A phosphotriesterase-like lactonase dubbed VmoLac isolated from the hyperthermophilic crenarchaeon V. moutnovskia was purified and crystallized. A diffraction data set was collected to 2.4 Å resolution.
Keywords: lactonases, quorum sensing, phosphotriesterases, organophosphorus, extremophiles, molecular promiscuity, Vulcanisaeta moutnovskia
Abstract
Phosphotriesterase-like lactonases (PLLs) are native lactonases that are capable of hydrolyzing lactones such as aliphatic lactones or acyl-homoserine lactones, which are involved in bacterial quorum sensing. Previously characterized PLLs are moreover endowed with a promiscuous phosphotriesterase activity and are therefore able to detoxify organophosphate insecticides. A novel PLL representative, dubbed VmoLac, has been identified from the hyperthermophilic crenarchaeon Vulcanisaeta moutnovskia. Because of its intrinsic high thermal stability, VmoLac may constitute an appealing candidate for engineering studies with the aim of producing an efficient biodecontaminant for organophosphorus compounds and a bacterial antivirulence agent. In combination with biochemical studies, structural information will allow the identification of the residues involved in substrate specificity and an understanding of the enzymatic catalytic mechanisms. Here, the expression, purification, crystallization and X-ray data collection at 2.4 Å resolution of VmoLac are reported.
1. Introduction
Organophosphates (OPs) are well known neurotoxic compounds that irreversibly inhibit acetylcholinesterase, a key enzyme of the central nervous system (Masson et al., 2009 ▶). These compounds, which are massively used as pesticides, are responsible for soil and water pollution, for which no satisfactory means of remediation are available (Singh, 2009 ▶). Indeed, existing methods for removing them are cost-prohibitive and cause environmental concerns (LeJeune et al., 1998 ▶). Moreover, before World War 2 these compounds were engineered as chemical warfare agents and were used in the Iran–Iraq war and in the terrorist attack on the Tokyo subway (Gupta, 2009 ▶). The use of enzymes that are capable of hydrolyzing these compounds represents an appealing alternative to chemical methods (Singh, 2009 ▶). Several organophosphate hydrolases (OPHs) belonging to different enzyme superfamilies have been identified and studied [e.g. organophosphate acid anhydrolase (OPAA; Vyas et al., 2010 ▶), OPHC2 (Gotthard et al., 2013 ▶) and paraoxonases (PONs; Ben-David et al., 2012 ▶)]. The most studied OPHs comprise the bacterial phosphotriesterases (PTEs) isolated from Brevundimonas diminuta (BdPTE; Benning et al., 1994 ▶) and Agrobacterium radiobacter (OpdA; Jackson et al., 2006 ▶). These enzymes, which exhibit near-diffusion-limit kinetic rates against the insecticide paraoxon (i.e. k cat/K M ≃ 108 M −1 s−1; Omburo et al., 1992 ▶), are believed to have emerged from the first uses of OPs as insecticides in the 1950s.
Recently, a protein family sharing ∼30% sequence identity with PTEs was identified by virtue of their ability to hydrolyze insecticides and were initially dubbed paraoxonases (Merone et al., 2005 ▶). A detailed biochemical and phylogenetic analysis later revealed that these proteins, named phosphotriesterase-like lactonases (PLLs), are in fact native lactonases endowed with promiscuous phosphotriesterase activity (Afriat et al., 2006 ▶; Elias & Tawfik, 2012 ▶). PLLs are likely to be the progenitors of PTEs (Afriat-Jurnou et al., 2012 ▶). PLLs hydrolyze various lactones, including aliphatic lactones and acyl-homoserine lactones (AHLs), that are involved in bacterial quorum sensing (Hiblot et al., 2012a ▶; Hawwa, Larsen et al., 2009 ▶). The ability of lactonases such as PLLs to hydrolyze AHLs enables these enzymes to interfere with bacterial communication (Dong et al., 2001 ▶) and they therefore offer interesting potentialities to develop new approaches in order to fight against several pathogens (Amara et al., 2011 ▶).
PTEs and PLLs, which belong to the amidohydrolase superfamily (Seibert & Raushel, 2005 ▶), share the same (α/β)8 topology in which a bimetallic centre is coordinated by four histidines, an aspartic acid and a carboxylated lysine (Elias et al., 2008 ▶; Del Vecchio et al., 2009 ▶). The bimetallic centre activates a water molecule into a hydroxide ion, which serves as a nucleophile for the hydrolysis of OPs or AHLs. The catalytic centre is surrounded by two loops involved in the substrate specificity: loops 7 and 8 (Elias et al., 2008 ▶; Jackson et al., 2009 ▶). PTEs and PLLs differ mainly in the relative size and conformations of these loops, which account for the different substrate specificity of these enzymes (Afriat-Jurnou et al., 2012 ▶). PLLs hydrolyze organophosphate compounds with poor to moderate efficiency (Hiblot et al., 2012b ▶; Hawwa, Larsen et al., 2009 ▶; Zhang et al., 2012 ▶) compared with PTEs (Omburo et al., 1992 ▶; Jackson et al., 2006 ▶; Donarski et al., 1989 ▶). Some PLL representatives, however, offer interesting biotechnological potentialities for engineering an efficient organophosphate biodecontaminant because of their high thermal stability (Hiblot et al., 2012a ▶,b ▶; Hawwa, Larsen et al., 2009 ▶; Hawwa, Aikens et al., 2009 ▶), as illustrated by several engineering studies on these enzymes (Merone et al., 2010 ▶; Hawwa, Larsen et al., 2009 ▶; Xue et al., 2013 ▶; Chow et al., 2010 ▶).
VmoLac (YP_004245953) is an enzyme identified from the recently sequenced crenarchaeon Vulcanisaeta moutnovskia strain 768-28 (Gumerov et al., 2011 ▶). This organism was isolated from solfataric fields close to the Moutnovsky volcano in Kamchatka, Russia. Its growth temperature ranges between 333 and 371 K (Gumerov et al., 2011 ▶). VmoLac shares ∼50% sequence identity with other PLLs. Here, we report the protein production, purification, crystallization and preliminary X-ray diffraction of VmoLac.
2. Cloning, expression and purification of VmoLac
The gene encoding VmoLac (YP_004245953) was optimized for Escherichia coli expression by the GeneArt service provider (Life Technologies, France). The optimized gene included N-terminal tags [a Strep-tag (MSAWSHPQFEK) for affinity chromatography purification and a TEV cleavage site (ENLYFQ/G) for removal of the tag; Gotthard et al., 2011 ▶] and was synthesized by GeneArt (Life Technologies, France). This construct leaves an N-terminal Gly residue after cleavage of the tag by TEV protease. The complete gene was subsequently cloned by the same provider into a custom version of pET-22b(+) (Novagen) using NdeI and XhoI as cloning sites. Recombinant VmoLac protein was overexpressed using a protocol that was previously used for another PLL (Gotthard et al., 2013 ▶; Hiblot et al., 2012a ▶). Briefly, recombinant VmoLac protein was overproduced in E. coli BL21 (DE3)-pGro7/GroEL strain (TaKaRa). Protein expression was performed in 4 l ZYP medium (100 µg ml−1 ampicillin, 34 µg ml−1 chloramphenicol) inoculated with a 50 ml overnight pre-culture. The culture was grown at 310 K until the OD600 nm reached 0.6. Induction of the protein was conducted by consumption of the lactose in the ZYP medium, a temperature transition to 298 K over 20 h and the addition of 0.2 mM CoCl2.
The cells were harvested by centrifugation (4500g, 277 K, 15 min). The pellets were resuspended in lysis buffer [50 mM HEPES pH 8, 150 mM NaCl, 0.2 mM CoCl2, 0.25 mg ml−1 lysozyme, 10 µg ml−1 DNAse I, 0.1 mM phenylmethylsulfonyl fluoride (PMSF)] and stored at 193 K for 2 h. The suspended frozen cells were thawed at 310 K for 15 min and disrupted by three sonication steps of 30 s (Branson Sonifier 450; 80% intensity and micro tip limit at 8). Cell debris was removed by centrifugation (14 500g, 277 K, 30 min). The cell lysate was then loaded onto a StrepTrap column (GE Healthcare) at a flow rate of 5 ml min−1 and protein elution was performed using elution buffer (50 mM HEPES pH 8, 150 mM NaCl, 0.2 mM CoCl2, 2.5 mM desthiobiotin). Because of the low binding capacity of the column, this step was repeated five times and all of the protein was then cleaved by TEV protease (1250 µg, 20 h, 310 K; van den Berg et al., 2006 ▶). Spontaneously precipitated TEV protease was harvested by centrifugation (12 000g, 277 K, 10 min). The protein was subsequently concentrated using a centrifugation device (Amicon Ultra MWCO 10 kDa; Millipore, Ireland) prior to a size-exclusion chromatography step (Superdex 75 16/60, GE Healthcare) in 50 mM HEPES pH 8, 150 mM NaCl, 0.2 mM CoCl2. Fractions containing pure protein were pooled and concentrated prior to crystallization trials using a centrifugation device (Amicon Ultra MWCO 10 kDa; Millipore, Ireland). The yield of production was about 5 mg per litre of culture.
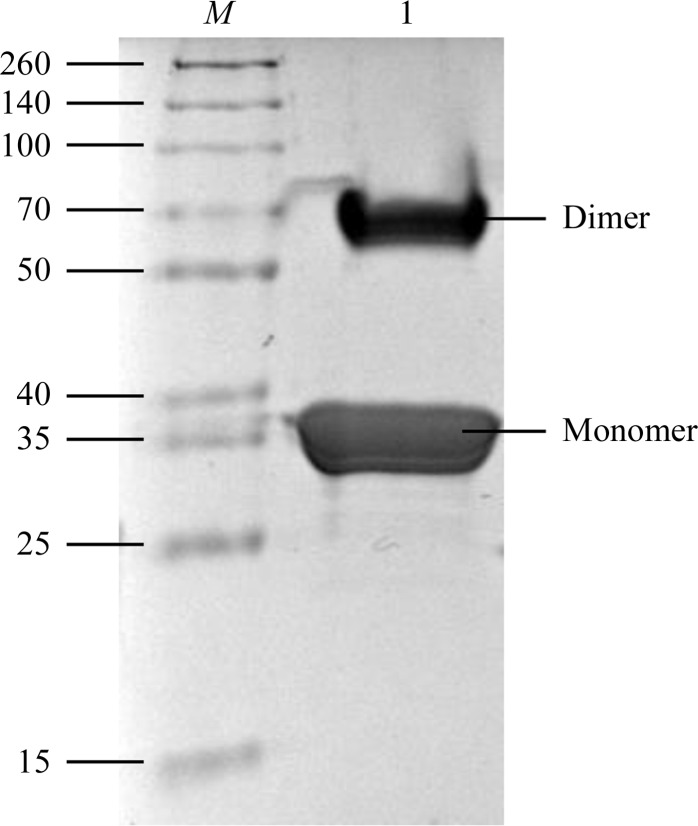
The purity of the protein was checked with Coomassie-stained 15% SDS–PAGE (Fig. 1 ▶), which revealed two main bands (35 and 70 kDa). Both bands were subjected to mass-spectrometric analysis (MS Platform Timone, Marseille, France) and both were identified as the VmoLac protein. The molecular mass of the VmoLac monomer being 35 548 Da, the two bands on the gel at 35 and 70 kDa were attributed to monomers and dimers of VmoLac, respectively. As the VmoLac dimer originates from an extremely thermophilic organism, its dimer may resist the denaturing conditions of sample preparation for SDS–PAGE experiments (368 K incubation for 10 min, 715 mM β-mercaptoethanol).
Figure 1.
15% SDS–PAGE of VmoLac protein performed under denaturing conditions and stained with Coomassie Blue. Lane M contains molecular-weight markers (Thermo Scientific Spectra Multicolor broad-range protein ladder; labelled in kDa). Lane 1 contains VmoLac protein.
3. Protein crystallization
VmoLac was concentrated to 20 mg ml−1 for crystallization trials. Crystallization assays were performed using the sitting-drop vapour-diffusion method setup in a 96-well plate and the commercial screen conditions Structure Screen 1 + 2 (Molecular Dimensions). The plate was incubated at 277 K. Crystals appeared after 1 d at 277 K in a condition consisting of 400 mM ammonium dihydrogen phosphate. Crystals grew in drops containing a 2:1 protein:reservoir ratio (Fig. 2 ▶).
Figure 2.

Crystals of VmoLac (average dimensions of 100 × 50 × 40 µm).
4. Data collection
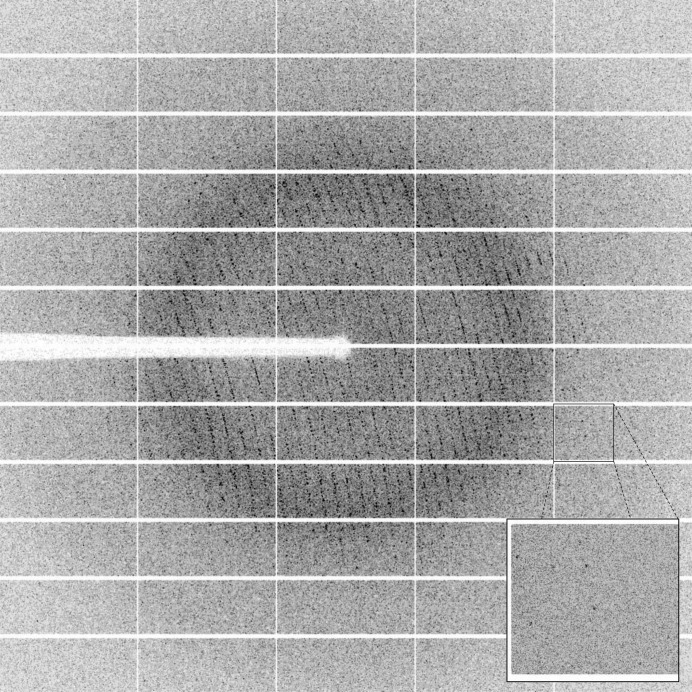
A cryoprotectant solution consisting of the crystallization solution supplemented with 30%(v/v) glycerol was added to the drop in order to exchange the solution containing the crystal. The crystal was subsequently mounted on a CryoLoop (Hampton Research) and flash-cooled in liquid nitrogen. X-ray diffraction intensities were collected on the ID29 beamline at the ESRF, Grenoble, France using a wavelength of 0.800 Å and a PILATUS 6M detector (DECTRIS, Switzerland) with 50 ms exposures. Diffraction data were collected using the fine-slicing method; individual frames consisted of 0.1° steps over a range of 100° (Fig. 3 ▶).
Figure 3.
A diffraction pattern from a crystal of VmoLac. The edge of the frame is at 2.0 Å resolution.
5. Results and conclusions
X-ray diffraction data were integrated and scaled using the XDS package (Kabsch, 1993 ▶; Table 1 ▶). The VmoLac crystals belonged to the hexagonal space group P64, with unit-cell parameters a = b = 174.06, c = 61.32 Å. VmoLac being a 35 kDa protein, the calculated Matthews coefficient (Matthews, 1968 ▶) suggests that between two and three monomers are present per asymmetric unit (3.78 and 2.52 Å3 Da−1, corresponding to 67.49 and 51.24% solvent content, respectively). An initial molecular replacement was performed using Phaser (McCoy et al., 2007 ▶) with the structure of SsoPox (52% sequence identity; PDB entry 2vc5; Elias et al., 2008 ▶) from which the fragment 265–277 was deleted as a starting model. Two molecules were initially placed in the asymmetric unit (R = 47.25%, R free = 48.1%). The initial solution was then submitted to ARP/wARP (Morris et al., 2003 ▶) for automated model construction. After 50 cycles of ARP/wARP and 20 cycles of refinement using REFMAC (Murshudov et al., 2011 ▶), the R and R free factor values were 18.95 and 22.38%, respectively. The electron-density maps revealed that, with the exception of some rotamers and loop conformations, the model is near final. The asymmetric unit contains one homodimer of VmoLac. The construction, refinement and interpretation of the structure are in progress.
Table 1. Data-collection statistics.
Values in parentheses are for the last bin.
| Beamline | ID29, ESRF |
| Wavelength (Å) | 0.800 |
| Detector | PILATUS 6M |
| Oscillation (°) | 0.1 |
| No. of frames | 1000 |
| Resolution (Å) | 47.6–2.4 (2.5–2.4) |
| Space group | P64 |
| Unit-cell parameters (Å) | a = b = 174.06, c = 61.32 |
| No. of observed reflections | 231397 (26406) |
| No. of unique reflections | 41676 (4740) |
| Completeness (%) | 99.6 (99.3) |
| R meas † (%) | 13.4 (63.5) |
| CC1/2 ‡ | 99.6 (81.8) |
| 〈I/σ(I)〉 | 14.55 (3.22) |
| Multiplicity | 5.55 (5.57) |
| Mosaicity (°) | 0.201 |
R
meas = 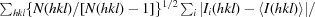
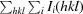 .
.
CC1/2 is the intra-data-set correlation coefficient calculated from the percentage of correlation between intensities (I 1 and I 2) from random half data sets (Karplus & Diederichs, 2012 ▶): CC1/2 = Corr(I 1, I 2).
Acknowledgments
This research was supported by a grant to EC from Délégation Générale pour l’Armement (REI #2009 34 0045). GG and JH are supported by Délégation Générale pour l’Armement.
References
- Afriat, L., Roodveldt, C., Manco, G. & Tawfik, D. S. (2006). Biochemistry, 45, 13677–13686. [DOI] [PubMed]
- Afriat-Jurnou, L., Jackson, C. J. & Tawfik, D. S. (2012). Biochemistry, 51, 6047–6055. [DOI] [PubMed]
- Amara, N., Krom, B. P., Kaufmann, G. F. & Meijler, M. M. (2011). Chem. Rev. 111, 195–208. [DOI] [PubMed]
- Ben-David, M., Elias, M., Filippi, J. J., Duñach, E., Silman, I., Sussman, J. L. & Tawfik, D. S. (2012). J. Mol. Biol. 418, 181–196. [DOI] [PubMed]
- Benning, M. M., Kuo, J. M., Raushel, F. M. & Holden, H. M. (1994). Biochemistry, 33, 15001–15007. [DOI] [PubMed]
- Berg, S. van den, Löfdahl, P. A., Härd, T. & Berglund, H. (2006). J. Biotechnol. 121, 291–298. [DOI] [PubMed]
- Chow, J. Y., Xue, B., Lee, K. H., Tung, A., Wu, L., Robinson, R. C. & Yew, W. S. (2010). J. Biol. Chem. 285, 40911–40920. [DOI] [PMC free article] [PubMed]
- Del Vecchio, P., Elias, M., Merone, L., Graziano, G., Dupuy, J., Mandrich, L., Carullo, P., Fournier, B., Rochu, D., Rossi, M., Masson, P., Chabriere, E. & Manco, G. (2009). Extremophiles, 13, 461–470. [DOI] [PubMed]
- Donarski, W. J., Dumas, D. P., Heitmeyer, D. P., Lewis, V. E. & Raushel, F. M. (1989). Biochemistry, 28, 4650–4655. [DOI] [PubMed]
- Dong, Y.-H., Wang, L.-H., Xu, J.-L., Zhang, H.-B., Zhang, X.-F. & Zhang, L.-H. (2001). Nature (London), 411, 813–817. [DOI] [PubMed]
- Elias, M., Dupuy, J., Merone, L., Mandrich, L., Porzio, E., Moniot, S., Rochu, D., Lecomte, C., Rossi, M., Masson, P., Manco, G. & Chabriere, E. (2008). J. Mol. Biol. 379, 1017–1028. [DOI] [PubMed]
- Elias, M. & Tawfik, D. S. (2012). J. Biol. Chem. 287, 11–20. [DOI] [PMC free article] [PubMed]
- Gotthard, G., Hiblot, J., Elias, M. & Chabrière, E. (2011). Acta Cryst. F67, 354–357. [DOI] [PMC free article] [PubMed]
- Gotthard, G., Hiblot, J., Gonzalez, D., Chabrière, E. & Elias, M. (2013). Acta Cryst. F69, 73–76. [DOI] [PMC free article] [PubMed]
- Gumerov, V. M., Mardanov, A. V., Beletsky, A. V., Prokofeva, M. I., Bonch-Osmolovskaya, E. A., Ravin, N. V. & Skryabin, K. G. (2011). J. Bacteriol. 193, 2355–2356. [DOI] [PMC free article] [PubMed]
- Gupta, R. C. (2009). Handbook of Toxicology of Chemical Warfare Agents London: Academic Press.
- Hawwa, R., Aikens, J., Turner, R. J., Santarsiero, B. D. & Mesecar, A. D. (2009). Arch. Biochem. Biophys. 488, 109–120. [DOI] [PMC free article] [PubMed]
- Hawwa, R., Larsen, S. D., Ratia, K. & Mesecar, A. D. (2009). J. Mol. Biol. 393, 36–57. [DOI] [PubMed]
- Hiblot, J., Gotthard, G., Chabriere, E. & Elias, M. (2012a). PLoS One, 7, e47028. [DOI] [PMC free article] [PubMed]
- Hiblot, J., Gotthard, G., Chabriere, E. & Elias, M. (2012b). Sci. Rep. 2, 779. [DOI] [PMC free article] [PubMed]
- Jackson, C. J., Carr, P. D., Kim, H.-K., Liu, J.-W., Herrald, P., Mitić, N., Schenk, G., Smith, C. A. & Ollis, D. L. (2006). Biochem. J. 397, 501–508. [DOI] [PMC free article] [PubMed]
- Jackson, C. J., Foo, J.-L., Tokuriki, N., Afriat, L., Carr, P. D., Kim, H.-K., Schenk, G., Tawfik, D. S. & Ollis, D. L. (2009). Proc. Natl Acad. Sci. USA, 106, 21631–21636. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (1993). J. Appl. Cryst. 26, 795–800.
- Karplus, P. A. & Diederichs, K. (2012). Science, 336, 1030–1033. [DOI] [PMC free article] [PubMed]
- LeJeune, K. E., Wild, J. R. & Russell, A. J. (1998). Nature (London), 395, 27–28. [DOI] [PubMed]
- Masson, P., Carletti, E. & Nachon, F. (2009). Protein Pept. Lett. 16, 1215–1224. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Merone, L., Mandrich, L., Porzio, E., Rossi, M., Müller, S., Reiter, G., Worek, F. & Manco, G. (2010). Bioresour. Technol. 101, 9204–9212. [DOI] [PubMed]
- Merone, L., Mandrich, L., Rossi, M. & Manco, G. (2005). Extremophiles, 9, 297–305. [DOI] [PubMed]
- Morris, R. J., Perrakis, A. & Lamzin, V. S. (2003). Methods Enzymol. 374, 229–244. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Omburo, G. A., Kuo, J. M., Mullins, L. S. & Raushel, F. M. (1992). J. Biol. Chem. 267, 13278–13283. [PubMed]
- Seibert, C. M. & Raushel, F. M. (2005). Biochemistry, 44, 6383–6391. [DOI] [PubMed]
- Singh, B. K. (2009). Nature Rev. Microbiol. 7, 156–164. [DOI] [PubMed]
- Vyas, N. K., Nickitenko, A., Rastogi, V. K., Shah, S. S. & Quiocho, F. A. (2010). Biochemistry, 49, 547–559. [DOI] [PubMed]
- Xue, B., Chow, J. Y., Baldansuren, A., Yap, L. L., Gan, Y. H., Dikanov, S. A., Robinson, R. C. & Yew, W. S. (2013). Biochemistry, 52, 2359–2370. [DOI] [PMC free article] [PubMed]
- Zhang, Y., An, J., Ye, W., Yang, G., Qian, Z.-G., Chen, H.-F., Cui, L. & Feng, Y. (2012). Appl. Environ. Microbiol. 78, 6647–6655. [DOI] [PMC free article] [PubMed]