spMCAT from Synechocystis sp. PCC 6803, the first MCAT counterpart from a cyanobacterium, has been successfully cloned, purified and crystallized. A high-quality crystal with good diffraction has been obtained.
Keywords: malonyl-CoA:acyl-carrier protein transacylase (MCAT), Synechocystis
Abstract
Malonyl-coenzymeA:acyl-carrier protein transacylase (MCAT), which catalyzes the transfer of the malonyl group from malonyl-CoA to acyl-carrier protein (ACP), is an essential enzyme in type II fatty-acid synthesis. The enzyme MCAT from Synechocystis sp. PCC 6803 (spMCAT), the first MCAT counterpart from a cyanobacterium, was cloned, purified and crystallized in order to determine its three-dimensional crystal structure. A higher-quality crystal with better diffraction was obtained by crystallization optimization. The crystal diffracted to 1.8 Å resolution and belonged to the orthorhombic space group P21212, with unit-cell parameters a = 43.22, b = 149.21, c = 40.59 Å. Matthews coefficient calculations indicated that the crystal contained one spMCAT molecule in the asymmetric unit with a Matthews coefficient of 2.18 Å3 Da−1 and a solvent content of 43.65%.
1. Introduction
As a consequence of the environmental problems associated with carbon dioxide emitted from burning of fossil fuels, energy production using biofuels and biochemicals is set to play an important role (Kerr, 2007 ▶). Recently, increasing attention has been paid to studies on engineering cyanobacteria because of their capability to convert atmospheric CO2 and sunlight directly into organic compounds, including carbon-based biofuels (Steuer et al., 2012 ▶). There are several advantages to their use such as their rapid growth rate, high lipid content, ability to thrive on non-arable land and ease of handling owing to the availability of genetic tools and sequenced genomes etc. (Herrera, 2006 ▶). In order to better understand and manipulate cyanobacterial lipid metabolism for the improvement of lipid production, an expanded knowledge of the mechanisms of some key enzymes involved in fatty-acid biosynthesis is needed. Synechocystis sp. PCC 6803 is a model cyanobacterium, making it ideal for the study of the mechanism governing fatty-acid synthesis.
The enzyme malonyl-coenzymeA:acyl-carrier protein transacylase (MCAT) serves to transfer malonate from malonyl-CoA to acyl-carrier protein (ACP), which plays an important role in type II fatty-acid biosynthesis (Ruch & Vagelos, 1973 ▶). Several MCAT crystal structures from different organisms have been published (Natarajan et al., 2012 ▶; Hong, Kim & Kim, 2010 ▶; Keatinge-Clay et al., 2003 ▶; Li et al., 2007 ▶; Oefner et al., 2006 ▶; Serre et al., 1995 ▶; Zhang et al., 2007 ▶). However, the structure of an MCAT from a cyanobacterium has not yet been reported. Sequence alignment indicated that the MCAT from Synechocystis sp. PCC 6803 has sequence identities of 40% to the MCAT from Escherichia coli (PDB entry 1mla; Serre et al., 1995 ▶), 36% to MCAT from Staphylococcus aureus (PDB entry 3im9; Hong, Kim, Park et al., 2010 ▶), 32% to MCAT from Mycobacterium tuberculosis (PDB entry 2qc3; Li et al., 2007 ▶) and 30% to MCAT from Helicobacter pylori (PDB entry 2h1y; Zhang et al., 2007 ▶). In order to determine the cyanobacterial MCAT structure, thereby enriching the structural information for the entire MCAT enzyme family, here we report the cloning, expression, purification, crystallization and preliminary X-ray crystallographic analysis of the MCAT encoded by fabD (slr2023) from Synechocystis sp. PCC 6803 (spMCAT). Elucidation of the structural information may provide us with a solid understanding of the theoretical background for higher biofuel production from microalgae and cyanobacteria.
2. Materials and methods
2.1. Cloning
Synechocystis sp. PCC 6803 was obtained from the Institute of Hydrobiology, Chinese Academy of Sciences. The DNA of Synechocystis sp. PCC 6803 was extracted as described by Xu et al. (1993 ▶). E. coli strains DH5α and BL21 were used for DNA cloning. Molecular manipulations were performed using standard methods. NdeI and XhoI were used according to the manufacturer’s instructions. The spMCAT-coding sequence of slr2023 (882 bp) was amplified from the Synechocystis sp. PCC 6803 DNA via PCR reaction using the forward primer 5′-CATATGAAAACTGCATGGGTA-3′ and the reverse primer 5′-CTCGAGCTACAGGGAATTCAAGTCG-3′ (NdeI and XhoI restriction sites are indicated in bold, respectively). The PCR fragment was ligated into the pMD18-T vector (TaKaRa) and confirmed by sequencing. The spMCAT fragment was then digested with NdeI and XhoI restriction endonucleases and ligated into digested pET-28a (Merck). The final construct encoded an N-terminally truncated spMCAT protein which had spMCAT fused to an N-terminal His tag.
2.2. Expression and purification
The pET28a-spMCAT expression vector containing an N-terminal hexahistidine tag was transformed into E. coli BL21 (DE3) cells. Cells were grown to an OD600 of 0.5 at 310 K in LB medium containing 34 µg ml−1 kanamycin. Protein expression was induced by the addition of 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and the cells were cultured at the same temperature for an additional 3.5 h. The cultured cells were harvested by centrifugation at 6500g for 20 min at 277 K. The cell pellet was then resuspended in lysis buffer [50 mM sodium phosphate pH 8.0, 300 mM NaCl, 5%(v/v) glycerol, 1 mM PMSF, 2 mM β-mercaptoethanol] and homogenized by ultrasonication on ice. The lysate was centrifuged for 30 min at 12 000g at 277 K. The supernatant containing soluble spMCAT protein was applied onto Ni–NTA resin (Qiagen) pre-equilibrated with buffer consisting of 50 mM sodium phosphate pH 8.0, 300 mM NaCl, 5%(v/v) glycerol, 5 mM imidazole. The column was washed with buffer consisting of 50 mM sodium phosphate pH 8.0, 300 mM NaCl, 5%(v/v) glycerol, 25 mM imidazole. The His-tagged spMCAT protein was then eluted using lysis buffer containing 300 mM imidazole. The eluate was further purified on a Superdex 200 column equilibrated with Tris–HCl buffer [50 mM Tris–HCl pH 7.8, 300 mM NaCl, 1 mM EDTA, 5%(v/v) glycerol, 2 mM β-mercaptoethanol] using an ÄKTAprime plus (GE Healthcare). The purity of the spMCAT was judged to be greater than 95% as shown by SDS–PAGE analysis (Fig. 1 ▶). The fraction containing spMCAT was concentrated to 30 mg ml−1 using a 10 kDa Amicon concentrator (Millipore) and the protein concentration was determined using the Bradford assay with BSA (bovine serum albumin) as a standard. Finally, the spMCAT was stored in storage buffer [50 mM Tris–HCl pH 7.8, 300 mM NaCl, 1 mM EDTA, 5%(v/v) glycerol, 2 mM β-mercaptoethanol] for 2 d at 193 K prior to crystallization attempts.
Figure 1.
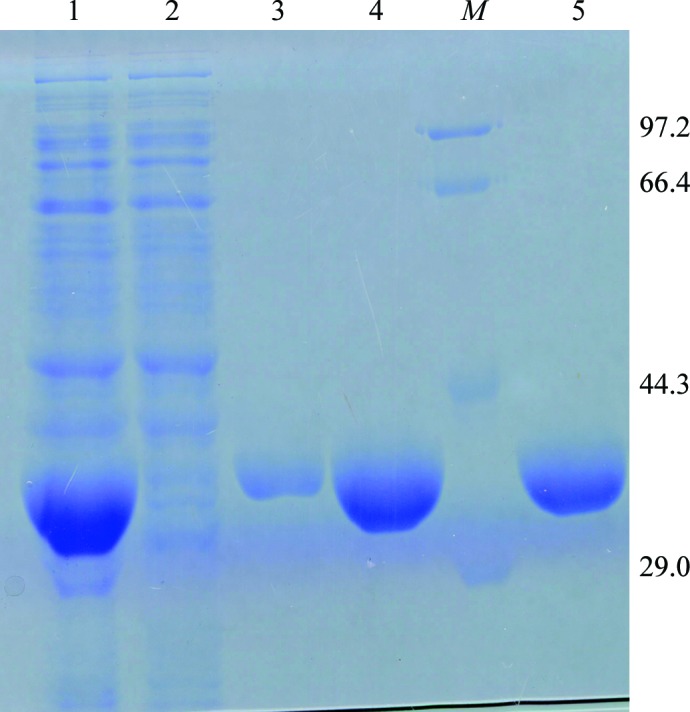
SDS–PAGE showing the purification of spMCAT. Lane 1, supernatant of sonicated E. coli containing spMCAT before loading onto the Ni2+-affinity column; lane 2, flowthrough of the sample, showing binding of spMCAT onto the column; lane 3, flowthrough from the washing buffer containing 25 mM imidazole; lane 4, spMCAT eluted from the Ni2+-affinity column using 300 mM imidazole; lane 5, spMCAT eluted from the Superdex 200 column, which was used for protein crystallization trials. Lane M, molecular-mass marker (labelled in kDa).
2.3. Crystallization
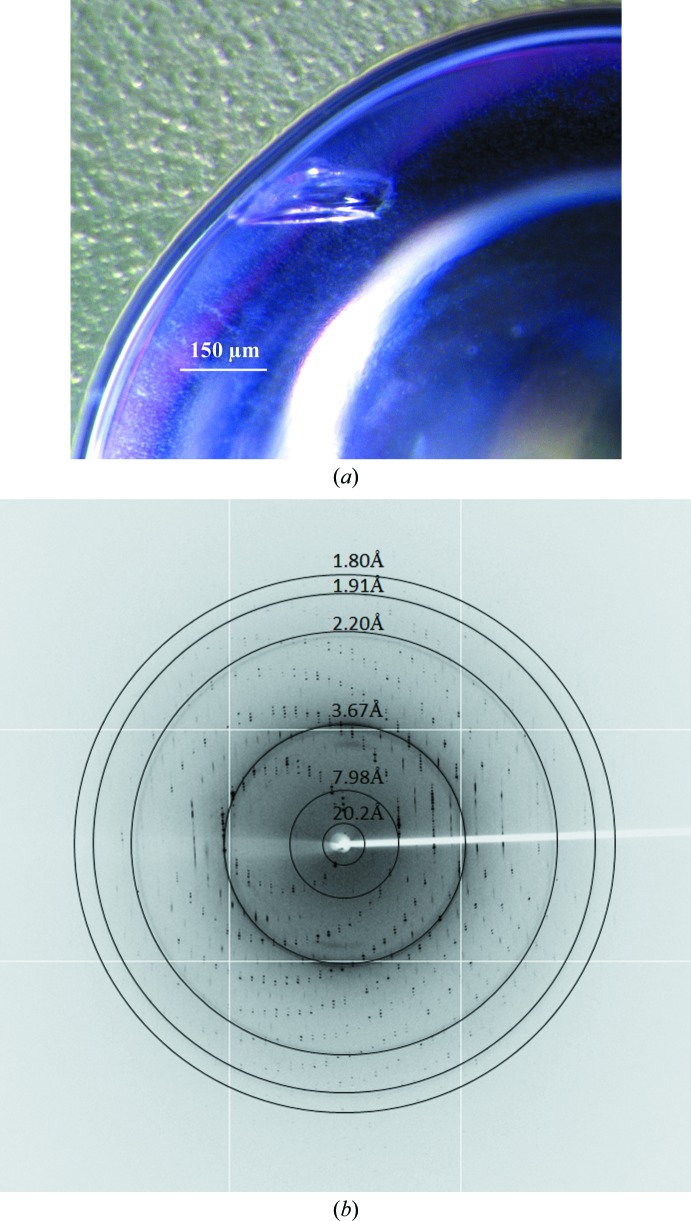
To obtain initial crystallization conditions, spMCAT was screened against 96 conditions from Crystal Screen and Crystal Screen 2 (Hampton Research) by the hanging-drop method. 1.2 µl spMCAT solution (25 or 10 mg ml−1) was mixed with an equal volume of reservoir solution on a siliconized cover slip and the mixture was equilibrated against 500 µl reservoir solution at 298 K. Needle or needle-cluster crystals of spMCAT appeared after 2 d using Crystal Screen condition No. 10 (0.2 M ammonium acetate, 0.1 M sodium acetate trihydrate pH 4.6, 30% PEG 4000) as the reservoir solution. Attempts to optimize the crystallization condition by altering the pH range and the PEG 4000 concentration or by using streak-seeding or additive screening (Hampton Research) were not successful. However, the crystal morphology was improved by changing the temperature, the addition of Al’s oil over the reservoir solution after the mixing of the drop according to Anandan et al. (2012 ▶) and an increase in the protein concentration. The spMCAT crystals with the best diffraction quality (Fig. 2 ▶ a) appeared after 2–3 d on mixing 2 µl protein solution (30 mg ml−1) in 50 mM Tris–HCl pH 7.8, 300 mM NaCl, 5%(v/v) glycerol, 1 mM EDTA, 2 mM β-mercaptoethanol with an equal volume of reservoir solution consisting of 0.2 M ammonium acetate, 0.1 M sodium acetate trihydrate pH 4.6, 30% PEG 4000 and equilibrating against 500 µl reservoir solution with the addition of 200 µl Al’s oil at 277 K.
Figure 2.
(a) Optimized crystal obtained after 2–3 d by mixing 2 µl protein solution (30 mg ml−1) at 50 mM Tris–HCl pH 7.8, 300 mM NaCl, 5%(v/v) glycerol, 1 mM EDTA, 2 mM β-mercaptoethanol with an equal volume of reservoir solution consisting of 0.2 M ammonium acetate, 0.1 M sodium acetate trihydrate pH 4.6, 30% PEG 4000 and equilibrating against 500 µl reservoir solution over which 200 µl Al’s oil was added at 277 K. (b) The X-ray diffraction image of an spMCAT crystal at ϕ = 90° collected at the Shanghai Synchrotron Radiation Facility (SSRF), China.
2.4. Data collection and processing
Optimized crystals, either preserved in liquid nitrogen after X-ray diffraction testing at an in-house facility (Agilent Gemini A Ultra dual-wavelength diffractometer) or transported directly in their original Linbro trays, were transported to the Shanghai Synchrotron Radiation Facility (SSRF) of China for X-ray data collection. Native spMCAT crystals were mounted directly on the goniometer using the mother liquor itself as cryoprotectant (as tested by in-house X-ray diffraction experiments). The problem of ice rings was partially solved by the simple annealing method (Hanson et al., 2003 ▶) when necessary. The crystal was flash-cooled on a CryoLoop (Hampton Research) in a liquid-nitrogen stream at 100 K. X-ray diffraction data were collected using an MX225 CCD detector at 100 K on beamline BL17U1 of the SSRF. The wavelength was 0.9793 Å and the exposure time was 1 s per image. The crystal was rotated through a total ϕ of 360° with an oscillation range of 1.0° per frame. The crystal-to-detector distance was set to 200 mm. The raw data were reduced and scaled using the HKL-2000 software package (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
spMCAT was successfully cloned from Synechocystis sp. PCC 6803 and expressed in E. coli with a fused hexahistidine tag. Homogeneous spMCAT protein was obtained after expression and purification by Ni–NTA and size-exclusion chromatography. Purified spMCAT showed a single band on SDS–PAGE with an apparent molecular weight of 32.2 kDa (Fig. 1 ▶), which is close to the value (31.5 kDa) estimated by ProtParam (http://www.expasy.org/tools/protparam.html). Initial crystallization attempts gave only needle-cluster crystals of spMCAT. To obtain well diffracting crystals, variations of the methods listed in §2.3 were performed. A satisfactory crystal (Fig. 2 ▶ a) was obtained by adding Al’s oil over the reservoir in combination with an increase in the protein concentration at 277 K.
The data-collection statistics after SCALEPACK are listed in Table 1 ▶. Even though a strategy of 360° crystal rotation was adopted to ensure data completeness, the merged data set after SCALEPACK was only 95.7% complete to 1.8 Å resolution (Fig. 2 ▶ b). Given that our data have only mild anisotropy as tested using the UCLA MBI Diffraction Anisotropy Server (http://services.mbi.ucla.edu/anisoscale/; Strong et al., 2006 ▶), this is most likely to be caused by the detrimental effect of the blind region on the completeness of our data; during our diffraction experiment one of the crystallographic twofold axes unfortunately happened to be aligned along the crystal rotation axis (Dauter, 1999 ▶). As a consequence of this data-completeness problem, the effective resolution cutoff was conservatively chosen at 1.8 Å in order to balance the data resolution and the data completeness, even though the I/σ(I) is over 11 in the last shell, the R merge residuals are lower than 18% (Table 1 ▶) and the outermost diffraction spots reached ∼1.5 Å resolution (Fig. 2 ▶ b). By doing this, the data completeness can be maintained at a level higher than 80% in the last resolution shell.
Table 1. Data-collection statistics for spMCAT.
Values in parentheses are for the outermost resolution shell.
| Source | Beamline BL17U1, SSRF |
| Wavelength (Å) | 0.9793 |
| Space group | P21212 |
| Unit-cell parameters (Å, o) | a = 43.22, b = 149.21, c = 40.59, α = β = γ = 90 |
| Temperature (K) | 100 |
| Multiplicity | 9.1 (9.7) |
| Resolution range (Å) | 50.0–1.8 (1.83–1.80) |
| Total/unique reflections | 219976/24117 |
| Completeness (%) | 95.7 (96.1) |
| No. of molecules in asymmetric unit | 1 |
| Criterion for observed reflections | I > −3σ(I) |
| Mean I/σ(I) | 34.0 (11.5) |
| R merge † (%) | 4.2 (17.8) |
R
merge = 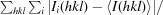
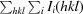 , where Ii(hkl) is the intensity of the ith measurement of reflection hkl and 〈I(hkl)〉 is the mean value of Ii(hkl) for all i measurements.
, where Ii(hkl) is the intensity of the ith measurement of reflection hkl and 〈I(hkl)〉 is the mean value of Ii(hkl) for all i measurements.
A total of 219 976 measured reflections were merged into 24 117 unique reflections with an R merge (on intensity) of 4.2% (Table 1 ▶). The crystal belonged to the orthorhombic space group P21212, with unit-cell parameters a = 43.22, b = 149.21, c = 40.59 Å, α = β = γ = 90°. Matthews coefficient calculations indicated that the crystal contains one spMCAT molecule per asymmetric unit with a Matthews coefficient of 2.18 Å3 Da−1 and a solvent content of 43.65% (the molecular mass of spMCAT is 31.5 kDa) (Matthews, 1968 ▶). The structural data for the spMCAT protein will provide an insight into its enzymatic mechanism and will be used to obtain a theoretical background for higher biofuel production from cyanobacteria.
Acknowledgments
We cordially thank the staff of beamline BL17U1 at the Shanghai Synchrotron Radiation Facility, People’s Republic of China for assistance in synchrotron X-ray data collection. This work was supported by the National Key Basic Research Program of China ‘973 Program’ (2011CBA00803) and the Hundred Talents Program of the Chinese Academy of Sciences (grant No. A1097).
References
- Anandan, A., Piek, S., Kahler, C. M. & Vrielink, A. (2012). Acta Cryst. F68, 1494–1497. [DOI] [PMC free article] [PubMed]
- Dauter, Z. (1999). Acta Cryst. D55, 1703–1717. [DOI] [PubMed]
- Hanson, B. L., Schall, C. A. & Bunick, G. J. (2003). J. Struct. Biol. 142, 77–87. [DOI] [PubMed]
- Herrera, S. (2006). Nature Biotechnol. 24, 755–760. [DOI] [PubMed]
- Hong, S. K., Kim, K. H. & Kim, E. E. (2010). Acta Cryst. F66, 20–22. [DOI] [PMC free article] [PubMed]
- Hong, S. K., Kim, K. H., Park, J. K., Jeong, K.-W., Kim, Y. & Kim, E. E. (2010). FEBS Lett. 584, 1240–1244. [DOI] [PubMed]
- Keatinge-Clay, A. T., Shelat, A. A., Savage, D. F., Tsai, S.-C., Miercke, L. J. W., O’Connell, J. D., Khosla, C. & Stroud, R. M. (2003). Structure, 11, 147–154. [DOI] [PubMed]
- Kerr, R. A. (2007). Science, 316, 188–190. [DOI] [PubMed]
- Li, Z., Huang, Y., Ge, J., Fan, H., Zhou, X., Li, S., Bartlam, M., Wang, H. & Rao, Z. (2007). J. Mol. Biol. 371, 1075–1083. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Natarajan, S., Kim, J.-K., Jung, T.-K., Doan, T. T. N., Ngo, H.-P.-T., Hong, M.-K., Kim, S., Tan, V. P., Ahn, S. J., Lee, S. H., Han, Y., Ahn, Y.-J. & Kang, L.-W. (2012). Mol. Cells, 33, 19–25. [DOI] [PMC free article] [PubMed]
- Oefner, C., Schulz, H., D’Arcy, A. & Dale, G. E. (2006). Acta Cryst. D62, 613–618. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Ruch, F. E. & Vagelos, P. R. (1973). J. Biol. Chem. 248, 8086–8094. [PubMed]
- Serre, L., Verbree, E. C., Dauter, Z., Stuitje, A. R. & Derewenda, Z. S. (1995). J. Biol. Chem. 270, 12961–12964. [DOI] [PubMed]
- Steuer, R., Knoop, H. & Machné, R. (2012). J. Exp. Bot. 63, 2259–2274. [DOI] [PubMed]
- Strong, M., Sawaya, M. R., Wang, S., Phillips, M., Cascio, D. & Eisenberg, D. (2006). Proc. Natl Acad. Sci. USA, 103, 8060–8065. [DOI] [PMC free article] [PubMed]
- Xu, X.-D., Wang, Y.-Q. & Li, S.-H. (1993). J. Graduate Sch. Acad. Sin. 10, 203–209.
- Zhang, L., Liu, W., Xiao, J., Hu, T., Chen, J., Chen, K., Jiang, H. & Shen, X. (2007). Protein Sci. 16, 1184–1192. [DOI] [PMC free article] [PubMed]