Abstract
Imprinted genes play important roles in placenta development and function. Parthenogenetic embryos, deficient in paternally expressed imprinted genes, lack extra-embryonic tissues of the trophoblast lineage. Parthenogenetic trophoblast stem cells (TSCs) are extremely difficult to derive, suggesting that an imprinted gene(s) is necessary for TSC establishment or maintenance. In a candidate study, we were able to narrow the list to one known paternally expressed gene, Sfmbt2. We show that mouse embryos inheriting a paternal Sfmbt2 gene trap null allele have severely reduced placentae and die before E12.5 due to reduction of all trophoblast cell types. We infected early embryos with lentivirus vectors expressing anti-Sfmbt2 shRNAs and found that TSC derivation was significantly reduced. Together, these observations support the hypothesis that loss of SFMBT2 results in defects in maintenance of trophoblast cell types necessary for development of the extra-embryonic tissues, the placenta in particular.
Keywords: Sfmbt2, Polycomb group protein, Trophoblast stem cell, Placenta, Loss of imprinting, Parthenogenesis, Mouse
INTRODUCTION
Waddington’s epigenetic landscape (Waddington, 1940), a theoretical approach to understanding development, has in recent years acquired significant mechanistic substance, even to the point of looking ‘hilly’ when data are displayed graphically. Genetic identification of various chromatin proteins and modifiers has led not just to expansion of the repertoire of molecular tools at our disposal, but to a redefinition of the term ‘epigenetics’, which now refers to a suite of biochemical processes that regulate how genomes are used during development and in response to environmental stress (Goldberg et al., 2007). Most of the biochemistry involves chromatin proteins and their wide ranging post-translational modifications, DNA modifications (cytosine methylation, hydroxymethylation) in some species, and the very ancient RNAi-based gene silencing strategies common to all life forms, including bacteria (Bhaya et al., 2011).
Among the chromatin modifiers that have received significant attention are the Polycomb Group genes (PcG), which encode components in a growing number of ‘complexes’ - PRC1, PRC2, PHO-RC, PR-Dub - the loss of which is exemplified in fruit flies by the classic polycomb phenotype, and in other organisms, from yeast to plants and animals, in sometimes catastrophic developmental failures (Bantignies and Cavalli, 2011). PcG genes, and their molecular antagonists, the Trithorax Group (TrxG) genes, regulate chromatin in a developmental context, acting to silence or activate specific suites of genes in different cell types or structures. Without them, organisms would be little more than puddles of cells.
Genomic imprinting, an epigenetic phenomenon in which alleles of some genes are silenced in a parent-of-origin fashion, has a special relationship with extra-embryonic tissues. The restriction of genomic imprinting in the vertebrate phylum to mammals has fueled speculation that it arose along with placentation, an idea that is reinforced by the observation that many imprinted genes are monoallelically expressed only in placenta and/or yolk sac (Lefebvre, 2012; Okae et al., 2012; Wang et al., 2011). Furthermore, among mammals, monotremes lack both genomic imprinting and placentae, and marsupials display a limited range of imprinted genes compared with eutherians (Hore et al., 2007; Renfree et al., 2008). What is so special about extra-embryonic tissues?
Extra-embryonic tissues play key roles in vertebrate development (Miri and Varmuza, 2009). For example, early signaling from extra-embryonic endoderm is required for formation of the body axes before gastrulation (Stern and Downs, 2012), and signaling from the chorion during gastrulation is necessary for establishment of the germline (de Sousa Lopes et al., 2007). In mammals, extra-embryonic tissues are also particularly important as the interface between the maternal and fetal circulatory systems that mediate gas, waste and nutrient exchange. Failure to form extra-embryonic tissues leads to early embryonic death. Parthenogenetic mouse embryos, which lack a paternal genome, typically fail to develop beyond early gastrulation, and have severely hypoplastic extra-embryonic tissues (Varmuza et al., 1993), suggesting that one or more paternally expressed imprinted genes may be required to sustain the trophoblast compartment, possibly through establishment and/or maintenance of trophoblast stem cells (TSCs).
TSCs can be derived from blastocyst outgrowths (Tanaka et al., 1998). Here, we demonstrate that the derivation of parthenogenetic trophoblast stem cells (PTSCs) is achievable but at a much lower frequency when compared with fertilized embryos. Previously reported reactivation of imprinted genes in embryos cultured in vitro (Rivera et al., 2008) prompted us to ask whether there are any maternally silenced genes that have lost their imprint, facilitating the derivation of PTSCs. Our results show that PTSCs reactivate several paternally expressed genes, including the PcG gene Sfmbt2, which is expressed robustly in extra-embryonic tissues (Kuzmin et al., 2008). Additional experiments revealed that, except for Sfmbt2, other candidate genes necessary for TSC establishment and maintenance could be discounted by virtue of their expression patterns in TSCs and uniparental blastocysts. We show that a gene trap null allele of Sfmbt2 is lethal when inherited paternally, and results in severely reduced extra-embryonic tissues at early post-implantation stages of development. Finally, to test if the observed defects are due to a diminished pool of TSCs, we show that lentiviral delivery of shRNA against Sfmbt2 to fertilized preimplantation embryos reduces TSC derivation significantly.
RESULTS
Parthenogenetic embryos are impaired in TSC production
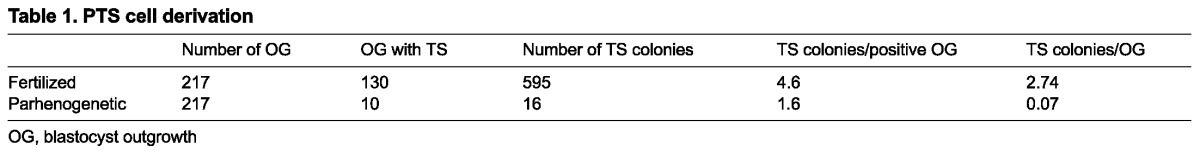
The failure of parthenogenetic embryos to sustain trophoblast development beyond early post-implantation stages (Varmuza et al., 1993) suggested that their TSC compartment may be impaired due to absence of one or more paternally expressed, early post-implantation-acting imprinted gene(s). To test this hypothesis, a series of paired experiments was performed to determine the frequency with which parthenogenetic TSCs (PTSC) could be generated in comparison with fertilized TSCs (FTSC) (Table 1). Fertilized blastocysts were efficient at producing TSCs; 60% of outgrowths generated at least one colony, and in most cases, more than one TSC colony was found in each well. Parthenogenetic embryos were 97.5% less efficient at generating TSCs than fertilized blastocysts, with only 4.6% of blastocysts yielding colonies, and fewer colonies per blastocyst, in most cases only one. Most parthenogenetic outgrowths contained only small clusters of giant cells after 3-4 days in culture.
Table 1.
PTS cell derivation
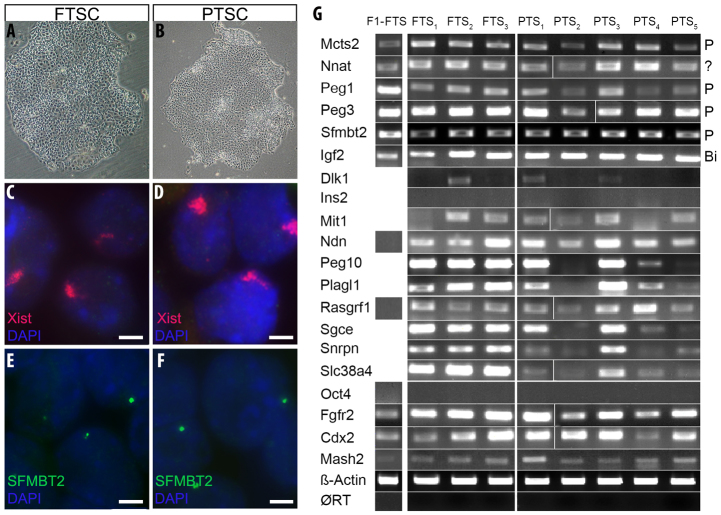
We compared five PTSC cell lines with four FTSC cell lines for a number of features. In addition to displaying a characteristic epithelial appearance with defined edges to the colonies (Fig. 1A,B), the identity of TSCs was confirmed by cell-type marker analysis, including the TSC-specific markers Cdx2, Fgfr2 and Mash2, and absence of the ES cell marker Oct4 (Fig. 1G). Removal of FGF4 resulted in differentiation of both FTSCs and PTSCs into trophoblast giant cells (not shown).
Fig. 1.
Fertilized and parthenogenetic TS cells show similarities in morphology and gene expression. (A,B) Phase-contrast images of fertilized (A) and parthenogenetic (B) TSC colonies. (C-F) FISH showing Xist (C,D) and Sfmbt2 (E,F) expression in FTSC (C,E) and PTSC (D,F) cells. Xist is labeled with Cy3-dCTP (red) and Sfmbt2 with FITC-dUTP (green). Nuclear staining is shown by DAPI (blue). Scale bar: 2.5 μm. (G) RT-PCR of various paternally expressed genes and cell type markers in four fertilized and five parthenogenetic TSC lines. Exclusion of genomic contamination is shown by ØRT in which reverse transcriptase was omitted during cDNA synthesis and PCR was performed using β-actin primers. P, paternally expressed; Bi, biallelic;?, unknown. Thin white lines mark separations between samples that were run on the same gel but not in adjacent lanes.
PTSCs reactivate some paternally expressed imprinted genes (PEGs)
Three PTSC lines derived from MI oocytes (PTS1-PTS3) and two lines derived from MII oocytes (PTS4, PTS5) were examined for expression of several known imprinted genes by RT-PCR. In addition, four FTSC lines, including F1-FTS, were assayed to evaluate consistency of expression across a range of cell lines (Fig. 1G). The analysis resulted in the categorization of these genes into two classes: those with consistent expression across all TSC lines, such as Mcts2, Nnat, Peg1 (Mest - Mouse Genome Informatics), Peg3, Sfmbt2 and Igf2, and those with inconsistent expression in TSC, such as Ndn, Rasgrf1, Mit1 (Copg2os2 - Mouse Genome Informatics), Mkrn3 (not shown), Peg10, Plagl1, Sgce, Snrpn, Slc38a4 and Dlk1. Ins2 was assayed but not detected in any TSC lines.
Genes with consistent expression across all PTSC lines were assayed for monoallelic expression in F1-FTS. SNP analysis in F1-FTS cell line revealed paternal expression of Mcts2, Peg1, Peg3 and Sfmbt2, confirming imprinted expression (supplementary material Fig. S2), and indicating that expression in PTSC is a function of loss of imprinting. Igf2 showed biallelic expression, indicating lack of imprinting of this gene in TSCs. We were unable to assess allelic expression of Nnat due to lack of polymorphisms.
Fluorescent in situ hybridization reveals that PTSCs undergo normal X-inactivation and express Sfmbt2 from one allele
The paternal X chromosome is preferentially inactivated in murine extra-embryonic tissues. Previous studies have shown that parthenogenetic embryos, despite lacking a paternal genome, appear to inactivate one of their X-chromosomes (Nesterova et al., 2001). We tested PTS2 and PTS5 cell lines for expression of Xist by fluorescent in situ hybridization (FISH). Both PTSC lines showed a distribution of Xist RNA characteristic of X chromosome silencing, indicating that X-inactivation can occur in PTSCs (Fig. 1C,D). In addition, only one allele of Sfmbt2 is active, as revealed by the presence of only one RNA FISH signal (Fig. 1E,F), indicating that reactivation is restricted to one of the grandparental alleles. Together, these results indicate that at least one maternal copy of normally silent imprinted genes can be reactivated in PTSCs.
Monoallelic expression of some PEGs does not start at the blastocyst stage
As TSC establishment commences at the blastocyst stage, we reasoned that the reduced efficiency of PTSC derivation must be due to the need to reactivate a normally silent maternal allele of a PEG in parthenogenetic blastocysts. The candidate PEG should therefore fulfill a number of criteria: (1) it should be expressed at the blastocyst stage; (2) it should be imprinted at the blastocyst stage; (3) it should be expressed in ALL TSC lines; (4) it should be imprinted in TSCs; (5) a knockout phenotype should have severe effects on trophoblast development commensurate with loss of the trophoblast stem cell compartment.
There are currently ∼50 PEGs identified (Miri and Varmuza, 2009). In order to assess the first two criteria, we examined the expression pattern of PEGs at the blastocyst stage using both published microarray data (GEO accession numbers GSE8163 and GSE1744) and qRT-PCR, discussed below. To ensure the integrity of identifiers as true representatives of annotated transcripts, the sequences for each transcript, as well as all splice variants, were individually examined for matching probe sets using NetAffx probe match software available at http://www.affymetrix.com/estore. A summary of the results can be found in supplementary material Tables S3 and S4. No detectable expression for genes such as Peg3os, Igf2os, Airn, Ins1, Ipw, Kcnq1ot1, Mas1, Nespos, Usp29 and Zdbf2 was found in the blastocyst microarray datasets.
We tested the expression and imprint status of 27 PEGs in androgenetic, gynogenetic (i.e. parthenogenetic) and biparental blastocysts by qRT-PCR (Fig. 2). Ten genes yielded no detectable expression signal [A19 (4930524O08Rik - Mouse Genome Informatics), Rasgrf1, Mit1, Dlk1, Slc38a4, Ins2, Ddc-exon1a, Magel2, Peg13 and Peg12; not shown]. The remaining 17 genes were categorized as biallelic or monoallelic based on the ratio of expression in uniparental blastocysts to fertilized blastocysts. Nap1l5, Mkrn3, Peg11 (Rtl1 - Mouse Genome Informatics), Impact, Igf2, Dio3 and Ndn showed expression at similar levels in both uniparental and biparental blastocyts, indicating their biallelism at this stage of development. Mcts2, Nnat, Peg1, Peg3, Peg10, Plagl1, Sfmbt2, Sgce, Snrpn and U2af1-rs1 (Zrsr1 - Mouse Genome Informatics), however, all showed similar levels of expression in androgenetic and biparental blastocysts compared to the marginal to no expression observed in gynogenetic blastocysts, indicating their monoallelism at this stage of development.
Fig. 2.
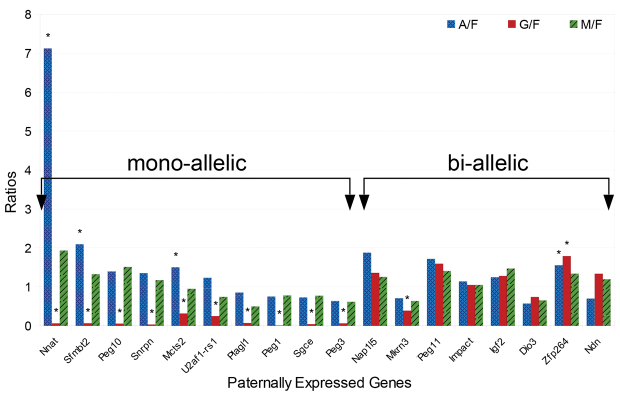
Survey of paternally expressed genes in biparental and uniparental blastocysts identifies genes with mono- or biallelic expression at the blastocyst stage. qRT-PCR results showing expression of known PEGs as ratios in androgenetic, gynogenetic or manipulated to fertilized (A/F, G/F and M/F, respectively) blastocysts. *P<0.05.
Criterion 3 was evaluated by assaying expression of selected genes in three FTSC and five PTSC lines. Several genes were found to display inconsistent activation, thus failing to pass criterion 3. The remaining genes were assessed for monoallelic expression in F1-FTS cell line (criterion 4); this test narrowed the list to a small number of PEGs (supplementary material Fig. S2). The latter observation was confirmed by the RNA-seq data published previously (Calabrese et al., 2012). A summary of these results can be found in supplementary material Table S4.
Using criterion 5, we provisionally excluded several genes as being required for TSC establishment and maintenance. Ddc, Dlk1, Gnasxl (Gnas - Mouse Genome Informatics), Igf2, Airn, Ins2, Kcnqlot1, Magel2, Mas1, MB11-85 (also known as Pwcr1 or Snord116), Ndn, Nespos, Peg1, Peg3, Peg10, Plagl1, Peg11, Peg3, Rasgrf1, Sgce, Snrpn, Snurf and U2af1-rs1 (Zrsr1 - Mouse Genome Informatics) have all been subjected to targeted mutagenesis in which the null phenotypes, with the exception of the Peg10 mutation, have no or very mild effects on placental development (see Miri and Varmuza, 2009). The most severe imprinted gene KO published to date is the Peg10 loss of function, which results in defects in the trophoblast derivatives of the placenta starting at E10.5 (Ono et al., 2006). However, Peg10 is not consistently reactivated in all PTSC, indicating that it is not required for TSC derivation/maintenance. The paternally expressed gene that fulfilled all criteria is Sfmbt2 (supplementary material Table S4), which we show below results in a severe reduction of trophoblast starting at E8.5 when it is missing.
PcG protein SFMBT2 displays nuclear staining in various cell types of trophoblast origin
The expression pattern of SFMBT2 was investigated at E10.5 by immunohistochemistry (IHC) of placenta sections. All trophoblast cell types were positive for SFMBT2 staining, although several different kinds of signal could be observed. Strong punctate nuclear staining can be seen in TGCs within the spongiotrophoblast (Fig. 3F), the labyrinth TGCs (Fig. 3G), as well as in some of the parietal trophoblast giant cells (PTGCs) juxtaposing the decidua (Fig. 3H), whereas some of the PTGCs displayed no staining (Fig. 3H; arrowheads). Spongiotrophoblast (SpT) showed weaker mainly cytoplasmic staining with occasional nuclear staining (Fig. 3F), and no staining (Fig. 3F, white arrowheads). The trophoblast cells of the labyrinthine layer displayed robust signal, whereas endothelial cells remained negative (Fig. 3G). SFMBT2 protein levels decline noticeably by E15.5 in all placenta layers (not shown); this observation is consistent with a drop in transcript levels recorded in microarray surveys of differentiating TSCs (Ralston et al., 2010); a summary graph can be found in supplementary material Fig. S3. The dynamic changes in SFMBT2 distribution are described below in a comparison of wild-type and mutant embryos.
Fig. 3.
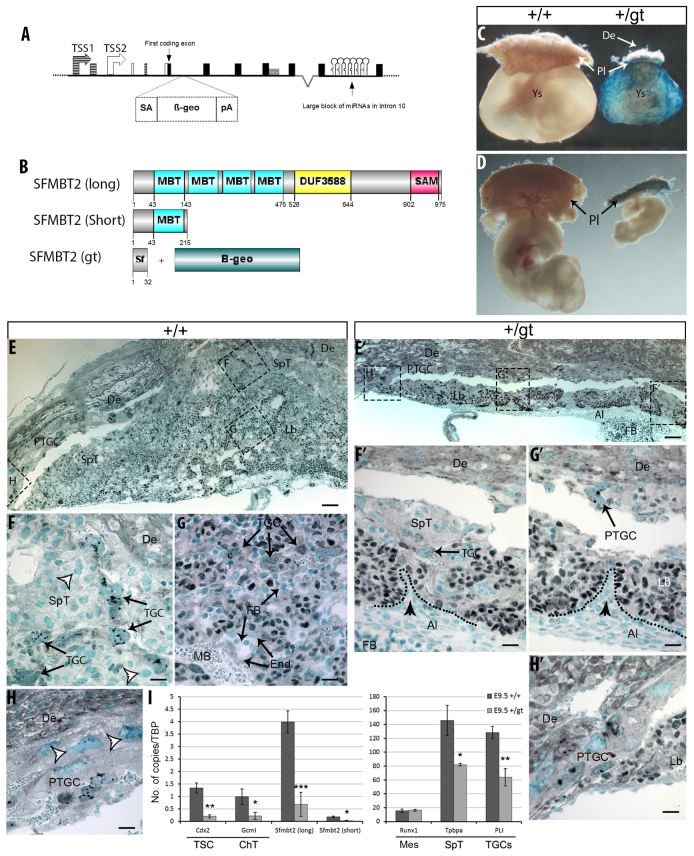
Paternal inheritance of a null Sfmbt2 allele is lethal due to defects in proper development of the extra-embryonic tissues. (A) Schematic representation of Sfmbt2 gene trap allele showing the insertion of a splice acceptor (SA) sequence followed by a β-geo cassette and a strong polyA signal (pA) downstream of the first coding exon. The two transcriptional start sites for Sfmbt2 are indicated by TSS1 and TSS2. (B) Schematic representation of wild-type (both long and short isoform) and truncated SFMBT2. Numbers correspond with the amino acid sequence. gt, genetrap; +, wild type. (C,D) Gross morphological comparison of E10.5 +/+ (lacZ negative) and +/gt (lacZ positive) fetuses with (C) and without (D) yolk sac. SFMBT2 IHC (E-H′) on E10.5 +/+ (E-H) and +/gt (E′-H′) placentae showing reduction in numbers of all trophoblast cell types, including SpT (F,F′), Lb (F′,G,G′) and PTGCs (H,H′). Methyl Green was used a counterstain. White arrowheads indicate unstained TGCs (F,H). Black arrowheads indicate allantoic protrusions into the chorion (F′,G′). (I) qRT-PCR results showing expression of various markers in +/gt and +/+ E9.5 placentae as a ratio to Tbp. Bars indicate s.d. *P≤0.05; **P≤0.01; ***P≤0.001. De, decidua; Ys, yolk sac; Lb, labyrinth; SpT, spongiotrophoblast; PTGC, parietal trophoblast giant cells; Al, allantois; FB, fetal blood; MB, maternal blood; Ch, chorion; TGC, trophoblast giant cells; Mes, allantoic mesothelium. Scale bars: 100 μm in E,E′; 25 μm in F-H′.
A null gene trap allele is lethal when transmitted paternally
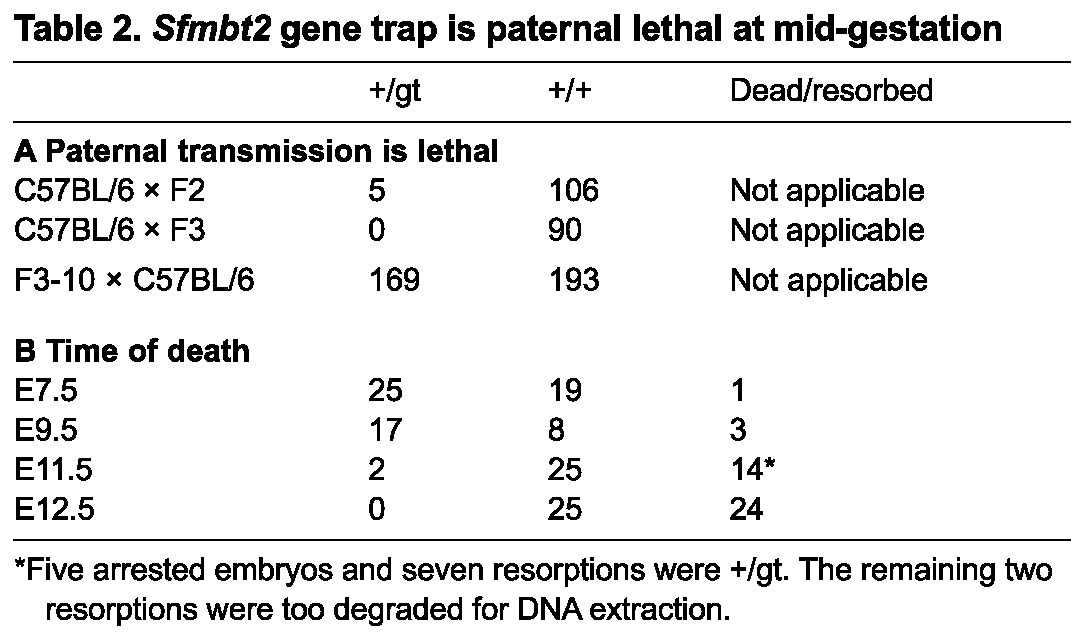
We obtained a mouse with a gene trap allele in which the first coding exon is trapped by a β-geo cassette; the fusion protein is predicted to contain only 32 amino acids of SFMBT2 at its N terminus (Fig. 3A,B). We reasoned that a null allele of Sfmbt2 should be lethal when transmitted paternally. The original gene trap cell line was from 129/Ola mice; a chimera was mated to a C57BL/6 female to produce a gene trap male whose sperm was archived. The male mouse we received, resuscitated by injection of archived gene trap sperm into C57BL/6 oocytes, was at backcross 2 on the C57BL/6 genetic background, and was able to sire only five gene trap offspring from 111 pups, one of which was a female. None of the male gene trap offspring has sired any live gene trap pups, and the litter sizes are approximately half those of wild-type C57BL/6 mice in our animal facility. The female, however, and her female offspring, produce gene trap offspring at Mendelian levels (Table 2A).
Table 2.
Sfmbt2 gene trap is paternal lethal at mid-gestation
Paternal inheritance of the gene trap allele of Sfmbt2 leads to severe defects in extra-embryonic tissues by E9.5
At E7.5, the time when parthenogenetic embryos begin to display severe phenotypes, Mendelian numbers of relatively normal looking egg cylinders can be recovered (Table 2B). By E9.5, although Mendelian numbers of embryos are recoverable, the +/gt conceptuses are readily recognizable by their small placentae and embryos. Although intact developmentally delayed embryos can be recovered at E10.5 (Fig. 3C,D), most embryos are dead by E11.5, and have been resorbed by E12.5 (Table 2B). Intercrosses of gt/+ males and females reveals that the time of death of null (gt/gt) embryos is similar, i.e. between E10.5 and E11.5.
Examination of +/gt extra-embryonic tissues at E10.5 revealed that, in comparison with wild-type littermates, all three trophoblast layers of the placenta are severely compromised (Fig. 3E-H′; supplementary material Fig. S4 shows lacZ images). The labyrinthine layer in +/gt placentae appears to have undergone the early stages of chorioallantoic branching morphogenesis, evident from villi projected into the chorion (Fig. 3E′-G′, arrowheads); however, in comparison with the wild-type littermates displaying extensive branching morphogenesis with the presence of both maternal and fetal blood (Fig. 3E,G), the +/gt labyrinth is severely underdeveloped. The spongiotrophoblast layer is also significantly reduced in +/gt placentae (Fig. 3F′) when compared with its wild-type counterpart (Fig. 3F), although expression of SFMBT2 in these cells is faint. Finally, all TGC compartments, including those associated with spongiotrophoblast (Fig. 3F,F′), labyrinth (Fig. 3F′,G,G′) and the region juxtaposing both the placenta (Fig. 3E,E′,G′,H,H′) and the parietal endoderm (supplementary material Fig. S4D,D′) are reduced. Interestingly, SFMBT2 protein levels in the remaining trophoblast cells in +/gt placentae appears to be the same as that in wild-type placentae (see below).
A qRT-PCR survey of genes expressed in various cell types of placenta was performed on both +/+ and +/gt placentae (Fig. 3I). To minimize loss of tissue during dissection and maximize the integrity of the placenta and PTGCs, the entire placental unit including the decidua was used for RNA extraction and cDNA synthesis. All trophoblast-specific markers, including Cdx2 (TSC marker), Gcm1 (chorionic trophoblast), Tpbpa (SpT) and Pl1 (Prl3d1 - Mouse Genome Informatics) (PTGC), show statistically significant reduction in +/gt placentae in comparison with wild-type counterparts. Runx1, however, expressed in the allantoic mesothelium (Mes) juxtaposing the chorionic trophoblast (Zeigler et al., 2006), did not show any reduction. In addition, qRT-PCR results show a significant reduction in both the long and the short isoforms of Sfmbt2, discussed below.
Reactivation of maternal allele
Given both the IHC and the qRT-PCR results showing a low level of expression of Sfmbt2 at E9.5 in +/gt placentae, we further examined E7.5 and E9.5 extra-embryonic tissues for expression of Sfmbt2. The possibility remained that alternative splicing around the gene trap insert might generate normal transcripts. Surprisingly, Sfmbt2 was found to be expressed in +/gt extra-embryonic tissues from the normally silent maternal allele (Fig. 4A). Quantitative RT-PCR at E7.5 and E9.5 of extra-embryonic tissues dissected out of the decidua reconfirmed that the levels of Sfmbt2 transcripts from the maternal allele were significantly lower than in wild-type littermates (Fig. 4B). Interestingly, analysis of Sfmbt2 expression in a placenta from a Chromosome 2 Maternal Duplication (MatDup) embryo (Cattanach et al., 2004) inheriting two maternal T(2;8)Wa chromosomes also displayed reactivation of the maternal allele (supplementary material Fig. S5).
Fig. 4.
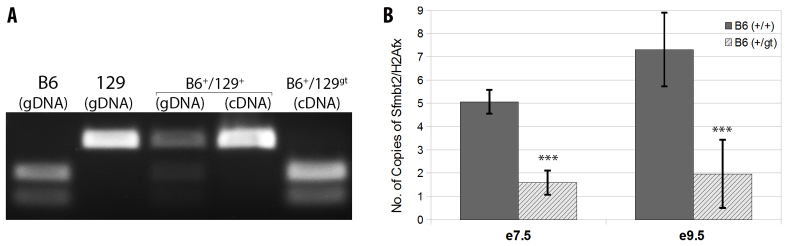
Maternal Sfmbt2 allele is reactivated in trans after fertilization. (A) Restriction fragment length polymorphism (RFLP) analysis of genomic DNA (gDNA) or cDNA derived from C57BL/6 (B6), 129, B6+/129+ and B6+/129gt. In each case, PCR product amplified with DCOSNP primers was digested with MspI. (B) qRT-PCR showing expression of Sfmbt2 in B6+/gt and their wild-type littermates B6+/+at E7.5 ectoplacental cone and E9.5 placentae as a ratio to H2Afx. Bars indicate s.d. ***P≤0.001.
Failure of trophoblast maintenance in SFMBT2-deficient embryos
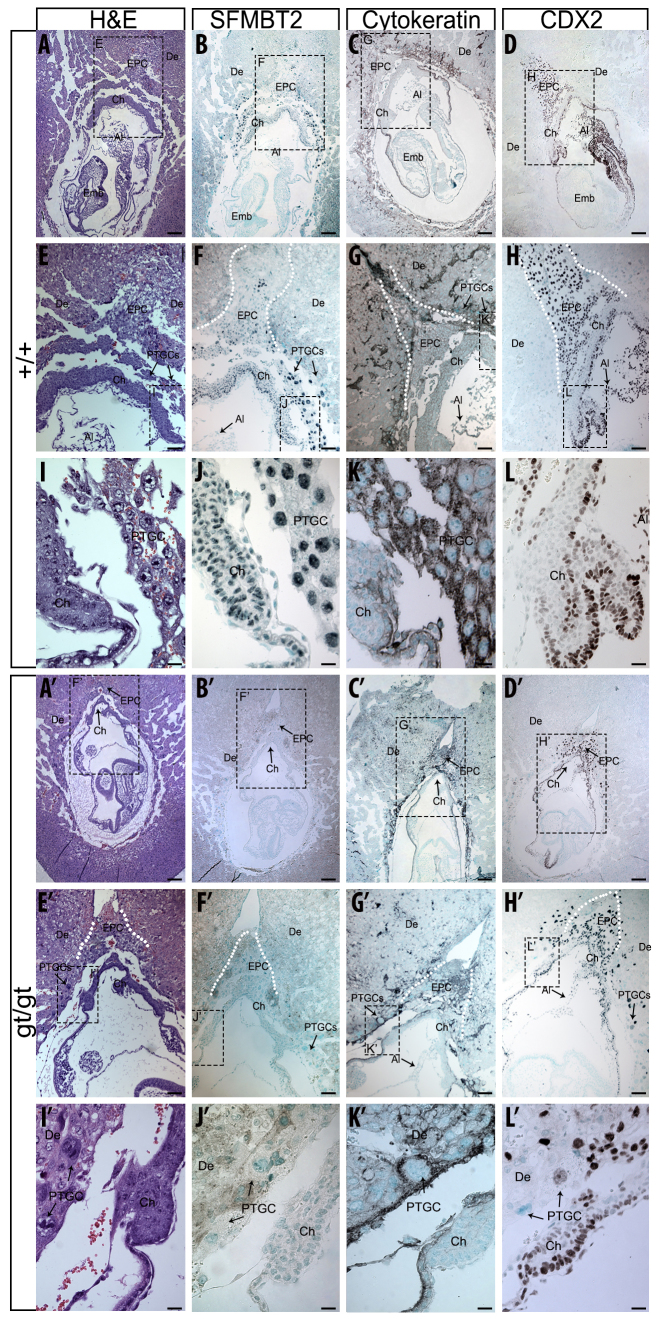
Reactivation of the maternal allele of Sfmbt2 in +/gt embryos created hypomorphs. What is the effect of nullizygosity for this gene? Histological analysis of gt/gt embryos at E8.5 after chorio-allantoic attachment (Fig. 5) revealed a reduction in the size of the epc (Fig. 5E,E′), the chorionic plate (Fig. 5I,I′) and the number of PTGCs (Fig. 5I,I′) (gt/gt embryos were readily identified by the complete lack of SFMBT2 signal following IHC). This observation was further confirmed by pan-cytokertin staining outlining the trophoblast cell types of the placenta (Fig. 5C,G,K, +/+; 5C′,G′,K′, +/gt). Despite the reduction in numbers, however, trophoblast cells of the gt/gt placenta display robust CDX2 expression, a marker of trophoblast, suggesting that SFMBT2 is required for maintenance rather than for establishment of trophoblast.
Fig. 5.
SFMBT2-null embryos display abnormalities in the extra-embryonic tissues as early as E8.5. Histology of E8.5 +/+ (A-L) and gt/gt (A′-L′) embryos. Hematoxylin and Eosin staining (A,E,I,A′,E′,I′); SFMBT2 IHC (B,F,J,B′,F′,J′); pan-cytokeratin IHC (C,G,K,C′,G′,K′) and CDX2 IHC (D,H,L,D′,H′,L′). Methyl Green was used as a counterstain. De, decidua; PTGC, Parietal trophoblast giant cells; Al; allantois; Ch, chorion; EPC, ectoplacental cone; Emb, embryo. Scale bars: 200 μm in A-D′; 100 μm in E-H′; 25 μm in I-L′.
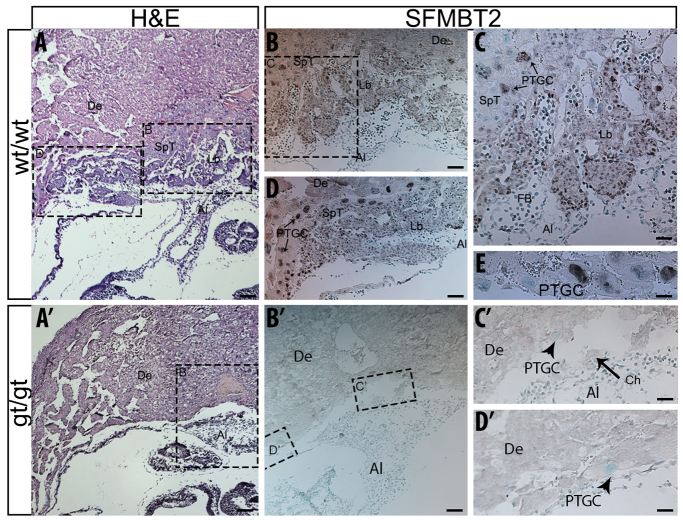
Histological analysis of gt/gt embryos at E9.25 and E9.75 was performed to refine the phenotypic analysis (supplementary material Fig. S6 and Fig. 6, respectively). Results indicate that despite the persistence of the chorionic plate by E9.25 of development (supplementary material Fig. S6), there was no evident advancement in the development of the placenta from E8.5 embryos (Fig. 5). By E9.75, when chorioallantoic branching morphogenesis is well under way in wild-type embryos (Fig. 6A-C), gt/gt embryos show a disintegrated chorionic plate with very few cells juxtaposing the allantois (Fig. 6A′-C′). In addition, at E9.75, unlike their wild-type littermates (Fig. 6D,E), gt/gt embryos retain only rare PTGCs (Fig. 6C′-D′). Taken together, these observations suggest that SFMBT2 plays a significant role in the maintenance of various trophoblast cell types, and is required for development of trophoblast derivatives beyond E8.5.
Fig. 6.
SFMBT2-null embryos fail to maintain tissues of trophoblast origin. Histology of E9.75 +/+ (A-E) and gt/gt (A′-D′) extra-embryonic tissues in decidua. Hematoxylin and Eosin staining (A,A′). SFMBT2 IHC (B-E,B′-D′); Arrowheads indicate PTGCs (C′,D). Methyl Green was used as a counterstain. De, decidua; Lb, labyrinth; SpT, spongiotrophoblast; PTGC, Parietal trophoblast giant cells; Al; allantois; Ch, chorion, FB, fetal blood; Emb, embryo. Scale bar: 200 μm in A,A′; 100 μm in B,B′,D; 50 μm in C,C′,D′; 25 μm in E.
ShRNA knockdown of Sfmbt2 impairs FTSC derivation and/or maintenance
The phenotype of Sfmbt2 mutant embryos suggested that the main defect was a failure to maintain a pool of trophoblast progenitors. In addition, Sfmbt2 was consistently reactivated in (surviving) PTSCs. We therefore tested the effect of reducing Sfmbt2 expression in preimplantation embryos on their ability to generate TSC in vitro. The approach we took was to infect morula stage embryos with lentiviruses-expressing shRNAs directed against Sfmbt2 prior to blastocyst outgrowth. The shRNAs we designed were first tested for their knockdown efficiency by infecting HEK293 cells that had been transiently transfected with myc-tagged Sfmbt2 expression vectors (Fig. 7A,B). Both shRNAs were able to reduce SFMBT2 protein significantly.
Fig. 7.
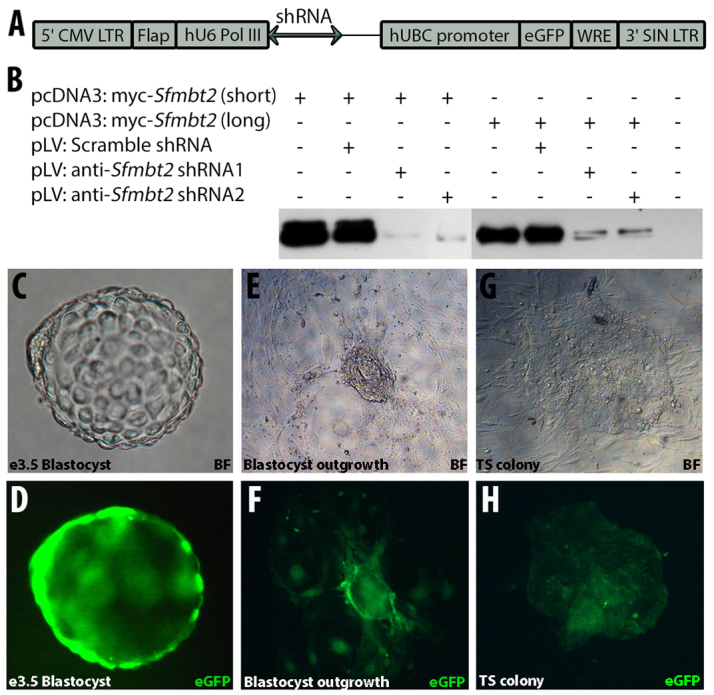
Sfmbt2 is required for the derivation of TS cells. (A) Map of shRNA-carrying lentiviral plasmid. ShRNA sequence was inserted downstream of hU6 Pol III promoter. Vectors also contain eGFP sequence driven by human UBC (hUBC) promoter. (B) Validation of anti-Sfmbt2 shRNAs shown by western blot analysis using anti-SFMBT2 antibody of whole protein lysate harvested from untransfected as well as transfected HEK293 cells with various combinations of plasmids as indicated. (C-H) Bright-field (C,E,G) and fluorescent (D,F,H) images of blastocysts (C,D) 36 hours post-infection, blastocyst outgrowths (E,F) and a TSC colony (G,H) derived from morulae infected with scrambled shRNA carrying lentivirus. (I) The frequency of TSC derivation after infection of morulae with various lentiviruses and controls.
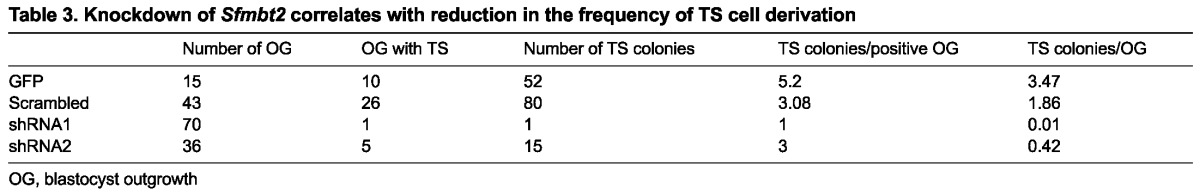
Infection of morulae with GFP-expressing lentiviruses is efficient, and leads to both blastocysts and outgrowths with clearly identifiable GFP-positive cells (Fig. 7C-F). TSC colonies derived from GFP expressing lentiviruses are similarly easy to identify visually (Fig. 7G,H). Infection with GFP-expressing lentiviruses had no obvious effect on TS cell derivation (Table 3). However, lentiviruses expressing shRNAs directed against Sfmbt2 severely reduced the frequency of TSC derivation. In particular, shRNA1 was very efficient at driving TSC derivation down to almost zero; in the entire series of experiments, only one TSC colony was generated, and expression of the GFP marker was very weak. ShRNA2 was less efficient at reducing TSC derivation. Interestingly, the scrambled shRNA also reduced TSC derivation somewhat, although not as well as the two Sfmbt2-specific shRNAs. Only GFP-positive TSC colonies were scored in these experiments. GFP-negative colonies were generated in all four groups involving infection with lentiviruses at rates between 0.1-0.4 per outgrowth, indicating that not all cells in each embryo were successfully infected.
Table 3.
Knockdown of Sfmbt2 correlates with reduction in the frequency of TS cell derivation
DISCUSSION
Early embryonic lethality of parthenogenetic embryos is likely to be due to lack of expression of one or more paternally expressed genes resulting in a hypoplastic trophoblast. This suggests that some paternally expressed imprinted genes are essential in the establishment and/or maintenance of a proliferative population of cells believed to give rise to the various compartments of the extra-embryonic tissues, including the placenta. Consistent with this hypothesis, we showed a 97.5% reduction in the frequency of PTS cell derivation compared with FTS cells. Several PEGs displayed consistent reactivation in the five PTS cell lines (Mcts2, Nnat, Peg1, Peg3 and Sfmbt2), suggesting that one or more PEGs, if reactivated, allows selection of TSCs in vitro. Our analysis of gene expression and survey of knockout phenotypes described in the literature led us to focus on Sfmbt2 as being essential for TSC and trophoblast development.
To test this candidate, we generated embryos with a paternal null allele of Sfmbt2 and were able to link the absence of the gene to severely reduced trophoblast and early lethality. By E9.5-E10.5, the mutant placentae are deficient in all trophoblast cell types, chorionic trophoblast, spongiotrophoblast and PTGCs. In addition, in comparison with wild-type littermates, the chorioallantoic branching morphogenesis is severely reduced in embryos with a paternally inherited gene trap allele. Complete null gt/gt embryos show similar trophoblast specific abnormalities including the thinning of the chorionic plate and PTGCs at E8.5, and appear to have almost entirely disintegrated trophoblast tissues by E9.75 with the exception of a small number of PTGCs.
Our data indicate that Sfmbt2 is the earliest acting imprinted gene analyzed to date. These phenotypes are consistent with reduction of the trophoblast progenitor pools of TSCs. Supporting this hypothesis is the observation that shRNA-mediated knockdown of Sfmbt2 severely reduced the ability of preimplantation embryos to support TSC derivation, reinforcing the idea that SFMBT2 is required for TSC establishment and/or maintenance. Given that parthenogenetic, gt/gt and shRNA-expressing blastocysts all make trophoblast, we favor the idea that SFMBT2 is required for maintenance of the progenitor pools.
Cell type specification is established by transcriptional control exerted by epigenetic modifications at both the DNA and histone levels. Polycomb group (PcG) proteins are highly conserved transcriptional repressors found in regulatory complexes that function by post-translational modification of histones to maintain cellular homeostasis, and have potential roles in stem cell identity and pluripotency. ES cells require SUZ12, a PcG protein involved in polycomb repressive complex 2 (PRC2), for their proper differentiation through repression of genes essential for ES cell self-renewal, such as Oct4 and Nanog. BMI1, another PcG protein, is also known for its involvement in stem cell self-renewal, and its dysregulation is linked with aberrant proliferation of cancer cells (Park et al., 2003). Consequently, the PcG protein SFMBT2 (Kuzmin et al., 2008) appears to be a strong candidate for a role in maintaining TSCs. Experiments in fruit flies revealed that the fly ortholog of SFMBT2, dSFMBT, along with the transcription factor PHO, forms the core of a third silencing complex, PHO-RC (Klymenko et al., 2006). Additionally, dSFMBT interacts physically and genetically with SCM, another PcG protein, but not under all conditions (Grimm et al., 2009). SFMBT2 also interacts with other chromatin proteins (Zhang et al., 2013); these experiments were conducted in HEK293 cells, which may lack some key trophoblast players.
Although Sfmbt2 is imprinted only in Old World rodents (mice and rats), it is expressed in the placentae of other mammals, including humans (Wang et al., 2011), is highly conserved at the amino acid level, and is therefore likely to play a central role in development of extra-embryonic lineages through its interactions with other chromatin proteins. Experiments are in progress to elaborate the molecular partners of SFMBT2 in the trophoblast stem cell lineage.
The maternal Sfmbt2 allele is reactivated in parthenogenetic embryos, but at a very low frequency compared with fertilized gene trap embryos. This observation is consistent with the requirement of another paternally expressed gene acting early that is a positive regulator of Sfmbt2. It is unlikely to be one of the known imprinted genes for which knockout data are available, as argued above; a novel paternally expressed gene with the capacity to be a transcriptional activator would be an appealing candidate. The observation that only one of the maternal alleles in PTSCs is reactivated, as demonstrated by FISH, is intriguing, and raises the issue of whether grandparental information is retained and passed through the oocyte. Other studies have yielded evidence for retention of grandparental effects (Davis et al., 2000; Han et al., 2008; Lucifero et al., 2004). Consistent with our observation of reactivation of the maternal allele in fertilized +/gt embryos, analysis of Sfmbt2 expression in a (fertilized) MatDup placenta from the T(2;8)Wa translocation line of mice revealed that reactivation of the maternal allele also occurred. This raises the possibility that the survival of some of the MatDup embryos in the study by Cattanach and colleagues may reflect reactivation of Sfmbt2 rather than the milder growth effects of Chr2 imprinted genes postulated by the authors (Cattanach et al., 2004).
We have shown that reducing expression of the PcG gene Sfmbt2 significantly abrogates the ability of mouse embryos to generate TSCs. This in vitro assay is consistent with the phenotype of a paternally inherited null allele of Sfmbt2, which results in major defects in placenta development. The role of SFMBT2 in maintaining the trophoblast cell types as a chromatin protein affords the possibility of establishing the gene networks involved in extra-embryonic tissues, and of defining important regulators of both fetal and maternal health.
MATERIALS AND METHODS
Sfmbt2 gene trap mutation
A male mouse with a gene trap insertion in the intron following the first coding exon of Sfmbt2 (derived from cell line DCO755) was obtained from the Mutant Mouse Regional Resource Centers (MMRRC, supported by the NIH, Office for Research Infrastructure Program, Comparative Medicine Branch) (see supplementary material Fig. S1 for insertion site mapping). This male was bred with C57BL/6 females and produced five gene trap offspring, one of which was the female founder of our colony. The colony is currently at backcross 10 on the C57BL/6 genetic background, and is maintained by breeding through the female germline, using standard protocols approved by the Canadian Council on Animal Care. Genotyping was performed by PCR analysis of tail biopsies with primers listed in supplementary material Table S1.
TSC derivation
Parthenogenetic embryos were generated by two methods. In the first method, MI oocytes from CD1 females were activated as described by Clarke et al. (Clarke et al., 1988). Activated oocytes were then transferred to the oviducts on pseudopregnant females and flushed on day 3 of pseudopregnancy. Blastocysts were distributed to feeder cell layers for the production of TSCs as described by Tanaka et al. (Tanaka et al., 1998). In the second method, MII oocytes retrieved from the oviducts of superovulated CD1 females were activated by brief exposure to 7% ethanol, exposed to cytochalasin D as described in Varmuza et al. (Varmuza et al., 1993) and cultured in vitro to the blastocyst stage before distribution to feeder cell layers for the production of TSCs. Normal fertilized TSCs (FTSC) were derived from CD1 blastocysts flushed from the uteri of mated females. A TSC line derived from mating a C57BL/6 female with a Mus castaneus male (F1-FTS) was generated for allelic expression analysis. In total, five PTSC lines (three from MI oocytes and two from MII oocytes), and four FTSC lines, including the F1-FTS line, were used in these experiments. For the experiment in which frequency of derivation was assessed, blastocysts were distributed individually to feeder cell layers in 96-well plates. Parthenogenetic embryos and fertilized embryos were assessed in parallel experiments to minimize confounding effects of culture conditions.
For lentiviral infections, CD1 superovulated female mice were mated with CD1 males, and E2.5 morulae were flushed from the genital tracts with M2 medium (Millipore; MR-015-D). Prior to viral infection, zonae pellucidae were removed with acid Tyrode’s solution (Millipore; MR-004-D). Embryos were cultured overnight in microdrops containing concentrated lentivirus, described below, diluted in CO2 equilibrated EmbryoMax KSOM Medium and 8 μg/ml of Polybrene (ABM; G062) under light mineral oil (Fisher Scientific) as described previously (Georgiades et al., 2007). Following incubation with virus, embryos were thoroughly washed with M2 medium and transferred individually to 96-well plates on a confluent layer of Mitomycin-C (R&D Systems; 3258/10)-treated mouse embryonic fibroblasts. Two days after infection, blastocyst outgrowths were scored for eGFP expression. Once the eGFP-positive outgrowths reached the appropriate size, they were disaggregated and cultured until TSC colonies emerged. Each experiment was performed in parallel with one or more of the controls to account for unspecific experimental effects.
RT-PCR and quantitative RT-PCR (qRT-PCR)
RNA extracted from tissues with Trizol reagent was converted to cDNA with AMV Reverse Transcriptase (Sigma) or Superscript III (Invitrogen) using buffers and protocols recommended by each supplier. Reactions were preceded by treatment of RNA with DNaseI for 30 minutes followed by heat inactivation at 65°C for 10 minutes. All cDNAs were tested for the presence of contaminating genomic DNA by amplification with either H2Afx or β-actin primers. No cDNA tested positive for contaminating genomic DNA. PCR amplification was performed with Takara Taq polymerase using reagents supplied by the manufacturer. Allelic expression was accomplished by either RFLP or capillary sequence analysis of cDNA from F1-FTS cells. qRT-PCR was performed in a Rotorgene light cycler, using SYBR Advantage qPCR Premix (Clontech; 639676). Standard curves, generated with cDNA product that was purified and quantitated on a Nanodrop spectrophotometer, were run with each set of samples. Three biological replicates were tested in each experiment. Data were collated and subjected to t-test analysis. Primer sequences are listed in the supplementary material Table S1.
qRT-PCR analysis of imprinted gene expression in blastocysts
Zygotes were isolated from superovulated B6D2 F1 hybrid females that had been mated to B6D2F1 males. Pronuclear transfer was performed as described previously (Han et al., 2010) to produce androgenetic and gynogenetic blastocysts, and sham-operated controls. Fertilized embryos and manipulated embryos were cultured to the blastocyst stage. The zonae pellucidae were removed using acid Tyrode’s solution. Total RNA was isolated from pools of blastocysts using the PicoPure RNA isolation kit (Life Technologies) according to the manufacturer’s instructions and stored at -70°C. RNA was reverse-transcribed into cDNA and amplified with SuperScript II Reverse Transcriptase (Life Technologies) and amplified using the Quantitect whole transcriptome kit (Qiagen). Quantitative RT-PCR analysis was performed using an ABI Prism 700 instrument (Life Technologies), and primer sets obtained for custom designed TaqMan assays (supplementary material Table S2). The mRNA abundance for each target gene was determined by normalizing to the signal obtained for the endogenous mitochondrial ribosomal protein S18C gene for sample to sample comparisons, and calculating relative expression by the comparative CT method (Livak and Schmittgen, 2001). Statistical analysis was performed between groups using Relative Expression Software Tool (REST) for the genes assayed by qRT-PCR method (Pfaffl et al., 2002).
FISH
RNA FISH was carried out as described previously using DNA probes generated from plasmids containing genomic Xist sequences, or Sfmbt2 cDNA (Mlynarczyk-Evans et al., 2006).
Western blots
Tissue or cells were homogenized in a lysis buffer containing 20 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol, 0.5 mM PMSF and a protease inhibitor cocktail (Sigma; P8340). Samples were electrophoresed on a conventional 8-10% SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked in 5% dried milk in PBS and 0.05% Tween followed by subsequent incubation in the primary antibody followed by HRP-conjugated secondary antibody and detected using enhanced ECL solution.
Histology and immunostaining
Tissues were immersed in formalin for 1 hour to overnight, depending on the size of the specimen, washed with PBS and dehydrated through an ethanol series before embedding in paraffin. Sections were stained with Hematoxylin and Eosin using standard histological procedures. In some cases, specimens were fixed briefly with formalin, stained for lacZ, and then post-fixed with formalin before embedding in paraffin. Immunostaining was performed following rehydration of sections and antigen retrieval. Antibody dilutions were as follows: 1:100 for anti-cytokeratin (Fitzgerald; 10R-C161a); 1:800 for anti-CDX2 (Novus Biologicals; EPR2764Y); 1:1000 for anti-SFMBT2 antibody (see Production of anti-SFMBT2 antisera, below). Secondary antibody used was from the VectaStain ABC kit (PK-4001; Vector Laboratories). Detection was performed with DAB substrate. Sections were subsequently counterstained with Methyl Green, dehydrated and mounted. Imaging was performed on an Olympus 1X71 inverted microscope, a Zeiss Lumar V12 stereomicroscope or an Olympus BX60 upright microscope.
Production of lentiviruses
Third-generation replication-deficient lentiviruses were generated using HEK293T cells as a packaging vehicle, as described previously with minor modifications (Tiscornia et al., 2006). Briefly, using CaPO4, HEK293T cells were transiently transfected with the appropriate lentiviral vectors [FUGW plasmid containing human Ubiquitin-C driven eGFP; FG12 plasmid containing UbiC driven eGFP, as well as either human U6-pol III driven anti-Sfmbt2-shRNA1, shRNA2 or scrambled shRNA (Fig. 7A), and three packaging plasmids encoding Gag-pol, Rev and an envelope protein VSV-G]. Culture medium was replaced with 30% heat-inactivated FBS (Gibco; 10437) in D-MEM (Gibco; 11965092), 7 to 11 hours after the initial transfection. Lentivirus-containing medium was harvested 36 hours later and concentrated using Lenti-X concentrator (Clontech; 631231) following the manufacturer’s protocol. Virus was diluted in TSC medium (Tanaka et al., 1998) and stored in small aliquots at -80°C. All lentiviruses contained a GFP marker used for visual confirmation of successful infection.
Production of anti-SFMBT2 antisera
A His-tagged Sfmbt2 cDNA encoding the short isoform was cloned in a bacterial expression vector (pET-15b) and used to transform STBL2 cells. Following induction with IPTG, protein was extracted, solubilized in urea and purified using Ni-NTA agarose beads (Invitrogen; R901-15). Purified protein was used for immunization of rabbits followed by serum collection (Hancock and O’Reilly, 2005; Tiscornia et al., 2006).
Supplementary Material
Acknowledgments
We thank Drs J. Rossant, J. Ellis and A. Jurisicova for sharing reagents; and Dr Jo Peters for donating the T(2;8)Wa MatDup placenta. We also thank Sangeetha Paramathas for assisting with some of the immunostaining experiments. Initial experiments with F1-FTS cells were performed on a cell line shared by Dr Mellissa Mann.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
K.L. and Z.Z. performed qRT-PCR on blastocysts (Fig. 2). B.P. and A.A. performed FISH (Fig. 1C-F), S.V. performed parthenogenetic TSC derivation (Table 1), genotyping (Table 2), reactivation analysis (Fig. 4), insertion site mapping supplementary material (Fig. S1), MatDup analysis supplementary material (Fig. S5). K.M. performed histology (Figs 3, 5, 6; supplementary material Figs S4, S6), allele-specific expression analysis (Fig. 1; supplementary material Fig. S2), lentivirus knockdown (Fig. 7), qRT-PCR (Fig. 3) and production of anti-SFMBT2 antisera. Manuscript was prepared by K.M. and S.V.
Funding
Funding for this work was provided by grants from the Natural Sciences and Engineering Research Council of Canada (S.V.) and the US National Institutes of Health [HD43092 to K.L.; GM088506 to B.P.]. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.096511/-/DC1
References
- Bantignies F., Cavalli G. (2011). Polycomb group proteins: repression in 3D. Trends Genet. 27, 454–464 [DOI] [PubMed] [Google Scholar]
- Bhaya D., Davison M., Barrangou R. (2011). CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 45, 273–297 [DOI] [PubMed] [Google Scholar]
- Calabrese J. M., Sun W., Song L., Mugford J. W., Williams L., Yee D., Starmer J., Mieczkowski P., Crawford G. E., Magnuson T. (2012). Site-specific silencing of regulatory elements as a mechanism of X inactivation. Cell 151, 951–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattanach B. M., Beechey C. V., Peters J. (2004). Interactions between imprinting effects in the mouse. Genetics 168, 397–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke H. J., Varmuza S., Prideaux V. R., Rossant J. (1988). The development potential of parthenogenetically derived cells in chimeric mouse embryos: implications for action of imprinted genes. Development 104, 175–182 [DOI] [PubMed] [Google Scholar]
- Davis T. L., Yang G. J., McCarrey J. R., Bartolomei M. S. (2000). The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum. Mol. Genet. 9, 2885–2894 [DOI] [PubMed] [Google Scholar]
- de Sousa Lopes S. M. C., Hayashi K., Surani M. A. (2007). Proximal visceral endoderm and extraembryonic ectoderm regulate the formation of primordial germ cell precursors. BMC Dev. Biol. 7, 140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiades P., Cox B., Gertsenstein M., Chawengsaksophak K., Rossant J. (2007). Trophoblast-specific gene manipulation using lentivirus-based vectors. Biotechniques 42, 317–325 [DOI] [PubMed] [Google Scholar]
- Goldberg A. D., Allis C. D., Bernstein E. (2007). Epigenetics: a landscape takes shape. Cell 128, 635–638 [DOI] [PubMed] [Google Scholar]
- Grimm C., Matos R., Ly-Hartig N., Steuerwald U., Lindner D., Rybin V., Müller J., Müller C. W. (2009). Molecular recognition of histone lysine methylation by the Polycomb group repressor dSfmbt. EMBO J. 28, 1965–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z., Mtango N. R., Patel B. G., Sapienza C., Latham K. E. (2008). Hybrid vigor and transgenerational epigenetic effects on early mouse embryo phenotype. Biol. Reprod. 79, 638–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z., Cheng Y., Liang C.-G., Latham K. E. (2010). Nuclear transfer in mouse oocytes and embryos. Methods Enzymol. 476, 171–184 [DOI] [PubMed] [Google Scholar]
- Hancock D. C., O’Reilly N. J. (2005). Production of polyclonal antibodies in rabbits. In Immunochemical Protocols (ed. Burns R.), pp. 27–39 Totowa, NJ: Humana Press; [PubMed] [Google Scholar]
- Hore T. A., Rapkins R. W., Graves J. A. M. (2007). Construction and evolution of imprinted loci in mammals. Trends Genet. 23, 440–448 [DOI] [PubMed] [Google Scholar]
- Klymenko T., Papp B., Fischle W., Köcher T., Schelder M., Fritsch C., Wild B., Wilm M., Müller J. (2006). A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 20, 1110–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin A., Han Z., Golding M. C., Mann M. R. W., Latham K. E., Varmuza S. (2008). The PcG gene Sfmbt2 is paternally expressed in extraembryonic tissues. Gene Expr. Patterns 8, 107–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre L. (2012). The placental imprintome and imprinted gene function in the trophoblast glycogen cell lineage. Reprod. Biomed. Online 25, 44–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- Lucifero D., Mann M. R. W., Bartolomei M. S., Trasler J. M. (2004). Gene-specific timing and epigenetic memory in oocyte imprinting. Hum. Mol. Genet. 13, 839–849 [DOI] [PubMed] [Google Scholar]
- Miri K., Varmuza S. (2009). Imprinting and extraembryonic tissues-mom takes control. Int Rev Cell Mol Biol 276, 215–262 [DOI] [PubMed] [Google Scholar]
- Mlynarczyk-Evans S., Royce-Tolland M., Alexander M. K., Andersen A. A., Kalantry S., Gribnau J., Panning B. (2006). X chromosomes alternate between two states prior to random X-inactivation. PLoS Biol. 4, e159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesterova T. B., Barton S. C., Surani M. A., Brockdorff N. (2001). Loss of Xist imprinting in diploid parthenogenetic preimplantation embryos. Dev. Biol. 235, 343–350 [DOI] [PubMed] [Google Scholar]
- Okae H., Hiura H., Nishida Y., Funayama R., Tanaka S., Chiba H., Yaegashi N., Nakayama K., Sasaki H., Arima T. (2012). Re-investigation and RNA sequencing-based identification of genes with placenta-specific imprinted expression. Hum. Mol. Genet. 21, 548–558 [DOI] [PubMed] [Google Scholar]
- Ono R., Nakamura K., Inoue K., Naruse M., Usami T., Wakisaka-Saito N., Hino T., Suzuki-Migishima R., Ogonuki N., Miki H., et al. (2006). Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat. Genet. 38, 101–106 [DOI] [PubMed] [Google Scholar]
- Park I. K., Qian D., Kiel M., Becker M. W., Pihalja M., Weissman I. L., Morrison S. J., Clarke M. F. (2003). Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423, 302–305 [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W., Horgan G. W., Dempfle L. (2002). Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30, e36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston A., Cox B. J., Nishioka N., Sasaki H., Chea E., Rugg-Gunn P., Guo G., Robson P., Draper J. S., Rossant J. (2010). Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development 137, 395–403 [DOI] [PubMed] [Google Scholar]
- Renfree M. B., Ager E. I., Shaw G., Pask A. J. (2008). Genomic imprinting in marsupial placentation. Reproduction 136, 523–531 [DOI] [PubMed] [Google Scholar]
- Rivera R. M., Stein P., Weaver J. R., Mager J., Schultz R. M., Bartolomei M. S. (2008). Manipulations of mouse embryos prior to implantation result in aberrant expression of imprinted genes on day 9.5 of development. Hum. Mol. Genet. 17, 1–14 [DOI] [PubMed] [Google Scholar]
- Stern C. D., Downs K. M. (2012). The hypoblast (visceral endoderm): an evo-devo perspective. Development 139, 1059–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Kunath T., Hadjantonakis A.-K., Nagy A., Rossant J. (1998). Promotion of trophoblast stem cell proliferation by FGF4. Science 282, 2072–2075 [DOI] [PubMed] [Google Scholar]
- Tiscornia G., Singer O., Verma I. M. (2006). Production and purification of lentiviral vectors. Nat. Protoc. 1, 241–245 [DOI] [PubMed] [Google Scholar]
- Varmuza S., Mann M., Rogers I. (1993). Site of action of imprinted genes revealed by phenotypic analysis of parthenogenetic embryos. Dev. Genet. 14, 239–248 [DOI] [PubMed] [Google Scholar]
- Waddington C. H. (1940). Organisers and Genes. Cambridge: Cambridge University Press; [Google Scholar]
- Wang Q., Chow J., Hong J., Smith A. F., Moreno C., Seaby P., Vrana P., Miri K., Tak J., Chung E. D., et al. (2011). Recent acquisition of imprinting at the rodent Sfmbt2 locus correlates with insertion of a large block of miRNAs. BMC Genomics 12, 204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeigler B. M., Sugiyama D., Chen M., Guo Y., Downs K. M., Speck N. A. (2006). The allantois and chorion, when isolated before circulation or chorio-allantoic fusion, have hematopoietic potential. Development 133, 4183–4192 [DOI] [PubMed] [Google Scholar]
- Zhang J., Bonasio R., Strino F., Kluger Y., Holloway J. K., Modzelewski A. J., Cohen P. E., Reinberg D. (2013). SFMBT1 functions with LSD1 to regulate expression of canonical histone genes and chromatin-related factors. Genes Dev. 27, 749–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.