To investigate the molecular basis of immune signalling initiated by the TIR domains of plant disease-resistance proteins, the crystallization and preliminary X-ray diffraction analyses of the TIR domains of three proteins involved in disease resistance in A. thaliana are reported.
Keywords: plant innate immunity, resistance proteins, Toll/interleukin-1 receptor domain, RPS4, RRS1, SNC1, hypersensitive response, Arabidopsis thaliana
Abstract
The Toll/interleukin-1 receptor (TIR) domain is a protein–protein interaction domain that is found in both animal and plant immune receptors. The N-terminal TIR domain from the nucleotide-binding (NB)–leucine-rich repeat (LRR) class of plant disease-resistance (R) proteins has been shown to play an important role in defence signalling. Recently, the crystal structure of the TIR domain from flax R protein L6 was determined and this structure, combined with functional studies, demonstrated that TIR-domain homodimerization is a requirement for function of the R protein L6. To advance the molecular understanding of the function of TIR domains in R-protein signalling, the protein expression, purification, crystallization and X-ray diffraction analyses of the TIR domains of the Arabidopsis thaliana R proteins RPS4 (resistance to Pseudomonas syringae 4) and RRS1 (resistance to Ralstonia solanacearum 1) and the resistance-like protein SNC1 (suppressor of npr1-1, constitutive 1) are reported here. RPS4 and RRS1 function cooperatively as a dual resistance-protein system that prevents infection by three distinct pathogens. SNC1 is implicated in resistance pathways in Arabidopsis and is believed to be involved in transcriptional regulation through its interaction with the transcriptional corepressor TPR1 (Topless-related 1). The TIR domains of all three proteins have successfully been expressed and purified as soluble proteins in Escherichia coli. Plate-like crystals of the RPS4 TIR domain were obtained using PEG 3350 as a precipitant; they diffracted X-rays to 2.05 Å resolution, had the symmetry of space group P1 and analysis of the Matthews coefficient suggested that there were four molecules per asymmetric unit. Tetragonal crystals of the RRS1 TIR domain were obtained using ammonium sulfate as a precipitant; they diffracted X-rays to 1.75 Å resolution, had the symmetry of space group P41212 or P43212 and were most likely to contain one molecule per asymmetric unit. Crystals of the SNC1 TIR domain were obtained using PEG 3350 as a precipitant; they diffracted X-rays to 2.20 Å resolution and had the symmetry of space group P41212 or P43212, with two molecules predicted per asymmetric unit. These results provide a good foundation to advance the molecular and structural understanding of the function of the TIR domain in plant innate immunity.
1. Introduction
Plants have developed a complex multilayered immune system to defend themselves against invading pathogens (Jones & Dangl, 2006 ▶). The first basal layer of immunity is termed PAMP (pathogen-associated molecular pattern)-triggered immunity (PTI) and involves the recognition of conserved PAMPs by pattern-recognition receptors (PRRs) on the exterior surface of the plant cell. The second layer involves the recognition of pathogen effector proteins by intracellular disease-resistance (R) proteins and is named effector-triggered immunity (ETI). ETI is often mediated by R proteins from the nucleotide-binding (NB) and leucine-rich repeat (LRR) class, which directly or indirectly recognize pathogen effector proteins. Once activated, NB–LRR proteins trigger defence responses that are often associated with localized cell death at infection sites through a process known as the hypersensitive response (HR; Chisholm et al., 2006 ▶). NB–LRR R proteins have either a coiled-coil (CC) or Toll/interleukin-1 receptor (TIR) domain at their amino-termini and are therefore grouped as CNL and TNL R proteins, respectively (Dodds & Rathjen, 2010 ▶). The central NB domain is believed to act as a molecular switch, utilizing the exchange of ADP and ATP to control activity (Lukasik & Takken, 2009 ▶; Williams et al., 2011 ▶), while the LRR domain has been shown to determine recognition specificity (Dodds et al., 2006 ▶; Padmanabhan et al., 2009 ▶; Ravensdale et al., 2012 ▶; Wang et al., 2007 ▶).
The N-terminal CC and TIR domains are believed to be involved in R-protein signalling. Recent reports of the crystal structures of the CC domain of the barley R protein MLA10 (Maekawa et al., 2011 ▶) and the TIR domain of the flax R protein L6 (Bernoux et al., 2011 ▶) helped to confirm this role. Maekawa and coworkers demonstrated that the CC domain of MLA10 forms a homodimer and mutational studies showed that this property is required for defence signalling (Maekawa et al., 2011 ▶). Similarly, the TIR domain of L6 has the capacity to homodimerize. Crystallographic and mutational studies identified a dimerization interface and a signalling interface in this domain. The TIR domain of L6 is also required and sufficient to induce cell death (Bernoux et al., 2011 ▶).
In Arabidopsis, RPS4 and RRS1 have been shown to cooperatively confer resistance to both fungal and bacterial pathogens (Colletotrichum higginsianum, Ralstonia solanacearum and Pseudomonas syringae pv. tomato strain DC3000 expressing avrRps4; Narusaka, Kubo et al., 2009 ▶; Narusaka, Shirasu et al., 2009 ▶; Birker et al., 2009 ▶). Several examples of paired NB–LRR genes acting cooperatively to confer resistance against a pathogen have been reported (Ashikawa et al., 2008 ▶; Lee et al., 2009 ▶; Sinapidou et al., 2004 ▶; Okuyama et al., 2011 ▶; Yuan et al., 2011 ▶). Both RPS4 and RRS1 belong to the TNL class of R proteins (Deslandes et al., 2002 ▶; Gassmann et al., 1999 ▶). Interestingly, RRS1 carries an additional C-terminal WRKY DNA-binding domain. RRS1 has been found to localize to the nucleus (Deslandes et al., 2003 ▶), while RPS4 is distributed both in the nucleus and cytoplasm, but nuclear localization is required for full pathogen resistance (Wirthmueller et al., 2007 ▶). While the genetic link between RPS4 and RRS1 in this cooperative dual-function resistance has been established, the molecular basis of this cooperation is largely unknown.
A mutation in the A. thaliana NB–LRR R-like protein-encoding gene SNC1 (suppressor of npr1-1, constitutive 1) leads to an autoactive phenotype (constitutive expression of defence genes and enhanced disease resistance against the virulent bacterial pathogen Pseudomonas syringae pv. maculicola ES4326 and the oomycete pathogen Hyaloperonospora arabidopsidis Noco2; Li et al., 2001 ▶). SNC1 has been reported to function through an association with the transcriptional corepressor protein Topless-related 1 (TPR1; Zhu et al., 2010 ▶). Overexpression of TPR1 activates SNC1-mediated immune responses, and GST pull-down assays have demonstrated that TPR1 associates with the SNC1 TIR domain in vitro (Zhu et al., 2010 ▶). However, the molecular basis of these associations remains unknown.
As a step towards further elucidating the mechanisms involved in R-protein signalling, we report the expression, purification and crystallization studies of the TIR domains of the A. thaliana proteins RPS4, RRS1 and SNC1. Because RPS4 and RRS1 function as a dual resistance-protein system, the study should reveal interesting differences in the molecular mechanism of TIR-domain signalling compared with the L6 protein. Comparative studies of different R-protein TIR domains will further shed light on the common features of the interfaces involved in TIR–TIR domain interactions.
2. Materials and methods
2.1. Protein production and purification
The cDNAs comprising the TIR domains, encoding residues 6–153 of RRS1 (designated RRS1TIR) and 10–178 of RPS4 (designated RPS4TIR), were amplified by PCR and inserted into the pMCSG7 vector using ligation-independent cloning (Stols et al., 2002 ▶) using the following primer combinations (RRS16FW, 5′-TACTTCCAATCCAATGCGAAGGATGAGGAATTCGTGTGCATCAGCTGCGTAG-3′; RRS1153RV, 5′-TATTCCACTTCCAATGTTATCCAATTCGTCCAACATAAAAGTGCGTCTCGTACACATC-3′; RPS410FW, 5′-TACTTCCAATCCAATGCGGAAGACAAGCCACCGCAGCATCAGGTG-3′; RPS4178RV, 5′-TTATCCACTTCCAATGTTATATTCCGGTCAACGCTGTCTTCACCGCC-3′). For SNC1, a synthetic gene, codon-optimized for expression in Escherichia coli, that encoded residues 1–184 was purchased from GeneArt (Life Technologies). The cDNA for the TIR domain-encoding residues 8–181 of SNC1 (designated SNC1TIR) was amplified by PCR and inserted into the pMCSG7 vector using ligation-independent cloning (Stols et al., 2002 ▶) using the primer combination SNC18FW, 5′-TACTTCCAATCCAATGCGGGTAGCCGTCGTTATGATGTTTTTCCGAG-3′; SNC1181RV, 5′-TATTCCACTTCCAATGTTAACCAAAATCATCACTCGGGGTCATGGTTT-3′. The resulting constructs contained an N-terminal His6 tag followed by a TEV (Tobacco etch virus) protease cleavage site. The integrity of the constructs was confirmed by sequencing. The expected molecular weights of RPS4TIR, RRS1TIR and SNC1TIR are 20, 17 and 19 kDa, respectively. The RRS1TIR and RPS4TIR constructs were expressed in E. coli Rosetta (DE3) cells, while SNC1TIR was expressed in E. coli BL21 (DE3) cells. For expression, the auto-induction method was used (Studier, 2005 ▶). In brief, cells were grown by continuous shaking at 310 K until the OD600 nm reached 0.6–0.8. The temperature was then lowered to 293 K for RPS4TIR and SNC1TIR and to 288 K for RRS1TIR and the cells were grown for a further 18 h before harvesting by centrifugation. Cells expressing RPS4TIR and SNC1TIR were resuspended in lysis buffer consisting of 50 mM HEPES pH 8.0, 500 mM NaCl, 1 mM DTT, while the cells expressing the RRS1TIR domain were resuspended in lysis buffer consisting of 50 mM HEPES pH 8.0, 150 mM NaCl, 1 mM DTT. The cells were lysed using sonication and were clarified by centrifugation (10 000g) for 40 min. The resulting supernatant was applied onto a 5 ml HisTrap column (GE Healthcare) pre-equilibrated with lysis buffer containing 30 mM imidazole. An ÄKTA FPLC system (GE Healthcare) was used for all chromatography steps. The column was washed with lysis buffer containing 30 mM imidazole to remove proteins interacting nonspecifically and the bound protein was eluted using a linear gradient of imidazole from 30 to 250 mM. Fractions containing the protein of interest, as determined by Coomassie-stained SDS–PAGE, were pooled and buffer-exchanged into a TEV protease-compatible buffer (100 mM Tris pH 8.0, 250 mM NaCl, 1 mM DTT, 0.05 mM EDTA). The N-terminal His6 tag was removed by overnight treatment with His6-tagged TEV protease at 277 K (100 µg per 5 mg of the protein of interest). The cleaved protein was reapplied onto the HisTrap column (pre-equilibrated with lysis buffer containing 30 mM imidazole) to remove the TEV protease and other contaminants. Unbound material that contained the tag-cleaved protein of interest was collected, concentrated and applied onto a Superdex 75 HiLoad 26/60 size-exclusion column (GE Healthcare) pre-equilibrated with gel-filtration buffer consisting of 10 mM HEPES pH 7.5, 150 mM NaCl, 1 mM DTT. The peak fractions were pooled and concentrated using Amicon Ultra-15 Centrifugal Filter Units (Millipore) to final concentrations of 10 mg ml−1 for RPS4TIR and SNC1TIR and 16 mg ml−1 for RRS1TIR. The purity of all of the purified proteins was estimated to be greater than 95% by Coomassie-stained SDS–PAGE. All proteins were stored in the gel-filtration buffer in aliquots at 193 K prior to setting up crystal trays.
2.2. Crystallization and X-ray data collection and processing
The optimal protein concentration for crystallization was determined using the PCT screen (Hampton Research). Initial screening was conducted in 96-well plates (Labtech) at 293 K using the hanging-drop vapour-diffusion method. Eight commercial screens were utilized: Index, PEG/Ion and PEGRx (Hampton Research), Morpheus, ProPlex, JCSG plus and PACT premier (Molecular Dimensions) and Precipitant Synergy (Emerald BioSystems). Hanging drops consisting of 100 nl protein solution and 100 nl reservoir solution were set up using a Mosquito robot (TTP LabTech, UK) and were equilibrated against 100 µl reservoir solution. The drops were monitored and imaged using a Rock Imager system (Formulatrix, USA).
Hits from the initial crystallization screens were optimized by varying the protein concentration, the precipitant concentration, the pH and the drop size and by using the Additive Screen (Hampton Research). Crystals were mounted in nylon loops and transferred into well solution containing 25%(v/v) glycerol as the final concentration prior to flash-cooling in liquid nitrogen.
For both RPS4TIR and SNC1TIR, an X-ray diffraction data set was collected from one single crystal in each case on the Australian Synchrotron MX2 beamline at a wavelength of 0.9537 Å (Table 1 ▶). Two data sets were collected for RRS1TIR at wavelengths of 0.9537 and 1.3776 Å (named data set 1 and 2, respectively) on the Australian Synchrotron MX2 beamline (Table 1 ▶). Data were collected using the Blu-Ice software (McPhillips et al., 2002 ▶), indexed and integrated using XDS (Kabsch, 2010 ▶) and scaled with AIMLESS (Evans & Murshudov, 2013 ▶) within the CCP4 suite (Winn et al., 2011 ▶).
Table 1. Crystallographic data collection and processing.
Values in parentheses are for the outer shell.
| RPS4 | RRS1 data set 1 | RRS1 data set 2 | SNC1 | |
|---|---|---|---|---|
| Detector | ADSC Quantum 315r CCD | ADSC Quantum 315r CCD | ADSC Quantum 315r CCD | ADSC Quantum 315r CCD |
| Wavelength (Å) | 0.9537 | 0.9537 | 1.3776 | 0.9537 |
| Temperature (K) | 100 | 100 | 100 | 100 |
| Crystal-to-detector distance (mm) | 320 | 180 | 170 | 310 |
| Rotation range per image (°) | 1 | 1 | 1 | 0.5 |
| Exposure time per image (s) | 1 | 1 | 1 | 1 |
| Space group | P1 | P41212 or P43212 | P41212 or P43212 | P41212 or P43212 |
| Unit-cell parameters | ||||
| a (Å) | 33.92 | 71.26 | 71.38 | 82.18 |
| b (Å) | 78.64 | 71.26 | 71.38 | 82.18 |
| c (Å) | 80.67 | 66.72 | 66.90 | 124.1 |
| α (°) | 65.63 | 90.00 | 90.00 | 90.00 |
| β (°) | 78.64 | 90.00 | 90.00 | 90.00 |
| γ (°) | 78.93 | 90.00 | 90.00 | 90.00 |
| Average mosaicity† (°) | 0.20 | 0.16 | 0.15 | 0.15 |
| Resolution range (Å) | 19.6–2.05 (2.11–2.05) | 71.24–1.75 (1.78–1.75) | 71.38–2.00 (2.11–2.00) | 19.68–2.20 (2.27–2.20) |
| Total No. of reflections | 165463 (10611) | 251704 (10757) | 929431 (130931) | 372024 (32369) |
| No. of unique reflections | 43623 (2762) | 17927 (955) | 12244 (1735) | 22223 (1888) |
| Completeness (%) | 94.0 (75.3) | 100.0 (100.0) | 100.0 (99.8) | 99.8 (99.6) |
| Multiplicity† | 3.8 (3.8) | 14.0 (11.3) | 75.9 (75.5) | 16.7 (17.1) |
| Mean I/σ(I) | 16.0 (5.9) | 32.3 (1.7) | 61.7 (11.5) | 17.8 (2.1) |
| R meas ‡ (%) | 7.0 (27.3) | 7.0 (167.3) | 7.0 (64.3) | 18.0 (184.9) |
| R p.i.m. § (%) | 3.6 (13.9) | 1.9 (49.4) | 0.8 (7.3) | 4.3 (44.1) |
| CC1/2 ¶ | 0.99 (0.95) | 0.99 (0.59) | 0.99 (0.98) | 0.99 (0.69) |
| Matthews coefficient†† (Å3 Da−1) | 2.44 | 2.49 | 2.51 | 2.63 |
| DelAnom correlation between half sets† | 0.43 (0.03) | |||
R
meas = 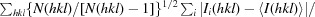
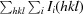 , where Ii(hkl) is the intensity of the ith measurement of an equivalent reflection with indices hkl.
, where Ii(hkl) is the intensity of the ith measurement of an equivalent reflection with indices hkl.
R
p.i.m. = 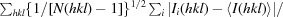
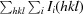 .
.
3. Results and discussion
The domain boundaries of the expression constructs were selected based on sequence alignments of RPS4, RRS1 and SNC1 with TIR domains of known structure, including those from flax L6 protein (Bernoux et al., 2011 ▶) and Arabidopsis NP_177436/At1g72930 (AtTIR; Chan et al., 2010 ▶). The sequence identities of RPS4TIR, RRS1TIR and SNC1TIR to L6TIR are 28, 16 and 36%, respectively. The sequence identities of RPS4TIR, RRS1TIR and SNC1TIR to AtTIR are 32, 23 and 37%, respectively. Four truncated variants each of the RRS1 TIR domain (residues 1–148, 1–153, 6–148 and 6–153) and the RPS4 TIR domain (residues 1–178, 1–183, 10–183 and 10–178) were generated. For the RRS1 TIR domain, the RRS16–153 variant (comprising residues 6–153; referred to here as RRS1TIR) was chosen for further study because of the superior protein-expression levels achieved in E. coli and the higher purity obtained after purification. For the RPS4 TIR domain, all four variants were soluble and could be purified to homogeneity. Both the RPS41–183 and the RPS410–178 variants yielded crystals in sparse-matrix screens. However, the crystals formed by RPS410–178 (referred to here as RPS4TIR) proved easier to optimize and diffraction-quality crystals were obtained. For the SNC1 TIR domain, the SNC18–181 variant (referred to here as SNC1TIR) was studied and could be produced in a soluble form in E. coli.
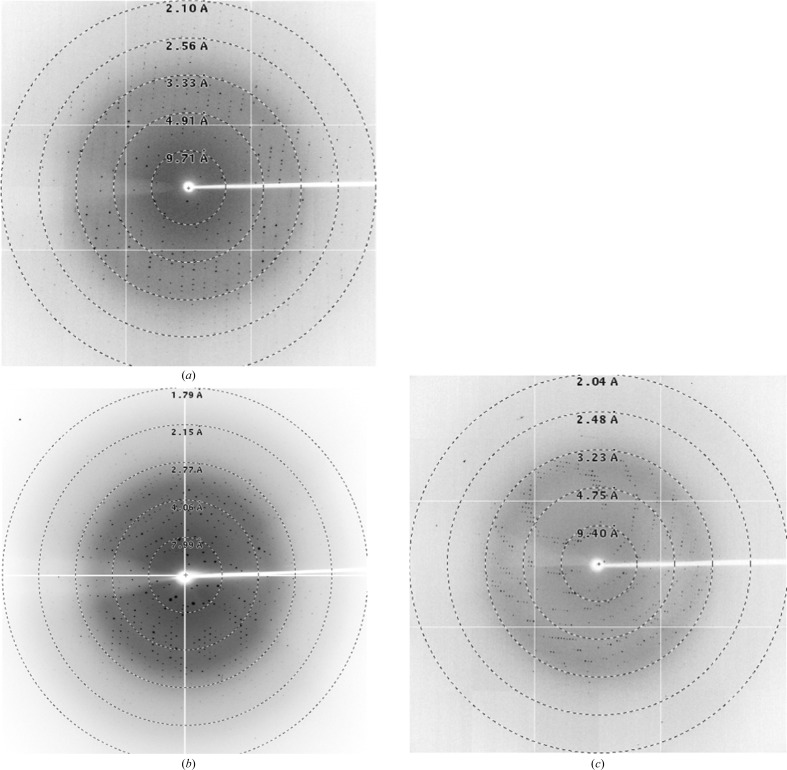
The optimal concentrations for the crystallization of RPS4TIR, RRS1TIR and SNC1TIR were determined to be 10, 2 and 5 mg ml−1, respectively. Initial crystallization screening was conducted at 293 K using 200 nl drops in 96-well plates. Crystals of RPS4TIR grew after 1 d in JCSG plus condition No. 3 consisting of 0.18 M tribasic ammonium citrate, 20%(w/v) PEG 3350. After initial optimization experiments in which the precipitant, salt and pH were manipulated, the crystals remained small and fragile. In an attempt to improve the crystal growth and morphology, we used the Additive Screen (Hampton Research). It was observed that the addition of numerous chloride-containing salts greatly improved the crystal morphology, with sodium chloride achieving the best morphological appearance. Long and thin plate-like crystals were obtained using a protein concentration of 10 mg ml−1 in a crystallization condition consisting of 23%(w/v) PEG 3350, 0.2 M ammonium citrate pH 6.5, 0.2 M sodium chloride at 293 K (Fig. 1 ▶ a). A data set was collected for RPS4TIR to 2.05 Å resolution (Table 1 ▶; Fig. 2 ▶ a). The crystals of RPS4TIR have the symmetry of space group P1 and are most likely to contain four molecules in the asymmetric unit, corresponding to a solvent content of 49% as calculated by MATTHEWS_COEF (Matthews, 1968 ▶; Kantardjieff & Rupp, 2003 ▶) within the CCP4 suite (Winn et al., 2011 ▶).
Figure 1.
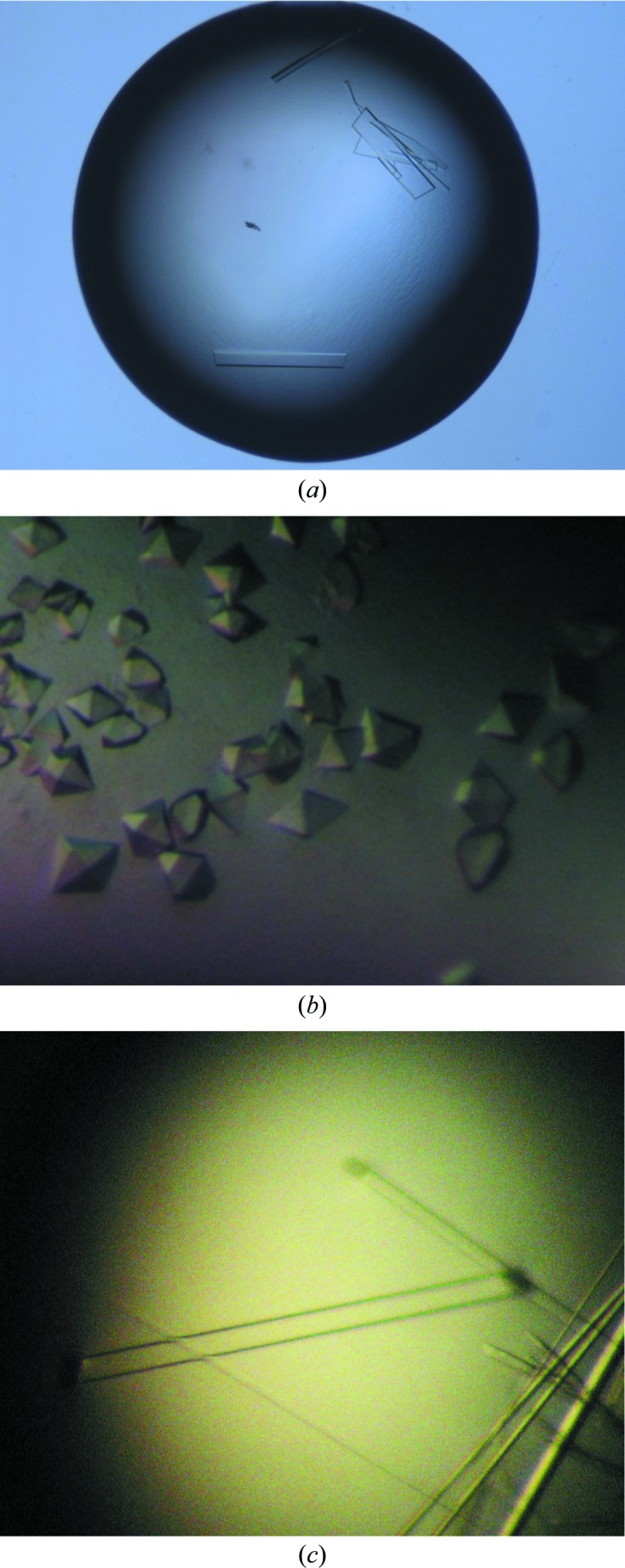
(a) Crystals of RPS4TIR grown after 1 d in 23%(w/v) PEG 3350, 0.2 M ammonium citrate pH 6.5, 0.2 M sodium chloride (approximate dimensions 150 × 150 × 10 µm). (b) Crystals of RRS1TIR grown after 1 d in 0.1 M bis-tris pH 7.0, 1.8 M ammonium sulfate (approximate dimensions 180 × 120 × 80 µm). (c) Crystals of SNC1TIR grown after 1 d in 18%(w/v) PEG 3350, 9%(w/v) glycerol, 0.1 M MMT buffer pH 7.5 (∼0.6 mm in the longest dimension).
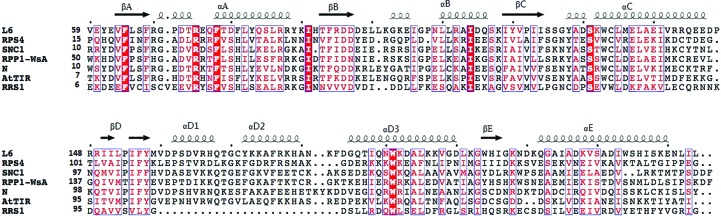
Figure 2.
Diffraction images of RPS4TIR, RRS1TIR and SNC1TIR crystals. The X-ray diffraction images were collected on the MX2 beamline of the Australian Synchrotron (see §2 and Table 1 ▶ for details). (a) Diffraction image of the RPS4TIR crystal, (b) diffraction image of the RRS1TIR crystal (from data set 1), (c) diffraction image of the SNC1TIR crystal.
Crystals of RRS1TIR appeared after 1 d in Index condition No. 4 consisting of 0.2 M ammonium sulfate pH 7.5, 0.1 M bis-tris pH 6.5. After optimization, tetragonal crystals were obtained in a buffer consisting of 0.1 M bis-tris pH 7.0, 1.8 M ammonium sulfate using a protein concentration of 2 mg ml−1 at 293 K (Fig. 1 ▶ b). A data set was collected at 1.75 Å resolution (Table 1 ▶; Fig. 2 ▶ b). The crystals of RRS1TIR have the symmetry of space group P41212 or P43212 and are most likely to contain one molecule in the asymmetric unit, corresponding to a solvent content of 50% as calculated by MATTHEWS_COEF (Matthews, 1968 ▶; Kantardjieff & Rupp, 2003 ▶) within the CCP4 suite (Winn et al., 2011 ▶).
The RRS1 TIR domain is an atypical plant TIR domain because it has a shorter αD helical region compared with other plant TIR domains (Fig. 3 ▶). AtTIR (Chan et al., 2010 ▶) represents the best sequence match for the RRS1 TIR domain to any protein with known structure, with a sequence identity of 21%. In light of the low sequence identity and the deletion within the αD helical region, we were unsure whether molecular replacement (MR) would be suitable for structure determination. To provide additional phase information based on sulfur single-wavelength anomalous diffraction (SAD), another data set was collected from the same crystal at a longer wavelength to maximize the anomalous signal from the S atoms (Table 1 ▶; the wavelength was chosen as a compromise to balance radiation damage and maximizing the anomalous signal). There are five S atoms in one molecule of the RRS1 TIR domain contributed by one methionine residue and four cysteine residues. We do not expect that the structure can be solved based solely on sulfur SAD data and intend to solve the structure by a combination of MR and SAD phasing.
Figure 3.
Multiple sequence alignment of TIR domains. Amino-acid sequences from the TIR domains of Arabidopsis RPS4 (residues 15–191), Arabidopsis SNC1 (10–180), Arabidopsis RPP1-WsA (50–229), tobacco N (10–191) and Arabidopsis RRS1 (6–153) were aligned with the sequences of TIR domains with known crystal structures, Arabidopsis AtTIR (PDB entry 3jrn; Chan et al., 2010 ▶) and flax L6 (PDB entry 3ozi; Bernoux et al., 2011), using MUSCLE (Edgar, 2004 ▶). The positions of the secondary-structure elements in L6 are shown at the top. The alignment was formatted using ESPript (Gouet et al., 2003 ▶).
The initial crystallization screens for SNC1TIR were set up with a protein concentration of 5 mg ml−1. Crystals of SNC1TIR appeared after 1 d under several different screening conditions with PEG 3350 as precipitant, including Index condition No. 44 consisting of 0.1 M HEPES pH 7.5, 37.5%(w/v) PEG 3350 and Index condition No. 45 consisting of 0.1 M Tris pH 8.5, 37.5%(w/v) PEG 3350. Optimization was carried out with PEG 3350 as precipitant and MMT (l-malic acid, MES, Tris) buffer to control the pH (Newman, 2004 ▶). The initial crystals were long and needle-like. Thicker orthorhombic crystals were obtained after glycerol was added to the screening solution. The best crystals grew in 18%(w/v) PEG 3350, 9%(w/v) glycerol, 0.1 M MMT buffer pH 7.5 with a protein concentration of 5 mg ml−1 at 293 K (Fig. 1 ▶ c). A data set was collected to 2.20 Å resolution (Fig. 2 ▶ c). The crystals of SNC1TIR have the symmetry of space group P41212 or P43212 and are most likely contain two molecules in the asymmetric unit, corresponding to a solvent content of 53% as calculated by MATTHEWS_COEF (Matthews, 1968 ▶; Kantardjieff & Rupp, 2003 ▶) within the CCP4 suite (Winn et al., 2011 ▶).
Structure determination of the RPS4, RRS1 and SNC1 TIR domains is currently under way. The crystal structures will provide new insights into the molecular details of the roles of TIR domains during plant immune signalling and in particular any differences between the RPS4/RRS1 dual resistance-protein system compared with the L6 protein. Comparative studies of different R-protein TIR domains will further shed light on the common features of the interfaces involved in TIR–TIR domain interactions.
Acknowledgments
We thank Daniel Ericsson for help and discussion. This work was supported by the Australian Research Council (ARC Discovery grant DP120100685 to BK, PND and JGE). BK is a National Health and Medical Research Council (NHMRC) Research Fellow (1003325).
References
- Ashikawa, I., Hayashi, N., Yamane, H., Kanamori, H., Wu, J., Matsumoto, T., Ono, K. & Yano, M. (2008). Genetics, 180, 2267–2276. [DOI] [PMC free article] [PubMed]
- Bernoux, M., Ve, T., Williams, S., Warren, C., Hatters, D., Valkov, E., Zhang, X., Ellis, J. G., Kobe, B. & Dodds, P. N. (2011). Cell Host Microbe, 9, 200–211. [DOI] [PMC free article] [PubMed]
- Birker, D., Heidrich, K., Takahara, H., Narusaka, M., Deslandes, L., Narusaka, Y., Reymond, M., Parker, J. E. & O’Connell, R. (2009). Plant J. 60, 602–613. [DOI] [PubMed]
- Chan, S. L., Mukasa, T., Santelli, E., Low, L. Y. & Pascual, J. (2010). Protein Sci. 19, 155–161. [DOI] [PMC free article] [PubMed]
- Chisholm, S. T., Coaker, G., Day, B. & Staskawicz, B. J. (2006). Cell, 124, 803–814. [DOI] [PubMed]
- Deslandes, L., Olivier, J., Peeters, N., Feng, D. X., Khounlotham, M., Boucher, C., Somssich, I., Genin, S. & Marco, Y. (2003). Proc. Natl Acad. Sci. USA, 100, 8024–8029. [DOI] [PMC free article] [PubMed]
- Deslandes, L., Olivier, J., Theulieres, F., Hirsch, J., Feng, D. X., Bittner-Eddy, P., Beynon, J. & Marco, Y. (2002). Proc. Natl Acad. Sci. USA, 99, 2404–2409. [DOI] [PMC free article] [PubMed]
- Dodds, P. N., Lawrence, G. J., Catanzariti, A. M., Teh, T., Wang, C.-I. A., Ayliffe, M. A., Kobe, B. & Ellis, J. G. (2006). Proc. Natl Acad. Sci. USA, 103, 8888–8893. [DOI] [PMC free article] [PubMed]
- Dodds, P. N. & Rathjen, J. P. (2010). Nature Rev. Genet. 11, 539–548. [DOI] [PubMed]
- Edgar, R. C. (2004). Nucleic Acids Res. 32, 1792–1797. [DOI] [PMC free article] [PubMed]
- Evans, P. R. & Murshudov, G. N. (2013). Acta Cryst. D69, 1204–1214. [DOI] [PMC free article] [PubMed]
- Gassmann, W., Hinsch, M. E. & Staskawicz, B. J. (1999). Plant J. 20, 265–277. [DOI] [PubMed]
- Gouet, P., Robert, X. & Courcelle, E. (2003). Nucleic Acids Res. 31, 3320–3323. [DOI] [PMC free article] [PubMed]
- Jones, J. D. G. & Dangl, J. L. (2006). Nature (London), 444, 323–329. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kantardjieff, K. A. & Rupp, B. (2003). Protein Sci. 12, 1865–1871. [DOI] [PMC free article] [PubMed]
- Karplus, P. A. & Diederichs, K. (2012). Science, 336, 1030–1033. [DOI] [PMC free article] [PubMed]
- Lee, S.-K., Song, M.-Y., Seo, Y.-S., Kim, H.-K., Ko, S., Cao, P.-J., Suh, J.-P., Yi, G., Roh, J.-H., Lee, S., An, G., Hahn, T.-R., Wang, G.-L., Ronald, P. & Jeon, J.-S. (2009). Genetics, 181, 1627–1638. [DOI] [PMC free article] [PubMed]
- Li, X., Clarke, J. D., Zhang, Y. & Dong, X. (2001). Mol. Plant Microbe Interact. 14, 1131–1139. [DOI] [PubMed]
- Lukasik, E. & Takken, F. L. (2009). Curr. Opin. Plant Biol. 12, 427–436. [DOI] [PubMed]
- Maekawa, T., Cheng, W., Spiridon, L. N., Töller, A., Lukasik, E., Saijo, Y., Liu, P., Shen, Q.-H., Micluta, M. A., Somssich, I. E., Takken, F. L. W., Petrescu, A. J., Chai, J. & Schulze-Lefert, P. (2011). Cell Host Microbe, 9, 187–199. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McPhillips, T. M., McPhillips, S. E., Chiu, H.-J., Cohen, A. E., Deacon, A. M., Ellis, P. J., Garman, E., Gonzalez, A., Sauter, N. K., Phizackerley, R. P., Soltis, S. M. & Kuhn, P. (2002). J. Synchrotron Rad. 9, 401–406. [DOI] [PubMed]
- Narusaka, M., Kubo, Y., Shiraishi, T., Iwabuchi, M. & Narusaka, Y. (2009). Plant Signal. Behav. 4, 954–955. [DOI] [PMC free article] [PubMed]
- Narusaka, M., Shirasu, K., Noutoshi, Y., Kubo, Y., Shiraishi, T., Iwabuchi, M. & Narusaka, Y. (2009). Plant J. 60, 218–226. [DOI] [PubMed]
- Newman, J. (2004). Acta Cryst. D60, 610–612. [DOI] [PubMed]
- Okuyama, Y., Kanzaki, H., Abe, A., Yoshida, K., Tamiru, M., Saitoh, H., Fujibe, T., Matsumura, H., Shenton, M., Galam, D. C., Undan, J., Ito, A., Sone, T. & Terauchi, R. (2011). Plant J. 66, 467–479. [DOI] [PubMed]
- Padmanabhan, M., Cournoyer, P. & Dinesh-Kumar, S. P. (2009). Cell. Microbiol. 11, 191–198. [DOI] [PMC free article] [PubMed]
- Ravensdale, M., Bernoux, M., Ve, T., Kobe, B., Thrall, P. H., Ellis, J. G. & Dodds, P. N. (2012). PLoS Pathog. 8, e1003004. [DOI] [PMC free article] [PubMed]
- Sinapidou, E., Williams, K., Nott, L., Bahkt, S., Tör, M., Crute, I., Bittner-Eddy, P. & Beynon, J. (2004). Plant J. 38, 898–909. [DOI] [PubMed]
- Stols, L., Gu, M., Dieckman, L., Raffen, R., Collart, F. R. & Donnelly, M. I. (2002). Protein Expr. Purif. 25, 8–15. [DOI] [PubMed]
- Studier, F. W. (2005). Protein Expr. Purif. 41, 207–234. [DOI] [PubMed]
- Wang, C.-I. A., Gunčar, G., Forwood, J. K., Teh, T., Catanzariti, A.-M., Lawrence, G. J., Loughlin, F. E., Mackay, J. P., Schirra, H. J., Anderson, P. A., Ellis, J. G., Dodds, P. N. & Kobe, B. (2007). Plant Cell, 19, 2898–2912. [DOI] [PMC free article] [PubMed]
- Williams, S. J., Sornaraj, P., deCourcy-Ireland, E., Menz, R. I., Kobe, B., Ellis, J. G., Dodds, P. N. & Anderson, P. A. (2011). Mol. Plant Microbe Interact. 24, 897–906. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Wirthmueller, L., Zhang, Y., Jones, J. D. G. & Parker, J. E. (2007). Curr. Biol. 17, 2023–2029. [DOI] [PubMed]
- Yuan, B., Zhai, C., Wang, W., Zeng, X., Xu, X., Hu, H., Lin, F., Wang, L. & Pan, Q. (2011). Theor. Appl. Genet. 122, 1017–1028. [DOI] [PubMed]
- Zhu, Z., Xu, F., Zhang, Y., Cheng, Y. T., Wiermer, M., Li, X. & Zhang, Y. (2010). Proc. Natl Acad. Sci. USA, 107, 13960–13965. [DOI] [PMC free article] [PubMed]