Abstract
The regulation of chromatin mobility in response to DNA damage is important for homologous recombination in yeast. Anchorage reduces rates of recombination, whereas increased chromatin mobility correlates with more efficient homology search. Here we tracked the mobility and localization of spontaneous S-phase lesions bound by Rad52, and find that these foci have reduced movement, unlike enzymatically induced double-strand breaks. Moreover, spontaneous repair foci are positioned in the nuclear core, abutting the nucleolus. We show that cohesin and nucleolar integrity constrain the mobility of these foci, consistent with the notion that spontaneous, S-phase damage is preferentially repaired from the sister chromatid.
Keywords: DNA damage, Rad52 foci, chromatin dynamics, cohesin, nucleolus
Introduction
Double-strand breaks (DSB) are particularly deleterious DNA lesions, as inappropriate repair can lead to translocations and genome instability [1]. Budding yeast efficiently repairs DSBs by homologous recombination (HR), which requires physical contact with a homologous template. Sister chromatids are the favoured template in S- and G2-phase cells, as they are exact copies of the damaged site, and are held together by cohesin. If a sister chromatid is unavailable or is similarly damaged, then a genome-wide search for a homologous template might take place [2].
In budding yeast, the Rad52 epistasis group composed of Mre11, Rad50, Xrs2, Rad52, Rad51 and Rad54, is responsible for catalyzing HR [3]. Each member is sequentially recruited to the DSB along with mediators of the DNA damage response, including the checkpoint kinase adaptor Rad9 (53BP1/BRCA1 in humans) [4]. These factors, when fused to a fluorescent protein, form microscopically discernible repair centres at the sites of damage [5], and are therefore convenient tools to study repair dynamics in living cells.
Most studies on mobility of repair foci have focused on Rad52, which accumulates at DSBs after resection [5] and disappears once pairing with the homologous template has occurred [6]. Rad52 foci form only in S-phase cells [7]. Quantitation of Rad52 focus mobility showed that enzymatically induced site-specific DSBs move within a larger volume than the same locus undamaged [6, 8]. The enhanced mobility of Rad52 foci requires Rad51 and Rad54, as well as the checkpoint kinase, Mec1 and the checkpoint adaptor protein Rad9 [6, 8]. Consistent with the notion that movement facilitates the homology search, increased mobility at DSBs correlates with faster production of repair intermediates and higher rates of recombination [8, 9]. Moreover, artificially tethered domains recombine less frequently [10].
Locus mobility might also be harnessed to relocate a DSB away from domains that are repressive for HR, such as heterochromatin or the nucleolus [11–13], and can shift DSBs that lack a homologous donor for HR towards nuclear pores [14]. The proposal that chromatin mobility promotes the efficiency of DSB repair, particularly when a homology search beyond the sister chromatid is required [15], is consistent with a recent study that examined the effect of nuclear architecture on ectopic recombination in yeast [10].
Importantly, DSBs are not the only lesions that generate Rad52 foci. For example, the repair of replication fork collision with a covalent DNA-protein adduct similarly recruits Rad52 [16], as do other spontaneous lesions in S-phase cells [7]. These spontaneous lesions generally do not activate the DNA damage checkpoint or arrest the cell cycle. Unlike enzymatically induced DSBs, they appear to reflect damage on one of the two sister chromatids, which are held together by cohesin following replication. Indeed, spontaneous Rad52 foci appear to stem largely from gaps behind the replication fork [17], making them structurally distinct from the breaks induced by enzymes that repeatedly cleave both sister chromatids. Intriguingly, these two types of lesions segregate spatially within the yeast nucleus: irreparable DSBs were shown to shift to the nuclear envelope [14, 18], whereas spontaneous Rad52 foci are predominantly found in the nuclear core [19].
Here we have explored the constraints that influence repair centre movement and position. Our results implicate cohesin and the nucleolus in constraining damage mobility, and support the hypothesis that reduced movement reflects the repair of spontaneous lesions by exchange with the sister chromatid.
Results and discussion
Mean square displacement (MSD) analysis robustly quantifies the mobility of diffusing, fluorescently tagged chromosomal loci and repair foci [6, 8, 9, 15]. In such analyses, the position of a moving spot is monitored from frame to frame, and the square of the distance travelled by the locus is plotted against increasing time intervals (Fig 1A). The resulting MSD curve, averaged over several independent time-lapse series, generally reaches a plateau proportional to the square of the radius of constraint (Rc). This is a useful measure that indicates the nuclear volume (nvol) within which a fluorescent spot can move. All movement parameters relevant for this study are compiled in supplementary Table S1 online.
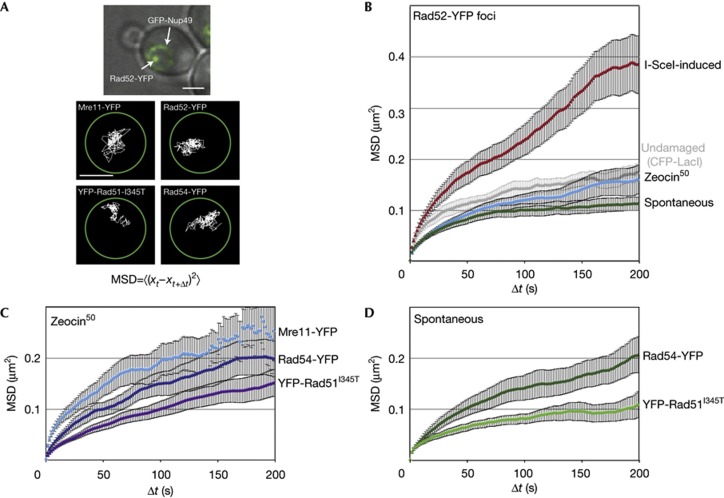
Figure 1.
The mobility of damaged DNA at various stages of repair. (A) Example of a Rad52-YFP focus in an S-phase cell (contrast-adjusted) in GA7997 (top). Scale bar, 2 μm. Bottom: Traces of Mre11-YFP (GA5832), Rad52-YFP (GA5820), YFP-Rad51I345T (GA6057) and Rad54-YFP (GA5833) on Zeocin addition. Scale bar, 1 μm. (B) MSD analysis of I-SceI-induced Rad52-YFP foci and the same undamaged locus (ZWF1; in grey; 17 movies) in GA6208, Zeocin-induced (50 μg/ml for 1 h; 28 movies) and spontaneous (21 movies) Rad52-YFP foci in GA5820. The data in this panel are from [8]. (C,D) MSD analysis of Mre11-YFP (12 movies), YFP-Rad51I345T (21 movies) and Rad54-YFP (10 movies) foci induced with 50 μg/ml Zeocin for 1 h (C) and for those that arose spontaneously (D; YFP-Rad51I345T: 16 movies; YFP-Rad54: 13 movies). All movies were taken in S-phase, except for Mre11-YFP, which is six from each S and G1 phases. Error bars represent the standard error. GFP, green fluorescent protein; MSD, mean square displacement; YFP, yellow fluorescent protein.
We have previously shown that the undamaged ZWF1 locus on chromosome 14, has an Rc of about 0.51 μm in S-phase cells, which is equivalent to ∼18% of the nvol (Fig 1B) [8]. Once an I-SceI-induced DSB is generated at this locus, the Rc value of the resulting Rad52-YFP focus increases to 0.7 μm (47% of nvol; Fig 1B) [8]. Not every Rad52 focus showed a similar mobility, however. For example, Rad52 foci induced by a limited dose of Zeocin (50 μg/ml for 1 h) have a mobility identical to that of an undamaged locus (Fig 1B). Similarly slow movement was detected for damage induced by the covalent binding of a mutated Flpase to DNA [8]. Surprisingly, spontaneous S-phase Rad52-YFP foci had an even lower Rc value (0.37 μm or 7% nvol; Fig 1B). We asked if spontaneous lesions were enriched in the ribosomal DNA (rDNA), but lesion localization by anti-Rad52 chromatin immunoprecipitation and deep sequencing failed to show significant enrichment for rDNA sequences (data not shown). Thus, we conclude that the type of DNA lesion determines the mobility of Rad52 foci.
In order to characterize the molecular constraints on spontaneously occurring S-phase damage, we asked whether the mobility of damaged DNA varies with different steps of the repair process. To this end, we fluorescently tagged several repair proteins besides Rad52, namely Mre11 (which binds transiently to the initial lesion in both S and G1 phases) and Rad51 and Rad54 (which both bind after 5′ end-resection). We determined the mobility of repair foci arising either spontaneously in S phase, or as a result of exposure to low-dose treatment with Zeocin, which induces single-strand nicks nine times more frequently than DSBs [20] (Fig 1C,D). As Rad51 is not functional when fused to YFP [5], we tracked both YFP-rad51 and a mutant of Rad51 that restores its activity in context of the YFP fusion, YFP-Rad51I345T [21].
Mre11 is part of the early-binding MRN complex that holds the two free ends of a DSB together, and has a role in end-resection [3, 22, 23]. Whereas spontaneous Mre11 foci were too short-lived to be imaged reliably, both Zeocin-induced and I-SceI-induced foci of Mre11 could be tracked in both G1- and S-phase cells. I-SceI-induced Mre11 foci showed a high degree of mobility in both stages of the cell cycle (Rc=0.81–0.82 μm; supplementary Fig S1A online), even though resection occurs only in S phase. Zeocin-induced Mre11-YFP foci had a lower Rc than I-SceI-induced foci (0.59 μm compared with 0.84 μm), yet was also unchanged between G1- and S-phase cells (supplementary Fig S1B online). Intriguingly, Zeocin-induced foci had a slightly higher mobility than undamaged loci in S phase, suggesting that the first response to damage is enhanced movement, which, for a short time, overrides the differences in constraint on chromatin mobility that correlate with stages of the cell cycle [24].
The tracking of spontaneous foci formed by a functional Rad51 fusion (YFP-Rad51I345T) showed behaviour similar to Rad52-YFP foci: spontaneous damage was more constrained than Zeocin-induced foci (7% versus ∼18% of nvol, respectively, Fig 1C,D). This was not the case for Rad54-YFP foci, which in both cases showed movement similar to that of an undamaged locus (∼18% of nvol). Given that Rad54 acts after Rad51 and Rad52, it might be that at late steps in the repair process, damage-induced changes in mobility are exhausted. Importantly, there was no instance in which either spontaneous or low-level Zeocin-induced foci showed the high level of mobility that is typical of an I-SceI-induced, irreparable DSB. Indeed, one major difference between I-SceI-induced damage and spontaneous Rad52 foci is that the latter do not activate the checkpoint kinase Mec1, which is required for enhanced DSB movement [8].
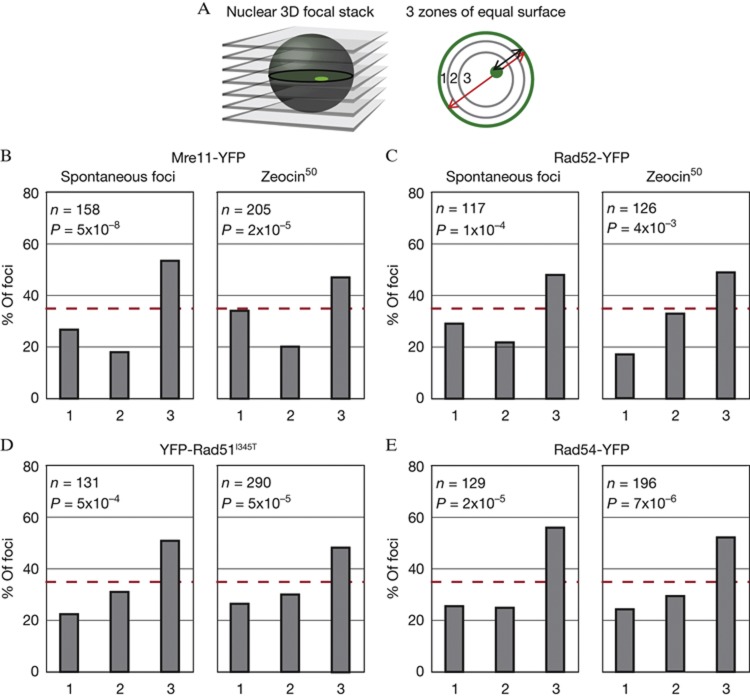
To test whether perinuclear anchoring accounts for the reduced mobility of spontaneous or low-level Zeocin-induced damage, we determined the radial position of the resulting repair foci using a well-characterized 3-zone assay [25, 26] (Fig 2A). We find that all types of spontaneous foci (that is, those scored in the mobility assays above) are enriched in the nuclear interior (Fig 2B–E). This resembles the distribution of Rad52 foci that arise from cleavage at the MAT locus, which is rapidly repaired by recombination with sequences at HMR or HML [19]. This result argues that the low mobility of spontaneous foci does not reflect their association with nuclear pores or other peripheral anchorage sites.
Figure 2.
Repair foci of tagged HR repair factors are enriched in the nuclear core. (A) Radial positions of repair foci were obtained by taking three-dimensional focal stacks of yeast nuclei and dividing the slice in which the spot is in focus into three zones of equal surface, with zone 1 being peripheral. (B–E) Zoning analysis of spontaneous and Zeocin-induced (50 μg/ml Zeocin for 1 h) Mre11-YFP foci (B; GA5832), Rad52-YFP (C; GA5820), YFP-Rad51I345T (D; GA6057), and Rad54-YFP (E; GA5833). Number of nuclei analysed (n) and the P-value for a χ2 test versus an expected random distribution are given. HR, homologous recombination; YFP, yellow fluorescent protein.
We next examined whether the increased mobility of Zeocin-induced damage over spontaneous foci reflects differential activation of the DNA damage checkpoint response, as appears to be the case for I-SceI-induced DSBs [8]. To this end, we monitored the mobility of spontaneous and Zeocin-induced Rad52-YFP foci, as well as that of an undamaged locus, in a rad9 mutant, which impairs checkpoint activation. While we found a slight increase in the mobility of undamaged DNA in rad9Δ cells, there was no longer a difference in the mobility of spontaneous and Zeocin-induced Rad52 foci (supplementary Fig S2 online). Thus, the slight increase in mobility of a Zeocin-induced focus over a spontaneous focus might indeed reflect activation of the DNA damage response.
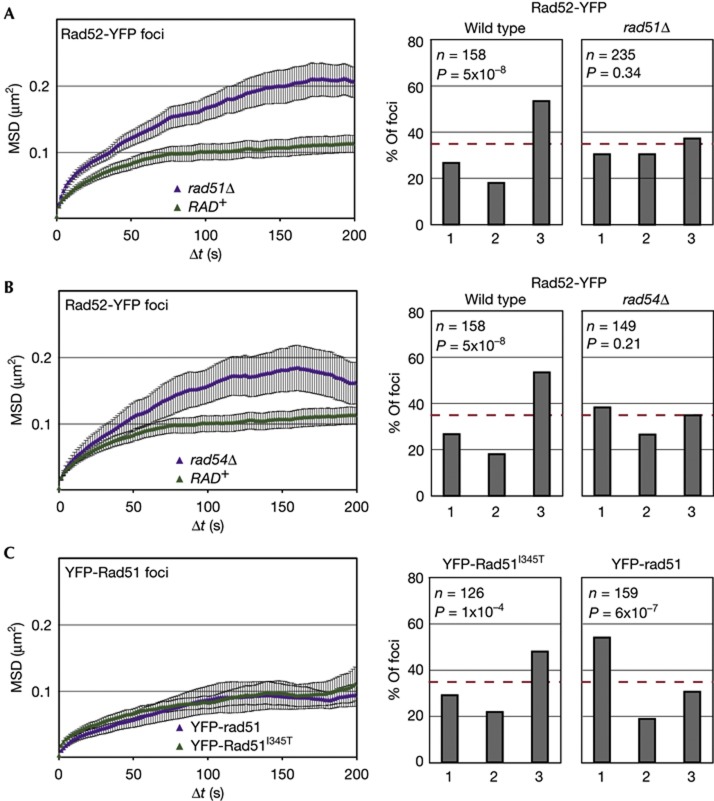
It remained to determine what elements restrict the mobility of spontaneous Rad52 foci. Given that Rad51 and Rad54 actively contribute to the enhanced movement of an I-SceI-induced DSB [8], we targeted these ATPases for deletion. Intriguingly, we found that loss of Rad51 or Rad54 partially relaxed the constraint on spontaneous Rad52-YFP foci (Fig 3A,B), which is the opposite of their effect on an induced DSB [8]. Nonetheless, in neither mutant did the Rad52 focus reach the mobility observed for an induced DSB.
Figure 3.
HR factors help constrain the mobility and non-peripheral enrichment of repair foci. (A–C) MSD analyses of spontaneous Rad52-YFP foci in rad51Δ (GA5836; 28 movies) and rad54Δ (GA6110; 13 movies) strains. MSD was also performed on foci of YFP-rad51 (GA6106; 9 movies) and YFP-Rad51I345T (GA6057; 16 movies). Spontaneous Rad52-YFP foci in wild type (GA5820) and YFP-Rad51I345T (GA6057) cells as in Fig 1. To the right are zoning assays of spontaneous Rad52 foci in wild type, rad51Δ or rad54Δ strains, or for YFP-rad51 (GA6106) and YFP-Rad51I345T (GA6057) in a wild-type background Number of nuclei analysed (n) and the P-value for a χ2 test versus an expected random distribution are given. The error bars on the MSD curves represent the s.e. HR, homologous recombination; MSD, mean square displacement; YFP, yellow fluorescent protein.
The subnuclear distribution of spontaneous Rad52-YFP foci was also altered in cells lacking Rad51 or Rad54, as spontaneous foci were now randomly distributed, rather than being internally enriched (Fig. 3A,B). This suggests that spontaneous damage becomes irreparable in these mutants, and moves, like an irreparable DSB, to the nuclear periphery. Consistent with this interpretation, we found that foci containing the non-functional YFP-rad51 fusion were enriched at the nuclear periphery, whereas functional YFP-Rad51I345T foci stayed internal (Fig 3C). In both strains, however, the foci showed constrained mobility (Fig 3C). Thus, the presence of Rad51 at spontaneous lesions seems to constrain mobility, and its ability to support HR determines whether the resulting repair focus will be internal (that is, HR positive) or peripheral (HR negative). In conclusion, both the mobility and localization of repair foci are influenced by HR proteins. In their absence, a default state might prevail, which confers mobility equivalent to the undamaged site with no preferential position within the nucleus. Alternatively, the nature of the underlying lesion might change fundamentally in rad51Δ or rad54Δ cells, which might also alter mobility or position of the repair foci.
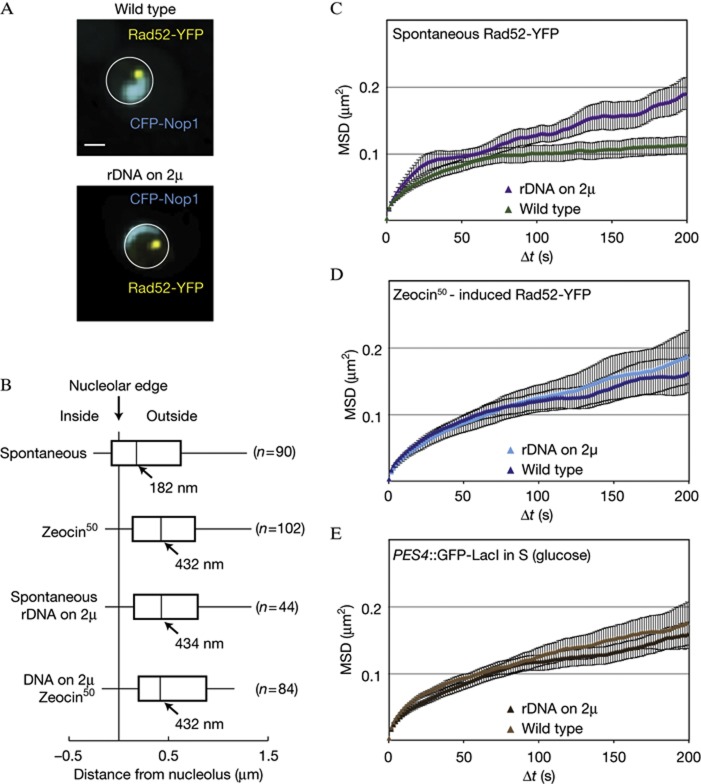
We next looked for structures that might constrain repair foci in the nuclear interior. One of the few identified substructures in the yeast nuclear core that could anchor foci is the nucleolus, which occupies about a third of the nvol. Indeed, mammalian chromosomal loci associated with the nucleolus are slow moving [27]. We scored the position of spontaneous and Zeocin-induced Rad52-YFP foci relative to the periphery of the nucleolus, which we tagged with a fluorescent marker (Fig 4A). The distances measured between the centre of each spontaneous Rad52-YFP focus and the nucleolar periphery yielded a median of 182 nm, whereas the more mobile Zeocin-induced foci were further away (median=432 nm; P=0.003; Fig 4B). These results indicate that the spontaneous repair foci are adjacent to the nucleolar surface, even though they are not enriched for rDNA lesions. Consistently, immunostaining of spontaneously occurring γH2A foci in S-phase cells also revealed strong enrichment at the nucleolar periphery (supplementary Fig S3 online).
Figure 4.
Spontaneous Rad52-YFP foci are constrained by the nucleolus. (A) Representative images of wild-type cells (GA5820) and those harbouring the rDNA on a multicopy plasmid (GA7997), both expressing CFP-Nop1. The CFP-Nop1 signal in the strain carrying the rDNA on a plasmid was ∼10-fold lower than in wild type, and thus brightness is adjusted. Scale bar, 1 μm. (B) Distances between Rad52-YFP foci and the nucleolus marked by CFP-Nop1 in GA5820 and GA7997. The number of nuclei analysed (n) is given for each condition. (C,D) MSD analyses of spontaneous (C) and Zeocin-induced (D) Rad52-YFP foci in both wild-type cells with (GA5820; 21 and 28 movies, respectively) or without the rDNA on a 2 μ plasmid (GA7997; 16 and 26 movies, respectively). 50 μg/ml Zeocin treatment was for 1 h. (E) MSD analysis of the LacO-tagged PES4 locus in wild type (GA1461; 20 movies) and in cells with the 5S and 35S on a 2 μ plasmid (GA8147; 22 movies). Analyses were done on S-phase cells. The error bars on the MSD curves represent the s.e. CFP, cyan fluorescent protein; MSD, mean square displacement; rDNA, ribosomal DNA; YFP, yellow fluorescent protein.
If spontaneous, slow-moving Rad52-YFP foci are juxtaposed to the nucleolus, then, we reasoned that changing the shape and size of the nucleolus should alter the organization and mobility of these repair foci. To test this, we obtained a strain that carries its only copies of the 5S and 35S rDNA genes on a multicopy plasmid [28]. In this strain, the nucleolus, detected by CFP-Nop1 fluorescence, is reduced by ∼10-fold (Fig 4A). In this strain, spontaneous foci no longer associate with the nucleolar periphery (Fig 4B) and their mobility increases slightly, reaching the mobility of Zeocin-induced foci (Fig 4C). In contrast, the mobility of Zeocin-induced Rad52-YFP foci is not different between the wild-type cells and the rDNA-plasmid strain, consistent with the fact that they are not associated with the nucleolar surface (Fig 4D). Similarly, there is no general increase in chromatin mobility: the MSD curve for the undamaged PES4 locus was unaltered (Fig 4E). We conclude that an intact nucleolar structure helps restrain the mobility of a spontaneous Rad52 focus.
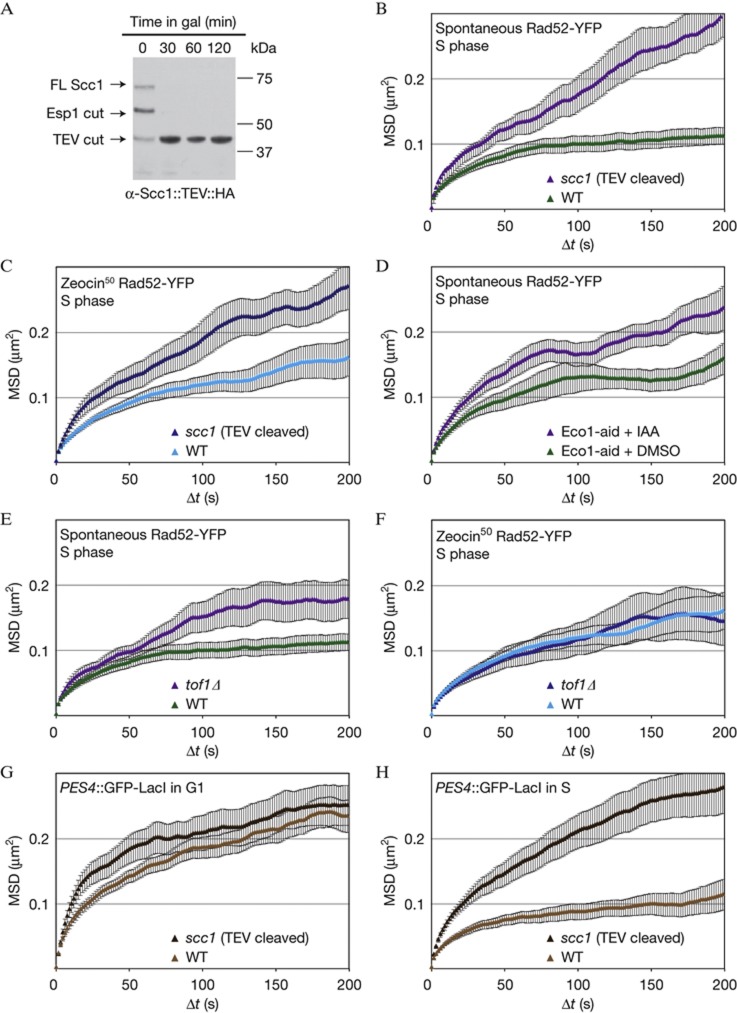
While constraint is reduced in the strain with defective nucleolar structure, the Rad52 foci in these cells do not become as mobile as an I-SceI-induced DSB. This argues that additional forces restrict the movement of spontaneous and Zeocin-induced foci. As these foci arise almost exclusively in S-phase cells [7], when the sister chromatid is synthesized, we surmised that the damaged site and its sister might be linked to each other by a ring-like complex called cohesin [29]. The cohesin ring is closed at one end by a protein called Scc1, whose cleavage releases the sister chromatids from each other in mitosis [30].
To see if the tethering of sister chromatids by cohesion confers constraint on spontaneous sites of damage, we took advantage of a Scc1 construct (Scc1::TEV) that contains a TEV protease cleavage site [30]. In a strain carrying a galactose-inducible TEV construct, the addition of galactose provokes complete cleavage of Scc1 within 30 min (Fig 5A). We found that Scc1::TEV cleavage led to increased Rc values for both spontaneous and Zeocin-induced Rad52-YFP foci (0.59 and 0.56 μm, respectively; Fig 5B,C).
Figure 5.
Sister chromatid cohesion constrains Rad52-YFP foci and undamaged chromatin. (A) Western blot showing efficient cleavage of Scc1::TEV on TEV induction in GA7757. (B,C) MSD analyses of spontaneous (B) and Zeocin-induced (C; 50 μg/ml for 1 h) Rad52-YFP foci after cleavage of cohesin (GA7757; 11 and 12 movies, respectively) compared with wild type (GA5820; 21 and 28 movies as in Fig 1). (D) MSD analysis of spontaneous Rad52-YFP foci in cells containing the Eco1-aid allele (GA8193) treated with DMSO (12 movies) or auxin (IAA; 14 movies). Only S-phase cells were analyzed. (E,F) MSD analysis of spontaneous and Zeocin-induced Rad52-YFP foci in tof1Δ cells (GA8160; 11 and 12 movies, respectively) compared with wild-type cells (GA5820; 21 and 28 movies, respectively). (G,H) MSD curves of LacO-tagged PES4 in GA1461 (wild type) and after cleavage of Scc1 (GA8112) in G1 (D; 26 and 22 movies, respectively) and S phase (E; 14 and 19 movies, respectively). The error bars on the MSD curves represent the s.e. MSD, mean square displacement; YFP, yellow fluorescent protein.
To confirm a role for sister chromatid cohesion (SCC) in constraining locus mobility, we exploited an allele of ECO1, which is fused to an auxin-inducible degron (Eco1-aid). Upon addition of auxin, Eco1 is rapidly degraded and sister chromatids separate [31]. Adding DMSO alone increased the mobility of both spontaneous Rad52 foci and undamaged loci slightly (Fig 5D, supplementary Fig S4A online), yet the addition of auxin led to an additional increase in mobility, exclusively in the Eco1-aid containing strain (Fig 5D, supplementary Fig S4A online). We therefore conclude that cohesin-dependent SCC restrains spontaneous Rad52-YFP foci in S-phase cells.
We further confirmed the role of SCC in constraining Rad52-YFP focus mobility by testing a tof1 mutant, which also compromises the establishment of cohesion at forks and damage [32]. We find that chromatin mobility at an undamaged locus is only slightly affected in this background (supplementary Fig S4B online). On the other hand, tof1Δ increased the mobility of spontaneous Rad52-YFP foci (Fig 5E), but not that of Zeocin-induced foci (Fig 5F). We propose that Tof1-established SCC specifically confers constraint at spontaneous Rad52-YFP foci.
Next, we asked whether the increase in mobility due to loss of cohesin was specific to repair foci. We tracked the undamaged LacO-tagged PES4 locus in G1- and S-phase cells, with and without Scc1 cleavage by TEV, because PES4, like most genomic loci, moves less in S-phase than in G1-phase cells [24]. Cleaving Scc1 did not increase chromatin mobility in G1, but led to a substantial increase in Rc in S-phase cells, suggesting that cohesin-mediated SCC is also responsible for cell cycle differences in mobility and not only for damage-specific constraint (Fig 5G,H). This is actually expected, as cohesin is not only loaded at sites of damage.
Finally, we tested whether contact with the nucleolar surface and SCC work together to constrain Rad52-YFP foci. Although Scc1 cleavage led to a dissolution of nucleolar structure (supplementary Fig S4C online), tof1Δ cells retain normal nucleolar shape, which we monitor by CFP-Nop1 (supplementary Fig S4C online). In the strain with plasmid-borne rDNA loci and reduced nucleolar structure (Fig 4A), we additionally deleted TOF1, yet this did not further increase in the mobility of either spontaneous or Zeocin-induced Rad52-YFP foci (supplementary Fig S4DE online). This argues that the effects of SCC and of nucleolar structure are epistatic with respect to their impact on the mobility of Rad52 foci.
Our work suggests that both cohesin-mediated SCC and some aspects of nucleolar structure constrain the movement of spontaneous Rad52-YFP foci. What could the biological implications of confining spontaneous repair centres be? We propose a model whereby the slow mobility of repair centres favours repair with the tethered sister chromatid, while reducing the likelihood of deleterious ectopic recombination. In support of this hypothesis, the scc1-73 mutant is defective in sister chromatid exchange, but has increased rates of ectopic recombination [33]. Moreover, several mutants defective in sister chromatid recombination, including those of H3K56 acetyltransferase Rtt109 and the repair factor Smc5/6, are defective in maintaining Rad52 foci outside of the nucleolus and have been associated with specific defects in sister chromatid recombination, while remaining competent for HR with ectopic donors [10, 11, 34, 35, 36, 37]. Together with these results, our study supports a model whereby both chromatin mobility and the molecules that constrain it contribute to genome stability in face of a diverse range of genetic insults.
Methods
Yeast strains and plasmids. The supplementary material online contains details about the strains and plasmids (supplementary Tables S2 and S3).
Microscopy. MSD and zoning analyses were done as described [8, 9, 19]. Time-lapse data for a given strain are based on several independent cultures or colonies of a given strain, filmed at different times. Details of the Zeocin treatment as well as the degradation of Scc1::TEV and Eco1-aid can be found in the supplementary information online.
Statistics. The P-values reported for the zoning assays represent a χ2 test with d.f.=2 comparing the experimental results to an expected random distribution. For the difference in the distance of the Rad52-YFP focus to the nucleolar periphery, we used a two-tailed Student’s t-test. The error on the Rc was determined as described [8, 9].
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank A. Ponti and R. Thierry for the Imaris add-on, R. Rothstein, O. Fritsch, F. Uhlmann, and L. Symington for strains and reagents. We are grateful to the FMI microscopy core for technical help, and F. Hamaratoglu and N. Hustedt for critical reading of the manuscript. This work has been supported by the Friedrich Miescher Institute for Biomedical Research, by a project grant from the Swiss National Foundation to S.M.G., the Swiss National Science Foundation "Frontiers in Genetics" programme, and a Terri Fox Foundation postdoctoral fellowship to V.D. (#19759).
Author contributions: V.D. and A.S. designed the experiments, carried them out, interpreted results and contributed to the writing; V.K. and T.S. carried out the experiments and interpreted the results, and S.M.G., helped to design the experiments, interpreted the results and contributed to writing.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alt FW, Zhang Y, Meng FL, Guo C, Schwer B (2013) Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell 152: 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehlen LR, Gasser SM, Dion V (2011) How broken DNA finds its template for repair: a computational approach. Prog Theoret Physics Suppl 191: 20–29 [Google Scholar]
- Heyer WD, Ehmsen KT, Liu J (2010) Regulation of homologous recombination in eukaryotes. Annu Rev Genet 44: 113–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn K, Lowndes NF, Grenon M (2012) Eukaryotic DNA damage checkpoint activation in response to double-strand breaks. Cell Mol Life Sci 69: 1447–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R (2004) Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713 [DOI] [PubMed] [Google Scholar]
- Mine-Hattab J, Rothstein R (2012) Increased chromosome mobility facilitates homology search during recombination. Nat Cell Biol 14: 510–517 [DOI] [PubMed] [Google Scholar]
- Lisby M, Rothstein R, Mortensen UH (2001) Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci USA 98: 8276–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion V, Kalck V, Horigome C, Towbin BD, Gasser SM (2012) Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat Cell Biol 14: 502–509 [DOI] [PubMed] [Google Scholar]
- Neumann FR, Dion V, Gehlen LR, Tsai-Pflugfelder M, Schmid R, Taddei A, Gasser SM (2012) Targeted INO80 enhances subnuclear chromatin movement and ectopic homologous recombination. Genes Dev 26: 369–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agmon N, Liefshitz B, Zimmer C, Fabre E, Kupiec M (2013) Effect of nuclear architecture on the efficiency of double-strand break repair. Nat Cell Biol 15: 694–699 [DOI] [PubMed] [Google Scholar]
- Torres-Rosell J et al. (2007) The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol 9: 923–931 [DOI] [PubMed] [Google Scholar]
- Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH (2011) Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell 144: 732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob B, Splinter J, Conrad S, Voss KO, Zink D, Durante M, Lobrich M, Taucher-Scholz G (2011) DNA double-strand breaks in heterochromatin elicit fast repair protein recruitment, histone H2AX phosphorylation and relocation to euchromatin. Nucleic Acids Res 39: 6489–6499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai S et al. (2008) Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 322: 597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion V, Gasser SM (2013) Chromatin movement in the maintenance of genome stability. Cell 152: 1355–1364 [DOI] [PubMed] [Google Scholar]
- Nielsen I, Bentsen IB, Lisby M, Hansen S, Mundbjerg K, Andersen AH, Bjergbaek L (2009) A Flp-nick system to study repair of a single protein-bound nick in vivo. Nat Methods 6: 753–757 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Prieto R, Munoz-Cabello AM, Cabello-Lobato MJ, Prado F (2013) Rad51 replication fork recruitment is required for DNA damage tolerance. EMBO J 32: 1307–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL (2009) Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev 23: 912–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystricky K, Van Attikum H, Montiel MD, Dion V, Gehlen L, Gasser SM (2009) Regulation of nuclear positioning and dynamics of the silent mating type loci by the yeast Ku70/Ku80 complex. Mol Cell Biol 29: 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povirk LF (1996) DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: bleomycin, neocarzinostatin and other enediynes. Mutat Res 355: 71–89 [DOI] [PubMed] [Google Scholar]
- Fung CW, Mozlin AM, Symington LS (2009) Suppression of the double-strand-break-repair defect of the Saccharomyces cerevisiae rad57 mutant. Genetics 181: 1195–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev K, Vitriol E, Stemple J, Resnick MA, Bloom K (2004) Chromosome fragmentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr Biol 14: 2107–2112 [DOI] [PubMed] [Google Scholar]
- Kaye JA, Melo JA, Cheung SK, Vaze MB, Haber JE, Toczyski DP (2004) DNA breaks promote genomic instability by impeding proper chromosome segregation. Curr Biol 14: 2096–2106 [DOI] [PubMed] [Google Scholar]
- Heun P, Laroche T, Shimada K, Furrer P, Gasser SM (2001) Chromosome dynamics in the yeast interphase nucleus. Science 294: 2181–2186 [DOI] [PubMed] [Google Scholar]
- Heun P, Laroche T, Raghuraman MK, Gasser SM (2001) The positioning and dynamics of origins of replication in the budding yeast nucleus. J Cell Biol 152: 385–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister P, Gehlen LR, Varela E, Kalck V, Gasser SM (2010) Visualizing yeast chromosomes and nuclear architecture. Methods Enzymol 470: 535–567 [DOI] [PubMed] [Google Scholar]
- Chubb JR, Boyle S, Perry P, Bickmore WA (2002) Chromatin motion is constrained by association with nuclear compartments in human cells. Curr Biol 12: 439–445 [DOI] [PubMed] [Google Scholar]
- Johzuka K, Horiuchi T (2009) The cis element and factors required for condensin recruitment to chromosomes. Mol Cell 34: 26–35 [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH (2009) Cohesin: its roles and mechanisms. Annu Rev Genet 43: 525–558 [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K (2000) Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103: 375–386 [DOI] [PubMed] [Google Scholar]
- Lopez-Serra L, Lengronne A, Borges V, Kelly G, Uhlmann F (2013) Budding yeast Wapl controls sister chromatid cohesion maintenance and chromosome condensation. Curr Biol 23: 64–69 [DOI] [PubMed] [Google Scholar]
- Mayer ML et al. (2004) Identification of protein complexes required for efficient sister chromatid cohesion. Mol Biol Cell 15: 1736–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tittel-Elmer M, Lengronne A, Davidson MB, Bacal J, Francois P, Hohl M, Petrini JH, Pasero P, Cobb JA (2012) Cohesin association to replication sites depends on rad50 and promotes fork restart. Mol Cell 48: 98–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Piccoli G et al. (2006) Smc5-Smc6 mediate DNA double-strand-break repair by promoting sister-chromatid recombination. Nat Cell Biol 8: 1032–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes-Ledesma F, Aguilera A (2006) Double-strand breaks arising by replication through a nick are repaired by cohesin-dependent sister-chromatid exchange. EMBO Rep 7: 919–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtele H et al. (2012) Histone H3 lysine 56 acetylation and the response to DNA replication fork damage. Mol Cell Biol 32: 154–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Galvan S, Jimeno S, Rothstein R, Aguilera A (2013) Histone H3K56 acetylation, Rad52, and non-DNA repair factors control double-strand break repair choice with the sister chromatid. PLoS Genet 9: e1003237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.