Abstract
Miro is a highly conserved calcium-binding GTPase at the regulatory nexus of mitochondrial transport and autophagy. Here we present crystal structures comprising the tandem EF hand and carboxy terminal GTPase (cGTPase) domains of Drosophila Miro. The structures reveal two previously unidentified ‘hidden’ EF hands, each paired with a canonical EF hand. Each EF hand pair is bound to a helix that structurally mimics an EF hand ligand. A key nucleotide-sensing element and a Pink1 phosphorylation site both lie within an extensive EF hand–cGTPase interface. Our results indicate structural mechanisms for calcium, nucleotide and phosphorylation-dependent regulation of mitochondrial function by Miro.
Keywords: EF hand, ELM domain, GTPase, Miro, mitochondria
Introduction
The outer mitochondrial membrane protein Miro (also called RhoT) is a highly conserved calcium (Ca2+)-binding GTPase that is a key player in a diverse set of mitochondrial processes, regulating mitochondrial shape, movement, organelle interactions and degradation. Relatively little is known about the molecular underpinnings of these processes and a structural understanding of the relevant protein machinery is lacking. Miro is notable for its domain architecture, as it is one of very few proteins to contain both EF hand and GTPase domains. It consists of two GTPase domains that flank a central, Ca2+-binding region containing two canonical EF hands [1] (cEF hands); a C-terminal transmembrane domain anchors Miro in the outer mitochondrial membrane [2].
Miro is essential for the axonal transport of mitochondria, as it attaches mitochondria to the microtubule-based motor protein kinesin-1 through the cargo-adaptor protein Milton [3–5]. Its cEF hands permit Miro to act as a Ca2+-dependent switch for mitochondrial movement, allowing transport at low, basal cytosolic Ca2+ concentrations but stopping mitochondria at higher levels of Ca2+ [6–8]. The mechanism underlying this Ca2+-dependent regulation is unclear [7, 8]. The GTPase domains of Miro influence mitochondrial morphology [1, 4], and both EF hand and GTPase domains regulate endoplasmic reticulum—mitochondrial connections [9, 10]. Miro is also a common substrate of the two Parkinson’s disease-related proteins Pink1 kinase and Parkin E3 ubiquitin ligase in a signalling cascade that facilitates mitochondrial degradation [11–13]. Phosphorylation of Miro by Pink1 kinase and its subsequent Parkin-mediated degradation leads to mitophagy of damaged mitochondria [12]. While it is clear that Pink1 is a necessary component in this Miro–Pink1–Parkin pathway, whether direct phosphorylation of Miro is required to trigger Parkin-mediated degradation is contested [12, 13]. Here, we present the first crystal structures of Miro and offer insights into the molecular mechanisms by which Miro might regulate such a wide variety of mitochondrial functions.
Results and Discussion
Overview of Miro structures
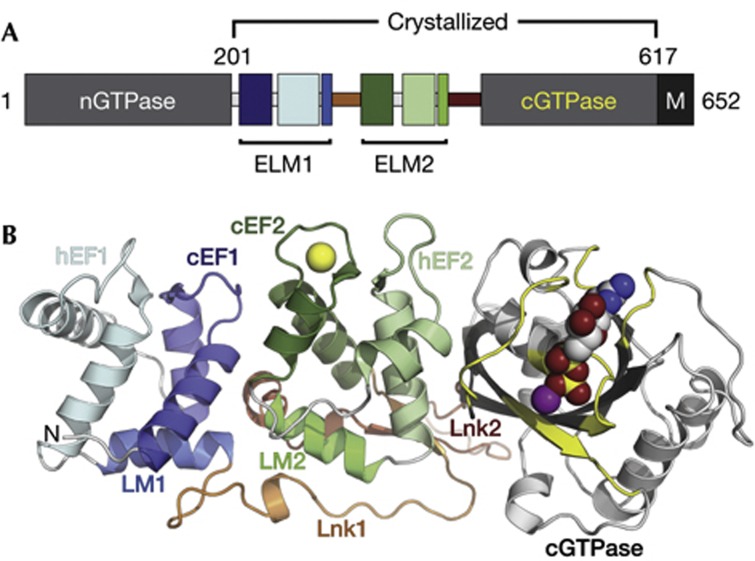
We determined the crystal structures of a Drosophila Miro (dMiro) fragment containing the EF hands and C-terminal GTPase (cGTPase) domain (Fig 1A; referred to as MiroS) in three states: apo (apo-MiroS at 2.82 Å resolution), Ca2+-bound (Ca-MiroS at 2.80 Å) and magnesium (Mg2+) and MgGDP-bound (MgGDP-MiroS at 3.00 Å). Supplementary Fig S1 online shows several views of the three structures, defined here as ‘side’, ‘top’ and ‘bottom’ views. A ribbon diagram highlighting the Ca2+ and nucleotide-binding sites is shown in the ‘side view’ in Fig 1B. The structure consists of three distinct domains that form a compact molecule ∼30 × 40 × 90 Å. The first two domains each contain paired EF hands and are related to each other by pseudo twofold symmetry (Fig 2A), whereas the third domain is the cGTPase. Two linker regions follow a circuitous path across the ‘bottom’ of the molecule to join the three domains. Crystallographic data are presented in supplementary Table S1 online.
Figure 1.
Domain architecture and structure of Miro. (A) Bar diagram of Miro domain structure. Numbering corresponds to dMiro isoform D, domain names are described in the text (B) Ribbon diagram of the MiroS structure in the ‘side view’. Note three distinct domains: ELM1 (blue), ELM2 (green), cGTPase (grey/yellow), joined by two linkers: Lnk1 (orange), and Lnk2 (red). The structure shown is MgGDP-MiroS, with the Ca2+ ion from the Ca-MiroS structure overlaid for orientation (Ca2+/Mg2+/GDP as yellow/purple/multicoloured spheres). cGTPase, C-terminal GTPase; dMiro, Drosophila Miro; ELM, EF hand pair with ligand mimic.
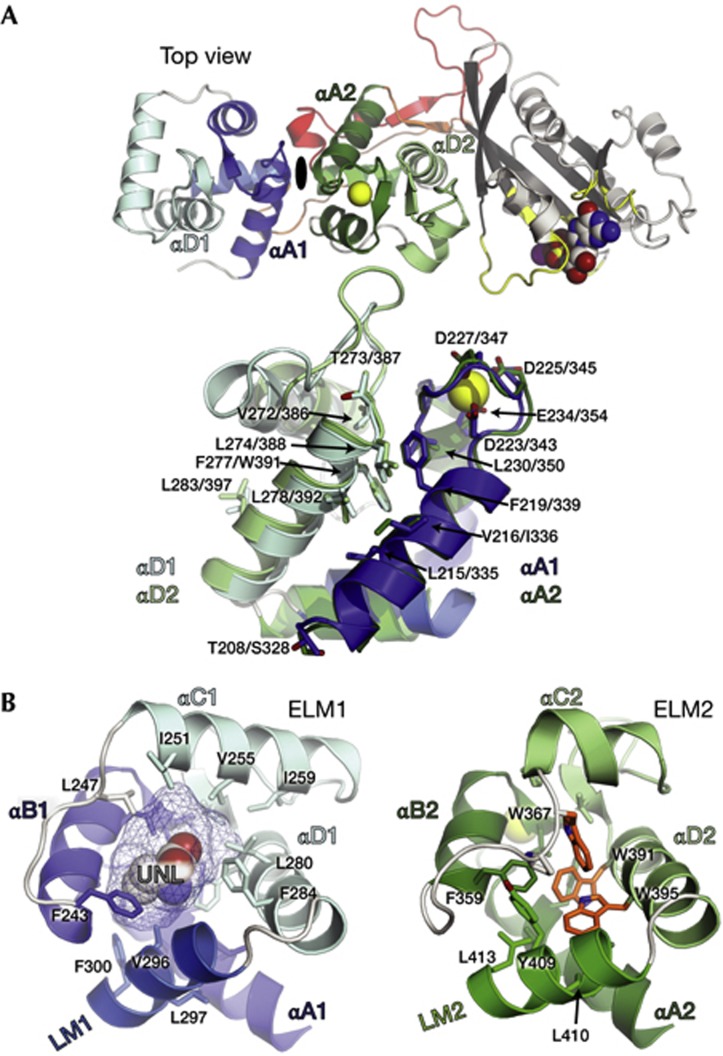
Figure 2.
Miro ELM domains. (A) Top, MiroS shown in the ‘top view’. ELM1 and ELM2 are related by pseudo twofold symmetry, represented by the dyad between them. Bottom, overlay of the two ELM domains (rmsd of 1.8 Å over 84 Cα positions). ELM1 (blue), ELM2 (green) and identical or similar conserved residues in identical positions are shown as sticks. (B) Comparison of the ELM domain hydrophobic cores. The ELM1 core contains the UNL, modelled here as homoserine shown in spheres, occupying a ∼442 Å3 cavity highlighted with mesh. The ELM2 core includes three tryptophans in close proximity (W367, W391, W395 in orange). Hydrophobic residues in the LM/EF hand pair interfaces are also shown in sticks. LM, ligand mimic; UNL, unidentified ligand.
Miro contains two EF hand pairs, such that each cEF hand is paired with a non-canonical ‘hidden’ EF hand [14] (hEF hand) that was not predicted by prior sequence analysis (Fig. 1; cEF1 and hEF1, cEF2 and hEF2). The hEF hands adopt a typical helix–loop–helix EF hand motif and stabilize the adjacent cEF hands through an antiparallel EF hand β-scaffold [15]. Each of the EF hand pairs is followed by a helix that structurally mimics an EF hand ligand, which we term the ‘ligand mimic’ [16] (LM) (Fig 1; LM1 and LM2). Each of the two LM helices is followed by a linker (Fig 1; Lnk1 and Lnk2) that enables a side-by-side orientation of the two cEF hands, positioning the EF hand pairs in-line with the cGTPase. The cGTPase exhibits a classic G-protein fold [17] comprising a six-stranded β-sheet core flanked by α-helices. An extensive interface between hEF2 and the cGTPase involves the nucleotide-responsive Switch I element, which is positioned away from the nucleotide-binding pocket in both the apo and MgGDP-bound structures. Sequence conservation between yeast, fly and human Miro suggests the linear arrangement of the two pseudosymmetric EF hand pairs and the cGTPase domain is likely to be a feature of all Miro proteins (supplementary Fig S1 online).
EF hand pair with ligand mimic domains
The LM helices, buried at the base of the four helices forming each EF hand pair, are a striking feature of the Miro structure, structurally reminiscent of several well-known EF hand proteins bound to their ligands, such as the Troponin C/Troponin I [18] and myosin essential light chain–heavy chain [19] complexes (supplementary Fig S2 online). Based on these comparisons, we refer to each Miro EF hand pair with LM helix arrangement as an ELM domain (Fig 1; ELM1 and ELM2). A handful of proteins contain ELM domains similar to Miro (supplementary Fig S2 online).
Miro’s two tandem ELM domains have similar folds, with 14 identical or similar residues at identical positions on the surface formed by the first (αA) and last (αD) helix of each EF hand pair (Fig 2A). The two ELM domains themselves do not form an extensive interface, but both LM helices bury over 1,000 Å2 of surface area within their respective EF hand pairs, and the linkers that follow them contribute several key polar contacts to the interface. The long linkers permit an ∼180° rotation of ELM2 with respect to ELM1, such that the C-termini of the LM helices point directly at one another and the two highly similar αA/αD surfaces are on opposite faces of Miro (Figs 1B and 2A).
Although the two ELM domains appear to be structurally similar, they have markedly different hydrophobic cores. The core of ELM1 core encloses an ∼442 Å3 cavity occupied in all of our structures by an unidentified ligand (UNL). In contrast, the ELM2 core is tightly packed and includes three conserved tryptophan residues in very close proximity (Fig 2B; supplementary Fig S3 online).
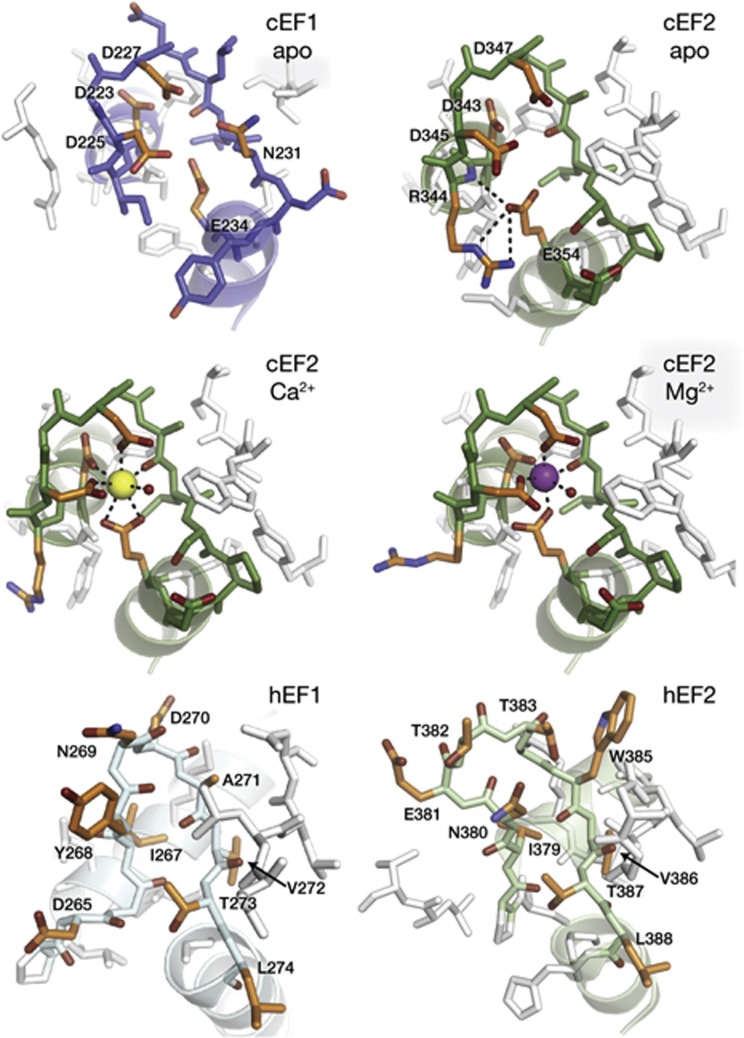
We observe Ca2+ or Mg2+ binding only to one EF hand site in the structures described here: cEF2 of ELM2. However, both cEF hand loops show a classic EF hand loop architecture and harbour the conserved glutamate residue previously shown to be critical for Ca2+-binding at cEF1 (E234) and cEF2 (E354) [7, 20] (Fig 3; supplementary Fig S4 online). The acidic residues of cEF2 coordinate Ca2+ in a classic pentagonal, bipyramidal fashion [15], and Mg2+-binding to cEF2 is readily accommodated by an octahedral coordination, as seen in other EF hands [21] (Fig 3). Unexpectedly, the overall fold of Miro is nearly identical in apo-MiroS, Ca-MiroS and MgGDP-MiroS (rmsd<0.7 Å over 404 Cα positions). The lack of Ca2+ or Mg2+ binding at cEF1 might be explained by the presence of the UNL at the core of ELM1. The αB1 helix is displaced relative to αB2 to accommodate the UNL, resulting in a 1.0 Å shift of E234 relative to E354 (supplementary Fig S5 online). In contrast to the cEF hands, the two hEF hand loops are likely incapable of Ca2+ binding, as they lack the typical EF hand loop structure and the negatively charged residues that typically coordinate Ca2+ (Fig 3).
Figure 3.
Details of the EF hand loops. All loops are shown in approximately the same orientation, with entering and exiting helices ghosted for clarity. Residues of note are coloured in orange. cEF1 is shown in its apo state. cEF2 is shown in its apo, Ca2+-bound and Mg2+-bound states. In the cEF2 apo state, E354 is stabilized by nearby loop residue R344. In the Ca2+-bound state, cEF2 loop residues coordinate Ca2+ in a pentagonal, bipyramidal arrangement, with E354 as a bidentate ligand. An ordered water molecule at the apex of one of the pyramids is represented as a small red sphere. In the Mg2+-bound state, the coordination is octahedral with E354 as a monodentate ligand. The hEF1 and hEF2 loops adopt a conformation quite different from the cEF hands, and also lack the acidic residues that typically coordinate cations. Note the conserved hEF1 272VTL274 and hEF2 386VTL388 residues, which overlay identically in Fig 2A. cEF, canonical EF hand; hEF, hidden EF hand.
EF hand/cGTPase interface
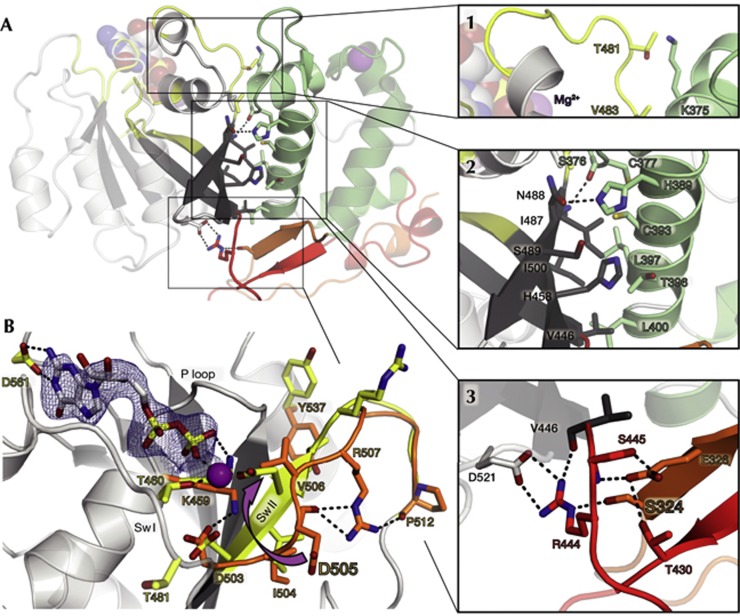
The MiroS structures presented here are, to our knowledge, the first reported structures showing a direct EF hand–GTPase interaction. The interface is amphiphilic, with a total buried surface area of ∼1,500 Å2 comprising three regions (Fig 4A, insets 1–3). In the first region, the hEF2 loop forms contacts with residues in and around Switch I of the cGTPase, including the Switch I threonine T481. The second region involves interdigitating hydrophobic residues in αD2 and the cGTPase β-sheet. The third region comprises an extensive network of polar contacts that includes residues from linkers Lnk1 and Lnk2 and features a salt bridge between highly conserved residues R444 and D521.
Figure 4.
The cGTPase and its interface. (A) The ELM2/cGTPase interface can be divided into three regions (insets 1–3). Select residues are shown as sticks, polar contacts as dashed lines, and Mg2+ and MgGDP as spheres. Region 1: αC2 and the hEF2 loop form both backbone and side-chain contacts with αG1 and Switch I in the cGTPase, including the Switch I threonine T481. Region 2: αD2 in hEF2 packs against the twisting cGTPase β-strands Gβ1-3. Region 3: the candidate Pink1 phosphorylation site residue S324 in Lnk1 hydrogen bonds with R444 in Lnk2, which also forms a salt bridge with D521, at the end of the Switch II loop. (B) cGTPase rearrangements in MgGDP binding. Displayed is the MiroS-MgGDP structure, with select residues from the aligned apo-MiroS structure shown in orange. GDP is shown as sticks (2Fo-Fc map, 1.2σ), and Mg2+ as a purple sphere. In the apo state, D505 in the Switch II 503DIDV506 motif is far from the nucleotide-binding pocket and is hydrogen bonded to R507, which is also stabilized by interactions with P512. In the MgGDP-bound state, the D505 side-chain rotates ∼7 Å to interact with the β-phosphate (arrow), locking the Switch II loop in a β-strand conformation and extending Gβ3. V506 rotates away from the nucleotide-binding pocket, and both R507 and Y537 become solvent exposed. cGTPase, C-terminal GTPase; Sw I/II, Switch I/II; P loop, phosphate-binding loop.
Both nucleotide binding and Pink1 phosphorylation could potentially modulate the ELM2/cGTPase interface of Miro. The Switch I threonine typically serves a γ-phosphate-sensing role in GTPases [17]. Although dMiro T481 is not conserved, any γ-phosphate-sensing mechanism enabling the Miro Switch I loop to access the nucleotide-binding pocket on MgGTP-binding would alter the first region of the ELM2/cGTPase interface (Fig 4). A Pink1 phosphorylation site lies at the heart of the network of polar contacts in the third region of the ELM2/cGTPase interface, in one of the three residues in Lnk1 (S323/S324/T325) [12]. Of these residues, S324 is most conserved and forms a hydrogen bond with R444 in Lnk2, coupling it to the cGTPase via the R444/D521 salt bridge (Fig 4A, inset 3). Phosphorylation of S324 would likely disrupt this network of interactions. Pink1 phosphorylation has been shown to trigger Parkin-dependent degradation of Miro [12]. A recent report [22] has identified several Parkin ubiquitination sites in human Miro, four of which map onto our structure on the same face as the S324 Pink1 site (Fig 5). Together, these findings might have mechanistic implications for Pink1-mediated recruitment of Parkin to the mitochondrial surface, a key step in the mitochondrial quality control pathway [23].
Figure 5.
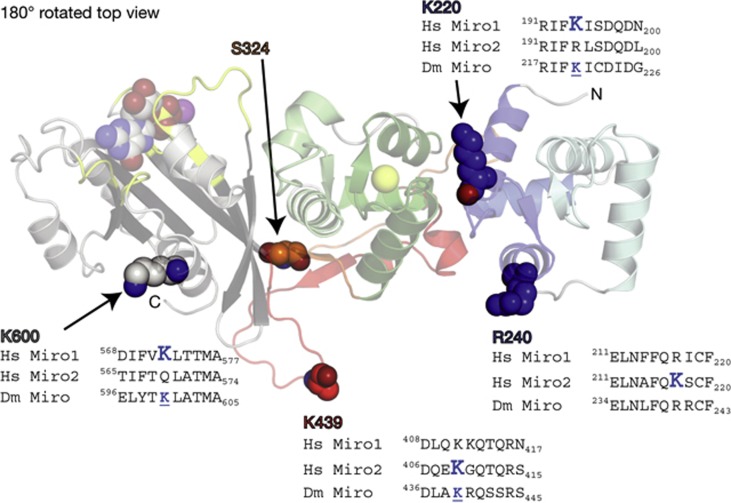
Location of known Parkin ubiquitination sites within the Miro structure. MiroS is shown in a 180°-rotated top view (relative to Fig 2). Four residues in dMiro correspond in sequence and structure to residues that are ubiquitinated by Parkin in human Miro1 (hMiro1) or human Miro2 (hMiro2) [22]. These are shown as spheres (K220, R240, K439 and K600; labelled with dMiro numbering). An alignment of a 10-aa segment of dMiro, hMiro1 and hMiro2 flanking each ubiquitinated site is shown. The large blue ‘K’ within each sequence alignment indicates the ubiquitinated residue in hMiro1 or hMiro2, and a smaller, underlined K indicates that dMiro has a lysine in the same position. Residue S324, a candidate Pink1 phosphorylation site, is also shown in spheres. Note the side-chain of K439 is disordered in the MiroS structure, so it is modelled as an alanine. dMiro/Dm Miro, Drosophila Miro; Hs Miro 1/2, human Miro1/2.
cGTPase structure is Ras-like
The Miro cGTPase is most structurally similar to the Ras homologue Rheb [24] (supplementary Fig S6 online). Both the N-terminal GTPase domain of Miro (nGTPase) and the cGTPase were originally classified as atypical Rho GTPases [1], as they depart from the Rho-conserved DxxG Switch II motif [17]. The structure of the MgGDP–MiroS complex shows that the atypical cGTPase Switch II motif 503DIDV506 is intimately involved in MgGDP binding. Comparison of the apo and MgGDP-bound cGTPase structures reveals a concerted rearrangement of residues in and around the Switch II and P loop motifs (Fig 4B). Upon MgGDP binding, Switch II undergoes a remarkable register shift that locks it into a β-strand conformation, extending the cGTPase β-sheet. Aspartate D505 rotates ∼7 Å (Fig 4B, arrow) to form an unusual, direct interaction with Mg2+, accommodated by shifts in R507 and Y537.
Solution studies of Miro
We evaluated the structure of Miro in solution using small-angle x-ray scattering (SAXS) and size-exclusion chromatography in-line with multiangle light scattering (SEC-MALS). Radii of gyration (Rg) and ab initio molecular envelope reconstructions from SAXS experiments indicate that the overall structural shape of MiroS bound to Ca2+ in solution corroborates the crystallographic data. The MiroS Rg remains similar under apo or Mg2+ conditions (Supplementary Figs S7,S8 online; Supplementary Table S2 online). The increased maximum diameter (Dmax) and Rg of MiroS-apo might indicate that Miro is less stably folded without Ca2+, like several other EF hand proteins [14, 21]. Notably, gross conformational rearrangements of MiroS into another stable form and/or oligomerization did not occur in the ion and nucleotide conditions tested. SEC-MALS shows that MiroS, and a longer Miro construct containing the nGTPase domain (aa 1-617, referred to as MiroL), are both monomers in solution regardless of Ca2+ concentration (supplementary Fig S9 online), consistent with the SAXS data for MiroS. Additionally, the respective elution profiles for both MiroS and MiroL±Ca2+ are superimposable, suggesting that the presence of the nGTPase does not greatly alter the behaviour of Miro in this assay. Taking these data together, neither MiroS nor MiroL appear to undergo a dramatic conformational rearrangement in response to Ca2+.
Conclusions
One of the clearest functions of Miro is the arrest of kinesin-1- and dynein-dependent mitochondrial movement in response to elevated Ca2+, a function that requires Ca2+ binding to the EF hands [8]. How do Miro’s ELM domains enable Ca2+-induced mitochondrial arrest? Although our structures illuminate Miro’s curious EF hand domain arrangement, the underlying mechanism is not yet readily apparent. Miro’s EF hand region is reminiscent of the Ca2+-myristoyl switch protein recoverin [25]. However, the Ca2+-induced structural changes that occur in recoverin [26], and in more conventional EF hand proteins [15], are in contrast with the minimal structural changes observed in our crystal and solution structures of Miro. Conformational changes in Miro might require the presence of an additional macromolecular binding partner, as seen in some other EF hand proteins [15]. Miro interacts with kinesin-1 [6–8], mitofusin [27], Milton [28–30], and Pink1 [11–13], all of which dimerize. The pseudosymmetric ELM domains might facilitate associations between the Miro monomer and dimeric binding partners. An appealing hypothesis is that Ca2+-binding primes Miro for interactions, permitting kinesin-1, for example, to bind Miro by displacing its LM helices, not unlike how rhodopsin kinase binds to recoverin by displacing its myristoylated N-terminal ‘LM’ helix [31] (supplementary Fig S2 online). Thus, the compact, linear MiroS conformation we have captured here likely reflects one key state of Miro, modulated by Ca2+, nucleotide and phosphorylation-dependent regulation in the context of mitochondrial function in the cell.
Methods
Crystallography. The dMiro1–617-6xHis isoform D (MiroL) was purified as described previously [12]. dMiro201–617-6xHis isoform D (MiroS) was expressed in Escherichia coli and purified by nickel-affinity and cation-exchange chromatography at 4 °C. Crystals were grown using the sitting-drop vapour diffusion method at 21 °C. Optimized conditions included 25 mM HEPES, 0.5 M NaCl, 0.5 mM TCEP, 1.5–1.9 M LiSO4, 0.1–0.2 M Bis-Tris at pH 7.4–8.0, 5 mM EGTA and 4.5–10.8 mg/ml protein. Ca-MiroS and MgGDP-MiroS crystals were subject to CaCl2 and MgCl2+GDP soaks for ∼72 h, respectively, before flash freezing directly in mother liquor (see supplementary Methods online). Diffraction data were measured at LS-CAT, Sector 21 of the Advanced Photon Source (APS) in the Argonne National Laboratory with an x-ray wavelength of 0.97872 Å. Data from two isomorphous selenomethionine (SeMet)-labelled crystals were merged, and phasing by single wavelength anomalous dispersion was carried out using Phenix. An initial model obtained using Phenix AutoBuild comprised 375 of an expected 417 residues and revealed the overall domain structure. After initial refinement, the selenomethionine structure was used as a search model in Phaser to obtain by molecular replacement the apo-MiroS, Ca-MiroS, and MgGDP-MiroS structures described here.
SAXS. Solution SAXS experiments were performed at BioCAT, beamline 18-ID-B at Sector 18 of the APS in the Argonne National Laboratory. MiroS solutions at 1.0 mg/ml in SAXS buffer (25 mM HEPES at pH 7.4,300 mM NaCl, 0.5 mM TCEP) supplemented with either EGTA, MgCl2, CaCl2, GDP and/or GTP were loaded into a capillary as a total of 20 exposures were measured (see supplementary Methods online). Buffer solutions matched to each Miro sample were measured in the same capillary before each experimental run. Images from buffer and sample sets were radially binned to obtain I versus q curves. Curves from each set were averaged and the resulting buffer curve was subtracted from the sample curve. The Rg was determined by Guinier analysis. Where possible, the particle distribution function P(r) was determined using the programme GNOM to confirm the Rg and to determine Dmax. Scattering data of apo-MiroS and MiroS in the presence of Mg2+ but not Ca2+ or nucleotide showed evidence of some unfolded and/or aggregated protein in the high and low Q range of the scattering curves. While these data were of sufficient quality to obtain Rg or Dmax, they were not used for SAXS reconstructions. Dummy atom modelling for SAXS reconstructions was done with the programme DAMMIF. Twofold, fivefold and 10-fold dilution of samples did not significantly change the Rg, Dmax or DAMMIF reconstructions of SAXS data. Comparisons of the SAXS reconstructions to the MiroS crystal structures were made with DAMAVER, SUPCOMB and CRYSOL.
SEC-MALS. Solution SEC-MALS experiments were conducted using Agilent Technologies 1200 LC HPLS system equipped with a Wyatt Dawn HeleosII 18-angle MALS light scattering detector. A total of 200 μl of MiroS or MiroL was injected onto a Superdex 200 column with a flow rate of 0.5 ml/min in SEC-MALS buffer (25 mM HEPES at pH 7.4, 300 mM NaCl, 0.5 mM TCEP, 0.5 mM EGTA, 1 mM MgCl2, 20 μM GTP). The same was repeated for the Ca2+-containing samples using SEC-MALS buffer+3 mM CaCl2. A void volume of 7.8 mL was determined using blue dextranReagents, constructs, references and details of protein expression, purification, crystallography, SAXS and SEC-MALS are described in the supplementary Methods online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank Y. Wong, M. Seeger, M. Gonzalez, C. Sato, A. Banks, K. Smith, S. Light, C. Janczak, A. Grigorescu and APS staff Z. Wawrzak, R. Graceffa and M. Vukonich for discussions and assistance; J. Waitzman, M. French, M. Wietecha, M. Cronin, T. Schwarz, Z. Grabarek for comments on the manuscript. This work was supported by NIH grants R01GM072656 (S.E.R.) and T32GM008382 (J.L.K.), the DePaul University College of Science and Health Faculty Research Grant (E.C.L.), and the ARCS Foundation, Kosciuszko Foundation, and PNA and PAMS scholarships (J.L.K.). This work used resources of the Northwestern University Structural Biology Facility and the Keck Biophysics Facility, supported by NCI-CCSG-P30-CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center. Use of the APS was supported by U.S. DOE contract DE-AC02-06CH11357. Use of the LS-CAT was supported by the Michigan Economic Development and the Michigan Technology Tri-Corridor (085P1000817). Use of BiocAT was supported by NIH grants 2 P41RR008630-17 and 9 P41 GM103622-17. See supplementary Methods online for more details.
Author contributions: S.E.R. supervised the research. J.L.K. initiated the project, expressed and purified Miro, performed crystallization trials and crystal soaks. J.L.K. and P.J.F. collected x-ray diffraction data and solved the structure. J.L.K., P.J.F., D.M.F. and S.E.R. refined and interpreted the structures. J.L.K., S.C., E.C.L. and S.E.R. designed, performed and analysed SAXS experiments. J.L.K. designed, performed and analysed SEC-MALS experiments. J.L.K. wrote the manuscript with D.M.F. and S.E.R; P.J.F., S.C., and E.C.L. assisted with manuscript preparation.
Footnotes
References
- Fransson A, Ruusala A, Aspenström P (2003) Atypical Rho GTPases have roles in mitochondrial homeostasis and apoptosis. J Biol Chem 278: 6495–6502 [DOI] [PubMed] [Google Scholar]
- Frederick RL, McCaffery JM, Cunningham KW, Okamoto K, Shaw JM (2004) Yeast Miro GTPase, Gem1p, regulates mitochondrial morphology via a novel pathway. J Cell Biol 167: 87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Macleod GT, Wellington A, Hu F, Panchumarthi S, Schoenfield M, Marin L, Charlton MP, Atwood HL, Zinsmaier KE (2005) The GTPase dMiro is required for axonal transport of mitochondria to Drosophila synapses. Neuron 47: 379–393 [DOI] [PubMed] [Google Scholar]
- Fransson S, Ruusala A, Aspenström P (2006) The atypical Rho GTPases Miro-1 and Miro-2 have essential roles in mitochondrial trafficking. Biochem Biophys Res Commun 344: 500–510 [DOI] [PubMed] [Google Scholar]
- Glater EE, Megeath LJ, Stowers RS, Schwarz TL (2006) Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J Cell Biol 173: 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saotome M, Safiulina D, Szabadkai G, Das S, Fransson A, Aspenström P, Rizzuto R, Hajnóczky G (2008) Bidirectional Ca2+-dependent control of mitochondrial dynamics by the Miro GTPase. Proc Natl Acad Sci 105: 20728–20733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAskill AF, Rinholm JE, Twelvetrees AE, Arancibia-Carcamo IL, Muir J, Fransson A, Aspenström P, Attwell D, Kittler JT (2009) Miro1 is a calcium sensor for glutamate receptor-dependent localization of mitochondria at synapses. Neuron 61: 541–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Schwarz TL (2009) The mechanism of Ca2+ -dependent regulation of kinesin-mediated mitochondrial motility. Cell 136: 163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Osman C, Walter P (2011) The conserved GTPase Gem1 regulates endoplasmic reticulum-mitochondria connections. Proc Natl Acad Sci 108: 14151–14156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murley A, Lackner LL, Osman C, West M, Voeltz GK, Walter P, Nunnari J (2013) ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. eLife 2: e00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihofen A, Thomas KJ, Ostaszewski BL, Cookson MR, Selkoe DJ (2009) Pink1 forms a multiprotein complex with Miro and Milton, linking Pink1 function to mitochondrial trafficking. Biochemistry 48: 2045–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, Rice S, Steen J, LaVoie MJ, Schwarz TL (2011) PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell 147: 893–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S et al. (2012) Parkinson’s disease–associated kinase PINK1 regulates Miro protein level and axonal transport of mitochondria. PLoS Genet 8: e1002537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopulos PB, Zheng L, Li G-Y, Plevin MJ, Ikura M (2008) Structural and mechanistic insights into STIM1-mediated initiation of store-operated calcium entry. Cell 135: 110–122 [DOI] [PubMed] [Google Scholar]
- Grabarek Z (2006) Structural basis for diversity of the EF-hand calcium-binding proteins. J Mol Biol 359: 509–525 [DOI] [PubMed] [Google Scholar]
- Heidarsson PO, Bjerrum-Bohr IJ, Jensen GA, Pongs O, Finn BE, Poulsen FM, Kragelund BB (2012) The C-Terminal tail of human neuronal calcium sensor 1 regulates the conformational stability of the Ca2+-activated state. J Mol Biol 417: 51–64 [DOI] [PubMed] [Google Scholar]
- Bourne H, Sanders D, McCormick F (1991) The GTPase superfamily: conserved structure and molecular mechanism. Nature 349: 117–127 [DOI] [PubMed] [Google Scholar]
- Vinogradova MV, Stone DB, Malanina GG, Karatzaferi C, Cooke R, Mendelson RA, Fletterick RJ (2005) Ca(2+)-regulated structural changes in troponin. Proc Natl Acad Sci USA 102: 5038–5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houdusse A, Cohen C (1996) Structure of the regulatory domain of scallop myosin at 2A resolution: implications for regulation. Structure 4: 21–32 [DOI] [PubMed] [Google Scholar]
- Koshiba T, Holman HA, Kubara K, Yasukawa K, Kawabata S-I, Okamoto K, MacFarlane J, Shaw JM (2011) Structure-function analysis of the yeast mitochondrial Rho GTPase, Gem1p: implications for mitochondrial inheritance. J Biol Chem 286: 354–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabarek Z (2011) Insights into modulation of calcium signaling by magnesium in calmodulin, troponin C and related EF-hand proteins. BBA—Mol Cell Res 1813: 913–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarraf SA, Raman M, Guarani-Pereira V, Sowa ME, Huttlin EL, Gygi SP, Harper JW (2013) Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 496: 372–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Walker JE, Youle R (2012) Mitochondrial quality control mediated by PINK1 and Parkin: links to Parkinsonism. Cold Spring Harb Perspect Biol 4: a011338–a011338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazhab-Jafari MT, Marshall CB, Ishiyama N, Ho J, Di Palma V, Stambolic V, Ikura M (2012) An autoinhibited noncanonical mechanism of GTP hydrolysis by Rheb maintains mTORC1 homeostasis. Structure 20: 1528–1539 [DOI] [PubMed] [Google Scholar]
- Tanaka T, Ames JB, Harvey TS, Stryer L, Ikura M (1995) Sequestration of the membrane-targeting myristoyl group of recoverin in the calcium-free state. Nature 376: 444–447 [DOI] [PubMed] [Google Scholar]
- Ames JB, Ishima R, Tanaka T, Gordon JI, Stryer L, Ikura M (1997) Molecular mechanics of calcium-myristoyl switches. Nature 389: 198–202 [DOI] [PubMed] [Google Scholar]
- Misko A, Jiang S, Wegorzewska I, Milbrandt J, Baloh RH (2010) Mitofusin 2 is necessary for transport of axonal mitochondria and interacts with the Miro/Milton complex. J Neurosci 30: 4232–4240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers RS, Megeath LJ, Górska-Andrzejak J, Meinertzhagen IA, Schwarz TL (2002) Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron 36: 1063–1077 [DOI] [PubMed] [Google Scholar]
- Koutsopoulos OS, Laine D, Osellame L, Chudakov DM, Parton RG, Frazier AE, Ryan MT (2010) Human Miltons associate with mitochondria and induce microtubule-dependent remodeling of mitochondrial networks. BBA—Mol Cell Res 1803: 564–574 [DOI] [PubMed] [Google Scholar]
- Brickley K, Stephenson FA (2011) Trafficking kinesin protein (TRAK)-mediated transport of mitochondria in axons of hippocampal neurons. J Biol Chem 286: 18079–18092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames JB, Levay K, Wingard JN, Lusin JD, Slepak VZ (2006) Structural basis for calcium-induced inhibition of rhodopsin kinase by recoverin. J Biol Chem 281: 37237–37245 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.