ALKBH5 catalyzes the demethylation of N 6-methyladenosine in RNA. The ALKBH5 protein was crystallized and X-ray diffraction data were collected to a resolution of 2.4 Å.
Keywords: N6-methyladenosine, ALKBH5
Abstract
N 6-Methyladenosine (m6A) is a ubiquitous modification found in mammalian mRNA and long noncoding RNA. ALKBH5 is a member of the iron(II)- and 2-oxoglutarate-dependent AlkB oxygenase family and has been shown to catalyze the oxidative demethylation of N 6-methyladenosine in RNA. The ALKBH5 protein was purified and crystallized using the hanging-drop vapour-diffusion method. The crystals diffracted to 2.4 Å resolution using synchrotron radiation. The crystals belonged to space group P212121, with unit-cell parameters a = 57.456, b = 83.406, c = 92.909 Å, α = β = γ = 90.00° and one molecule in the asymmetric unit.
1. Introduction
N 6-Methyladenosine (m6A) is one of the most common and abundant modifications of RNA molecules in eukaryotes (Bokar, 2005 ▶), with each RNA molecule having three to five m6A bases. About 7000 mRNAs have been found to possess m6A sites in both human and mouse cells (Dominissini et al., 2012 ▶; Meyer et al., 2012 ▶). m6A modification can affect diverse biological processes including mRNA splicing, nuclear export, stability and translational efficiency (Bokar, 2005 ▶). The m6A modification has been shown to function as a tag in cellular mRNAs to prevent recognition and initiation of the host cellular immune response (Karikó et al., 2005 ▶), whereas the human m6A methyltransferase MTA70 is a subunit of a nuclear-localized complex, MT-A (Bokar et al., 1997 ▶). Consistently, RNA interference by MTA70 results in cell death through apoptosis in HeLa cells. Recently, FTO and ALKBH5 have been demonstrated to function as RNA m6A demethylases (Jia et al., 2011 ▶; Zheng et al., 2013 ▶). Several potential m6A-binding proteins have been identified such as ELAV1, YTHDF2 and YTHDF3 (Dominissini et al., 2012 ▶). ELAV1 is highly expressed in cancer cells and is involved in inflammation through the regulation of mRNA stability, splicing and translation (Filippova et al., 2012 ▶), consistent with the roles proposed for m6A modification. YTHDF2 is a translocation partner of RUNX1 in acute myeloid leukaemia patients (Nguyen et al., 2006 ▶). Functional studies on methyltransferase, demethylase and potential m6A-binding proteins reveal that m6A in RNA may be involved in adipogenesis, spermatogenesis, development and carcinogenesis, as well as other as yet unidentified life processes. The reversible m6A modification in mammalian mRNA represents another novel epigenetic marker, a finding that has recently established the concept of RNA epigenetics as another layer of epigenetic regulation (He, 2010 ▶).
Iron(II)- and 2-oxoglutarate (2OG)-dependent dioxygenases play important roles in a wide range of biological processes, including hypoxic response, nucleic acid repair and modification and chromatin modification (Loenarz & Schofield, 2008 ▶). These enzymes are characterized by their catalytic requirement for ferrous iron as well as the co-substrate 2OG. The Escherichia coli AlkB protein is an iron(II)/2OG dioxygenase that is involved in DNA and RNA repair. AlkB can demethylate 1-methyladenine (1-meA) and 3-methylcytosine (3-meC) in DNA and RNA (Aas et al., 2003 ▶). Nine AlkB homologues have been identified in mammals: ALKBH1–8 and FTO. ALKBH1, the mammalian homologue with the highest similarity to AlkB, is a mitochondrial protein that demethylates 3-meC in DNA and RNA (Aas et al., 2003 ▶). ALKBH2 and ALKBH3 are functionally similar to AlkB. ALKBH2 is more active towards dsDNA, while ALKBH3 preferentially demethylates ssDNA and ssRNA (Yang et al., 2008 ▶; Sundheim et al., 2006 ▶). Unlike the other proteins, ALKBH8 catalyzes the hydroxylation of a modified uridine (mcm5U) in the wobble position of the anticodon stem loop of a specific tRNA (van den Born et al., 2011 ▶). FTO was identified as an obesity susceptibility factor in genome-wide association studies (Frayling et al., 2007 ▶; Scott et al., 2007 ▶; Dina et al., 2007 ▶). FTO is able to catalyze the demethylation of 3-methylthymine (3-meT) and 3-methyluracil (3-meU) in ssDNA and ssRNA (Jia et al., 2008 ▶). Structural investigations confirm that single-stranded nucleic acids are the preferred substrate of FTO (Han et al., 2010 ▶). The latest research results show that m6A in nuclear RNA may be a physiological substrate of FTO (Jia et al., 2011 ▶).
Similar to FTO, ALKBH5 has been identified as a mammalian m6A RNA demethylase in vitro and in vivo (Zheng et al., 2013 ▶). ALKBH5 is a direct transcriptional target of hypoxia-inducible factor 1 (HIF-1), which is induced by low concentrations of oxygen (Thalhammer et al., 2011 ▶). ALKBH5 is ubiquitously expressed in the testicle and lung. Knockdown of ALKBH5 leads to increased amounts of m6A in mRNA, whereas overexpression of ALKBH5 results in decreased amounts of m6A in human cells. The ALKBH5 protein, which colocalizes with nuclear speckles, significantly affects nuclear mRNA export and RNA metabolism when disturbed. ALKBH5 deficiency results in testicular atrophy and reduction in the number and motility of sperm (Zheng et al., 2013 ▶).
The available data have unambiguously established the link between ALKBH5 and RNA demethylation. However, how ALKBH5 specifically selects its substrates remains unknown. Structural study will provide insight into the underlying mechanism. Here, we report the cloning, expression, purification, crystallization and preliminary X-ray analysis of ALKBH5.
2. Experimental procedures
2.1. Cloning
A region encoding ALKBH5 residues 67–352 (NCBI Reference Sequence NP_060228.3) was PCR-amplified from a human heart cDNA library using Pfu DNA polymerase. The forward oligonucleotide primer was 5′-GGAATTCCATATGGAGCGCAGCGACTATGAGGAG-3′ and the reverse oligonucleotide primer was 5′-CCGCTCGAGCAGCACCGAGCGCCGGTT-3′. NdeI and XhoI restriction-enzyme recognition sites (bold) were added to the forward primer and the reverse primer, respectively. The ALKBH567–352 fragment was cloned into the pET-30a vector, resulting in the pET-30a-ALKBH567–352 plasmid encoding a C-terminally His6-tagged sequence.
2.2. Protein expression and purification
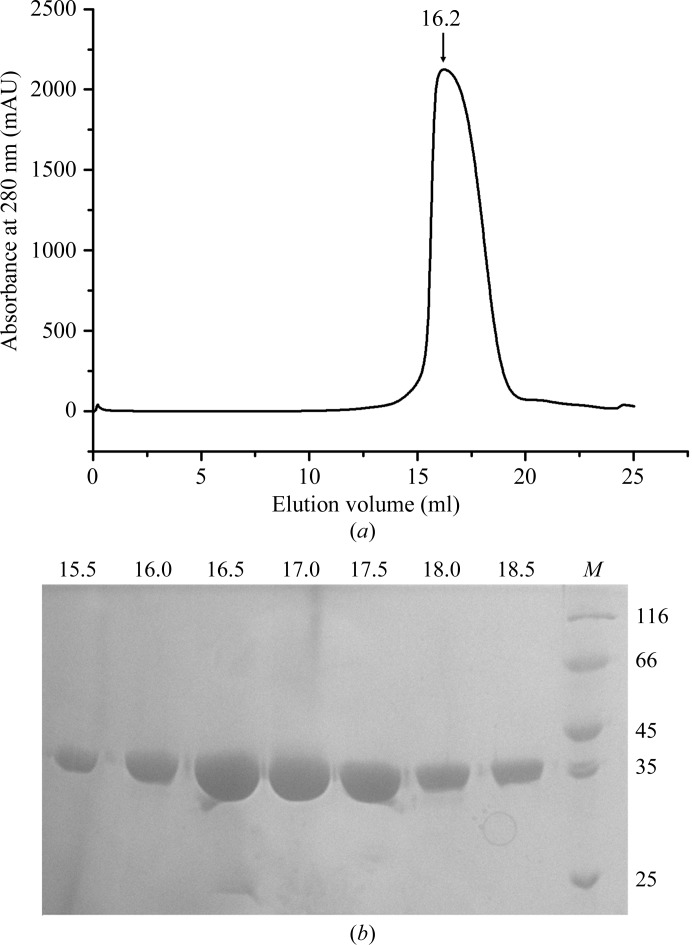
Escherichia coli BL21 (DE3) cells containing the pET-30a-ALKBH567–352 plasmid were grown in Luria broth (LB) medium with 50 mg ml−1 kanamycin at 310 K. When the OD600 of the cells reached 0.6, the temperature was decreased to 291 K and isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to a final concentration of 0.1 mM to induce protein expression. The cells were harvested by centrifugation at 4000 rev min−1 for 20 min after 16 h induction. The cells were resuspended in buffer A (150 mM NaCl, 25 mM Tris pH 7.2, 15 mM imidazole) and lysed by sonication. All subsequent purification steps were performed at 293 K. The crude lysate was centrifuged at 16 000 rev min−1 for 30 min and the supernatant was loaded onto Ni–NTA His-Bind resin columns (Novagen). The affinity columns were washed with buffer A. The protein was eluted with buffer B (25 mM Tris pH 7.2, 50 mM NaCl, 250 mM imidazole). The diluted sample in buffer A was applied onto a Source 15S cation-exchange column (GE Healthcare Biosciences) pre-equilibrated with buffer C (25 mM Tris pH 7.2, 3 mM DTT). The protein was eluted with a linear gradient of 0–0.3 M sodium chloride in buffer C. The next step was size-exclusion chromatography on a Superdex 200 10/300 GL column (GE Healthcare Biosciences) employing elution buffer D (10 mM Tris–HCl pH 7.2, 100 mM sodium chloride, 3 mM DTT). All fractions containing the ALKBH5 protein were pooled and concentrated to ∼10 mg ml−1. The protein was flash-frozen and stored at 193 K for subsequent crystallization experiments. SDS–PAGE with Coomassie Blue staining was used to monitor the purity of the protein at each step and to verify that the final protein was >95% pure (Fig. 1 ▶).
Figure 1.
(a) Purification of ALKBH5 protein by size-exclusion chromatography (Superdex 200). The elution volume of the protein is indicated. (b) SDS–PAGE. The numbers at the top correspond to the elution volumes in (a). Lane M, protein molecular-weight marker (labelled in kDa).
2.3. Crystallization
2OG was added to the protein stock solution to a final concentration of 1 mM. Initial crystallization screening was carried out at 293 K using the hanging-drop vapour-diffusion method in 24-well VDX plates (Hampton Research). The reagents screened were from Index, Crystal Screen, Crystal Screen 2, PEG/Ion and PEG/Ion 2 from Hampton Research. Each drop consisted of 1 µl protein solution plus 1 µl reservoir solution. The total volume of solution in each well was 0.4 ml. The ALKBH5 protein crystallized using PEG/Ion condition No. 48 [0.2 M ammonium citrate dibasic pH 5.1, 20%(w/v) PEG 3350].
2.4. X-ray data collection
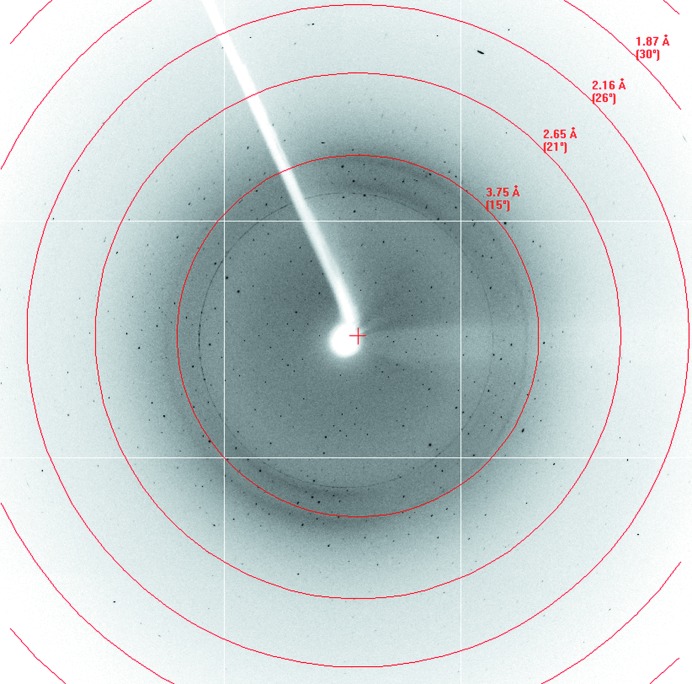
Prior to data collection, a single crystal of ALKBH567–352 was transferred to a CryoLoop and soaked in a cryoprotectant consisting of 0.2 M ammonium citrate dibasic, 20%(w/v) PEG 3350, 10%(v/v) glycerol before being cooled to 100 K in a stream of nitrogen gas. Crystal diffraction data were collected using a MAR 345 CCD detector at a crystal-to-detector distance of 350 mm on beamline BL17U at the Shanghai Synchrotron Radiation Facility (SSRF). The data were indexed and integrated using the HKL-2000 processing software (Otwinowski & Minor, 1997 ▶). Data-collection and processing statistics are summarized in Table 1 ▶.
Table 1. Data-collection and processing statistics.
Values in parentheses are for the outer shell.
| Wavelength (Å) | 0.9792 |
| Oscillation angle (°) | 1 |
| No. of frames | 180 |
| Space group | P212121 |
| Unit-cell parameters (Å, °) | a = 57.456, b = 83.406, c = 92.909, α = β = γ = 90.00 |
| Observed reflections | 126210 |
| Unique reflections | 18112 |
| Resolution range (Å) | 25.00–2.40 (2.44–2.40) |
| R merge † | 0.144 (0.515) |
| Average I/σ(I) | 20.5 (5.3) |
| Multiplicity | 7.0 (7.3) |
| Completeness (%) | 100.0 (100.0) |
R
merge = 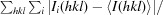
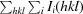 , where I(hkl) is the intensity of reflection hkl,
, where I(hkl) is the intensity of reflection hkl, 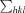 is the sum over all reflections and
is the sum over all reflections and 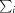 is the sum over i measurements of reflection hkl.
is the sum over i measurements of reflection hkl.
3. Results and discussion
In this study, the ALKBH5 protein was successfully overexpressed in E. coli. The expressed protein was purified using Ni–NTA affinity chromatography followed by cation-exchange and size-exclusion chromatography steps, resulting in a yield of 30 mg purified protein from 12 l of culture. SDS–PAGE analysis revealed a single band corresponding to a molecular weight of about 35 kDa, which is close to the calculated molecular weight of 33.75 kDa (Fig. 1 ▶). Although mRNA is a substrate of ALKBH5, we did not identify the presence of mRNA in the purified protein from E. coli. This could reflect the requirement for a modified mRNA as a substrate of ALKBH5. After purification and screening of crystallization conditions, crystals of ALKBH5 were obtained using a reservoir solution consisting of 0.2 M ammonium citrate dibasic pH 5.1, 20%(w/v) PEG 3350 (Fig. 2 ▶).
Figure 2.
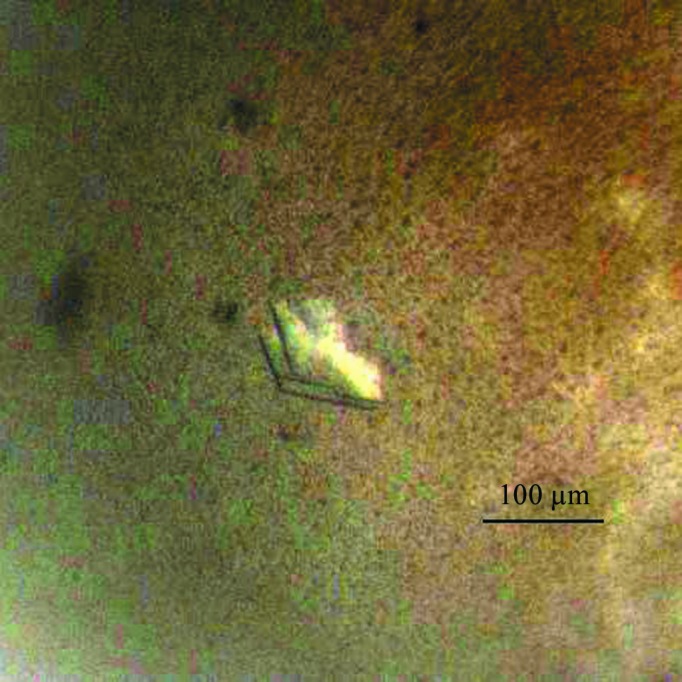
Crystals of ALKBH567–352. The crystals were grown by the hanging-drop vapour-diffusion method using a solution consisting of 0.2 M ammonium citrate dibasic pH 5.1, 20%(w/v) PEG 3350.
The crystals of ALKBH567–352 diffracted to high resolution (2.4 Å; Fig. 3 ▶) at a synchrotron source. The ALKBH5 crystals belonged to space group P212121, with unit-cell parameters a = 57.456, b = 83.406, c = 92.909 Å, α = β = γ = 90.00°. The R merge of the overall data set was 0.144. Calculation of the Matthews coefficient (Matthews, 1968 ▶) suggested the presence of one molecule in the asymmetric unit (V M = 3.30 Å3 Da−1, 62.73% solvent content). Additional data-set statistics and information are given in Table 1 ▶.
Figure 3.
A diffraction image of an ALKBH5 crystal collected on a CCD detector.
Molecular replacement (MR) was attempted using three ALKBH5 homologue structures (ALKBH8, PDB entry 3thp, Pastore et al., 2012 ▶; ALKBH3, PDB entry 2iuw, Sundheim et al., 2006 ▶; ALKBH2, PDB entry 3buc, Yang et al., 2008 ▶) with Phaser (McCoy et al., 2007 ▶). However, Phaser gave no correct solution. Multiple structure alignments of the three AlkB members showed that they share a highly conserved core domain. We built a model using the data for this core domain and successfully solved the phase by MR with Phaser.
Acknowledgments
We thank the staff of beamline BL17U at the SSRF for their help during data collection. We thank Dr Shilong Fan for assistance with the X-ray diffraction experiment. This work was funded by the National Outstanding Young Scholar Science Foundation of China (20101331722) and the State Key Program of National Natural Science of China (31130063) to Jijie Chai.
References
- Aas, P. A., Otterlei, M., Falnes, P. O., Vågbø, C. B., Skorpen, F., Akbari, M., Sundheim, O., Bjørås, M., Slupphaug, G., Seeberg, E. & Krokan, H. E. (2003). Nature (London), 421, 859–863. [DOI] [PubMed]
- Bokar, J. A. (2005). Top. Curr. Genet. 12, 141–177.
- Bokar, J. A., Shambaugh, M. E., Polayes, D., Matera, A. G. & Rottman, F. M. (1997). RNA, 3, 1233–1247. [PMC free article] [PubMed]
- Born, E. van den, Vågbø, C. B., Songe-Møller, L., Leihne, V., Lien, G. F., Leszczynska, G., Malkiewicz, A., Krokan, H. E., Kirpekar, F., Klungland, A. & Falnes, P. Ø. (2011). Nature Commun. 2, 172. [DOI] [PubMed]
- Dina, C. et al. (2007). Nature Genet. 39, 724–726. [DOI] [PubMed]
- Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., Cesarkas, K., Jacob-Hirsch, J., Amariglio, N., Kupiec, M., Sorek, R. & Rechavi, G. (2012). Nature (London), 485, 201–206. [DOI] [PubMed]
- Filippova, N., Yang, X., King, P. & Nabors, L. B. (2012). J. Biol. Chem. 287, 32277–32287. [DOI] [PMC free article] [PubMed]
- Frayling, T. M. et al. (2007). Science, 316, 889–894. [DOI] [PMC free article] [PubMed]
- Han, Z., Niu, T., Chang, J., Lei, X., Zhao, M., Wang, Q., Cheng, W., Wang, J., Feng, Y. & Chai, J. (2010). Nature (London), 464, 1205–1209. [DOI] [PubMed]
- He, C. (2010). Nature Chem. Biol. 6, 863–865. [DOI] [PubMed]
- Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., Yi, C., Lindahl, T., Pan, T., Yang, Y.-G. & He, C. (2011). Nature Chem. Biol. 7, 885–887. [DOI] [PMC free article] [PubMed]
- Jia, G., Yang, C.-G., Yang, S., Jian, X., Yi, C., Zhou, Z. & He, C. (2008). FEBS Lett. 582, 3313–3319. [DOI] [PMC free article] [PubMed]
- Karikó, K., Buckstein, M., Ni, H. & Weissman, D. (2005). Immunity, 23, 165–175. [DOI] [PubMed]
- Loenarz, C. & Schofield, C. J. (2008). Nature Chem. Biol. 4, 152–156. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E. & Jaffrey, S. R. (2012). Cell, 149, 1635–1646. [DOI] [PMC free article] [PubMed]
- Nguyen, T. T., Ma, L. N., Slovak, M. L., Bangs, C. D., Cherry, A. M. & Arber, D. A. (2006). Genes Chromosomes Cancer, 45, 918–932. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 277, 307–326. [DOI] [PubMed]
- Pastore, C., Topalidou, I., Forouhar, F., Yan, A. C., Levy, M. & Hunt, J. F. (2012). J. Biol. Chem. 287, 2130–2143. [DOI] [PMC free article] [PubMed]
- Scott, L. J. et al. (2007). Science, 316, 1341–1345. [DOI] [PMC free article] [PubMed]
- Sundheim, O., Vågbø, C. B., Bjørås, M., Sousa, M. M., Talstad, V., Aas, P. A., Drabløs, F., Krokan, H. E., Tainer, J. A. & Slupphaug, G. (2006). EMBO J. 25, 3389–3397. [DOI] [PMC free article] [PubMed]
- Thalhammer, A., Bencokova, Z., Poole, R., Loenarz, C., Adam, J., O’Flaherty, L., Schödel, J., Mole, D., Giaslakiotis, K., Schofield, C. J., Hammond, E. M., Ratcliffe, P. J. & Pollard, P. J. (2011). PLoS One, 6, e16210. [DOI] [PMC free article] [PubMed]
- Yang, C.-G., Yi, C., Duguid, E. M., Sullivan, C. T., Jian, X., Rice, P. A. & He, C. (2008). Nature (London), 452, 961–965. [DOI] [PMC free article] [PubMed]
- Zheng, G. et al. (2013). Mol. Cell, 49, 18–29. [DOI] [PMC free article] [PubMed]