Abstract
Smoking influences body weight such that smokers weigh less than non-smokers and smoking cessation often leads to weight increase. The relationship between body weight and smoking is partly explained by the effect of nicotine on appetite and metabolism. However, the brain reward system is involved in the control of the intake of both food and tobacco. We evaluated the effect of single-nucleotide polymorphisms (SNPs) affecting body mass index (BMI) on smoking behavior, and tested the 32 SNPs identified in a meta-analysis for association with two smoking phenotypes, smoking initiation (SI) and the number of cigarettes smoked per day (CPD) in an Icelandic sample (N=34 216 smokers). Combined according to their effect on BMI, the SNPs correlate with both SI (r=0.019, P=0.00054) and CPD (r=0.032, P=8.0 × 10−7). These findings replicate in a second large data set (N=127 274, thereof 76 242 smokers) for both SI (P=1.2 × 10−5) and CPD (P=9.3 × 10−5). Notably, the variant most strongly associated with BMI (rs1558902-A in FTO) did not associate with smoking behavior. The association with smoking behavior is not due to the effect of the SNPs on BMI. Our results strongly point to a common biological basis of the regulation of our appetite for tobacco and food, and thus the vulnerability to nicotine addiction and obesity.
Keywords: addiction, body mass index, nicotine dependence, obesity, smoking
Introduction
Smoking and obesity are major risk factors for many serious diseases.1, 2 Eating and smoking are behavioral traits that are at least in part controlled by the same reward mechanisms.3 Genome-wide association studies (GWAS) have yielded 32 single-nucleotide polymorphisms (SNPs) associated with body mass index (BMI).4 Smoking and SNPs associated with increased smoking quantity have been shown to correlate with lower BMI.5, 6
According to the World Health Organization (WHO), more than one billion people smoke and over 400 million people are obese (BMI >30 kg m−2), with both prevalences rising (see url section). Eating can become compulsive, and the neurobiological processes relating to overindulgence in food overlap with those involved in substance abuse and addiction.3 All drugs of abuse have been shown to increase dopamine in the mesolimbic reward system, and studies of both human brain images3 and animal brains7 have revealed that similar neurocircuits are involved in the regulation of rewarding and reinforcement in drug addiction and compulsive eating. Based on the many similarities between hyperphagia and excessive drug use in addiction, it has even been suggested that some forms of obesity should be included as a diagnosis in future editions of the Diagnostic and Statistical Manual of Mental Disorders.8, 9
Smoking influences body weight, such that smokers weigh less than non-smokers, and smoking cessation is often accompanied by an increase in weight.5 These effects have been largely attributed to nicotine that increases the metabolic rate and suppresses appetite. Although increased food intake upon smoking cessation is partly explained by a reward substitution mechanism, as food intake is increased to make up for the lack of nicotine, the absence of nicotine has also been shown to increase the reward value of certain foods.10 At the molecular level, these effects are most likely achieved through activation of the nicotinic acetylcholine receptors. The melanocortin (MC) system has a key role in regulating body weight,11 and nicotine was recently shown to interact directly with the MC system in the brain through activation of α3β4 nicotinic acetylcholine receptors on pro-opiomelanocortin (POMC) neurons12 in the arcuate nucleus of the hypothalamus. The POMC neurons project to secondary neurons influencing appetite, and nicotine activation leads to the release of melanocortin-4 agonists activating MC4 receptors in the paraventricular nucleus producing appetite suppression, an effect that is absent from POMC KO mice.12
However, the relationship between smoking phenotypes and obesity is more complicated than can be accounted for by the known effects of nicotine on appetite and metabolism. This is evident from the fact that the number of cigarettes smoked per day (CPD) correlates with elevated BMI.13, 14 Thus, although smokers weigh less than non-smokers, heavy smokers indeed weigh more than light smokers.
BMI and smoking data are widely available from various studies and large sample sizes have been obtained for GWAS of BMI4 and some smoking phenotypes,15, 16, 17 and these studies have uncovered a number of variants associating with obesity (BMI) and with smoking behavior. The variant most strongly correlating with CPD,15, 16, 17 rs1051730-A/rs16969968-A, correlates with reduced BMI both in current and former smokers, but does not have an impact on the BMI of never smokers.6 This observation is consistent with the notion that smoking influences body weight through nicotine's effects on body and brain, the increase of metabolic rate and suppression of appetite. Here we report how variants correlating with BMI influence smoking behavior.
Materials and methods
Study subjects
Written informed consent was obtained from all subjects. Inclusion in the study required the availability of genotypes from ongoing SNP array typing in Iceland or previous GWAS,15, 16, 17 and the study populations have all been described previously.15, 16, 17 The GWAS of smoking initiation (SI) involved comparison of ever smokers and never smokers, and the studies of smoking quantity probed CPD as a quantitative trait among smokers only. The definitions of smokers and never smokers varied somewhat between studies,15, 16, 17 as questions addressing smoking behavior varied with most studies probing for regular smoking over a certain period of time. Questions probing for smoking quantity also varied between studies, and for analysis of smoking quantity we used CPD data for smokers in categories with each category representing 10 CPD (effect size of 0.1=1 CPD).15, 16, 17 CPD at the time of smoking was used for past smokers, and never smokers were excluded from analysis of CPD. All subjects were of European descent. The total sample sizes were N=100 860 and N=161 490 for CPD and SI, respectively.
Icelandic study design
A generalized form of linear regression was used to test the correlation between quantitative traits (BMI and height) and smoking phenotypes (CPD and SI) in Iceland. The generalized form assumes that the smoking behavior of related individuals is correlated proportional to the kinship between them rather than assuming that the smoking phenotypes of all individuals are independent. Let y be the vector of smoking behavior measurements, and let x be the vector of BMI or height measurements. We assume that the expectation of the smoking behavior depends linearly on BMI or height, Ey=α+βx, and that the variance–covariance matrix of the smoking behavior depends only on the pairwise kinship between the study participants, Var(y)=2σ2Φ, where
is based on the kinship between individuals as estimated from the Icelandic genealogical database (kij) and an estimate of the heritability of the trait (ρ). Assuming normally distributed errors, the maximum likelihood method gives estimates for β, which will asymptotically follow a normal distribution and can be used to estimate the correlation between height and BMI on the one side and CPD and SI on the other.
In order to test the correlation between the set of 32 BMI SNPs or the set of 180 height SNPs and smoking behavior, the same type of analysis was performed replacing the observed BMI and height with the BMI and height predicted based on the sets of 32 and 180 SNPs. We shall describe how this was achieved for BMI, the analysis for height being conceptually identical. For each of the 32 SNPs reported to associate with BMI, let fi be its minor allele frequency and γi be its published effect on BMI. For an individual with gi minor alleles at SNP i, the set of 32 BMI SNPs predict a BMI of
Conditional independence
We observe a correlation between the 32 BMI SNPs and smoking behavior. The 32 BMI SNPs associate with BMI and BMI associates with CPD. The question then arises of whether the correlation between the 32 BMI SNPs and CPD is all going through BMI. In other words, are the 32 BMI SNPs and CPD correlated conditional on BMI? Assuming that the 32 BMI SNPs and CPD are independent conditional on BMI, then the correlation between the 32 BMI SNPs and CPD will be the product of the correlation between the 32 BMI SNPs and BMI and the correlation between BMI and CPD. Denoting the estimator for the correlation between the 32 BMI SNPs and BMI with 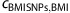 , and the variance of the estimator with
, and the variance of the estimator with 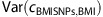 , and similarly for the correlation between BMI and CPD. Then,
, and similarly for the correlation between BMI and CPD. Then, 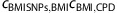 is an estimator of the correlation between the 32 BMI SNPs and CPD, assuming conditional independence, and
is an estimator of the correlation between the 32 BMI SNPs and CPD, assuming conditional independence, and 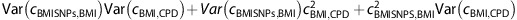 gives an estimate of the variance of the estimator. A standard test for the mean of two samples can now be applied to test the difference between the observed correlation between the 32 BMI SNPs and CPD and the correlation predicted based on the 32 BMI SNPs and CPD being independent conditional on BMI.
gives an estimate of the variance of the estimator. A standard test for the mean of two samples can now be applied to test the difference between the observed correlation between the 32 BMI SNPs and CPD and the correlation predicted based on the 32 BMI SNPs and CPD being independent conditional on BMI.
Replication outside of Iceland
The non-Icelandic studies shared only summary results from the genome-wide smoking behavior association scans in the form of effect sizes, P-values and allele frequencies. The ∼2.5 million SNPs from the HapMap dataset were imputed and tested for association within each study population.15, 16, 17 The significance levels of each study population were adjusted individually using the method of genomic control.18 We used standard fixed-effects additive meta-analysis to combine the results for each SNP. After combining the results from all the populations, we again applied the method of genomic control and adjusted both smoking phenotypes accordingly (λGC=1.10 and λGC=1.06 for SI and CPD, respectively).
As data were not available on the individual level, we could not predict SI and CPD on the individual level as was done in Iceland. In order to test for the association of the 32 SNPs associating with BMI and the 180 SNPs associating with height with smoking behavior, we weighted the combined significance over all the populations of each SNP by the expected z-score associated with the SNP, assuming that the effect on smoking behavior was proportional to the effect on BMI or height as follows. Again let us take BMI as an example. For each of the 32 SNPs reported to associate with BMI, let fi be its minor allele frequency and γi be its published effect on BMI. We denote the unknown effect of each SNP on smoking behavior with βi and our assumption about the SNP's effect on smoking behavior being proportional to the SNP's effect on BMI can be stated as βi=kγi for some constant k. Quantifying the signifiance of the association of each SNP with smoking behavior by its z-score zi, maximal power is achieved by weighing the SNPs according to the expected z-score. The expected z-score for the ith SNP is proportional to 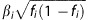 , which we assume is proportional to
, which we assume is proportional to 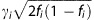 , which we will refer to as wi and use to weigh the smoking behavior z-scores of the 32 BMI SNPs together:
, which we will refer to as wi and use to weigh the smoking behavior z-scores of the 32 BMI SNPs together: 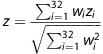 .
.
Results and discussion
To study the correlation between obesity variants and smoking phenotypes, we focused on the 32 SNPs associating with BMI described in a recent report of a study of 249 796 subjects.4 We weighted the 32 SNPs together based on their published effect on BMI and tested the correlation with both CPD and SI in 49 565 chip-typed Icelanders (Table 1). We also tested the correlation between the actual measured BMI and the smoking phenotypes in a slightly larger set of Icelanders. For comparison, we performed a corresponding study using Icelandic data on human height and 180 SNPs reported to influence human height in a recent study of 183 731 individuals19 (Table 1).
Table 1. Association of BMI, height and SNPs associating with BMI and height with smoking phenotypes in Iceland.
| |
CPD |
|
|
Smoking |
|
|
|---|---|---|---|---|---|---|
| From | N | Correlation (95% CI) | P | N | Correlation (95% CI) | P |
| BMI | 33 620 | 0.095 (0.085, 0.106) | 2.5 × 10−68 | 49 565 | −0.005 (−0.014, 0.004) | 0.29 |
| 32 BMI SNPs | 24 618 | 0.032 (0.019, 0.045) | 8.0 × 10−7 | 34 216 | 0.019 (0.008, 0.030) | 0.00054 |
| Height | 33 875 | −0.004 (−0.015, 0.007) | 0.46 | 49 931 | −0.012 (−0.021, −0.002) | 0.013 |
| 180 Height SNPs | 24 630 | 0.001 (−0.011, 0.014) | 0.84 | 34 231 | 0.004 (−0.007, 0.015) | 0.44 |
Abbreviations: BMI, body mass index; CI, confidence interval; SNP, single-nucleotide polymorphism.
BMI associated with CPD (r=0.095, P=2.5 × 10−68) but not SI (r=−0.005, P=0.29), whereas height did not associate with CPD (r=−0.004, P=0.46) and showed only weak association with SI (r=−0.012, P=0.013). The set of 32 BMI SNPs associated with both CPD (r=0.032, P=8.0 × 10−7) and SI (r=0.019, P=0.00054), whereas the set of 180 height SNPs associated with neither smoking behavior (P=0.84 and 0.44 for CPD and SI, respectively).
The correlation between the set of 32 BMI SNPs and BMI and the correlation between BMI and CPD predict a correlation between the 32 BMI SNPs and CPD of 0.013, which is significantly lower than the observed correlation of 0.032 between the set of 32 BMI SNPs and CPD (P=0.0033). The correlation between BMI and SI is negative so that the predicted correlation between the 32 BMI SNPs and SI is also negative and even more significantly different from the observed correlation of 0.019 than from 0. Hence, the observed associations between the BMI variants and the smoking phenotypes are not explained by the direct phenotypic correlations between BMI and smoking behavior.
To investigate the contributions of individual SNPs and to replicate our observations in other populations, we looked up the correlations of each of the 32 SNPs with CPD and SI, using data from our previous studies outside of Iceland15, 16, 17 (N=76 242 for CPD, and N=127 274 for SI). For these studies, we utilized the fixed-effect additive meta-analysis results for ∼2 500 000 SNPs obtained using the inverse-variance method for each of the two smoking phenotypes. Before conducting the meta-analysis, we performed a genomic control correction of each study.18 The combined χ2-test statistics were still somewhat inflated by a factor of λGC=1.10 (SI) and λGC=1.06 (CPD). The correlations between the set of 32 BMI SNPs and the two smoking variables were significant in this replication sample with P=1.2 × 10−5 and 9.3 × 10−5, for SI and CPD, respectively. Combined with Iceland, the association between the 32 BMI SNPs and SI and CPD reached a significance of P=1.2 × 10−7 and P=1.6 × 10−9, respectively.
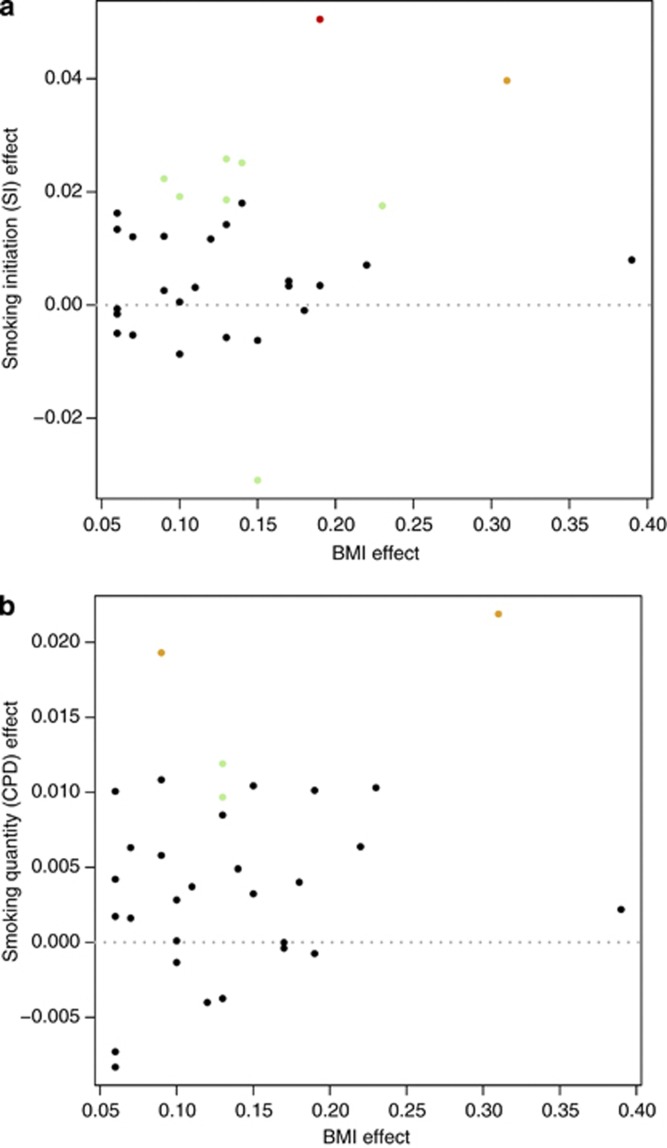
As expected, based on the correlations observed between the combined set of the 32 BMI SNPs (Table 1), we observe congruence in the effects that these SNPs have on BMI and smoking behavior. For most of the SNPs, the allele that associates with increased BMI also associates with both increased probability of SI and higher CPD (Figure 1). We note that the effect sizes are small and although the markers as a group clearly associate with the smoking behaviors, further studies are required to determine unequivocally which of the markers have an impact on smoking behavior. The SNP by far most strongly associated with BMI (rs1558902-A in FTO) represents a notable exception from the trend observed and shows no evidence for association with either CPD or SI.
Figure 1.
Association of obesity variants with smoking initiation (SI) and CPD. The effects on smoking behaviors are depicted vs the effects on BMI from a large meta-analysis.4 (a) The effect on smoking initiation vs the effect on BMI. (b) The effect on CPD vs the effect on BMI. The BMI effect is in standard units, and the effects on SI and CPD were obtained using a standard fixed-effects additive meta-analysis to combine the results for each SNP from Iceland with additional data from three large GWAS.15, 16, 17 The effects on SI are the β-values from logistic regression treating ever smoking as the response and the allele counts as covariates, and the GWAS of CPD used smoking quantity in categories with each category representing 10 CPD (effect size of 0.1=1 CPD). The dots representing each data point are color coded to indicate the p-value obtained as red (P<0.0001), yellow (P<0.001), green (P<0.05) and black (P≥0.05) and the input data are provided in (Supplementary Table 1).
Considering the 11 BMI SNPs most strongly associated with smoking (P<0.05), 9 SNPs associate with smoking initiation and 4 with CPD (Supplementary Table 1 and Figure 1). For smoking initiation the most significant associations were to rs10767664-A (effect=0.050495, P=1.14 × 10−6) in the Brain Neurotrophin Factor gene (BDNF) and rs2867125-C (effect=0.0397, P=0.000021) 45 kb upstream of the Transmembrane protein 18 gene (TMEM18), and for CPD the most significant associations were with rs2867125-C (effect=0.286, P=0.000346) (TMEM18) and rs4771122-G (effect=0.0193, P=0.00048) in the mitochondrial translational initiation factor 3 gene (MTIF3). In addition to rs286125-C (TMEM18), rs2815752-A (NEGR1) is among the top markers (P<0.05) for both SI (effect=0.186, P=0.0244) and CPD (effect=0.0097, P=0.0305). A SNP within the BDNF gene has previously been shown to associate with smoking initiation (rs6265-C).16 This SNP is in linkage disequilibrium with the BMI-associated rs10767664 (r2=0.85 in Iceland). The association with SI remains significant after removing rs10767664 (P=1.3 × 10−5).
In summary, we have demonstrated that as a group, the 32 common variants identified in GWAS of BMI4 also have an impact on the smoking behavior. A variant within the nAChR gene cluster at chrs 15q25 (rs1051730-A) was discovered in GWAS of smoking behavior20, 21 and subsequently shown to correlate with reduced BMI in smokers without an effect on the BMI of never smokers,6 thus most likely influencing BMI mainly through its effect on smoking behavior. The variants studied here represent a different class of SNPs affecting both BMI and smoking: They were found in GWAS of BMI and influence BMI in both smokers and never smokers, and the alleles correlating with elevated BMI tend to increase the propensity to smoke and/or associate with increased cigarette intake. We note that, in Iceland, the correlation between the predicted BMI and observed BMI is similar for smokers (0.15, P=3.0 × 10−97, N=20 462) and never smokers (0.13, P=7.2 × 10−33, N=7910). The direction of this trend is opposite to what would be expected based on the known effects of nicotine on BMI, and inconsistent with an effect rooted in nicotine-mediated increase of metabolic rate and suppression of appetite. That the majority of variants known to associate with elevation of BMI correlate with smoking behaviors in this manner points to a common biological basis to regulation of the intake of food and tobacco.
Acknowledgments
We thank the participants in the genetic studies whose contributions made this work possible. This work was supported in part by NIH (R01-DA017932 and R01-DA022522) and the European Commission's Sixth Framework Programme, Integrated Project GENADDICT (LSHM-CT-2004-005166). The ENGAGE smoking consortium was formed through a component of the Integrated Project ENGAGE, supported by the European Commission's Seventh Framework Program, grant agreement HEALTH-F4–2007–201413. SB was funded by the FP7-PEOPLE-2009-IAPP 251592 grant (NextGene).
Author Contributions
TET, DFG, and KS wrote the manuscript. The study was designed by and the results interpreted by TET, DFG, PS, SB, UT and KS. The meta-analyses of smoking GWAS data were performed by DFG. TET, DFG, PS, SB,US, GT, BW and VS worked on data management and analysis. Smoking GWAS consortia were coordinated by HF (TAG), PFS(TAG) JM (OX-GSK) and MIM (ENGAGE). All authors contributed to the final version of the paper.
CONSORTIA
The data utilized came from three large GWAS done by the ENGAGE, TAG, and OX-GSK consortia (references 15–17). The additional collaborators from these three consortia are listed below.
ENGAGE Consortium—Ida Surakka8,9, Jacqueline M Vink10, Najaf Amin11, Frank Geller12, Thorunn Rafnar1, Tõnu Esko13,14, Stefan Walter11, Christian Gieger15, Rajesh Rawal15, Massimo Mangino16, Inga Prokopenko5,6, Reedik Mägi5,6,13, Kaisu Keskitalo19, Iris H. Gudjonsdottir1, Solveig Gretarsdottir1, Hreinn Stefansson1, Yurii S Aulchenko11, Mari Nelis13,14, Katja K Aben21,22, Martin den Heijer21,23, Nicole Soranzo16,24, Ana M Valdes16, Claire Steves16, André G Uitterlinden11,25, Albert Hofman6, Anke Tönjes26,27, Peter Kovacs28, Jouke Jan Hottenga10, Gonneke Willemsen10, Nicole Vogelzangs29, Angela Döring15, Norbert Dahmen30, Barbara Nitz15, Samuli Ripatti8,9, Markus Perola9,13, Johannes Kettunen24, Anna-Liisa Hartikainen30, Anneli Pouta31, Jaana Laitinen32, Matti Isohanni30, Shen Huei-Yi8,9, Maxine Allen5, Maria Krestyaninova33, Alistair S Hall34, John R Thompson35, Hogni Oskarsson36, Thorarinn Tyrfingsson37, Lambertus A Kiemeney21,22,38, Marjo-Riitta Järvelin31,39,40,41, Veikko Salomaa9, Michael Stumvoll26, Tim D Spector16, H-Erich Wichmann15,42,43, Andres Metspalu13,14, Nilesh J Samani44, Brenda W Penninx29, Ben A Oostra45, Dorret I Boomsma10, Henning Tiemeier11, Cornelia M van Duijn11, Jaakko Kaprio8,19,46, Jeffrey R Gulcher1
1Decode genetics/AMGEN, Sturlugata 8, Reykjavik, Iceland. 5Wellcome Trust Center of Human Genetics, Oxford, UK 6Oxford Centre for Diabetes, Endocrinology and Metabolism, University of Oxford, Oxford, UK. 8Institute for Molecular Genetics Finland, FIMM, University of Helsinki, Finland. 9National Institute for Health and Welfare, Helsinki, Finland. 10Department of Biological Psychology, VU University Amsterdam, Amsterdam, The Netherlands. 11Department of Epidemiology, Erasmus University Medical Center, Rotterdam, The Netherlands. 12Department of Epidemiology Research, Statens Serum Institut, Copenhagen, Denmark. 13Estonian Genome Center, University of Tartu, Riia 23b, Tartu 51010, Estonia. 14IMCB of University of Tartu and Estonian Biocentre, Riia str 23, Tartu 51010, Estonia. 15Institute of Epidemiology, Helmholtz Zentrum München, Ingolstaedter Landstr. 1, 85764 Munich/Neuherberg, Germany. 16Department of Twin Research and Genetic Epidemiology, King's College London, St Thomas' Hospital Campus, London SE17EH, UK. 19Department of Public Health, University of Helsinki, Helsinki, Finland. 21Radboud University Nijmegen Medical Centre, Department Of Epidemiology, Biostatistics and HTA, Nijmegen, The Netherlands. 22Comprehensive Cancer Centre East, Nijmegen, The Netherlands. 23Radboud University Nijmegen Medical Centre, Department of Endocrinology, Nijmegen, The Netherlands. 24Wellcome Trust Sanger Institute, Hinxton, UK. 25Department of Internal Medicine, Erasmus University Medical Center, Rotterdam, The Netherlands. 26Department of Medicine, University of Leipzig, Liebigstr. 18, 04103, Leipzig, Germany. 27Coordination Centre for Clinical Trials, University of Leipzig, Härtelstr. 16–18, 04103, Leipzig, Germany. 28Interdisciplinary Centre for Clinical Research, University of Leipzig,Inselstr. 22, 04103, Leipzig, Germany. 29EMGO Institute/Department of Psychiatry, VU University Medical Center, Amsterdam, The Netherlands. 30Department of Psychiatry, University of Mainz, Mainz, Germany. 30Institute of Clinical Medicine, University of Oulu, Oulo, Finland. 31Lifecourse and service Department, National Institute of Health and Welfare, Oulu, Finland. 32Finnish Institute of Occupational Health, Oulo, Finland. 33European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, CB10 1 SD, UK. 34Multidisciplinary Cardiovascular Research Centre (MCRC), Leeds Institute of Genetics, Health and Therapeutics (LIGHT), University of Leeds, Leeds, LS2 9JT, UK. 35Department of Health Sciences and Genetics, University of Leicester, LE1 7RH Leicester, UK. 36Therapeia, 101 Reykjavik, Iceland. 37Vogur SAA Addiction Treatment Center, Reykjavik, Iceland. 38Radboud University Nijmegen Medical Centre, Department. of Urology, Nijmegen, The Netherlands. 39Department of Epidemiology and Public Health, Imperial College, Faculty of Medicine, London, UK. 40Institute of Health Sciences, University of Oulu, Oulu, Finland. 41Biocenter Oulu, University of Oulu, Oulu, Finland. 42Institute of Medical Informatics, Biometry and Epidemiology, Ludwig-Maximilians-Universität, Munich, Germany. 43Klinikum Grosshadern, Munich, Germany.44Department of Cardiovascular Sciences, University of Leicester, Clinical Sciences Wing, Glenfield Hospital, Groby Road, Leicester, LE3 9QP, UK. 45Department of Clinical Genetics, Erasmus University Medical Center, Rotterdam, The Netherlands. 46Department of Mental Health and Alcohol Abuse Services, National Institute for Health and Welfare, Helsinki, Finland.
The Tobacco and Genetics Consortium (TAG)–Yun Jung Kim1, Jennifer Dackor1, Eric Boerwinkle3, Nora Franceschini4, Diego Ardissino5, Luisa Bernardinelli6,7, Pier M Mannucci8, Francesco Mauri9, Piera A Merlini9, Devin Absher10, Themistocles L Assimes11, Stephen P Fortmann12, Carlos Iribarren13, Joshua W Knowles11, Thomas Quertermous11, Luigi Ferrucci14, Toshiko Tanaka15, Joshua C Bis16,17, Curt D Furberg18, Talin Haritunians19, Barbara McKnight16,20, Bruce M Psaty16,17,21,22, Kent D Taylor19, Evan L Thacker16,23, Peter Almgren24, Leif Groop24, Claes Ladenvall24, Michael Boehnke25, Anne U Jackson25, Karen L Mohlke1,2, Heather M Stringham25, Jaakko Tuomilehto26–28, Emelia J Benjamin29,30, Shih-Jen Hwang31, Daniel Levy32, Sarah Rosner Preis31, Ramachandran S Vasan29,32, Jubao Duan33, Pablo V Gejman33, Douglas F Levinson34, Alan R Sanders33, Jianxin Shi35, Esther H Lips36, James D McKay36, Antonio Agudo37, Luigi Barzan38, Vladimir Bencko39, Simone Benhamou40,41, Xavier Castellsagué37, Cristina Canova42, David I Conway43, Eleonora Fabianova44, Lenka Foretova45, Vladimir Janout46, Claire M Healy47, Ivana Holcátová39, Kristina Kjaerheim48, Pagona Lagiou49, Jolanta Lissowska50, Ray Lowry51, Tatiana V Macfarlane52, Dana Mates53, Lorenzo Richiardi54, Peter Rudnai55, Neonilia Szeszenia-Dabrowska56, David Zaridze57, Ariana Znaor58, Mark Lathrop59,60, Paul Brennan36, Stefania Bandinelli61, Timothy M Frayling62, Jack M Guralnik63, Yuri Milaneschi64, John R B Perry62, David Altshuler65–70, Roberto Elosua71, Sek Kathiresan65,68,72, Gavin Lucas71, Olle Melander73, Christopher J O'Donnell74, Veikko Salomaa75, Stephen M Schwartz16, Benjamin F Voight76, Brenda W Penninx77,78, Johannes H Smit77,78, Nicole Vogelzangs77,78, Dorret I Boomsma79, Eco J C de Geus79, Jacqueline M Vink79, Gonneke Willemsen79, Stephen J Chanock80, Fangyi Gu81, Susan E Hankinson82, David J Hunter81, Albert Hofman83, Henning Tiemeier83,84, Andre G Uitterlinden83,85, Cornelia M van Duijn83,86, Stefan Walter83,87, Daniel I Chasman88, Brendan M Everett88,89, Guillaume Paré88, Paul M Ridker88,89, Ming D Li90, Hermine H Maes91,92, Janet Audrain-McGovern93, Danielle Posthuma94,95, Laura M Thornton96, Caryn Lerman93,97, Jaakko Kaprio26,75,98, Jed E Rose99, John P A Ioannidis100–102, Peter Kraft81 and Dan-Yu Lin103.
1Department of Genetics, University of North Carolina, Chapel Hill, North Carolina, USA. 2University of North Carolina Lineberger Comprehensive Cancer Center, University of North Carolina, Chapel Hill, North Carolina, USA. 3Human Genetics Center and Institute for Molecular Medicine, University of Texas Health Science Center, Houston, Texas, USA. 4Department of Epidemiology, University of North Carolina, Chapel Hill, North Carolina, USA. 5Division of Cardiology, Azienda Ospedaliero-Universitaria di Parma, Parma, Italy. 6Statistical Laboratory, Centre for Mathematical Sciences, University of Cambridge, Cambridge, UK. 7Department of Applied Health Sciences, University of Pavia, Pavia, Italy. 8Department of Internal Medicine and Medical Specialties, Fondazione Istituto di Ricovero e Cura a Carattere Scientifico, Ospedale Maggiore, Mangiagalli e Regina Elena, University of Milan, Milan, Italy. 9Department of Cardiology, Azienda Ospedaliera Niguarda Ca' Granda, Milan, Italy. 10HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA. 11Cardiovascular Medicine, Stanford University, Stanford, California, USA. 12Stanford Prevention Research Center, Stanford University, Stanford, California, USA. 13Kaiser Permanente Northern California Division of Research, Oakland, California, USA. 14National Institute on Aging, Baltimore, Maryland, USA. 15Medstart Research Institute, National Institute on Aging, Baltimore, Maryland, USA. 16Cardiovascular Health Research Unit, University of Washington, Seattle, Washington, USA. 17Department of Medicine, University of Washington, Seattle, Washington, USA. 18Division of Public Health Sciences, Wake Forest University Health Sciences, Winston-Salem, North Carolina, USA. 19Medical Genetics Institute, Cedars-Sinai Medical Center, Los Angeles, California, USA. 20Department of Biostatistics, University of Washington, Seattle, Washington, USA. 21Department of Epidemiology and Health Services, University of Washington, Seattle, Washington, USA. 22Group Health Research Institute, Seattle, Washington, USA. 23Department of Epidemiology, University of Washington, Seattle, Washington, USA. 24Department of Clinical Sciences, Diabetes and Endocrinology Unit, Lund University, Malmö, Sweden. 25Department of Biostatistics, School of Public Health, University of Michigan, Ann Arbor, Michigan, USA. 26Hjelt Institute, Department of Public Health, University of Helsinki, Helsinki, Finland. 27Diabetes Prevention Unit, National Institute for Health and Welfare, Helsinki, Finland. 28Finland South Ostrobothnia Central Hospital, Seinäjoki, Finland. 29Boston University School of Medicine, Boston, Massachusetts, USA. 30Boston University School of Public Health, Boston, Massachusetts, USA. 31Center for Population Studies, National Heart, Lung and Blood Institute, Bethesda, Maryland, USA. 32Department of Medicine, Sections of Preventive Medicine and Cardiology, Boston University School of Medicine, Boston, Massachusetts, USA. 33Center for Psychiatric Genetics, NorthShore University HealthSystem Research Institute, Evanston, Illinois, USA. 34Department of Psychiatry and Behavioral Sciences, Stanford University, Stanford, California, USA. 35Biostatistics Branch, Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, Maryland, USA. 36International Agency for Research on Cancer (IARC), Lyon, France. 37Institut Català d'Oncologia, Barcelona, Spain. 38General Hospital, Pordenone, Italy. 39Institute of Hygiene and Epidemiology, First Faculty of Medicine, Charles University, Prague, Czech Republic. 40Institut National de la santé et de la Recherche Medicalé (INSERM) U794, Paris, France. 41Institut Gustave Roussy, Villejuif, France. 42Department of Environmental Medicine and Public Health, University of Padua, Padua, Italy. 43University of Glasgow Medical Faculty Dental School, Glasgow, UK. 44Specialized Institute of Hygiene and Epidemiology, Banska Bystrica, Slovakia. 45Department of Cancer Epidemiology and Genetics, Masaryk Memorial Cancer Institute, Brno, Czech Republic. 46Palacky University, Olomouc, Czech Republic. 47Trinity College School of Dental Science, Dublin, Ireland. 48Cancer Registry of Norway, Oslo, Norway. 49University of Athens School of Medicine, Athens, Greece. 50Department of Cancer Epidemiology and Prevention, Maria Sklodowska-Curie Cancer Center and Institute of Oncology, Warsaw, Poland. 51University of Newcastle Dental School, Newcastle, UK. 52University of Aberdeen School of Medicine, Aberdeen, UK. 53Institute of Public Health, Bucharest, Romania. 54Center for Experimental Research and Medical Studies, University of Turin, Turin, Italy. 55National Institute of Environmental Health, Budapest, Hungary. 56Department of Epidemiology, Institute of Occupational Medicine, Lodz, Poland. 57Institute of Carcinogenesis, Cancer Research Centre, Moscow, Russia. 58Croatian National Cancer Registry, Zagreb, Croatia. 59Centre National de Genotypage, Institut Genomique, Comissariat à l'énergie Atomique, Evry, France. 60Fondation Jean Dausset-Centre d‘Étude du Polymorphisme Humain (CEPH), Paris, France. 61Geriatric Unit, Azienda Sanitaria di Firenze, Florence, Italy. 62Genetics of Complex Traits, Peninsula Medical School, The University of Exeter, Exeter, UK. 63Laboratory of Epidemiology, Demography and Biometry, National Institute on Aging, Bethesda, Maryland, USA. 64Tuscany Health Regional Agency, Florence, Italy. 65Broad Institute of Harvard and Massachusetts Institute of Technology, Cambridge, Massachusetts, USA. 66Department of Molecular Biology, Massachusetts General Hospital, Boston, Massachusetts, USA. 67Diabetes Unit, Massachusetts General Hospital, Boston, Massachusetts, USA. 68Center for Human Genetics Research, Massachusetts General Hospital, Boston, Massachusetts, USA. 69Department of Genetics, Harvard Medical School, Boston, Massachusetts, USA. 70Department of Medicine, Harvard Medical School, Boston, Massachusetts, USA. 71Cardiovascular Epidemiology and Genetics, Institut Municipal d'Investigacio Medica, Barcelona, Spain. 72Harvard Medical School, Boston, Massachusetts, USA. 73Department of Clinical Sciences, Hypertension and Cardiovascular Diseases, University Hospital Malmö, Lund University, Malmö, Sweden. 74National Heart, Lung, and Blood Institute's Framingham Heart Study, Framingham, Massachusetts, USA. 75National Institute for Health and Welfare (THL), Helsinki, Finland. 76Program in Medical and Population Genetics, Broad Institute of Harvard and Massachusetts Institute of Technology, Cambridge, Massachusetts, USA. 77EMGO Institute, Vrije Universiteit (VU) Medical Center, Amsterdam, The Netherlands. 78Department of Psychiatry, VU University Medical Center, Amsterdam, The Netherlands. 79Biological Psychology, VU University Amsterdam, Amsterdam, The Netherlands. 80Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, Maryland, USA. 81Program in Molecular and Genetic Epidemiology, Department of Epidemiology, Harvard University, Boston, Massachusetts, USA. 82Channing Laboratory, Department of Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, Massachusetts, USA. 83Department of Epidemiology, Erasmus Medical Center, Member of the Netherlands Consortium on Healthy Aging, Rotterdam, The Netherlands. 84Department of Child and Adolescent Psychiatry, Erasmus Medical Center, Rotterdam, The Netherlands. 85Department of Internal Medicine, Erasmus Medical Center, Rotterdam, The Netherlands. 86Centre for Medical Systems Biology, Erasmus Medical Center, Rotterdam, The Netherlands. 87Department of Public Health, Erasmus Medical Center, Rotterdam, The Netherlands. 88Division of Preventive Medicine, Department of Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, Massachusetts, USA. 89Division of Cardiovascular Medicine, Department of Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, Massachusetts, USA. 90Department of Psychiatry and Neurobehavioral Sciences, University of Virginia, Charlottesville, Virginia, USA. 91Virginia Institute for Psychiatric and Behavioral Genetics, Virginia Commonwealth University, Richmond, Virginia, USA. 92Massey Cancer Center, Virginia Commonwealth University, Richmond, Virginia, USA. 93Department of Psychiatry, University of Pennsylvania, Philadelphia, Pennsylvania, USA. 94Department of Functional Genomics, VU Amsterdam, Amsterdam, The Netherlands. 95Department of Medical Genomics, VU University Medical Center Amsterdam, Amsterdam, The Netherlands. 96Department of Psychiatry, University of North Carolina, Chapel Hill, North Carolina, USA. 97Abramson Cancer Center, University of Pennsylvania, Philadelphia, Pennsylvania, USA. 98Institute for Molecular Medicine, University of Helsinki, Helsinki, Finland. 99Department of Psychiatry and Behavioral Sciences, Duke University Medical Center, Durham, North Carolina, USA. 100Department of Hygiene and Epidemiology, University of Ioannina School of Medicine, Ioannina, Greece. 101Tufts Clinical and Translational Science Institute, Tufts University School of Medicine, Boston, Massachusetts, USA. 102Center for Genetic Epidemiology and Modeling, Institute for Clinical Research and Health Policy Studies, Tufts Medical Center, Boston, Massachusetts, USA. 103Department of Biostatistics, University of North Carolina, Chapel Hill, North Carolina, USA.
Oxford-GSK Consortium—Jason S Liu1, Federica Tozzi2,3, Dawn M Waterworth4, Sreekumar G Pillai5, Pierandrea Muglia6, Lefkos Middleton7, Wade Berrettini8, Christopher W Knouff9, Xin Yuan4, Gérard Waeber10,11, Peter Vollenweider10,11, Martin Preisig10,12, Nicholas J Wareham13, Jing Hua Zhao13, Ruth JF Loos13, Inês Barroso14, Kay-Tee Khaw15, Scott Grundy16, Philip Barter17, Robert Mahley18,19, Antero Kesaniemi20, Ruth McPherson21,22, John Vincent23, John Strauss23, James Kennedy23, Anne Farmer24, Peter McGuffin24, Richard Day25, Keith Matthews26, Per Bakke26, Amund Gulsvik26, Susanne Lucae27, Marcus Ising27, Tanja Brueckl27, Sonja Horstmann27, Joachim Heinrich28,29,30, Rajesh Rawal28, Norbert Dahmen31, Claudia Lamina28,32, Ozren Polasek33, Lina Zgaga34, Jennifer Huffman35, Susan Campbell35, Jaspal Kooner36, John C Chambers37, Mary Susan Burnett38, Joe Devaney38, Augusto D Pichard38, Kenneth M Kent38, Lowell Satler38, Joseph M Lindsay38, Ron Waksman38, Stephen Epstein38, Jim F Wilson39, Sarah H Wild39, Harry Campbell39, Veronique Vitart3, Muredach P Reilly40,41, Mingyao Li42, Liming Qu42, Robert Wilensky40, William Matthai40, Hakon H Hakonarson43, Daniel J Rader40,41, Andre Franke44, Michael Wittig44, Arne Schäfer44, Manuela Uda45, Antonio Terracciano46, Xiangjun Xiao47, Fabio Busonero45, Paul Scheet47, David Schlessinger46, David St Clair48, Dan Rujescu49, Gonçalo R Abecasis50, Hans Jörgen Grabe51, Alexander Teumer52, Henry Völzke53, Astrid Petersmann54, Ulrich John55, Igor Rudan39, Caroline Hayward35, Alan F Wright35, Ivana Kolcic33, Benjamin J Wright56, John R Thompson56, Anthony J Balmforth57, Alistair S Hall57, Nilesh J Samani58, Carl A Anderson14, Tariq Ahmed59, Christopher G Mathew60, Miles Parkes61, Jack Satsangi62, Mark Caulfield63, Patricia B Munroe63, Martin Farrall 64, Anna Dominiczak65, Jane Worthington66,67, Wendy Thomson66,67, Steve Eyre66,67, Anne Barton66,67, Vincent Mooser68, Clyde Francks69.
1Department of Statistics, University of Oxford, 1 South Parks Road, Oxford OX1 3TG, UK. 2Clinical Sciences-Aptuit Medicines Reasearch Center, Verona, Italy. 3Department of Psychiatry, School of Medicine, University of North Carolina, Chapel Hill, North Carolina, USA. 4Genetics Division, GlaxoSmithKline, Upper Merion, Pennsylvania, USA. 5Roche Pharmaceuticals, Nutley, New Jersey, USA. 6Neurosearch Denmark and Department of Psychiatry, University of Toronto, Toronto, Canada. 7Division of Neurosciences and Mental Health, Imperial College London, UK. 8Department of Psychiatry, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA. 9Genetics Division, GlaxoSmithKline, Research Triangle Park, North Carolina, USA. 10University Hospital Center, University of Lausanne, Lausanne, Switzerland. 11Department of Internal Medicine, University of Lausanne, Lausanne, Switzerland. 12Department of Psychiatry, University of Lausanne, Lausanne, Switzerland. 13MRC Epidemiology Unit, Institute of Metabolic Science, Cambridge, UK. 14Wellcome Trust Sanger Institute, Hinxton, UK. 15Department of Public Health and Primary Care, University of Cambridge, Cambridge, UK. 16Center for Human Nutrition, University of Texas Southwestern Medical Center, Dallas, Texas, USA. 17The Heart Research Institute, Sydney, New South Wales, Australia. 18Gladstone Institute of Cardiovascular Disease, University of California, San Francisco, California, USA. 19American Hospital, Istanbul, Turkey. 20Department of Internal Medicine, University of Oulu, Oulu, Finland. 21Division of Cardiology, University of Ottawa Heart Institute, Ottawa, Ontario, Canada. 22Biocenter Oulu, University of Oulu, Oulu, Finland. 23Centre for Addiction and Mental Health, University of Toronto, ON, Canada. 24Medical Research Council Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, King's College London, UK. 25Center for Neuroscience, Division of Medical Sciences, University of Dundee, Dundee, UK. 26Institute of Medicine, University of Bergen, Bergen, Norway. 27Max-Planck Institute of Psychiatry, Munich, Germany. 28Institute of Epidemiology, Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany. 29Institute of Medical Informatics, Biometry and Epidemiology, Ludwig-Maximilians-Universität, Munich, Germany. 30Klinikum Grosshadern, Munich, Germany. 31Psychiatrische Klinik und Poliklinik University of Mainz, Germany. 32Division of Genetic Epidemiology, Department of Medical Genetics, Molecular and Clinical Pharmacology, Innsbruck Medical University, Innsbruck, Austria. 33Medical School, University of Split, Split, Croatia. 34Centre for Population Health Sciences, University of Edinburgh, Edinburgh, UK. 35Institute of Genetics and Molecular Medicine, MRC Human Genetics Unit, Edinburgh, UK. 36National Heart and Lung Institute, Imperial College London, UK. 37Division of Epidemiology, Imperial College London, UK. 38Cardiovascular Research Institute, MedStar Health Research Institute, Washington Hospital Center, Washington, District of Columbia, USA. 39Centre for Population Health Sciences, University of Edinburgh, UK. 40The Cardiovascular Institute, University of Pennsylvania, Philadelphia, Pennsylvania, USA. 41The Institute for Translational Medicine and Therapeutics, School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA. 42Biostatistics and Epidemiology, University of Pennsylvania, Philadelphia, Pennsylvania, USA. 43The Center for Applied Genomics, Children's Hospital of Philadelphia, Philadelphia, Pennsylvania, USA. 44Institute of Clinical Molecular Biology, Christian-Albrechts-University, Kiel, Germany. 45Istituto di Neurogenetica e Neurofarmacologia, CNR, Monserrato, Cagliari, Italy. 46National Institute on Aging, Baltimore, Maryland, USA. 47Department of Epidemiology, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 48Department of Mental Health, University of Aberdeen, Aberdeen, UK. 49Department of Psychiatry, University of Halle, Halle, Germany. 50Center for Statistical Genetics, Department of Biostatistics, University of Michigan, Ann Arbor, Michigan, USA. 51Department of Psychiatry and Psychotherapy, University of Greifswald, Greifswald, Germany. 52Interfacultary Institute for Genetics and Functional Genomics, University of Greifswald, Greifswald, Germany. 53Institute for Community Medicine, University of Greifswald, Greifswald, Germany. 54Institute of Clinical Chemistry and Laboratory Medicine, University of Greifswald, Greifswald, Germany. 55Department of Social Medicine and Epidemiology, University of Greifswald, Greifswald, Germany. 56Department of Health Sciences, University of Leicester, Leicester, UK. 57Mulitdisciplinary Cardiovascular Research Centre (MCRC), Leeds Institute of Genetics, Health and Therapeutics (LIGHT), University of Leeds, Leeds, UK. 58Department of Cardiovascular Sciences, University of Leicester, Glenfield Hospital, Leicester, UK. 59Peninsula College of Medicine and Dentistry, Exeter, UK. 60Department of Medical and Molecular Genetics, King's College London School of Medicine, Guy's Hospital, London, UK. 61Gastroenterology Research Unit, Addenbrooke's Hospital, Cambridge, UK. 62Gastrointestinal Unit, Molecular Medicine Centre, University of Edinburgh, Western General Hospital, Edinburgh, UK. 63Clinical Pharmacology and Barts and the London Genome Centre, William Harvey Research Institute, Barts and the London School of Medicine, Queen Mary University of London, London, UK. 64Department of Cardiovascular Medicine, University of Oxford, Wellcome Trust Centre for Human Genetics, Oxford, UK. 65BHF Glasgow Cardiovascular Research Centre, Division of Cardiovascular and Medical Sciences, University of Glasgow, Western Infirmary, Glasgow, UK. 66Arthritis Research UK Epidemiology Unit, Musculoskeletal Research Group, University of Manchester, Manchester Academic Health Sciences Centre, Manchester, UK. 67NIHR Manchester Musculoskeletal Biomedical Research Unit, Central Manchester NHS Foundation Trust, Manchester, UK. 68Department of Pathology and Laboratory Medicine, Centre Hospitalier Universitaire Vaudois, Lausanne, Switzerland. 69Max Planck Institute for Psycholinguistics.
URLS
http://www.who.int/tobacco/mpower/mpower_report_full_2008.pdf
http://www.who.int/mediacentre/factsheets/fs311/en/index.html
Authors whose affiliations are listed as Decode genetics/AMGEN are employees of Decode genetics/AMGEN.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- Haslam DW, James WP. Obesity. Lancet. 2005;366:1197–1209. doi: 10.1016/S0140-6736(05)67483-1. [DOI] [PubMed] [Google Scholar]
- Jha P. Avoidable global cancer deaths and total deaths from smoking. Nat Rev Cancer. 2009;9:655–664. doi: 10.1038/nrc2703. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Wang GJ, Fowler JS, Telang F. Overlapping neuronal circuits in addiction and obesity: evidence of systems pathology. Philos Trans R Soc Lond B Biol Sci. 2008;363:3191–3200. doi: 10.1098/rstb.2008.0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audrain-McGovern J, Benowitz NL. Cigarette smoking, nicotine, and body weight. Clin Pharmacol Ther. 2011;90:164–168. doi: 10.1038/clpt.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freathy RM, Kazeem GR, Morris RW, Johnson PC, Paternoster L, Ebrahim S, et al. Genetic variation at CHRNA5-CHRNA3-CHRNB4 interacts with smoking status to influence body mass index. Int J Epidemiol. 2011;40:1617–1628. doi: 10.1093/ije/dyr077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny PJ. Reward mechanisms in obesity: new insights and future directions. Neuron. 2011;69:664–679. doi: 10.1016/j.neuron.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND, O'Brien CP. Issues for DSM-V: should obesity be included as a brain disorder. Am J Psychiatry. 2007;164:708–710. doi: 10.1176/ajp.2007.164.5.708. [DOI] [PubMed] [Google Scholar]
- Devlin MJ. Is there a place for obesity in DSM-V. Int J Eat Disord. 2007;40:S83–S88. doi: 10.1002/eat.20430. [DOI] [PubMed] [Google Scholar]
- Lerman C, Berrettini W, Pinto A, Patterson F, Crystal-Mansour S, Wileyto EP, et al. Changes in food reward following smoking cessation: a pharmacogenetic investigation. Psychopharmacology. 2004;174:571–577. doi: 10.1007/s00213-004-1823-9. [DOI] [PubMed] [Google Scholar]
- Schwartz MW, Woods SC, Porte D, Jr., Seeley RJ, Baskin DG. Central nervous system control of food intake. Nature. 2000;404:661–671. doi: 10.1038/35007534. [DOI] [PubMed] [Google Scholar]
- Mineur YS, Abizaid A, Rao Y, Salas R, DiLeone RJ, Gundisch D, et al. Nicotine decreases food intake through activation of POMC neurons. Science. 2011;332:1330–1332. doi: 10.1126/science.1201889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamia C, Trichopoulou A, Lenas D, Trichopoulos D. Tobacco smoking in relation to body fat mass and distribution in a general population sample. Int J Obes Relat Metab Disord. 2004;28:1091–1096. doi: 10.1038/sj.ijo.0802697. [DOI] [PubMed] [Google Scholar]
- Chiolero A, Jacot-Sadowski I, Faeh D, Paccaud F, Cornuz J. Association of cigarettes smoked daily with obesity in a general adult population. Obesity. 2007;15:1311–1318. doi: 10.1038/oby.2007.153. [DOI] [PubMed] [Google Scholar]
- Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42:448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The-Tobacco-and-Genetics-Consortium Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42:436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.