An alanine dehydrogenase from B. pseudofirmus OF4 was expressed in E. coli and purified. Crystallization and preliminary X-ray crystallographic analysis of the recombinant enzyme were performed.
Keywords: alanine dehydrogenase, Bacillus pseudofirmus
Abstract
Alanine dehydrogenase (OF4Ald) from the alkaliphilic Bacillus pseudofirmus OF4 was expressed and purified with a His6 tag in a form suitable for X-ray crystallographic analysis. Crystals were grown by the hanging-drop vapour-diffusion method at 289 K using a solution consisting of 0.1 M Tris–HCl pH 8.0, 0.2 M LiSO4, 22%(w/v) PEG 3350. X-ray diffraction data were collected to 2.8 Å resolution. The crystal belonged to the triclinic space group P1, with unit-cell parameters a = 88.04, b = 105.59, c = 120.53 Å, α = 88.37, β = 78.77, γ = 82.65°.
1. Introduction
Alanine dehydrogenase (Ald; l-alanine:NAD+ oxidoreductase, deaminating; EC 1.4.1.1), which catalyzes the NADH-dependent interconversion of pyruvate, ammonia and l-alanine (Gallagher et al., 2004 ▶), plays an important role in the carbon and nitrogen metabolism of microorganisms. Since the first alanine dehydrogenase was reported from Mycobacterium tuberculosis H37Ra (Andersen et al., 1992 ▶), a number of Ald enzymes have been purified and characterized from a variety of bacteria such as Bacillus stearothermophilus (Ohshima et al., 1990 ▶), Rhizobium leguminosarum (Allaway et al., 2000 ▶), Archaeoglobus fulgidus (Smith et al., 2003 ▶) and Mycobacterium smegmatis (Sakamoto et al., 1990 ▶). Based on their apparent molecular masses analyzed by gel filtration and SDS–PAGE and their crystal structures, alanine dehydrogenases are classified into four types of subunit structure: homohexameric in Phormidium lapideum (Baker et al., 1998 ▶), M. tuberculosis (Agren et al., 2008 ▶) and Thermus thermophilus (Váli et al., 1980 ▶), homotetrameric in Mycobacterium strain HE5 (Schuffenhauer et al., 1999 ▶) and soybean nodule bacteroids (Smith & Emerich, 1993 ▶), homooctameric in Streptomyces aureofaciens (Vančurová et al., 1988 ▶) and homodimeric in the archaeon A. fulgidus (Gallagher et al., 2004 ▶).
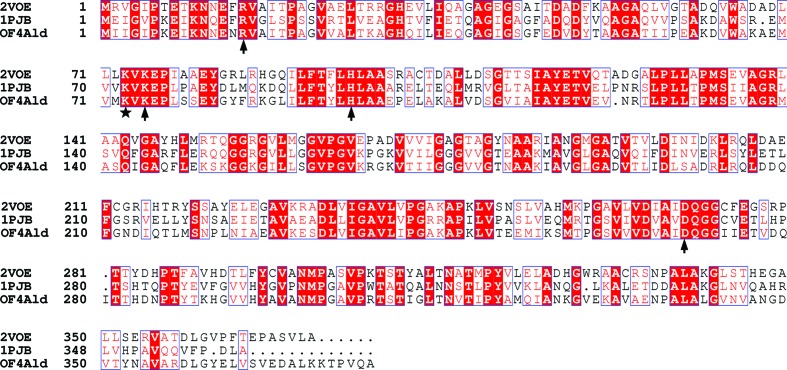
Alkaliphilic B. pseudofirmus OF4 are obligately aerobic spore-forming Gram-positive motile rods with an optimal growth pH of 10.5 and an optimal growth temperature of 303 K (Guffanti et al., 1986 ▶; Takami & Krulwich, 2000 ▶). Genetic data analysis of B. pseudofirmus OF4 (GenBank accession No. CP001878.2) indicates that an open reading frame encodes an alanine dehydrogenase protein (OF4Ald; GenBank accession No. ADC50010.1) which is located beside a gene encoding alanine racemase, and together they form an operon (Ju et al., 2009 ▶). OF4Ald shares 56% sequence homology with the alanine dehydrogenase from P. lapideum (PDB entry 1pjb; Baker et al., 1998 ▶). According to an analysis of the crystal structures and catalytic mechanisms of the alanine dehydrogenases from P. lapideum and M. tuberculosis (53% identity; PDB entry 2voe; Tripathi & Ramachandran, 2008 ▶), two conserved residues His96 and Asp270 are potential acid–base catalysts in the reaction and pyruvate is anchored to the active site through interaction with the side chains of the conserved residues Arg15 and Lys75 (Tripathi & Ramachandran, 2008 ▶). However, a multiple sequence alignment and biochemical analysis lead to conjecture that Lys73 in OF4Ald (Fig. 1 ▶) is also involved in the catalytic reaction.
Figure 1.
Multiple sequence alignment of alanine dehydrogenases from B. pseudofirmus OF4 (OF4Ald), P. lapideum (PDB entry 1pjb) and M. tuberculosis (PDB entry 2voe). The arrows and stars indicate the potential active-site residues.
In order to elucidate the biochemical properties of OF4Ald and the role of Lys73 in substrate recognition, a structural investigation was initiated. Here, we report our progress on crystallization and preliminary X-ray diffraction data analysis of recombinant OF4Ald protein.
2. Materials and methods
2.1. Cloning, expression and purification
Genomic DNA of B. pseudofirmus OF4 was isolated as described previously (Ju et al., 2009 ▶) and used as a template. The of4ald gene (GenBank accession No. ADC50010.1) was amplified using the primers 5′-CACGCATATGATTATCGGTATTCCA-3′ and 5′-AGCCTCGAGTGCTTGAACAGGTGTTTTC-3′; bold sequences represent NdeI and XhoI restriction sites, respectively. PCR was carried out with Ex Taq DNA polymerase (TaKaRa, Japan) for 25 cycles of 368 K for 45 s, 328 K for 1 min and 345 K for 2 min. The PCR products were digested with NdeI and XhoI and the fragments were then inserted into the corresponding sites of plasmid pET-22b(+) (Novagen, USA). The recombinant pET-OF4Ald, which was confirmed by DNA sequencing, was introduced into Escherichia coli BL21(DE3) cells (Novagen, USA). The E. coli BL21(DE3) transformants were grown in 800 ml LB medium containing 100 µg ml−1 ampicillin at 310 K until the OD600 reached 0.5. Expression of the recombinant OF4Ald protein was induced by the addition of 1 mM IPTG (final concentration) and growth of the culture continued at 310 K for 12 h.
The cells were harvested by centrifugation (7656g, 10 min, 277 K) and the cell pellets were resuspended in 30 ml lysis buffer (50 mM Tris–HCl pH 7.5, 200 mM NaCl, 5% glycerol) and disrupted by sonication. Cell debris was removed by centrifugation (15 000g, 30 min, 277 K). The supernatant was loaded onto an Ni2+-chelating affinity chromatography column (10 ml containing 2 ml Ni2+ beads; GE Healthcare, USA) and the column was rinsed with 30 ml washing buffer (50 mM Tris–HCl pH 7.5, 200 mM NaCl, 5% glycerol, 20 mM imidazole) and then eluted with 5 ml elution buffer (50 mM Tris–HCl pH 7.5, 200 mM NaCl, 5% glycerol, 200 mM imidazole). Subsequent purification was performed on FPLC by size-exclusion chromatography at 277 K on a Superdex 200 10/300 GL column (GE Healthcare, USA) equilibrated with 50 mM Tris–HCl pH 7.5, 50 mM NaCl, 5% glycerol (flow rate of 0.5 ml min−1). The protein fraction was pooled, concentrated and dialyzed at 277 K against 10 mM Tris–HCl buffer pH 7.5 containing 10 µM NAD+ using an Amicon Ultra-15 Centrifugal Filter Device (30 kDa molecular-weight cutoff; Millipore). The molecular mass of OF4Ald was determined by gel-filtration chromatography as described previously (Ju et al., 2005 ▶). Cytochrome c (12.4 kDa), carbonic anhydrase (29 kDa), bovine serum albumin (66 kDa), alcohol dehydrogenase (150 kDa) and β-amylase (200 kDa) were used as molecular-mass standards (Sigma, USA). The purity of the OF4Ald protein was analyzed by 12%(w/v) SDS–PAGE and the protein concentration was determined using a NanoDrop device at 280 nm. (The extinction coefficient of OF4Ald is 20 400 M −1 cm−1 in water; Thermo Fisher, USA.)
2.2. Crystallization
The purified OF4Ald protein with a His6 tag at the C-terminus (LEHHHHHH) was used for crystallization screening. Initially, crystallization screening was carried out using Crystal Screen, Crystal Screen 2, Index and PEG/Ion from Hampton Research at 289 K by the hanging-drop vapour-diffusion method. Drops consisting of 1 µl protein solution (10 mg ml−1) mixed with an equal volume of reservoir solution were equilibrated against 300 µl reservoir solution. The crystallization conditions were optimized based on the initial screening through adjustment of the precipitant concentration, the protein concentration and the buffer pH.
2.3. X-ray data collection and processing
For X-ray diffraction experiments, the crystals were first soaked in reservoir solution containing 15%(v/v) glycerol for 30 s. The soaked crystals were then mounted in nylon loops and flash-cooled in liquid nitrogen at 100 K (Parkin & Hope, 1998 ▶).
Diffraction data were collected at 100 K using an in-house X-ray source (Rigaku MicroMax-007HF desktop rotating-anode X-ray generator with a Cu target operated at 40 kV and 30 mA) and a Saturn 944+ detector with a 70 mm crystal-to-detector distance at a wavelength of 1.5418 Å. 720 frames were collected with 0.5° oscillation per image at 100 K. The collected intensities were indexed, integrated, corrected for absorption, scaled and merged using HKL-2000 (Otwinowski & Minor, 1997 ▶).
3. Results
A 1131 bp DNA fragment from B. pseudofirmus OF4 was amplified and cloned into the expression vector pET-22b(+) for construction of the plasmid pET-OF4Ald. The recombinant OF4Ald protein with a His6 tag was successfully expressed and purified to electrophoretic homogeneity. The final yield of OF4Ald was 30 mg of protein per litre of culture. The molecular mass of OF4Ald was estimated to be about 40 kDa by SDS–PAGE (Fig. 2 ▶), coinciding with the predicted molecular mass of 40.5 kDa for recombinant OF4Ald with a His6 tag at the C-terminus. The OF4Ald protein purified by size-exclusion chromatography eluted as a single Gaussian-shaped peak. The molecular mass of OF4Ald was calculated to be 166.1 kDa, indicating that OF4Ald is a tetramer.
Figure 2.
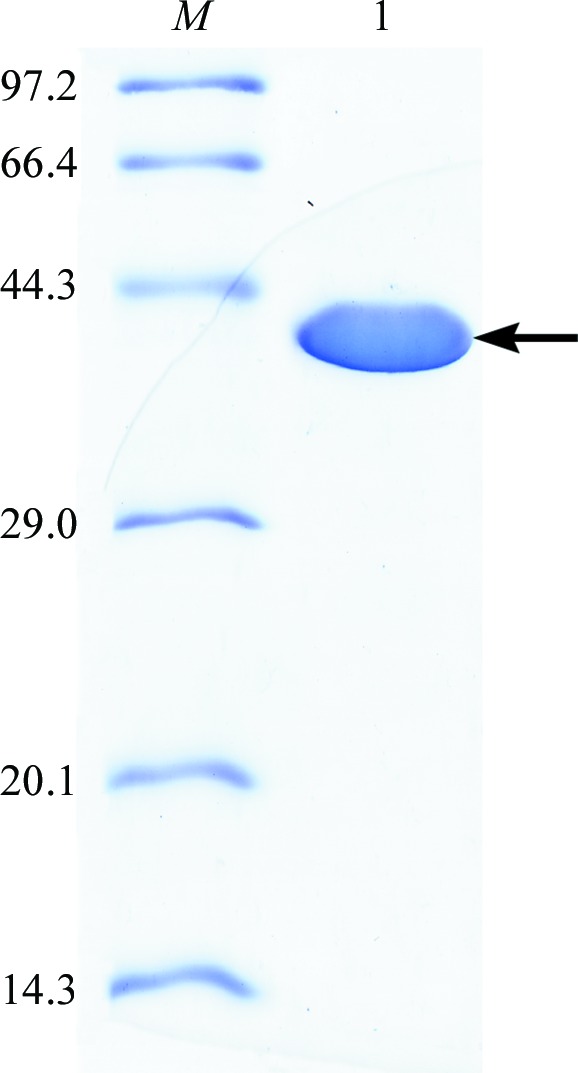
12% SDS–PAGE analysis of OF4Ald. Lane M, molecular-mass standards (labelled in kDa); lane 1, purified OF4Ald protein (40 kDa).
Initial crystallization screening was performed at 289 K using 16-well tissue-culture plates. After 2–5 d incubation, crystals of different shapes were obtained in condition Nos. 6 (fine needles; 0.2 M NaCl, 20% PEG 3350), 14 (irregular tetrahedra; 0.2 M KSCN, 20% PEG 3350) and 18 (irregular sheets; 0.2 M KNO3, 20% PEG 3350) of the PEG/Ion kit and condition Nos. 77 (small cubes; 0.2 M Li2SO4.H2O, 0.1 M Tris–HCl pH 8.5, 25% PEG 3350) and 93 [small prisms; 0.2 M Zn(CH3COO)2.2H2O, 20% PEG 3350] of the Index kit. A well diffracting crystal was obtained from condition No. 77 of the Index kit using a protein concentration of 10 mg ml−1. Optimization of this condition through adjustment of the precipitant concentration, the buffer pH and the protein concentration resulted in fewer nucleation sites and yielded crystals of identical size and morphology. Crystals sufficient for X-ray data collection of about 310 × 280 × 130 µm in size (Fig. 3 ▶) were obtained using a solution consisting of 0.1 M Tris–HCl pH 8.0, 0.2 M LiSO4.H2O, 22% PEG 3350 at a protein concentration of 15 mg ml−1.
Figure 3.
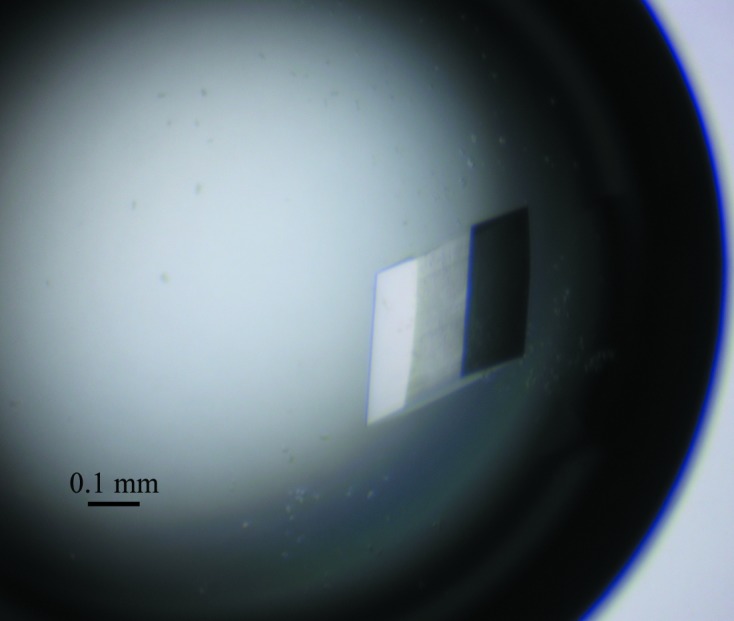
Typical crystals of OF4Ald. The optimized crystals (about 310 × 280 × 130 µm) were grown in 0.1 M Tris–HCl pH 8.0, 0.2 M LiSO4.H2O, 22%(w/v) PEG 3350.
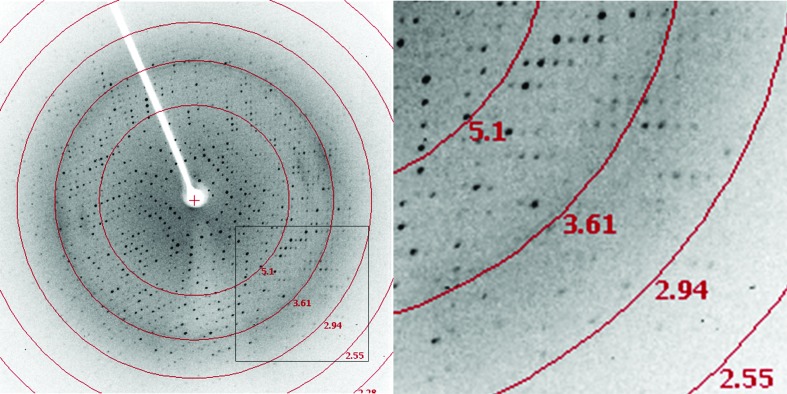
Crystallographic data statistics are shown in Table 1 ▶. A diffraction data set was collected from the recombinant OF4Ald crystal to 2.8 Å resolution with an overall R merge of 11.9% (Fig. 4 ▶). This data set has 97.4% overall completeness and 94.6% completeness in the highest resolution shell. The crystal belonged to the triclinic space group P1, with unit-cell parameters a = 88.04, b = 105.59, c = 120.53 Å, α = 88.37, β = 78.77, γ = 82.65°. Matthews parameter calculations indicate the presence of eight to 12 protein molecules in the asymmetric unit (Matthews, 1968 ▶).
Table 1. Diffraction data statistics.
Values in parentheses are for the outer shell.
| Space group | P1 |
| Wavelength (Å) | 1.5418 |
| Unit-cell parameters (Å, °) | a = 88.04, b = 105.59, c = 120.53, α = 88.37, β = 78.77, γ = 82.65 |
| Resolution range (Å) | 50.00–2.80 (2.85–2.80) |
| Total No. of reflections | 377621 (15533) |
| No. of unique reflections | 99374 (4854) |
| Average multiplicity | 3.8 (3.2) |
| Completeness (%) | 97.4 (94.6) |
| R merge † (%) | 11.9 (43.2) |
| Average I/σ(I) | 9.9 (1.8) |
R
merge = 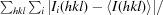
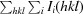 , where Ii(hkl) is the intensity of the ith measurement of reflection hkl and 〈I(hkl)〉 is the mean intensity of all symmetry-related reflections.
, where Ii(hkl) is the intensity of the ith measurement of reflection hkl and 〈I(hkl)〉 is the mean intensity of all symmetry-related reflections.
Figure 4.
Diffraction pattern of an OF4Ald crystal.
The molecular-replacement (MR) method using AMoRe (Navaza, 1994 ▶) was employed to determine the initial phases using the structure with PDB code 1pjb as a search model. After a rotational and translational search, we failed to find a promising MR solution. We therefore labelled the protein with selenomethionine in an attempt to obtain the initial phases. We hope that the three-dimensional structure of OF4Ald will provide insights into the biochemical properties of alanine dehydrogenase from B. pseudofirmus OF4, in particular the role of residue Lys73.
Acknowledgments
This work was supported by grants from the Natural Science Foundation of Hebei Province (C2011205045), Hebei Science and Technology Development Foundation (10205521D), Hebei Province Foundation for Returnees (20100705), the Youth Foundation of Hebei Educational Committee (2011102) and Funding of Hebei Normal University (L2009B13 and L2012Z12).
References
- Agren, D., Stehr, M., Berthold, C. L., Kapoor, S., Oehlmann, W., Singh, M. & Schneider, G. (2008). J. Mol. Biol. 377, 1161–1173. [DOI] [PubMed]
- Allaway, D., Lodwig, E. M., Crompton, L. A., Wood, M., Parsons, R., Wheeler, T. R. & Poole, P. S. (2000). Mol. Microbiol. 36, 508–515. [DOI] [PubMed]
- Andersen, A. B., Andersen, P. & Ljungqvist, L. (1992). Infect. Immun. 60, 2317–2323. [DOI] [PMC free article] [PubMed]
- Baker, P. J., Sawa, Y., Shibata, H., Sedelnikova, S. E. & Rice, D. W. (1998). Nature Struct. Biol. 5, 561–567. [DOI] [PubMed]
- Gallagher, D. T., Monbouquette, H. G., Schröder, I., Robinson, H., Holden, M. J. & Smith, N. N. (2004). J. Mol. Biol. 342, 119–130. [DOI] [PubMed]
- Guffanti, A. A., Finkelthal, O., Hicks, D. B., Falk, L., Sidhu, A., Garro, A. & Krulwich, T. A. (1986). J. Bacteriol. 167, 766–773. [DOI] [PMC free article] [PubMed]
- Ju, J., Xu, S., Wen, J., Li, G., Ohnishi, K., Xue, Y. & Ma, Y. (2009). J. Biosci. Bioeng. 107, 225–229. [DOI] [PubMed]
- Ju, J., Yokoigawa, K., Misono, H. & Ohnishi, K. (2005). J. Biosci. Bioeng. 100, 409–417. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Navaza, J. (1994). Acta Cryst. A50, 157–163.
- Ohshima, T., Sakane, M., Yamazaki, T. & Soda, K. (1990). Eur. J. Biochem. 191, 715–720. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Parkin, S. & Hope, H. (1998). J. Appl. Cryst. 31, 945–953.
- Sakamoto, Y., Nagata, S., Esaki, N., Tanaka, H. & Soda, K. (1990). J. Ferment. Bioeng. 69, 154–158.
- Schuffenhauer, G., Schräder, T. & Andreesen, J. R. (1999). Arch. Microbiol. 171, 417–423. [DOI] [PubMed]
- Smith, M. T. & Emerich, D. W. (1993). J. Biol. Chem. 268, 10746–10753. [PubMed]
- Smith, N., Mayhew, M., Robinson, H., Héroux, A., Charlton, D., Holden, M. J. & Gallagher, D. T. (2003). Acta Cryst. D59, 2328–2331. [DOI] [PubMed]
- Takami, H. & Krulwich, T. A. (2000). Extremophiles, 4, 19–22. [DOI] [PubMed]
- Tripathi, S. M. & Ramachandran, R. (2008). Proteins, 72, 1089–1095. [DOI] [PubMed]
- Váli, Z., Kilár, F., Lakatos, S., Venyaminov, S. A. & Závodszky, P. (1980). Biochim. Biophys. Acta, 615, 34–47. [DOI] [PubMed]
- Vančurová, I., Vančura, A., Volc, J., Neužzil, J., Flieger, M., Basařová, G. & Běhal, V. (1988). Arch. Microbiol. 150, 438–440.