KatB, a manganese catalase from Anabaena PCC7120 was crystallized and preliminary diffraction data obtained. Crystal belonged to space group P41212 (or enantiomer) and diffracted to 2.2 Å resolution.
Keywords: manganese catalases, Anabaena, KatB
Abstract
Catalases are enzymes that play an important role in the detoxification of hydrogen peroxide (H2O2) in aerobic organisms. Among catalases, haem-containing catalases are ubiquitously distributed and their enzymatic mechanism is very well understood. On the other hand, manganese catalases that contain a bimanganese core in the active site have been less well characterized and their mode of action is not fully understood. The genome of Anabaena PCC 7120 does not show the presence of a haem catalase-like gene; instead, two ORFs encoding manganese catalases (Mn-catalases) are present. Here, the crystallization and preliminary X-ray crystallographic analysis of KatB, one of the two Mn-catalases from Anabaena, are reported. KatB was crystallized using the hanging-drop vapour-diffusion method with PEG 400 as a precipitant and calcium acetate as an additive. Diffraction data were collected in-house on an Agilent SuperNova system using a microfocus sealed-tube X-ray source. The crystal diffracted to 2.2 Å resolution at 100 K. The tetragonal crystal belonged to space group P41212 (or enantiomer), with unit-cell parameters a = b = 101.87, c = 138.86 Å. Preliminary X-ray diffraction analysis using the Matthews coefficient and self-rotation function suggests the presence of a trimer in the asymmetric unit.
1. Introduction
Catalases are enzymes that break down hydrogen peroxide (H2O2) directly into O2 and H2O (Bernroitner et al., 2009 ▶). H2O2 is an important reactive oxygen species (ROS) generated in the living cell as a byproduct of aerobic respiration or by the action of certain enzymes such as superoxide dismutases and oxidases. In the presence of the Fe2+ ion, H2O2 undergoes the Fenton reaction to generate a hydroxyl radical (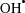 ), the most deleterious ROS that can damage all cellular macromolecules (Halliwell & Gutteridge, 1986 ▶). Therefore, it is important to detoxify H2O2 for survival of the cell and catalases play a crucial role in this process. Three classes of catalases have been identified: (i) monofunctional catalases, (ii) bifunctional catalase–peroxidases and (iii) binuclear manganese catalases (Zamocky et al., 2008 ▶). The first two categories include haem-containing catalases that are ubiquitously distributed among aerobic organisms and have been very well characterized. Several crystal structures of these from different organisms are available in the Protein Data Bank. Manganese catalases (Mn-catalases), in contrast, lack haem and are restricted to prokaryotes and archaea (Amo et al., 2002 ▶). They have been relatively less well characterized and the crystal structures of only two Mn-catalases, i.e. those from Thermus thermophilus (Antonyuk et al., 2000 ▶) and Lactobacillus plantarum (Barynin et al., 2001 ▶), are known.
), the most deleterious ROS that can damage all cellular macromolecules (Halliwell & Gutteridge, 1986 ▶). Therefore, it is important to detoxify H2O2 for survival of the cell and catalases play a crucial role in this process. Three classes of catalases have been identified: (i) monofunctional catalases, (ii) bifunctional catalase–peroxidases and (iii) binuclear manganese catalases (Zamocky et al., 2008 ▶). The first two categories include haem-containing catalases that are ubiquitously distributed among aerobic organisms and have been very well characterized. Several crystal structures of these from different organisms are available in the Protein Data Bank. Manganese catalases (Mn-catalases), in contrast, lack haem and are restricted to prokaryotes and archaea (Amo et al., 2002 ▶). They have been relatively less well characterized and the crystal structures of only two Mn-catalases, i.e. those from Thermus thermophilus (Antonyuk et al., 2000 ▶) and Lactobacillus plantarum (Barynin et al., 2001 ▶), are known.
Cyanobacteria were the first organisms to evolve oxygen and are responsible for the subsequent oxygenation of the earth’s atmosphere. It is very likely that cyanobacteria have developed intricate mechanisms to protect themselves from ROS (Banerjee et al., 2012a ▶). The genome of the heterocystous, filamentous, nitrogen-fixing cyanobacterium Anabaena PCC 7120 (Kaneko et al., 2001 ▶) shows the presence of two Mn-catalases encoded by ORFs alr0998 and alr3090. Interestingly, a haem catalase-like gene is absent in Anabaena PCC 7120 (Banerjee et al., 2012b ▶). The alr3090 ORF has been shown to be important for survival under desiccation (Katoh, 2012 ▶). Increased production of the Alr3090 protein (KatB) under arsenic stress and iron stress has been shown by proteomic analysis (Narayan et al., 2011 ▶; Pandey et al., 2012 ▶). These data suggest that KatB may play a crucial role in the stress tolerance of Anabaena PCC 7120.
The Anabaena PCC 7120 Mn-catalase KatB is smaller (230 amino acids) when compared with the Mn-catalases from both L. plantarum (273 amino acids) and T. thermophilus (302 amino acids). Pairwise sequence alignment shows that KatB shares ∼28% and ∼26% identity with the Mn-catalases from L. plantarum and T. thermophilus, respectively. Based on a BLAST search against the nonredundant database and distance-based analysis, KatB is clustered in a group along with Mn-catalases from other cyanobacteria. This cluster is quite distant from the Mn-catalase clusters of Lactobacillus sp. and Thermus sp. Here, we report the crystallization and preliminary X-ray diffraction analysis of KatB from Anabaena. The three-dimensional structure of KatB will help in understanding the basic mechanism of catalase action and will aid in establishing the evolutionary relationship within the Mn-catalase family.
2. Materials and methods
2.1. Cloning, overexpression and purification
The ORF alr3090 was PCR-amplified employing Anabaena PCC 7120 genomic DNA as a template. The following primers were used: forward primer, 5′-GGACCATGGTTTTTCACAAAGAAAGAACCGATTC-3′, and reverse primer, 5′-GGGGATCCTCGAGTTAGT GATGGTGATGGAATGTTTTTGTAGTGGGTTAG-3′. Restriction-enzyme sites for NcoI and BamHI (underlined) were incorporated in the forward and the reverse primer, respectively. The reverse primer also had six His codons (shown in bold) followed by a stop codon. The PCR product was purified, digested and ligated to NcoI/BamHI double-digested pET-16b vector. The recombinant plasmid (pETKatB) was sequenced to confirm the nucleotide identity of the cloned alr3090 ORF. For overexpression, pETKatB was transformed into Escherichia coli BL21 (DE3) pLysS strain. E. coli pET-KatB cells were grown at 310 K and 180 rev min−1 in Luria–Bertani (LB) medium supplemented with 100 µg ml−1 carbenicillin and 34 µg ml−1 chloramphenicol. At an OD600 of ∼0.6, 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and 100 µM MnCl2 were added to the medium and the cells were incubated for a further 16 h at 293 K.
After 16 h, the cells were harvested by centrifugation and resuspended in cold lysis buffer (50 mM Tris pH 8.0, 200 mM NaCl, 5 mM imidazole). Cell lysis was performed on ice by sonication. The supernatant obtained by centrifuging the cell lysate at 13 000 rev min−1 for 30 min at 277 K was allowed to bind Ni2+–NTA (nitrilotriacetic acid) agarose with gentle shaking at 277 K for 2.5 h. The slurry was thoroughly washed with lysis buffer supplemented with increasing concentration of imidazole (10, 20 and 30 mM). The bound protein was eluted by increasing the imidazole concentration in the lysis buffer to 100 mM. The eluted protein was dialyzed against 20 mM Tris buffer pH 8.0 overnight. Subsequently, the dialyzed fraction containing the His-tagged KatB protein was resolved on SDS–PAGE and visualized by staining with Coomassie Brilliant Blue G-250 (Fig. 1 ▶).
Figure 1.

Coomassie Brilliant Blue-stained SDS–polyacrylamide gel showing the purified His-tagged KatB protein (indicated by an arrow) along with protein marker (lane M; labelled in kDa).
2.2. Crystallization
The KatB protein was concentrated to 4–5 mg ml−1 in 20 mM Tris buffer pH 8.0 using 10 kDa cutoff centrifugal filters. Initial crystallization screening was performed by the sitting-drop vapour-diffusion method in 96-well crystallization plates (three-well, Greiner) using a CyBio HTPC robot. Crystallization drops were prepared by mixing protein solution (4–5 mg ml−1) with a varying volume of reservoir solution and were equilibrated against 75 µl reservoir solution. Commercially available crystallization screens (Qiagen, Germany) were used for initial screening. Crystallization plates were incubated at 293 K in an incubator/imager (Formulatrix). Plates were analyzed immediately after setup, once every day for the first week and then once a week. Tetragonal crystals of the purified protein appeared in various conditions from the initial crystallization screens. Most of these conditions contained polyethylene glycol (PEG) as a precipitant. Optimization of the initial hits was performed manually by the hanging-drop vapour-diffusion method using 24-well plates. PEG 8000 was found to be the most effective precipitant for crystallization. Crystals grew within hours but showed cracks and a loss of morphology with time. Lower molecular weight PEGs were found to reduce the growth rate of crystals, thereby improving the quality of the crystals (Fig. 2 ▶). Crystals appeared in 1 d and continued to grow for 15 d. The final crystallization condition consisted of 18–25% PEG 400, 100 mM imidazole pH 8.0, 200 mM calcium acetate.
Figure 2.

A typical crystal of KatB.
2.3. Data collection and processing
For X-ray diffraction data collection, crystals were soaked in cryosolution for 30–60 s and flash-cooled in liquid nitrogen. The cryosolution was the same as the crystallization solution but with an increased concentration of PEG 400 (30–35%). X-ray diffraction data were collected in-house on an Agilent SuperNova system using a microfocus sealed-tube X-ray source with multilayer optics operated at 50 kV and 0.8 mA (Cu Kα; λ = 1.5406 Å). A total of 218 frames were collected at a crystal-to-detector distance of 61 mm with 0.5° oscillation per frame and an exposure time of 120 s (Table 1 ▶). The diffraction data were processed using the CrysAlisPro software suite (v.1.171.36.28c; Agilent Technologies UK Ltd, Oxford, England). Scaling and merging were performed using SCALA as implemented in the CCP4 suite (Winn et al., 2011 ▶).
Table 1. Data-collection statistics.
Values in parentheses are for the outer shell.
| Wavelength (Å) | 1.54056 |
| Temperature (K) | 100 |
| Detector | Titan CCD |
| Crystal-to-detector distance (mm) | 61 |
| Rotation range per image (°) | 0.5 |
| Total rotation range (°) | 109 |
| Exposure time per image (s) | 120 |
| Space group | P41212 or P43212 |
| Unit-cell parameters (Å, °) | a = b = 101.87, c = 138.86, α = β = γ = 90 |
| Mosaicity (°) | 0.9 |
| Resolution range (Å) | 15.64–2.2 (2.32–2.20) |
| Total No. of reflections | 313446 (45243) |
| No. of unique reflections | 37668 (5440) |
| Completeness (%) | 99.7 (100) |
| Mean I/σ(I) | 14.3 (4.6) |
| Multiplicity | 8.3 (8.3) |
| No. of molecules in the asymmetric unit | 3 |
| R merge (%) | 11.7 (42.6) |
| Overall B factor from Wilson plot (Å2) | 15.8 |
3. Results and discussion
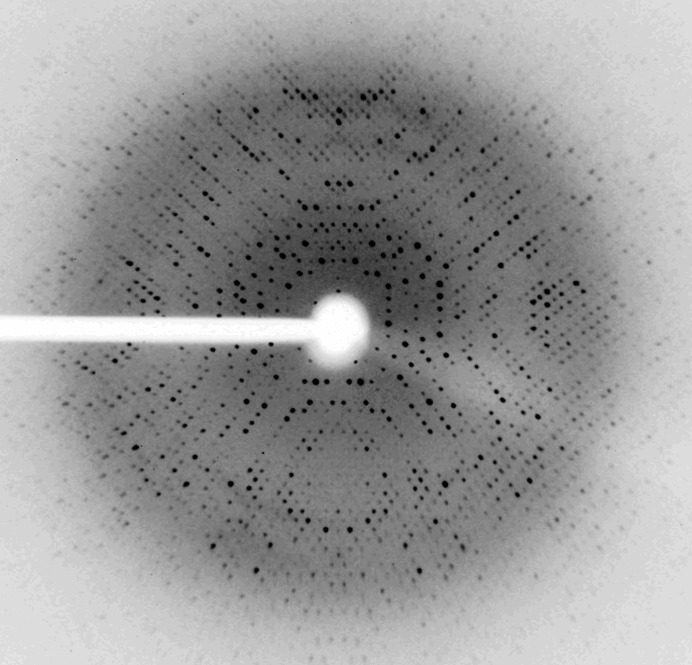
The KatB protein from Anabaena PCC 7120 was overexpressed in E. coli and purified using Ni–NTA affinity chromatography. The KatB was concentrated to 4–5 mg ml−1 in 20 mM Tris buffer pH 8.0 for crystallization. The crystal used for diffraction data collection was obtained after 15 d and was approximately 0.2 × 0.2 × 0.15 mm in size. The crystal diffracted to a resolution of 2.2 Å at 100 K (Fig. 3 ▶). The crystal belonged to space group P41212 (or enantiomer), with unit-cell parameters a = b = 101.87, c = 138.86 Å. Data statistics are shown in Table 1 ▶. Based on the molecular mass of the protein (26 488 Da) deduced from the nucleotide sequence and the volume of the unit cell (1 440 931 Å3), a Matthews parameter (Matthews, 1968 ▶) of 2.27 Å3 Da−1 and a solvent content of 45.8% suggested that the highest normalized probability (P tot of ∼0.86) was for the presence of three monomers in the asymmetric unit. To check for noncrystallographic symmetry (NCS) within the asymmetric unit, a fast self-rotation function was calculated using the program POLARRFN from the CCP4 suite (Winn et al., 2011 ▶). Self-rotation functions were computed for κ = 0–180° in the resolution range 8–2.5 Å with a radius of integration of 20 Å. Peaks related to crystallographic symmetry and threefold noncrystallographic symmetry were clearly observed. Plots of the rotation function calculated using κ = 30, 60, 120 and 180° are shown in Fig. 4 ▶. The presence of a strong peak at ω, ϕ = 0, 37° in the κ = 120° section suggests the presence of threefold noncrystallographic symmetry. The presence of threefold NCS is confirmed by the presence of symmetry-related peaks in other κ sections (Fig. 4 ▶). The presence of threefold NCS in the tetragonal asymmetric unit suggests that the oligomeric assembly of KatB is likely to be a homohexamer with 32 point symmetry, as similar biological assemblies are present in the other two Mn-catalases.
Figure 3.
Diffraction image of KatB crystal from data obtained using an in-house Agilent SuperNova X-ray diffractometer system with a Titan CCD detector.
Figure 4.
Plots of self-rotation functions of the KatB crystals calculated using data between 8.0 and 2.5 Å resolution and a radius of integration of 20 Å. Different κ sections (κ = 30, 60, 120 and 180°) are shown. A strong peak (90% of the height of origin peak) at ω, ϕ = 0, 37° in the κ = 120° section suggests the presence of a threefold noncrystallographic axis. The presence of symmetry-related NCS peaks in other κ sections confirms the presence of threefold NCS. Noncrystallographic symmetry peaks are marked with an asterisk in all of the sections.
Acknowledgments
We thank the National Facility for Macromolecular Crystallography, Solid State Physics Division, BARC for the X-ray diffractometer equipment. We also thank Dr Vinay Kumar, Dr Mukesh Kumar, Dr Vishal Prashar, Dr Gagandeep Gupta and Dr Manisha Banerjee for useful discussions and S. R. Jadhav for technical help.
References
- Amo, T., Atomi, H. & Imanaka, T. (2002). J. Bacteriol. 184, 3305–3312. [DOI] [PMC free article] [PubMed]
- Antonyuk, S. V., Melik-Adamyan, V. R., Popov, A. N., Lamzin, V. S., Hempstead, P. D., Harrison, P. M., Artymyuk, P. J. & Barynin, V. V. (2000). Crystallogr. Rep. 45, 105–116.
- Banerjee, M., Ballal, A. & Apte, S. K. (2012a). Biochem. J. 442, 671–680. [DOI] [PubMed]
- Banerjee, M., Ballal, A. & Apte, S. K. (2012b). Environ. Microbiol. 14, 2891–2900. [DOI] [PubMed]
- Barynin, V. V., Whittaker, M. M., Antonyuk, S. V., Lamzin, V. S., Harrison, P. M., Artymiuk, P. J. & Whittaker, J. W. (2001). Structure, 9, 725–738. [DOI] [PubMed]
- Bernroitner, M., Zamocky, M., Furtmüller, P. G., Peschek, G. A. & Obinger, C. (2009). J. Exp. Bot. 60, 423–440. [DOI] [PubMed]
- Halliwell, B. & Gutteridge, J. M. C. (1986). Arch. Biochem. Biophys. 246, 501–514. [DOI] [PubMed]
- Kaneko, T. et al. (2001). DNA Res. 8, 205–213. [DOI] [PubMed]
- Katoh, H. (2012). Biochim. Biophys. Acta, 1817, 1263–1269. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Narayan, O. P., Kumari, N. & Rai, L. C. (2011). J. Microbiol. Biotechnol. 21, 136–146. [DOI] [PubMed]
- Pandey, S., Rai, R. & Rai, L. C. (2012). J. Proteomics, 75, 921–937. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Zamocky, M., Furtmüller, P. G. & Obinger, C. (2008). Antioxid. Redox Signal. 10, 1527–1548. [DOI] [PMC free article] [PubMed]