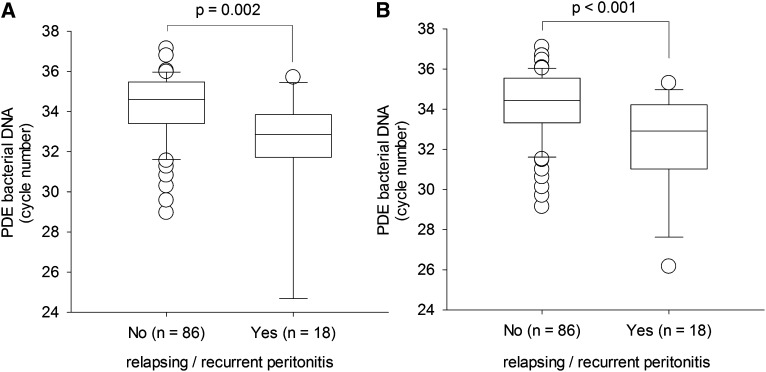
Patients with relapsing or recurrent peritonitis episodes had significantly higher levels of bacterial DNA fragments in PD effluent than those without relapsing or recurrent peritonitis, both 5 days before (31.9±3.4 versus 34.3±3.0 cycles; Mann-Whitney U test, P=0.002) and on the day of (32.3±2.6 versus 34.1±1.7 cycles; P<0.001) completion of antibiotic treatment (Figure 2).
Figure 2.
Comparison of bacterial DNA fragment levels in peritoneal dialysis effluent (PDE). (A) Five days before and (B) on the day of completion of antibiotic therapy, between patients with and without relapsing or recurrent peritonitis episodes. Bacterial DNA fragment levels are expressed as the number of polymerase chain reaction (PCR) cycles; a lower cycle number indicates a higher level of bacterial DNA level. In whisker-box plots, boxes indicate median, 25th and 75th percentiles, and whiskers indicate 5th and 95th percentiles. Data are compared by Mann-Whitney U test.
Accuracy of Prediction
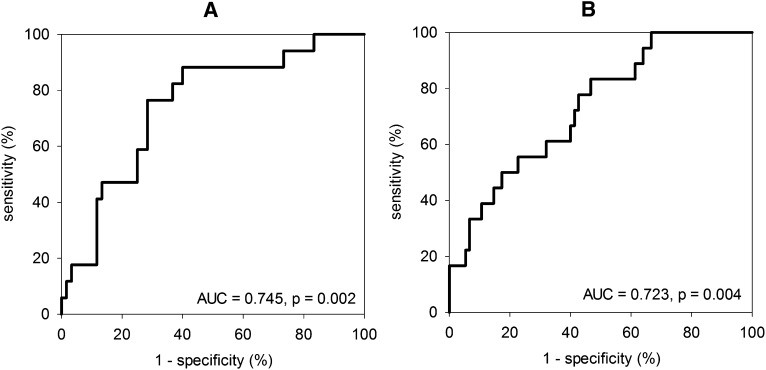
We further explored the performance of bacterial DNA fragment levels in PD effluent 5 days before and on the day of completion of antibiotic therapy for the prediction of relapsing or recurrent peritonitis episodes by constructing two ROC curves (Figure 3). In essence, the areas under the ROC curve (AUC) at both time points are statistically significant, with the AUC for the bacterial DNA fragment levels 5 days before the completion of antibiotic therapy being slightly higher. Further analysis showed that when bacterial DNA fragment detectable by 34 RT-QPCR cycles 5 days before the completion of antibiotic therapy is used as the cutoff, it has a sensitivity of 88.9% and specificity of 60.5% for the prediction of relapsing or recurrent peritonitis.
Figure 3.
Receiver-operating characteristic (ROC) curves indicating the performance of bacterial DNA fragment levels in peritoneal dialysis effluent (PDE). (A) Five days before and (B) on the day of completion of antibiotic therapy, for the prediction of relapsing or recurrent peritonitis episodes. AUC, area under curve.
Discussion
In this study, we found that bacterial DNA fragment levels in PD effluent are significantly higher, both 5 days before and on the date of completion of antibiotic therapy, among patients who subsequently developed relapsing or recurrent peritonitis episodes than among those without relapsing or recurrent peritonitis. On the basis of our data, bacterial DNA fragment detectable by 34 RT-QPCR cycles 5 days before the completion of antibiotic therapy is a sensitive and fairly specific cutoff for the prediction of relapsing or recurrent peritonitis.
At present, there is no accurate laboratory test to determine which patient is going to develop relapsing or recurrent peritonitis episodes, important causes of peritoneal failure in PD patients (5). Previous studies showed that the white blood cell count in dialysate 3 days after antibiotic treatment is a reliable predictor of immediate treatment failure (catheter removal or peritonitis-related death) (20), while patients with relapsing peritonitis tend to be younger, have a lower Charlson comorbidity score, and have peritonitis caused by S. aureus or Pseudomonas species (5–7). However, these studies did not identify any diagnostic test for the prediction of relapsing or recurrent peritonitis; our study represents an early effort in this area.
In general, the distribution of causative organisms of the peritonitis episodes is similar to that in our previous study (4), and the incidence of concomitant exit site infections is also consistent with our previous in-house data. The proportion of culture-negative cases in this study is apparently higher than that in our previous report (21), probably because the index episodes of our present study exclude repeat and recurrent episodes, as well as peritonitis caused by Mycobacterium or fungal species. Although we did not exclude more episodes caused by Pseudomonas species, the incidence of Pseudomonas peritonitis in this study is lower than in our previous report (6), so much so that Pseudomonas species do not appear to be a common cause of relapsing peritonitis.
It is important to note that the presence of bacterial DNA fragment in PD effluent does not indicate the presence of living bacteria capable of causing an active infection. Because standard PD effluent bacterial culture is negative, it seems unlikely to represent the persistence of viable bacteria in the effluent. However, bacterial DNA fragment per se, irrespective of the presence of viable intact bacteria, could induce local inflammatory response, although we do not have data on the PD effluent levels of inflammatory cytokines. An elevated PD effluent bacterial DNA fragment level may imply inadequate antibiotic treatment or the presence of viable bacteria in a protected site (e.g., biofilm on the PD catheter). On the other hand, intraperitoneal sequestration of bacteria is also possible. A previous study showed that mesothelial cells can ingest S. aureus, and the ingested bacteria proliferated within mesothelial cells, which may be released subsequently (22). Another study found that after host cell invasion, S. aureus resided within phagocytic vacuoles, and a high percentage of the ingested bacteria remained alive within mesothelial cells and might be released after host cell death, resulting in relapsing peritonitis (23).
In the present study, bacterial DNA fragment levels in PD effluent are presented as the number of RT-QPCR cycles. Our preliminary study showed that bacterial DNA fragment is marginally detectable in PD effluent from stable PD patients without peritonitis, as well as from PD effluent 1 month after antibiotic treatment for an episode of peritonitis has been completed (Szeto CC, unpublished data). Ideally, control samples with a known amount of bacterial DNA could be used to construct a standard curve, and the DNA fragment level could then be calculated as the number of copies per volume of sample. However, because of the limitations in our original study design and limited volume of archive samples, we cannot determine and report the exact amount of bacterial DNA in terms of copies per milliliter. We believe it is reasonable to report the RT-QPCR cycle number rather than the exact DNA copy number and standardize the sample volume and experimental procedure, although the new-generation technology of PCR in combination with electrospray ionization/mass spectrometry would greatly facilitate the precise quantification of bacterial DNA fragments in biologic samples.
We found that PD effluent bacterial DNA levels 5 days before and on the day of completion of antibiotics were both useful for the prediction of relapsing or recurrent peritonitis. It should be noted that in order to conform with the latest treatment guideline, the duration of antibiotic treatment was not identical for all recruited patients. We favor the use of DNA levels 5 days before antibiotic treatment is stopped because it is shorter than the turnaround time for the test. If bacterial DNA fragment levels are validated for clinical use, the test could be performed a few days before treatment is planned to stop; if the result suggests a high risk of relapsing or recurrent peritonitis, a prolonged course of antibiotic or other therapy (e.g., intraperitoneal fibrinolytic agent) could be used for prevention. This hypothesis, however, needs to be tested by further study. On the basis of our result, when bacterial DNA fragment detectable by 34 RT-QPCR cycles 5 days before the completion of antibiotic is used as the cutoff, the positive predictive value is 32%. If a therapeutic intervention is 50% effective, one episode of recurrent or relapsing peritonitis could be prevented for eight patients being treated; a randomized controlled trial with a sample size 135 cases (allowing for 10% dropout) would be needed to test this hypothesis.
This study had several limitations. First, the sample size is small, which reduces the significance and clinical importance of our conclusion. The study would need many more cases in order to be large enough to allow elaborated multivariate analysis to adjust for other confounding clinical factors, such as the presence of exit site infection or S. aureus carrier status. In theory, our technique may be particularly useful for guiding the management of culture-negative peritonitis, but the number of such cases was small in our study and no definite conclusion could be made in this regard. For the same reason, we cannot detect possible difference in PD effluent bacterial DNA levels between patients with and without exit site infection, different antibiotic protocols, or different bacteriologic causes. Similarly, we cannot perform reliable subgroup analysis for relapsing and recurrent episodes or for whether the prognostic role of PD effluent bacterial DNA fragment level is generally applicable or is being confined to peritonitis episodes caused by specific organisms (for example, S. aureus).
Because all patients in this study came from a single PD center, external validity needs to be substantiated before the result can be extrapolated to other centers. Moreover, the accuracy of PD effluent bacterial DNA fragment level may not be ideal. In general, the AUC <0.8 may not be helpful for clinical use. Further studies are urgently needed for a more accurate quantification of bacterial DNA fragment level in PD effluent so that a reliable diagnostic cutoff can be identified. In addition, a second set of patients is also needed for validation before bacterial DNA fragment levels in PD effluent can be applied for routine clinical use.
Disclosures
C.C.S. reported receiving research grant and consultancy from Baxter Healthcare.
Acknowledgments
This study was supported in part by the Research Fund for the Control of Infectious Disease (RFCID) of the Food and Health Bureau, Government of Hong Kong, and the Chinese University of Hong Kong (CUHK) research account 6901031. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
References
- 1.Szeto CC, Wong TY, Leung CB, Wang AY, Law MC, Lui SF, Li PK: Importance of dialysis adequacy in mortality and morbidity of chinese CAPD patients. Kidney Int 58: 400–407, 2000 [DOI] [PubMed] [Google Scholar]
- 2.Szeto CC, Wong TY, Chow KM, Leung CB, Li PK: Are peritoneal dialysis patients with and without residual renal function equivalent for survival study? Insight from a retrospective review of the cause of death. Nephrol Dial Transplant 18: 977–982, 2003 [DOI] [PubMed] [Google Scholar]
- 3.Szeto CC, Chow KM, Wong TY, Leung CB, Wang AY, Lui SF, Li PK: Feasibility of resuming peritoneal dialysis after severe peritonitis and Tenckhoff catheter removal. J Am Soc Nephrol 13: 1040–1045, 2002 [DOI] [PubMed] [Google Scholar]
- 4.Szeto CC, Leung CB, Chow KM, Kwan BC, Law MC, Wang AY, Lui SF, Li PK: Change in bacterial aetiology of peritoneal dialysis-related peritonitis over 10 years: Experience from a centre in South-East Asia. Clin Microbiol Infect 11: 837–839, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szeto CC, Kwan BC, Chow KM, Law MC, Pang WF, Chung KY, Leung CB, Li PK: Recurrent and relapsing peritonitis: Causative organisms and response to treatment. Am J Kidney Dis 54: 702–710, 2009 [DOI] [PubMed] [Google Scholar]
- 6.Szeto CC, Chow KM, Leung CB, Wong TY, Wu AK, Wang AY, Lui SF, Li PK: Clinical course of peritonitis due to Pseudomonas species complicating peritoneal dialysis: A review of 104 cases. Kidney Int 59: 2309–2315, 2001 [DOI] [PubMed] [Google Scholar]
- 7.Szeto CC, Chow KM, Kwan BC, Law MC, Chung KY, Yu S, Leung CB, Li PK: Staphylococcus aureus peritonitis complicates peritoneal dialysis: Review of 245 consecutive cases. Clin J Am Soc Nephrol 2: 245–251, 2007 [DOI] [PubMed] [Google Scholar]
- 8.Schindler R, Beck W, Deppisch R, Aussieker M, Wilde A, Göhl H, Frei U: Short bacterial DNA fragments: Detection in dialysate and induction of cytokines. J Am Soc Nephrol 15: 3207–3214, 2004 [DOI] [PubMed] [Google Scholar]
- 9.Gomila M, Gascó J, Busquets A, Gil J, Bernabeu R, Buades JM, Lalucat J: Identification of culturable bacteria present in haemodialysis water and fluid. FEMS Microbiol Ecol 52: 101–114, 2005 [DOI] [PubMed] [Google Scholar]
- 10.Gomila M, Gascó J, Gil J, Bernabeu R, Iñigo V, Lalucat J: A molecular microbial ecology approach to studying hemodialysis water and fluid. Kidney Int 70: 1567–1576, 2006 [DOI] [PubMed] [Google Scholar]
- 11.Bruns T, Sachse S, Straube E, Assefa S, Herrmann A, Hagel S, Lehmann M, Stallmach A: Identification of bacterial DNA in neutrocytic and non-neutrocytic cirrhotic ascites by means of a multiplex polymerase chain reaction. Liver Int 29: 1206–1214, 2009 [DOI] [PubMed] [Google Scholar]
- 12.Rogers GB, Russell LE, Preston PG, Marsh P, Collins JE, Saunders J, Sutton J, Fine D, Bruce KD, Wright M: Characterisation of bacteria in ascites—reporting the potential of culture-independent, molecular analysis. Eur J Clin Microbiol Infect Dis 29: 533–541, 2010 [DOI] [PubMed] [Google Scholar]
- 13.Li PKT, Szeto CC, Piraino B, Bernardini J, Figueiredo AE, Gupta A, Johnson DW, Kuijper EJ, Lye WC, Salzer W, Schaefer F, Struijk DG, International Society for Peritoneal Dialysis : Peritoneal dialysis-related infections recommendations: 2010 update. Perit Dial Int 30: 393–423, 2010 [DOI] [PubMed] [Google Scholar]
- 14.Kwan BC, Chow KM, Leung CB, Law MC, Cheng PM, Yu V, Li PK, Szeto CC: Circulating bacterial-derived DNA fragments as a marker of systemic inflammation in peritoneal dialysis [published online ahead of print June 5, 2013]. Nephrol Dial Transplant doi: 10.1093/ndt/gft100 [DOI] [PubMed]
- 15.Bossola M, Sanguinetti M, Scribano D, Zuppi C, Giungi S, Luciani G, Torelli R, Posteraro B, Fadda G, Tazza L: Circulating bacterial-derived DNA fragments and markers of inflammation in chronic hemodialysis patients. Clin J Am Soc Nephrol 4: 379–385, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weisburg WG, Barns SM, Pelletier DA, Lane DJ: 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173: 697–703, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sleigh J, Cursons R, La Pine M: Detection of bacteraemia in critically ill patients using 16S rDNA polymerase chain reaction and DNA sequencing. Intensive Care Med 27: 1269–1273, 2001 [DOI] [PubMed] [Google Scholar]
- 18.Warwick S, Wilks M, Hennessy E, Powell-Tuck J, Small M, Sharp J, Millar MR: Use of quantitative 16S ribosomal DNA detection for diagnosis of central vascular catheter-associated bacterial infection. J Clin Microbiol 42: 1402–1408, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothman RE, Majmudar MD, Kelen GD, Madico G, Gaydos CA, Walker T, Quinn TC: Detection of bacteremia in emergency department patients at risk for infective endocarditis using universal 16S rRNA primers in a decontaminated polymerase chain reaction assay. J Infect Dis 186: 1677–1681, 2002 [DOI] [PubMed] [Google Scholar]
- 20.Chow KM, Szeto CC, Cheung KK, Leung CB, Wong SS, Law MC, Ho YW, Li PK: Predictive value of dialysate cell counts in peritonitis complicating peritoneal dialysis. Clin J Am Soc Nephrol 1: 768–773, 2006 [DOI] [PubMed] [Google Scholar]
- 21.Szeto CC, Wong TY, Chow KM, Leung CB, Li PK: The clinical course of culture-negative peritonitis complicating peritoneal dialysis. Am J Kidney Dis 42: 567–574, 2003 [DOI] [PubMed] [Google Scholar]
- 22.Visser CE, Brouwer-Steenbergen JJ, Schadee-Eestermans IL, Meijer S, Krediet RT, Beelen RH: Ingestion of Staphylococcus aureus, Staphylococcus epidermidis, and Escherichia coli by human peritoneal mesothelial cells. Infect Immun 64: 3425–3428, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haslinger-Löffler B, Wagner B, Brück M, Strangfeld K, Grundmeier M, Fischer U, Völker W, Peters G, Schulze-Osthoff K, Sinha B: Staphylococcus aureus induces caspase-independent cell death in human peritoneal mesothelial cells. Kidney Int 70: 1089–1098, 2006 [DOI] [PubMed] [Google Scholar]