Abstract
C1q is the first subcomponent of the classical pathway of the complement system and a major connecting link between innate and acquired immunity. As a versatile charge pattern recognition molecule, C1q is capable of engaging a broad range of ligands via its heterotrimeric globular domain (gC1q) which is composed of the C-terminal regions of its A (ghA), B (ghB) and C (ghC) chains. Recent studies using recombinant forms of ghA, ghB and ghC have suggested that the gC1q domain has a modular organization and each chain can have differential ligand specificity. The crystal structure of the gC1q, molecular modeling and protein engineering studies have combined to illustrate how modular organization, charge distribution and the spatial orientation of the heterotrimeric assembly offer versatility of ligand recognition to C1q. Although the biochemical and structural studies have provided novel insights into the structure-function relationships within the gC1q domain, they have also raised many unexpected issues for debate.
Keywords: Complement, C1q, Crystal, Immunoglobulin, Innate immunity, C-reactive protein
1. Introduction
C1q is the first subcomponent of the classical pathway of complement activation and a major connecting link between classical pathway-driven innate immunity and IgG- or IgM-mediated acquired immunity. The C1q molecule is made up of six copies of three polypeptide chains: the A-chain (223 residues), the B-chain (226 residues) and the C-chain (217 residues) [1–4]. Each of these chains has a short three to nine residue amino-terminal region, containing a cysteine residue, followed by a collagen-like region (CLR) of ~81 residues and a carboxy-terminal globular head module (ghA, ghB or ghC) of ~135 residues [5]. The globular head modules from the different chains combine to form a heterotrimeric globular head domain (gC1q) via conserved hydrophobic patches. The gC1q domain, which is believed to be the nucleation point in the formation of the C1q molecule, is further stabilized by the CLR before a combination of intra-chain and A to B and C to C inter-chain disulfide bonds yield the structural unit: six chains forming two gC1q domains covalently linked by the inter C-chain disulphide bond. Three of these structural units then associate via strong non-covalent bonds in the fibril-like central portion to yield the hexameric C1q molecule that has a tulip-like structure [6,7].
In the classical complement pathway, the binding of C1q to IgG- or IgM-containing immune complex (IC) leads to the auto-activation of C1r, which, in turn, activates C1s. C1r and C1s, the two serine protease proenzymes, together with C1q constitute C1, the first component of the classical complement pathway [2]. The activation of the C1 complex (C1q + C1r2 + C1s2) subsequently leads to the activation of the C2–C9 components of the classical pathway and formation of the terminal membrane attack complex (MAC) [3].
2. C1q is a versatile innate immune molecule
In addition to being the target recognition protein of the classical complement pathway, C1q is also involved in a number of other immunological processes (Table 1), including maintenance of immune tolerance via clearance of apoptotic cells, phagocytosis of bacteria, neutralization of virions, cell adhesion, and modulation of dendritic cells (DC), B cells and fibroblasts. Its ability to carry out such diverse functions is aided by its capacity to engage a broad range of ligands, such as envelope proteins of certain retroviruses, β-amyloid fibrils, lipopolysaccharides (LPS), porins from Gram-negative bacteria, phospholipids (PL), apoptotic cells and some acute phase reactants including pentraxins (Table 2). The polyanionic nature of these ligands appears to suggest that C1q recognizes a pattern of charged residues or groups [7].
Table 1.
Immune functions of C1q
| Biological processes | Comments | References |
|---|---|---|
| Activation of classical pathway of complement | The classical pathway of complement activation is triggered by either IgG or IgM antibody bound to antigen. The binding of the antibody to antigen exposes a site on the antibody which is a binding site for the first complement component C1 (comprising C1q, C1r and C1s). The classical pathway usually culminates in the formation of MAC on the target cell which causes its lysis. | [10,19,56,57] |
| IC clearance/solubilization | An IC is a cluster of interlocking antigens and antibodies (antigen–antibody complex). The level of circulating ICs is a measure of immune system function. The formation of such complexes is a natural and successful attempt to neutralize antigens (exogenous or endogenous). ICs can cause an exaggerated immune response. C1q plays a vital role in solubilization and clearance of ICs. C1q deficiency can lead to IC accumulation, leading to glomerulonephritis. There is a strong association between C1q deficiency and SLE. | [58–60] |
| Bacterial clearance | C1q is involved in bacterial clearance. It is an opsonin and coats bacteria and enhances uptake of bacteria by phagocytic cells. C1q interaction with its receptor on neutrophils induces the super-oxidative burst. This activity is attributed to the CLR. | [61,62] |
| Virus binding/inactivation | C1q can bind to various viruses, including retroviruses and influenza viruses. The globular head is believed to be involved in the binding viral envelope proteins, e.g. C1q binds to the envelope protein p15e of Type C retroviruses leading to the classical pathway mediated lysis of the virus. | [63,64] |
| Induction of pro-inflammatory cytokines | C1q triggers the production of IL-8, IL-6 and MCP-1 by human umbilical vein endothelial cells. C1q interaction with its receptor calreticulin–CD91 complex operates via CLR. This interaction enhances p38 MAPK activation, NF-κB activity and production of pro-inflammatory cytokines in macrophages. Both gC1q and CLR can suppress LPS and CpG-induced MyD88-dependent pathway and production of TNF-α and IL-12 p40 in bone marrow-derived DC. | [65–67] |
| Clearance of apoptotic/necrotic cells (via PTX3, CRP, SAP) | Dying cells must be cleared effectively by phagocytic cells in order to avoid chronic inflammation. Autoimmunity can be the result of presence of autoantigens from apoptotic or necrotic cells. The pentraxins CRP, SAP and PTX3 all bind nuclear debris and are all C1q ligands. C1q is involved in the clearance of apoptotic cells either directly or via CRP, SAP or PTX3. This is quite crucial to preventing pathological conditions like SLE. | [8,45,49,68–72] |
| Induction of apoptosis | C1q can induce mitotic arrest in G1 stage in fibroblasts. Increased p38 MAPK activation has been shown in C1q treated proliferating fibroblasts. P38 MAPK activation leads to mitotic arrest and subsequent apoptosis of fibroblasts. | [73,74] |
| Modulation of dendritic cells | C1q is known to be involved in capturing IC in lymphoid tissues. Immature DC are a good source of C1q. DC are known to accumulate in atherosclerotic lesions. IC are also detected in such lesions and C1q is considered to bind and trap these IC. Ox-LDL is known to associate with IC which is then ingested by DC through the Fc-γ receptor. C1q captures IC like Fc-γ receptor and in atherogenesis, C1q may be involved in binding ox-LDL within the IC. | [75–77] |
| Chemotaxis | Being apotent chemoattractant, C1q can recruit neutrophils and eosinophils to the sites of infection and inflammation. | [78–80] |
| Proliferative/anti-proliferative effects | C1q CLR has been shown to have anti-proliferative effect on fibroblasts. C1q inhibits the growth of a wide variety of cultured somatic cells. The effect of C1q is not cytotoxic, but cytostatic. It is proposed that the interaction of C1q with C1q receptor can lead to suppression of events necessary for cell proliferation. | [73,81–83] |
| Adhesion and coagulation via platelets | Platelets are known to be involved in inflammatory processes and contribute to the development of thrombosis, atherosclerosis and vasculitis. C1q has been shown to modulate platelet interactions with collagen and ICs. Expression of αIIb/β3 integrins and P-selectin on C1q exposure also contributes to the coagulation events associated with complement activation and inflammation. | [84,85] |
| Isotype switching | C1q knock-out mice show significantly reduced production of T cell dependent antigen-specific IgG2a and IgG3 isotypes owing to reduced IFN-γ production by T cells. C1q, therefore, may be crucial to antigen delivery to follicular DCs and subsequent generation of normal secondary antibody response. | [86] |
Table 2.
Ligands of C1q which interact with its gC1q domain
| Ligands | Interacting sites/motifs | Implications/comments | References |
|---|---|---|---|
| IgG | The ghB is the principal IgG-binding module of C1q. Residues Arg114, Arg129 and Glu162 are important in binding IgG. A modeling exercise of binding of IgG to gC1q also identified ArgB161 as important in forming salt bridges with Glu195 and Glu287. Other residues, e.g. ArgA162 also play complementary role. Murine and human IgG bind differently to gC1q, illustrating species specific differences in C1q binding by IgG. Glu318, Lys320, and Lys322 in mouse IgG2b, which is highly conserved in different IgG isotypes, are the main residues in gC1q binding. In human IgG1 Asp270, Lys322, Pro329, Pro331, and residues at 326 and 333 are implicated. | C1q also interacts with the Fab (CL domain), supporting observations that Fab-Fc flexibility has a crucial role in C1q binding. | [4,26–28] |
| IgM | Hexameric and pentameric IgM both can bind gC1q via the Cμ3 domain involving His, Asp/Glu and Pro residues at 430–434. However, ghC module is the most specific in binding IgM although ghA module can bind both IgG and IgM. The residues in gC1q involved in binding IgM have not been identified. | Monomeric IgM, fixed to antigen, does not bind C1q. Hexameric IgM is >100 times more efficient at activating complement than the pentameric form, possibly reflecting better symmetry for binding one entire C1q molecule. | [87] |
| CRP | Binding of gC1q to CRP is achieved by a fitting of the globular C1q head in the pore of the pentameric CRP. However, as CRP has pseudo five fold symmetry, the binding of gC1q to CRP is understood to involve not a symmetrical binding but an asymmetric positioning of the gC1q such that the two Tyr175 of CRP subunits A and D are within hydrogen bonding distances from C1q residues TyrB175 and LysA200. | CRP, like SAP, binds nuclear debris and has a major role in clearing chromosomal material from necrotic cells, via C1q binding. | [4,31,32,45] |
| SAP | Since SAP is a homologue of CRP, binding may be similar. However, CRP Asp112 is not conserved in SAP. | SAP is a highly conserved plasma protein. It is responsible for controlling chromatin degradation and prevention of antinuclear autoimmunity. | [71,88] |
| PTX3 | CRP Asp112 is not conserved in PTX3. PTX3 is able to interact with C1q that is complexed with C1r2C1s2 tetramer. PTX3 binds via gC1q. | PTX3 is structurally related to CRP and SAP and shares similarities with them in its C terminal half but not in the N terminus. PTX3 was shown to enhance the deposition of both C1q and C3 on the surface of apoptotic cells and thus facilitating uptake of apoptotic cells by phagocytes. PTX3 can both activate and inhibit C1q. C1 complex can bind to immobilized PTX3 and subsequently activate C4 and induce complement activation. However, fluid-phase PTX3 inhibits complement activation by blocking the binding of C1q to its ligand (antibody sensitized erythrocytes) PTX3 is believed to play a dual role in innate immunity, supporting safe clearance of damaged self-material and amplifying innate immunity against fungal pathogens. | [49,69,89] |
| Decorin | Decorin (a proteoglycan) may bind to the “reck” region of C1q (between gC1q and CLR), or to both domains via decorin core protein. Decorin also binds collagen I and V. | May modulate classical pathway activation in the tissue as it can inhibit the hemolytic activity of purified C1 as well as C1 in normal human serum. | [90–92] |
| HTLV-I gp21 | C1q binding site has been mapped to residues 400–429 and 426–460 of gp21 which lies between the anchorage domain and the fusion domain of gp21. The ghC module is the principal module involved in binding gp21. | C1q has been shown to inhibit the infectivity of the cell free HTLV-I virion via its interaction with the extramembrane region of gp21, a transmembrane protein considered responsible for fusion between the virus and target cell membrane. | [11,93–95] |
| HIV-1 gp41 | C1q has been shown to interact with residues 601–613 of the envelope protein gp41 of HIV-1, which facilitates infection of complement-receptor bearing cells, instead of viral lysis. Structurally the gp41 loop structure exhibits an absence of charged resides and an abundance of exposed hydrophobic side chains which are characteristic of protein–protein interaction sites. The Ala substitution of all hydrophobic residues (including the 4-carbon aliphatic moiety of the Lys608 side chain) within the loop abolishes C1q–gp41 interaction. When modeled on the gp41 structure of simian immunodeficiency virus, the HIV-1 gp41 loop region forms a cleft, which is likely to be complementary to the binding of gC1q | C1q binding to viruses may result in virus neutralization. HIV-1 infected B cells, which may serve as an alternative reservoir for HIV-1, also activate the classical complement pathway in a gp41 dependent manner. | [11,96–104] |
| p15E | Binding sites not identified. | C1q binds to p15E, the envelope protein from the Moloney virus, which leads to classical pathway-mediated virolysis in human serum. | [63,64] |
| Aβ1–42 | C1q globular heads bind to the acidic N-terminal 1–11 region of the β-amyloid peptide. The ghB module has been shown to interact with the peptide. | β-amyloid fibrils are the major protein components of the neuritic plaques in the brain of Alzheimer's disease and activate the complement in an antibody-independent manner. | [105–107] |
| ABri in FBD and ADan in FDD | The gC1q domain interacts with the N-terminal region of familial dementia peptides. The ghB module binds both these peptides specifically (Bonifati, unpublished data) | Familial British dementia (FBD) and familial Danish dementia (FDD) (collectively known as chromosome 13 dementias) have striking neuropathological similarities to Alzheimer's disease. ABri and ADan can fully activate complement similar to Aβ1–42. Both peptides are degradation products of the same precursor molecule BriPP. | [108,109] |
| Prion protein | Any specific interaction site is not known, it is suspected that it involves the gC1q domain. | Mice genetically deficient in C1q show a significant delay in the onset of scrapie. C1q probably contributes to the early localization of the transmissible spongiform encephalopathies in the lymphoid tissue before neuroinvasion. | [110,111] |
| Apoptotic cells | C1q can bind to the surface blebs of apoptotic cells. All modules can independently bind to apoptotic cells. C1q can also bind via pentraxins. | Surface blebs of apoptotic keratinocytes are concentrated source of autoantigens, which have to be properly cleared before they present a challenge to immune tolerance. C1q may serve as an opsonin, mediating efficient recognition and clearance of apoptotic cells to protect against autoimmunity. C1q deficiency is almost invariably associated with SLE as a result of impaired clearance of apoptotic cells. C1q knock-out mice have glomerulonephritis with immune deposits and show presence of numerous apoptotic bodies in affected glomeruli. | [11,58,60,112,113] |
| PL | C1q binds cardiolipin and other anionic PL. | Possible role in clearance of apoptotic and necrotic cells. | [114] |
| Omp from Gram–ve bacteria, LPS | C1q binds directly to the surfaces of many Gram-negative bacteria in an antibody-independent manner. The gC1q can bind lipid A, LPS and porins. OmpK36 porin from Klebsiella pneumoniae competes directly with IgG for binding to C1q. Binding to lipid A and LPS is mainly via the phosphate groups of lipid A. | Many pathogenic Gram-negative bacteria evade C1q fixation by steric hindrance of C1q binding. | [104,114] |
| HRG | Binding involves both gC1q and CLR domains. Binding site within HRG is unknown. | HRG is an acute phase protein and can regulate/inhibit the formation of insoluble immune complexes. | [115] |
| NIA | The ghB and ghC are the main modules which bind NIA. The gC1q-binding site overlaps with OmpK36-binding site | These are highly glycosylated proteins, forming large aggregates and agglutinating a large variety of bacteria. NIA and gp340 (a glycoprotein that binds C1q, SP-A and SP-D) have identical sequences. NIA/gp340 act as scavenger molecules. | [104,116–118] |
| C1q BP/gC1qR/p33 | Conserved acidic protein, found on a large variety of cells with a unique “donut”-shaped structure. Proposed involvement of ghA (155–164 residues). C1q-binding site is localized on its “S”-surface of gC1qR. Putative C1q binding motifin ligand is located on the A chain between 189 and 201 amino acid residues. (EDEVGQEDEAES). Residues 76 and 93 may also participate in binding C1q. | gC1qR was originally described as a receptor for gC1q domain, which subsequently turned out to be a multi-ligand and multifunctional molecule. gC1qR has recently been shown to be the receptor for internalin B, the invasion protein of Listeria monocytogenes. | [119–125] |
| Heparin | The C1q-binding site for heparin, heparan sulphate, dermatansulfate, keratansulpate, dextran-sulphate, chondroitin 4 sulphate and chondroitin-6-sulphate is partially overlapping with the C1r2s2-binding site; another ligand-binding site within the gC1q domain. | B cells possibly produce a soluble CSPG (proteochon droitin sulfate) that closely resembles the serum-derived C1q inhibitor (C1qI) in structure, which may act as a C1q inhibitor under physiological conditions. | [92,126–129] |
| HNP 1, 2, 3 | Defensins are short cationic peptides from the neutrophil granules. Two possible binding sites are proposed, one in the CLR and another in gC1q. The defensins have Glu–X–Arg–X–Arg motif similar to Gly–X–Lys–X–Lys from IgG. | Defensins are able to modulate complement by both C1q dependent and independent mechanisms. Inhibitory activity of HNP-1 may be a mechanism to control the inflammatory response. The binding site for HNP-1 is probably in the CLR. | [130–132] |
| Fibronectin, fibrin, fibrinogen | The proposed ligand-binding site for fibronectin lies between the CLR and gC1q. On fibronectin, the binding site is the gelatin-binding domain. Both CLR and gC1q domains are involved in binding fibrinogen. | C1q enhances the binding of plasma fibronectin to bacteria. C1q and fibrinogen are capable of high-affinity interactions that may be important to sequester these complexes in tumors and wounds. | [133–136] |
C1q is typically a very efficient scavenging molecule, which mostly modulates pro-inflammatory responses. It has been shown to be involved in neurodegenerative diseases such as Alzheimer's disease, familial dementia and prion disease [8], while its deficiency has been linked to systemic lupus erythematosus (SLE) and glomerulonephritis [9].
3. The ligand recognition domain, gC1q, has a modular organization
As a charge pattern recognition molecule of innate immunity, C1q interacts with the majority of its known ligands via the gC1q domain. Given the heterotrimeric organization of the gC1q, it has been debated whether the carboxy-terminal globular head modules of the human C1q A, B and C chains are functionally autonomous, with ghA, ghB and ghC having distinct binding properties, or that the ability of C1q to bind its ligands is dependent upon a combined, globular structure [10,11]. To address this question, recombinant forms of ghA, ghB and ghC modules have been expressed in bacteria and examined for their specific interactions with ligands known to bind C1q. These include aggregated IgG and IgM, human immunodeficiency virus-1 (HIV-1) transmembrane envelope glycoprotein gp41 peptide 601–613, human T cell lymphtropic virus-I (HTLV-I) gp21 peptide 400–429, β-amyloid peptide (Aβ1–42), and apoptotic cells [11]. The ghA can bind heat-aggregated IgG and IgM, in addition to binding specifically to HIV-1 gp41-derived loop peptide; the ghB prefers aggregated IgG to IgM, in addition to binding βA1–42; whereas the ghC shows preference for IgM as well as HTLV-I gp21 peptide. Both ghA and ghB can independently inhibit C1q-dependent hemolysis of IgG- and IgM-sensitized sheep erythrocytes, ghC being a better inhibitor than ghB in case of IgM-coated erythrocytes [12–14]. Thus, each module within the gC1q domain appears to fold and function with some degree of autonomy, suggesting that they probably evolved as functionally specialized modules.
4. There is a structural basis to the modular organization of the gC1q domain
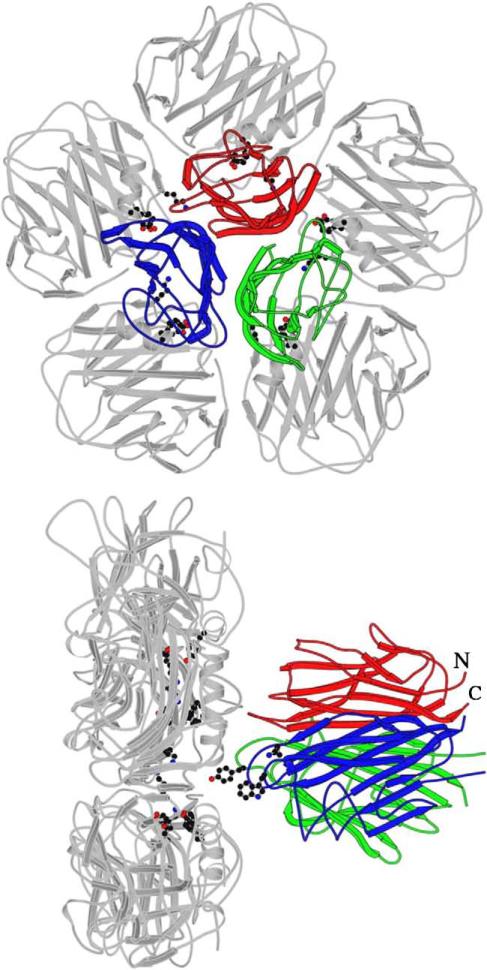
The X-ray crystal structure of the gC1q domain of human C1q (1.9 Å resolution; PDB code: 1PK6), which was based on the structure of adipocyte-specific complement-related protein of 30 kDa (ACRP30) [15] and collagen X [16], has confirmed the modular organization of the domain. The gC1q structure revealed a compact, spherical heterotrimer (50 Å diameter) with a non-crystallographic pseudo-three-fold symmetry. Each of the individual globular head modules, with their N- and C-termini emerging at the base of the trimer, has a jellyroll topology consisting of a 10-stranded β sandwich made up of two 5-stranded anti-parallel β sheets (Fig. 1; ribbon diagram shown in color) [4]. The gC1q is held together predominantly by non-polar interactions, with contributions from a series of interactions along its three-fold axis that include hydrogen bonds, a well-exposed Ca2+ ion located near the apex and main-chain polar interactions. Additional lateral interactions, which are hydrophobic at the base, and polar and hydrophilic towards the apex, further stabilize the heterotrimeric assembly.
Fig. 1.
Schematic illustration of the overall fit between gC1q [4] and one pentameric molecule of CRP [31]. (a) The separation and relative orientation of the two molecules are arbitrary with no specific interactions between the two implied. Highlighted on the CRP molecule are the key residues (Asp112, Lys114, Tyr175) shown by mutagenesis to be involved in the interaction with gC1q [45]. (b) Highlighted on the gC1q molecule (ghA shown in blue, ghB in green and ghC in red) are the residues which interact with CRP following modeling procedures. The starting CRP–gC1q model was designed to facilitate exploration of the potential gC1q interaction with CRP Asp112 and Tyr175 [4]. The gC1q residues shown are ghB–Tyr175, ghA–Lys200 and ghA–Trp147. Figure drawn using MOLSCRIPT [137].
The three modules within gC1q, ghA, ghB and ghC, show clear differences in their electrostatic surface potentials, which in part explains their modularity in terms of ligand recognition [4]. Modules ghA and ghC both show a combination of basic and acidic residues scattered on their external face, whereas module ghB shows a predominance of positive charges, especially a continuous patch of arginine residues (Arg101, Arg114 and Arg129), which have been implicated in the C1q–IgG interaction [17,18]. Several hydrophobic residues are exposed on the external face of each module, but only ghC displays solvent-accessible aromatic residues. Thus, the modular organization of the heterotrimeric assembly, together with different surface charge patterns and the spatial orientation of individual modules, enables gC1q to interact with a diverse range of ligands (Fig. 2) [4,10,11].
Fig. 2.
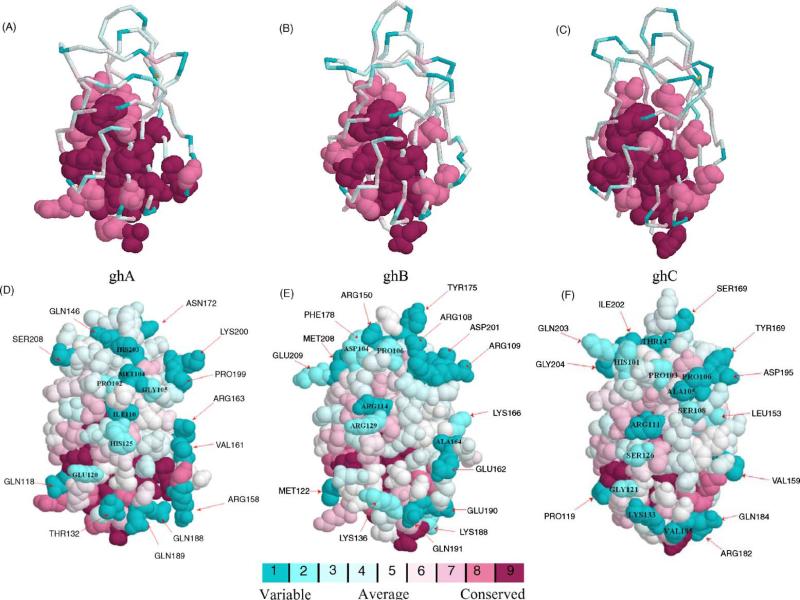
Conserved and variable residues in the gC1q domain. The most conserved residues in the modules ghA, ghB and ghC appear to lie in the lower half of the gC1q domain and towards the subunit–subunit interface (A–C). This core region is shared by all three modules. This is consistent with the role of the lower region of the gC1q domain in being the nucleating center for heterotrimer assembly. The variable residues are markedly different in each module (D–F). Several highly variable residues are labeled. The color-coding for the residues is given below the images. Each color corresponds to a particular rank given by ConSurf. The figure also illustrates how the gC1q domain family has used a common structural core supplanted with variable loop regions to achieve functional diversity.
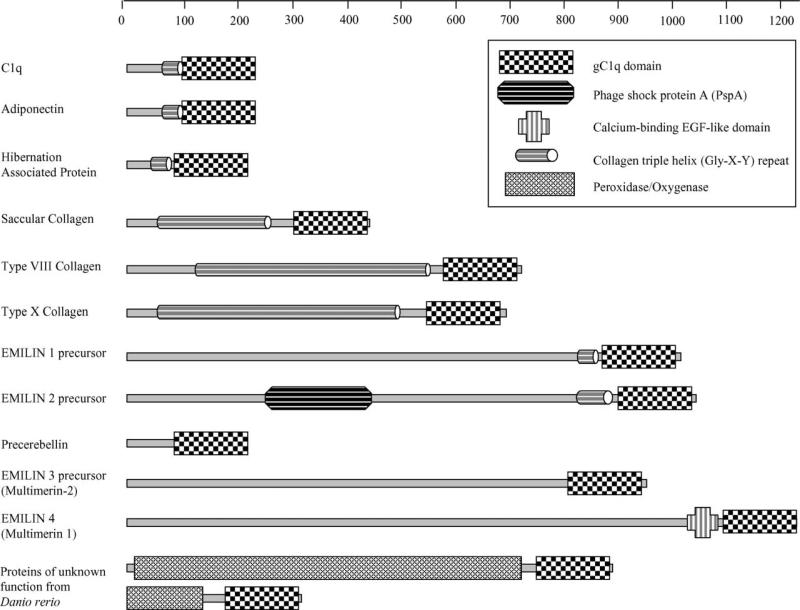
In contrast to this divergence in the individual modules, their core β-sandwich scaffold is well conserved and is also found in a variety of functionally diverse non-complement proteins including collagen VIII and X, precerebellin, hibernation proteins, multimerin, ACRP30/adiponectin, saccular collagen, and elastin microfibril interface located protein (EMILIN) (Fig. 3) [6,7,19], as well as in members of the tumor necrosis factor (TNF) ligand superfamily [15,20,21]. This suggests that although the individual modules of the heterotrimeric gC1q may have evolved with different functional capabilities, the core gC1q three-dimensional structure has been conserved to coordinate the diverse functions of the individual modules and those of other proteins in the C1q family.
Fig. 3.
Domain architecture of proteins containing a gC1q signature domain. The gC1q signature domains are also found in variety of collagen and non-collagen containing proteins, which constitute a novel C1q family. The gC1q are either homotrimeric (type VIII and X collagen, multimerin, ACRP30, and saccular collagen), or heterotrimeric (C1q and hibernation proteins, and probably precerebellin) structures. The crystal structure of the homotrimeric adiponectin (ACRP30/adipoQ), an adipocyte secreted adipokine which is involved in insulin resistance and fatty acid homeostasis, revealed the typical jellyroll fold which is also shared by TNF ligand superfamily members [15]. Three hibernation proteins (HP-20, -25 and -27), which disappear from serum prior to chipmunk hibernation, are also considered to regulate energy homeostasis [138]. Saccular collagen/Otolin, an inner ear structural protein in fish, may be an epicenter for otolith calcification [139–141]. Precerebellin is remarkably similar to ghB module [142]. Type VIII collagen, composed of two gene products (α1 and α2), occurs as homotrimers [143]. Type X collagen is a homotrimer of three α1 (X) chains. EMILIN 1 and 2 are secretory proteins associated with elastic fibres [144]. Multimerin/EMILIN 4, found in platelets and blood vessel endothelium, can form large, variably sized homomultimers. Most members of the C1q family have a triple helical Gly–X–Y collagen repeat, except precerebellin, EMILIN 3 and 4 [145]. EMILIN 2 has a striking PspA (phage shock protein A) domain which is the major antigen of S. pneumoniae [146]. Multimerin 1/EMILIN 4 has an EGF_CA (calcium binding EGF-like domain) which are known to be involved in protein–protein interactions [147]. Two proteins of unknown function from Danio rerio having the gC1q domain also have a peroxidase/oxygenase domain. The domain architectures were obtained from the CDART database [148]. Two additional members, CORS26 and CTRP5 have been described recently. CORS26, which has been implicated in embryonic skeletal development and bore tumors, is considered a candidate susceptibility gene in the development of arthritis [149,150]. The gC1a domain of CTRP5 is considered to be involved in the formation of an extracellular hexagonal lattice between retiral pigment epithelium and Bruch's membrane [151]. A substitution mutation within the gC1q domain of CTRP5 has been linked to late-onset retinal degeneration [151].
5. The ghB module appears to have a central role in the C1q–IgG interaction
Human C1q shows only weak binding to the Fc regions of non-aggregated IgG (functional affinity constant, K = 4 × 103 M–1 to 5 × 104 M–1) [22–24]. However, when presented as multiple, closely spaced Fc regions, as found in IC, the strength of binding of the hexameric C1q to IgG increases dramatically (K = 107 M–1 to 108 M–1) [24]. This interaction is predominantly ionic in nature [25].
Various research groups have attempted to identify the IgG residues involved in binding gC1q. Initially, three residues from murine IgG2b, Glu318, Lys320, and Lys322, which are conserved across the different IgG isotypes, were proposed as being important, but not all isotypes fix complement [26]. Studies by Idusogie et al. using a chimeric monoclonal antibody rituximab, which contains a human IgG1 constant region, have highlighted that the region containing Glu318, Lys320 and Lys322 is likely to be one of several possible C1q binding sites. However, alanine substitutions at positions Glu318 and Lys320 in rituximab had no serious effect on C1q binding or complement activation. Instead, alanine substitution at positions Asp270, Lys322, Pro329 and Pro331 significantly reduced the ability of the chimera to bind C1q and activate complement [27,28]. Gaboriaud et al. [4] have proposed that the gC1q may also bind to the Fab region through interactions with the light chains. Consequently, apart from identifying some of the critical IgG residues involved in the C1q–IgG interaction, all of these studies imply that the structure of the hinge region in the various IgG isotypes, which provides them with different levels of flexibility in the region near the Cγ2 domain, may affect access of the C1q molecule to the IgG binding site.
The complementary IgG binding sites on the gC1q domain have recently been identified via mutational analysis [18] and molecular modeling based on crystallographic data [4]. Experiments involving chemical modification of arginine and histidine residues and subsequent cross-linking to heterologous IgG have implicated Arg114, Arg129 and Arg163 in ghB, Arg162 in ghA and Arg156 in ghC in this interaction [17]. Site-directed mutagenesis studies, however, have given more precise clues on the nature of the gC1q–IgG interaction [18]. These involved engineering a series of single residue mutations in individually expressed modules, such as substituting specific arginine (R) residues with either alanine (A), glutamate (E) or glutamine (Q), or replacing histidine (H) residues with either alanine (A) or aspartate (D), and then comparing the ability of these mutant modules to compete with their wild-type counterparts in binding heat aggregated IgG. Substitutions with negatively charged glutamate or aspartate residues led to a larger decrease in the IgG binding by the mutants compared to their corresponding alanine substitutions, confirming the ionic nature of this interaction. Substitution of Arg114 of ghB with either alanine (ghB-R114A) or glutamate (ghB-R114E) caused maximum reduction in IgG binding (~50%), highlighting a major role for Arg114 of the human C1q B chain in the C1q–IgG interaction. A further mutation of Arg114 to glutamine confirmed that the reduction in binding was due to the removal of the arginine residue rather than the physical properties of either the alanine or the glutamate residues. Wild-type ghA and ghC modules both significantly inhibit IgG binding by C1q. The substitution of Arg162 of ghA and Arg156 of ghC with alanine (ghA-R162A, ghC-R156A), or glutamate (ghA-R162E, ghC-R156E), led to ~20% reduction in this inhibition, which indicates that although they are probably involved in the IgG–ghA and IgG–ghC binding regions, they may not be the critical IgG binding residues of these modules. Thus, these mutational studies not only suggest that the ghB module has a dominant role in the gC1q–IgG interaction, but that Arg114 of ghB is the key residue, and Arg129, Arg163 and His117 of the same module have complementary roles.
The crystal structure of the gC1q of human C1q has revealed that Arg162 of ghA and Arg156 of ghC are engaged in internal salt bridges with Asp191 of ghA and Glu187 of ghC, respectively, and hence are unlikely contributors to the C1q–IgG interaction within a heterotrimer [4]. However, the ghB module, which is the most accessible of the three modules, has a predominantly positively charged outer surface distinguished by the presence of the three basic amino acids: Arg101, Arg114 and Arg129, two of which have already been described as important in the IgG–C1q interaction [17]. Even though in the crystal structure Arg114 and Arg129 are shown to have ordered structures, this is probably due to the stabilizing effects of crystal contacts and like Arg163 they are likely to be available for ligand binding in solution. Thus, the most attractive structural model which attempts to describe C1q–IgG interaction positions the two molecules in such a way that Asp270 and Lys322 of IgG form salt bridges with Arg129 and Glu162 of the ghB, respectively, with additional ionic interactions provided by Arg114 and Arg161 of ghB. In this orientation, the Arg129 appears to ‘act like a wedge’ between the Cγ2 and the light chain constant (CL) domains [4]. Gaboriaud et al. [4] have contemplated that the gC1q may bind to the Fab region as well through interactions with the CL domain. Thus, Fab/Fc orientation may be a critical factor in dictating access of the ghB module to Cγ2 domain. This is consistent with the biochemical studies which suggest a limiting role of the hinge region, and hence isotype specificity in the C1q–IgG interaction.
6. The interaction between gC1q and C-reactive protein-complexes involves multiple modules
The versatile nature of C1q is further demonstrated by its interaction with the complexes of human C-reactive protein (CRP), a major acute phase protein, which activates the classical complement pathway [29,30]. The CRP molecule has five identical, 206 amino acid long subunits, held together through non-covalent interactions and arranged with pentameric symmetry around a central pore (Fig. 1). The crystal structures of the protein reveal a flattened jellyroll appearance for each subunit (A, B, C, D and E) that is made up of two anti-parallel β-sheets and a single short α-helix [31,32]. CRP readily forms complexes, in a Ca2+-dependent manner, to substances with exposed phosphocholine (PCh) groups, such as C-polysaccharide (CPS) of the cell wall of Streptococcus pneumoniae, oxidized low-density lipoprotein (oxLDL), and apoptotic cells [33–37]. The PCh-binding site, located at the Ca2+-binding site, is present on each CRP subunit. All subunits have the same relative orientation in the pentamer and all five PCh-binding sites fall on the same pentameric face, commonly known as the ‘recognition face’. The pentameric surface opposite to the recognition face of CRP is designated as the ‘effector face’ of the protein [29–32].
Activation of the classical complement pathway initiated by the binding of C1q to ligand-bound CRP is reported to participate in several proposed anti-inflammatory actions of CRP such as phagocytosis of apoptotic cells and protection from S. pneumoniae infection [35,38–40]. However, the mechanism of action of CRP following C1q binding is not completely understood, as CRP–C1q-mediated complement activation does not proceed to completion. The process generates C3 convertase but does not result in the formation of an effective C5 convertase. Therefore, pro-inflammatory molecules like C5a are not produced and assembly of the MAC of the complement pathway is prevented [41–43]. Presumably, the ligands bound to CRP and opsonized by C3 fragments alone under the actions of C3 convertase are targeted for opsonophagocytosis. Thus, C1q assists CRP in playing its opsonic role and in acting as an anti-inflammatory acute phase protein of the innate immune system.
While no structural detail for CRP–ligand complexes, which effect C1q-binding and complement activation, has yet been reported, the C1q-binding region on CRP has been defined. Site-directed mutagenesis initially identified residues Asp112 and Lys114 as important in the CRP–C1q interaction [44]. Subsequently, the crystal structure [31] revealed a striking extended cleft on the effector face of CRP, which starts at about the center of each subunit and extends its edge to the central pore of the pentamer. The cleft, which is deep and narrow at its origin but opens up and becomes wider and shallow towards the center of the pentameric ring, incorporates Asp112 and was proposed as the possible C1q binding site [31]. Subsequent site-directed mutagenesis of amino acids located in the CRP cleft revealed that the wider and shallow end of the cleft, which is close to the pore, provided accommodation for C1q [45]. The wider portion of the cleft is bound by the 112–114 loop on one side, the 86–92 loop, the C-terminus, and Tyr175 of the helix 169–176 on the other. Tyr175, Asp112, Glu88, His38, and Asn158 in each CRP subunit contribute to the structure and topology of the C1q-binding site [45]. Asp112 and Tyr175 appear to be the contact residues and participate directly in CRP–C1q interactions. Glu88 influences the conformational change of C1q necessary for complement activation, while His38 and Asn158 probably contribute to the geometry of the binding site. In addition, Lys114, possibly from two adjacent subunits, also affects binding of CRP to C1q with the positive charge of Lys114 hindering binding of C1q [44,45]. Molecular modeling has suggested that this may be due to the C1q Lys residues directed towards the negatively charged residues of the CRP pore [4]. The conformation of the CRP cleft is not changed by the presence or absence of Ca2+ at the recognition face [31,46] consistent with reports that CRP-polycation complexes, that do not require either Ca2+ or the critical residues from the PCh-binding site, are avidly recognized by C1q [37,47].
Deposition of CRP on an appropriate multivalent ligand-bearing surface through the recognition face will present the effector face for C1q recognition, and it is now clear that the gC1q, rather than the CLR [48], is recognized and bound [45]. Further evidence is provided by long pentraxin 3 (PTX3), a protein containing a CRP homology domain, which has recently been shown to bind the gC1q domain as well as recombinant ghA, ghB and ghC modules [49]. Agrawal et al. [45] suggested that only one gC1q could be accommodated per CRP pentamer and that the binding site involved more than one CRP subunit. Assuming that efficient CRP–C1q binding and complement activation requires multiple gC1q interactions for each C1q molecule, the presence of five equivalent gC1q binding sites on each of an array of CRP pentamers will provide the opportunity for multiple interactions without the need for structural rearrangement [45]. C1q binding by CRP is effected only by the binding of CRP to an appropriate physiological ligand and it has been suggested that this results in a conformational change in the CRP structure, either within each subunit or in the form of a change in their relative disposition within the pentameric ring [31,45]. This conformational change may fully open the C1q recognition site facilitating relaxed binding of C1q for further interactions with the components of the complement pathway.
The recent determination of the structure of gC1q has provided further evidence for the proposed gC1q–CRP interaction both directly and in the form of a model of CRP complexed with gC1q [4]. The features of this model are informative with regard to the structure and topology of the CRP-binding site on gC1q. The effector face of one pentamer of CRP can accommodate a single gC1q domain with its crown docked within the negatively-charged central pore and its surrounds, resulting in contact between key CRP residues and the predominantly basic crown of gC1q (Fig. 1) [45]. Since CRP has a five-fold axis of symmetry, gC1q can interact with CRP in one of five equivalent orientations. The starting point for the refinement of the model of the CRP–gC1q interaction aligned basic residues of gC1q with the CRP cleft region and CRP Asp112 and Tyr175 in particular. In the resulting best-fit model Gaboriaud et al. [4] report that CRP Tyr175 (subunits A and D) is within H-bond distance of gC1q residues Tyr175 (ghB) and Lys200 (ghA) respectively, with CRP Tyr175 (subunit E) at 4Å from the gC1q Trp147 (ghA). The proposed model was found to be sterically restrained, consistent with the requirement for ligand-bound CRP and associated conformational change within either the protomer or the pentameric ring [4,31,45].
7. Highly variable residues on the gC1q surface may confer differential ligand specificity to each module
The ability of C1q to bind a large variety of ligands is the key factor influencing the range of biological processes affected by it (Tables 1 and 2). This, together with the specific binding properties of the ghA, ghB and ghC modules, gives another dimension to the functional versatility of the C1q molecule. However, the basis for binding all these ligands is still unclear, except for IgG and CRP. Since the X-ray crystal structure of the native gC1q domain of human C1q is available [4], we analyzed modules ghA, ghB and ghC individually, using a computer program called ‘ConSurf’, in order to identify functionally critical residues. Given the three-dimensional structure of a protein domain as an input, ConSurf [50] carries out a search for close homologues of the protein in the SWISS-PROT database [51] using PSI-BLAST [52]. Once the homologues have been identified, a multiple sequence alignment is created using CLUSTALW [53]. A phylogenetic tree for the homologous sequences is built using this alignment. ConSurf then calculates conservation scores for each residue position. These conservation scores are subsequently mapped onto the three-dimensional structure in order to provide a view of the conserved and variable residues of the protein.
ConSurf is usually used to identify residues that are highly conserved within a protein family. However, we believe that in a group of proteins that have a conserved three-dimensional structure but have distinct functions, the functionally important residues will be the most different. Therefore, we used ConSurf to identify the most variable residues within each C1q module compared to a set of other gC1q containing proteins (Fig. 3). We concentrated on the regions that were more variable in the alignment of each module with the rest of the members of the C1q family (Table 3). In order to ensure reliable identification of functionally important residues, several runs with increasing number of homologues were performed using a high E-value cut-off of 0.001. A separate run using a curated multiple sequence alignment for the gC1q domain with the 12 homologues available from the PFAM database of protein families was also used [54].
Table 3.
Most variable residues within gC1q identified by ConSurf
| ghA | ghB | ghC | ||||
|---|---|---|---|---|---|---|
| 1 | ● PRO2 | 102 | ●ASN2 | 104 | ● HIS 2 | 101 |
| 2 | ● MET1 | 104 | ● PRO 2 | 106 | ● PRO 2 | 103 |
| 3 | ●GLY2 | 105 | ● ARG 1 | 108 | ● ALA 1 | 105 |
| 4 | ILE 1 | 110 | ● ARG 1 | 109 | ● PRO 1 | 106 |
| 5 | ○GLN1 | 118 | ARG 1 ★ | 114 | ●SER2 | 108 |
| 6 | ○GLU2 | 120 | MET 1 | 122 | ● ARG 1 | 111 |
| 7 | HIS 2 | 125 | ARG 2 ★ | 129 | ○ PRO 1 | 119 |
| 8 | THR1 | 132 | LYS 2 | 136 | ○GLY2 | 121 |
| 9 | GLN1 | 146 | ARG 1 | 150 | SER2 | 126 |
| 10 | SER2 | 152 | ●GLU1★ | 162 | LYS 1 | 133 |
| 11 | ● ARG 1 | 158 | ● ALA 1 | 164 | THR1 | 147 |
| 12 | ● VAL 1 | 161 | ● LYS 2 | 166 | LEU 2 | 153 |
| 13 | ● ARG 1 | 163 | CYS 2 | 171 | VAL 1 | 159 |
| 14 | CYS 1 | 168 | ● TYR 1 ◀ | 175 | ● CYS 1 | 165 |
| 15 | ASN1 | 172 | ●PHE2 | 178 | ●SER1 | 169 |
| 16 | ●GLN2 | 186 | ○ LYS 2 | 188 | ●ASN2 | 172 |
| 17 | ●GLN1 | 188 | ○GLU1 | 190 | ○ ARG 1 | 182 |
| 18 | ●GLN1 | 189 | ○GLN1 | 191 | ○GLN1 | 184 |
| 19 | ○ PRO 1 | 199 | ●ASP1 | 201 | ○ VAL 1 | 185 |
| 20 | ○ LYS 1 ◀ | 200 | ● LYS 1 | 202 | ●ASP1 | 195 |
| 21 | ○ HIS 1 | 203 | ○ MET 1 | 208 | ● TYR 1 | 196 |
| 22 | SER1 | 208 | ○GLU2 | 209 | ○GLY2 | 201 |
| 23 | ○GLY1 | 210 | ○ ILE 1 | 202 | ||
| 24 | ○GLN2 | 203 | ||||
| 25 | ○GLY1 | 204 |
(★) Residues known to be involved in IgG binding; (◀) residues proposed to be involved in CRP binding. Residues are sorted on the basis of position. (●) and (○) indicate possible important residue clusters for ligand binding. Clusters are defined wherever residues are at/within 3-residue distance from each other. Hydrophobic and ring residues are underlined. Arginine and lysine residues are shown in boldface. Numbers in superscript indicate the “rank” of the residue given by ConSurf (1–9, most variable to most conserved, see also Fig. 2). The most variable residues (ranks 1 and 2) are shown here. Only the gC1q domain was used in the analysis.
As expected, nearly all the residues identified as highly variable appeared to lie on the surface of gC1q heterotrimer (Fig. 2). ConSurf identified ghA–Lys200 and ghB–Tyr175 as highly variable residues, which have been proposed to be involved in the gC1q–CRP interaction [4]. The ghB–Arg114, ghB–Arg129 and ghB–Glu162, which are considered central to the gC1q–IgG interaction, were also identified as highly variable. The ConSurf program concurs with different experimental and computational studies that suggest that the ghA–Arg162 has only a complementary role in binding IgG as a soluble monomer. However, two adjoining residues (ghA–Val161 and ghA–Arg163) were identified as functionally significant. The binding site of HTLV-I gp21 syncytium peptide is believed to be hydrophobic in nature. The ghC–His101, ghC–Pro103, ghC–Ala105 and ghC–Pro106 form a hydrophobic patch on the surface of the ghC module. It is possible that these residues are important in binding HTLV-I gp21 peptide. It is interesting to note that there are five arginine and four lysine residues identified by ConSurf as most variable in the ghB module compared to two arginine and one lysine residues in each of the ghA and ghC modules. It is likely that the ability of C1q to bind polyanions is largely dependent on the ghB module.
8. Conclusions and perspectives
It has long been debated whether the heterotrimeric globular head region of C1q needed a combined effort from all three chains (ghA, ghB and ghC) for its recognition properties. It is increasingly becoming evident that each module within the gC1q domain has a fair degree of structural and functional autonomy. The concept of modularity within the gC1q domain, initially considered provocative, has recently been supported by the gC1q crystal structure and molecular modeling. The spatial orientation of the three modules appears to offer flexibility of ligand recognition to the heterotrimeric gC1q domain. This is illustrated by the equatorial position of the ghB module, which seems best placed to interact with IgG. The proposed involvement of the Fab/Fc interface and a direct access of IgG-bound antigen to C1q are exciting additions to the knowledge of gC1q–IgG interaction.
The structural and functional anatomy of gC1q, combined with the studies on C1q–CRP interactions, provides important clues on the functional consequences of binding of PCh-containing substances (bacteria, modified LDL, apoptotic, damaged and necrotic cells) to CRP. The multiple binding of PCh, as part of a physiological ligand, to CRP may bring CRP pentamers close together, thus facilitating C1q binding and complement activation. If a slight structural change in CRP is necessary as proposed, it may occur simultaneously with the binding of multiple gC1q domains from one molecule of C1q to multiple CRP molecules arranged on the PCh-rich ligand.
The C1q–IgG interaction activates the classical complement pathway leading to the generation of MAC, whereas interaction between C1q and ligand-bound CRP activates complement to form C3 convertases with no generation of MAC. The circumstances for the failure of PCh–CRP–C1q complex to lead to the formation of MAC are unresolved, although a Factor H-dependent mechanism has been indicated [35,42]. Here we present two possible scenarios. In the first, a PCh-containing target such as a damaged cell, which is decorated with bound CRP, is unlikely to offer enough space on its surface for the assembly of the multi-protein complex MAC because of its huge molecular architecture. Indeed, it has been shown that CRP protects assembly of MAC on the targets bound to CRP [35,55]. The extent of complement activation by CRP complexed with a target with an irregular distribution of fewer PCh moieties has yet to be determined. The second possibility revolves around the hypothesis that C1q recognizes CRP and IgG through different mechanisms, which is supported by the recent modeling data [4]. In the two proposed recognition modes of gC1q, IgG is recognized by the equatorial region of a single chain of gC1q (ghB) while CRP is recognized by the top of the gC1q involving all three chains (ghA + ghB + ghC). It is possible that the orientation of gC1q on ligand-bound CRP is central to the inability of CRP-activated complement to proceed through C3 to MAC.
The availability of the recombinant forms of the wild-type and mutant ghA, ghB and ghC modules has made it possible to examine their interactions with a range of ligands which are known to bind via gC1q. Site directed mutagenesis of highly variable residues identified using ConSurf may lead to a better definition of ligand binding sites in each of the modules. A tentative assembly of C1q homotrimers in silico has been shown to result in severe steric hindrances, especially at the level of lateral contacts, providing a structural basis for the assembly of protomers only as heterotrimers [4]. This raises the possibility that denaturation and renaturation procedures using equimolar concentrations of ghA, ghB and ghC modules could yield a recombinant heterotrimeric gC1q domain. Generation of such a molecule could allow inclusion of the point mutants of ghA, ghB or ghC modules individually. This would facilitate the study of the effects of single residue mutations on the structure-function relationship, and structure determination of the gC1q complexed with a range of ligands.
Acknowledgements
The authors are funded by the European Commission (PW, UK), the German National Genome Network (NGFN; TC, UK, RG), the Alexander van Humboldt Foundation (UK), the University of Padova (DMB), Bulgarian National Scientific Foundation (MSK; Grant 2349), the Wellcome Trust (TJG, AKS), and the National Institutes of Health (AA; Grant RO1HL071233). RG is supported by a grant from the Deutsche Forschungsgemeinschaft through the Graduierteu Kolleg Gk370.
Abbreviations
- gC1q
globular domain of C1q and similar signature domains of members of the C1q family
- ghA, ghB, ghC
the carboxy-terminal, globular head regions of C1q A, B and C chains, respectively
- CLR
collagen-like region
- MAC
membrane attack complex
- DC
dendritic cell
- LPS
lipopolysaccharides
- PL
phospholipids
- SLE
systemic lupus erythematosus
- HIV
human immunodeficiency virus
- HTLV
human T-cell lymphotrophic virus
- Aβ1–42
β-amyloid peptide residues 1–42
- ACRP30
adipocyte-specific complement-related protein of 30 kDa
- EMILIN
elastin microfibril interface located protein
- TNF
tumor necrosis factor
- IC
immune complex
- CL
light chain constant region
- CRP
human C-reactive protein
- PCh
phosphocholine
- CPS
C-polysaccharide
- oxLDL
oxidized low-density lipoprotein
- PTX3
pentraxin 3
- PspA
phage shock protein A
- EGF_CA
epidermal growth factor like module
- MCP
monocyte chemoattractant peptide
- MAPK
mitogen-activated protein kinase
- SAP
serum amyloid protein
- FBD
familial British dementia
- FDD
familial Danish dementia
- HRG
histidine-rich glycoprotein
- NIA
non-immunoglobulin salivary agglutinin
- Omp
outer membrane protein
- HNP
human neutrophils peptide
References
- 1.Arlaud GJ, Gaboriaud C, Thielens NM, Rossi V, Bersch B, Hernandez JF, Fontecilla-Camps JC. Structural biology of C1: dissection of a complex molecular machinery. Immunol Rev. 2001;180:136–45. doi: 10.1034/j.1600-065x.2001.1800112.x. [DOI] [PubMed] [Google Scholar]
- 2.Arlaud GJ, Gaboriaud C, Thielens NM, Budayova-Spano M, Rossi V, Fontecilla-Camps JC. Structural biology of the C1 complex of complement unveils the mechanisms of its activation and proteolytic activity. Mol Immunol. 2002;39:383–94. doi: 10.1016/s0161-5890(02)00143-8. [DOI] [PubMed] [Google Scholar]
- 3.Arlaud GJ, Gaboriaud C, Thielens NM, Rossi V. Structural biology of C1. Biochem Soc Trans. 2002;30:1001–6. doi: 10.1042/bst0301001. [DOI] [PubMed] [Google Scholar]
- 4.Gaboriaud C, Juanhuix J, Gruez A, Lacroix M, Darnault C, Pignol D, Verger D, Fontecilla-Camps JC, Arlaud GJ. The crystal structure of the globular head of complement protein C1q provides a basis for its versatile recognition properties. J Biol Chem. 2003;278:46974–82. doi: 10.1074/jbc.M307764200. [DOI] [PubMed] [Google Scholar]
- 5.Sellar GC, Blake DJ, Reid KBM. Characterization and organization of the genes encoding the A-B- and C-chains of human complement subcomponent C1q. The complete derived amino acid sequence of human C1q. Biochem J. 1991;274:481–90. doi: 10.1042/bj2740481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kishore U, Reid KBM. Modular organization of proteins containing C1q-like globular domain. Immunopharmacology. 1999;42:15–21. doi: 10.1016/s0162-3109(99)00011-9. [DOI] [PubMed] [Google Scholar]
- 7.Kishore U, Gaboriaud C, Waters P, Shrive AK, Greenhough TJ, Sim RB, Arlaud GJ, Reid KBM. C1q and Tumor Necrosis Factor Superfamily: Modularity and Versatility. Trend Immunol. 2004 doi: 10.1016/j.it.2004.08.006. In Press. [DOI] [PubMed] [Google Scholar]
- 8.van Beek J, Elward K, Gasque P. Activation of complement in the central nervous system: roles in neurodegeneration and neuroprotection. Ann NY Acad Sci. 2003;992:56–71. doi: 10.1111/j.1749-6632.2003.tb03138.x. [DOI] [PubMed] [Google Scholar]
- 9.Botto M, Walport MJ. C1q, autoimmunity and apoptosis. Immuno-biology. 2002;205:395–406. doi: 10.1078/0171-2985-00141. [DOI] [PubMed] [Google Scholar]
- 10.Kishore U, Kojouharova MS, Reid KBM. Recent progress in the understanding of the structure-function relationships of the globular head regions of C1q. Immunobiology. 2002;205:355–64. doi: 10.1078/0171-2985-00138. [DOI] [PubMed] [Google Scholar]
- 11.Kishore U, Gupta SK, Perdikoulis MV, Kojouharova MS, Urban BC, Reid KBM. Modular organization of the carboxyl-terminal, globular head region of human C1q A, B, and C chains. J Immunol. 2003;171:812–20. doi: 10.4049/jimmunol.171.2.812. [DOI] [PubMed] [Google Scholar]
- 12.Kishore U, Leigh LE, Eggleton P, Strong P, Perdikoulis MV, Willis AC, Reid KBM. Functional characterization of a recombinant form of the C-terminal, globular head region of the B-chain of human serum complement protein C1q. Biochem J. 1998;333:27–32. doi: 10.1042/bj3330027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kojouharova MS, Panchev ID, Tchorbadjieva MI, Reid KBM, Hoppe HJ. Differential binding of IgG and of a HIV gp41 peptide by the B chain and A chain globular head sequences of C1q, respectively. J Immunol. 1998;161:4325–31. [PubMed] [Google Scholar]
- 14.Kishore U, Strong P, Perdikoulis MV, Reid KBM. A recombinant homotrimer, composed of the α-helical neck region of human surfactant protein D and C1q B chain globular domain, is an inhibitor of the classical complement pathway. J Immunol. 2001;166:559–65. doi: 10.4049/jimmunol.166.1.559. [DOI] [PubMed] [Google Scholar]
- 15.Shapiro L, Scherer PE. The crystal structure of a complement-1q family protein suggests an evolutionary link to tumor necrosis factor. Curr Biol. 1998;8:335–8. doi: 10.1016/s0960-9822(98)70133-2. [DOI] [PubMed] [Google Scholar]
- 16.Bogin O, Kvansakul M, Rom E, Singer J, Yayon A, Hohenester E. Insight into Schmid metaphyseal chondrodysplasia from the crystal structure of the collagen X NC1 domain trimer. Structure (Camb) 2002;10:165–73. doi: 10.1016/s0969-2126(02)00697-4. [DOI] [PubMed] [Google Scholar]
- 17.Marques G, Anton LC, Barrio E, Sanchez A, Ruiz S, Gavilanes F, Vivanco F. Arginine residues of the globular regions of human C1q involved in the interaction with immunoglobulin G. J Biol Chem. 1993;268:10393–402. [PubMed] [Google Scholar]
- 18.Kojouharova MS, Gadjeva MG, Tsacheva IG, Zlatarova A, Roumenina LT, Tchorbadjieva MI, Atanasov BP, Waters P, Urban BC, Sim RB, Reid KBM, Kishore U. Mutational analyses of the recombinant globular regions of human C1q A, B, and C chains suggest an essential role for arginine and histidine residues in the C1q–IgG interaction. J Immunol. 2004;172:4351–8. doi: 10.4049/jimmunol.172.7.4351. [DOI] [PubMed] [Google Scholar]
- 19.Kishore U, Reid KBM. C1q: structure, function, and receptors. Immunopharmacology. 2000;49:159–70. doi: 10.1016/s0162-3109(00)80301-x. [DOI] [PubMed] [Google Scholar]
- 20.Jones EY, Stuart DI, Walker NP. Structure of tumour necrosis factor. Nature. 1989;338:225–8. doi: 10.1038/338225a0. [DOI] [PubMed] [Google Scholar]
- 21.Eck MJ, Beutler B, Kuo G, Merryweather JP, Sprang SR. Crystallization of trimeric recombinant human tumor necrosis factor (cachectin). J Biol Chem. 1988;263:12816–9. [PubMed] [Google Scholar]
- 22.Sledge CR, Bing DH. Binding properties of the human complement protein Clq. J Biol Chem. 1973;248:2818–23. [PubMed] [Google Scholar]
- 23.Schumaker VN, Calcott MA, Spiegelberg HL, Muller-Eberhard HJ. Ultracentifuge studies of the binding of IgG of different subclasses to the Clq subunit of the first component of complement. Biochemistry. 1976;15:5175–81. doi: 10.1021/bi00668a035. [DOI] [PubMed] [Google Scholar]
- 24.Hughes-Jones NC, Gardner B. The reaction between the complement subcomponent C1q. IgG complexes and polyionic molecules. Immunology. 1978;34:459–63. [PMC free article] [PubMed] [Google Scholar]
- 25.Burton DR, Boyd J, Brampton AD, Easterbrook-Smith SB, Emanuel EJ, Novotny J, Rademacher TW, van Schravendijk MR, Sternberg MJ, Dwek RA. The Clq receptor site on immunoglobulin G. Nature. 1980;288:338–44. doi: 10.1038/288338a0. [DOI] [PubMed] [Google Scholar]
- 26.Duncan AR, Winter G. The binding site for C1q on IgG. Nature. 1988;332:738–40. doi: 10.1038/332738a0. [DOI] [PubMed] [Google Scholar]
- 27.Idusogie EE, Presta LG, Gazzano-Santoro H, Totpal K, Wong PY, Ultsch M, Meng YG, Mulkerrin MG. Mapping of the C1q binding site on rituxan, a chimeric antibody with a human IgG1 Fc. J Immunol. 2000;164:4178–84. doi: 10.4049/jimmunol.164.8.4178. [DOI] [PubMed] [Google Scholar]
- 28.Idusogie EE, Wong PY, Presta LG, Gazzano-Santoro H, Totpal K, Ultsch M, Mulkerrin MG. Engineered antibodies with increased activity to recruit complement. J Immunol. 2001;166:2571–5. doi: 10.4049/jimmunol.166.4.2571. [DOI] [PubMed] [Google Scholar]
- 29.Volanakis JE. Human C-reactive protein: expression, structure, and function. Mol Immunol. 2001;38:189–97. doi: 10.1016/s0161-5890(01)00042-6. [DOI] [PubMed] [Google Scholar]
- 30.Hirschfield GM, Pepys MB. C-reactive protein and cardiovascular disease: new insights from an old molecule. QJM. 2003;96:707–93. doi: 10.1093/qjmed/hcg134. [DOI] [PubMed] [Google Scholar]
- 31.Shrive AK, Cheetham GM, Holden D, Myles DA, Turnell WG, Volanakis JE, Pepys MB, Bloomer AC, Greenhough TJ. Three dimensional structure of human C-reactive protein. Nat Struct Biol. 1996;3:346–54. doi: 10.1038/nsb0496-346. [DOI] [PubMed] [Google Scholar]
- 32.Thompson D, Pepys MB, Wood SP. The physiological structure of human C-reactive protein and its complex with phosphocholine. Structure Fold Des. 1999;7:169–77. doi: 10.1016/S0969-2126(99)80023-9. [DOI] [PubMed] [Google Scholar]
- 33.Bhakdi S, Torzewski M, Klouche M, Hemmes M. Complement and atherogenesis: binding of CRP to degraded, nonoxidized LDL enhances complement activation. Arterioscler Thromb Vasc Biol. 1999;19:2348–54. doi: 10.1161/01.atv.19.10.2348. [DOI] [PubMed] [Google Scholar]
- 34.Chang MK, Binder CJ, Torzewski M, Witztum JL. C-reactive protein binds to both oxidized LDL and apoptotic cells through recognition of a common ligand: Phosphorylcholine of oxidized phospholipids. Proc Natl Acad Sci USA. 2002;99:13043–8. doi: 10.1073/pnas.192399699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gershov D, Kim S, Brot N, Elkon KB. C-Reactive protein binds to apoptotic cells, protects the cells from assembly of the terminal complement components, and sustains an antiinflammatory innate immune response: implications for systemic autoimmunity. J Exp Med. 2000;192:1353–64. doi: 10.1084/jem.192.9.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nauta AJ, Daha MR, Van Kooten C, Roos A. Recognition and clearance of apoptotic cells: a role for complement and pentraxins. Trend Immunol. 2003;24:148–54. doi: 10.1016/s1471-4906(03)00030-9. [DOI] [PubMed] [Google Scholar]
- 37.Claus DR, Siegel J, Petras K, Skor D, Osmand AP, Gewurz H. Complement activation by interaction of polyanions and polycations. III. Complement activation by interaction of multiple polyanious and polycations in the presence of C-reactive protein. J Immunol. 1977;118:83–7. [PubMed] [Google Scholar]
- 38.Szalai AJ, Briles DE, Volanakis JE. Role of complement in C-reactive-protein-mediated protection of mice from Streptococcus pneumoniae. Infect Immun. 1996;64:4850–3. doi: 10.1128/iai.64.11.4850-4853.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown JS, Hussell T, Gilliland SM, Holden DW, Paton JC, Ehrenstein MR, Walport MJ, Botto M. The classical pathway is the dominant complement pathway required for innate immunity to Streptococcus pneumoniae infection in mice. Proc Natl Acad Sci USA. 2002;99:16969–74. doi: 10.1073/pnas.012669199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mold C, Rodic-Polic B, Du Clos TW. Protection from Streptococcus pneumoniae infection by C-reactive protein and natural antibody requires complement but not Fcγ receptors. J Immunol. 2002;168:6375–81. doi: 10.4049/jimmunol.168.12.6375. [DOI] [PubMed] [Google Scholar]
- 41.Volanakis JE. Complement activation by C-reactive protein complexes. Ann N Y Acad Sci. 1982;389:235–50. doi: 10.1111/j.1749-6632.1982.tb22140.x. [DOI] [PubMed] [Google Scholar]
- 42.Mold C, Gewurz H, Du Clos TW. Regulation of complement activation by C-reactive protein. Immunopharmacology. 1999;42:23–30. doi: 10.1016/s0162-3109(99)00007-7. [DOI] [PubMed] [Google Scholar]
- 43.Berman S, Gewurz H, Mold C. Binding of C-reactive protein to nucleated cells leads to complement activation without cytolysis. J Immunol. 1986;136:1354–9. [PubMed] [Google Scholar]
- 44.Agrawal A, Volanakis JE. Probing the C1q-binding site on human C-reactive protein by site-directed mutagenesis. J Immunol. 1994;152:5404–10. [PubMed] [Google Scholar]
- 45.Agrawal A, Shrive AK, Greenhough TJ, Volanakis JE. Topology and structure of the C1q-binding site on C-reactive protein. J Immunol. 2001;166:3998–4004. doi: 10.4049/jimmunol.166.6.3998. [DOI] [PubMed] [Google Scholar]
- 46.Ramadan MA, Shrive AK, Holden D, Myles DA, Volanakis JE, DeLucas LJ, Greenhough TJ. The three-dimensional structure of calcium-depleted human C-reactive protein from perfectly twinned crystals. Acta Crystallogr D Biol Crystallogr. 2002;58:992–1001. doi: 10.1107/s0907444902005693. [DOI] [PubMed] [Google Scholar]
- 47.Agrawal A, Simpson MJ, Black S, Carey MP, Samols D. A C-reactive protein mutant that does not bind to phosphocholine and pneumococcal C-polysaccharide. J Immunol. 2002;169:3217–22. doi: 10.4049/jimmunol.169.6.3217. [DOI] [PubMed] [Google Scholar]
- 48.Jiang HX, Siegel JN, Gewurz H. Binding and complement activation by C-reactive protein via the collagen-like region of C1q and inhibition of these reactions by monoclonal antibodies to C-reactive protein and C1q. J Immunol. 1991;146:2324–30. [PubMed] [Google Scholar]
- 49.Nauta AJ, Bottazzi B, Mantovani A, Salvatori G, Kishore U, Schwaeble WJ, Gingras AR, Tzima S, Vivanco F, Egido J, Tijsma O, Hack EC, Daha MR, Roos A. Biochemical and functional characterization of the interaction between pentraxin 3 and C1q. Eur J Immunol. 2003;33:465–73. doi: 10.1002/immu.200310022. [DOI] [PubMed] [Google Scholar]
- 50.Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben Tal N. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–4. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 51.Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, Phan I, Pilbout S, Schneider M. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in. 2003;31:365–70. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR. The Pfam protein families database. Nucleic Acids Res. 2004;32(Database issue):D138–141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bhakdi S, Torzewski M, Paprotka K, Schmitt S, Barsoom H, Suriyaphol P, Han SR, Lackner KJ, Husmann M. Possible protective role for C-reactive protein in atherogenesis: complement activation by modified lipoproteins halts before detrimental terminal sequence. Circulation. 2004;109:1870–6. doi: 10.1161/01.CIR.0000124228.08972.26. [DOI] [PubMed] [Google Scholar]
- 56.Roos A, Nauta AJ, Broers D, Faber-Krol MC, Trouw LA, Drijfhout JW, Daha MR. Specific inhibition of the classical complement pathway by C1q-binding peptides. J Immunol. 2001;167:7052–9. doi: 10.4049/jimmunol.167.12.7052. [DOI] [PubMed] [Google Scholar]
- 57.Sim RB, Reid KBM. C1: molecular interactions with activating systems. Immunol Today. 1991;12:307–11. doi: 10.1016/0167-5699(91)90004-D. [DOI] [PubMed] [Google Scholar]
- 58.Botto M, Dell'Agnola C, Bygrave AE, Thompson EM, Cook HT, Petry F, Loos M, Pandolfi PP, Walport MJ. Homozygous C1q deficiency causes glomerulonephritis associated with multiple apoptotic bodies. Nat Genet. 1998;19:56–9. doi: 10.1038/ng0598-56. [DOI] [PubMed] [Google Scholar]
- 59.Korb LC, Ahearn JM. C1q binds directly and specifically to surface blebs of apoptotic human keratinocytes: complement deficiency and systemic lupus erythematosus revisited. J Immunol. 1997;158:4525–8. [PubMed] [Google Scholar]
- 60.Navratil JS, Korb LC, Ahearn JM. Systemic lupus erythematosus and complement deficiency: clues to a novel role for the classical complement pathway in the maintenance of immune tolerance. Immunopharmacology. 1999;42:47–52. doi: 10.1016/s0162-3109(99)00018-1. [DOI] [PubMed] [Google Scholar]
- 61.Bobak DA, Washburn RG, Frank MM. C1q enhances the phagocytosis of Cryptococcus neoformans blastospores by human monocytes. J Immunol. 1988;141:592–7. [PubMed] [Google Scholar]
- 62.Goodman EB, Tenner AJ. Signal transduction mechanisms of C1q-mediated superoxide production. Evidence for the involvement of temporally distinct staurosporine-insensitive and sensitive pathways. J Immunol. 1992;148:3920–8. [PubMed] [Google Scholar]
- 63.Rother RP, Fodor WL, Springhorn JP, Birks CW, Setter E, Sandrin MS, Squinto SP, Rollins SA. A novel mechanism of retrovirus inactivation in human serum mediated by anti-α-galactosyl natural antibody. J Exp Med. 1995;182:1345–55. doi: 10.1084/jem.182.5.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thielens NM, Tacnet-Delorme P, Arlaud GJ. Interaction of C1q and mannan-binding lectin with viruses. Immunobiology. 2002;205:563–74. doi: 10.1078/0171-2985-00155. [DOI] [PubMed] [Google Scholar]
- 65.van den Berg RH, Faber-Krol MC, Sim RB, Daha MR. The first subcomponent of complement, C1q, triggers the production of IL-8, IL-6, and monocyte chemoattractant peptide-1 by human umbilical vein endothelial cells. J Immunol. 1998;161:6924–30. [PubMed] [Google Scholar]
- 66.Yamada M, Oritani K, Kaisho T, Ishikawa J, Yoshida H, Takahashi I, Kawamoto S, Ishida N, Ujiie H, Masaie H, Botto M, Tomiyama Y, Matsuzawa Y. Complement C1q regulates LPS-induced cytokine production in bone marrow-derived dendritic cells. Eur J Immunol. 2004;34:221–30. doi: 10.1002/eji.200324026. [DOI] [PubMed] [Google Scholar]
- 67.Gardai SJ, Xiao YQ, Dickinson M, Nick JA, Voelker DR, Greene KE, Henson PM. By binding SIRPα or calreticulin/CD91, lung collectins act as dual function surveillance molecules to suppress or enhance inflammation. Cell. 2003;115:13–23. doi: 10.1016/s0092-8674(03)00758-x. [DOI] [PubMed] [Google Scholar]
- 68.Bottazzi B, Vouret-Craviari V, Bastone A, De Gioia L, Matteucci C, Peri G, Spreafico F, Pausa M, D'Ettorre C, Gianazza E, Tagliabue A, Salmona M, Tedesco F, Introna M, Mantovani A. Multimer formation and ligand recognition by the long pentraxin PTX3. Similarities and differences with the short pentraxins C-reactive protein and serum amyloid P component. J Biol Chem. 1997;272:32817–23. doi: 10.1074/jbc.272.52.32817. [DOI] [PubMed] [Google Scholar]
- 69.Mantovani A, Garlanda C, Bottazzi B. Pentraxin 3, a non-redundant soluble pattern recognition receptor involved in innate immunity. Vaccine. 2003;21(Suppl. 2):43–7. doi: 10.1016/s0264-410x(03)00199-3. [DOI] [PubMed] [Google Scholar]
- 70.Roos A, Xu W, Castellano G, Nauta AJ, Garred P, Daha MR, Van Kooten C. A pivotal role for innate immunity in the clearance of apoptotic cells. Eur J Immunol. 2004;34:921–9. doi: 10.1002/eji.200424904. [DOI] [PubMed] [Google Scholar]
- 71.Sorensen IJ, Nielsen EH, Andersen O, Danielsen B, Svehag SE. Binding of complement proteins C1q and C4bp to serum amyloid P component (SAP) in solid contra liquid phase. Scand J Immunol. 1996;44:401–7. doi: 10.1046/j.1365-3083.1996.d01-326.x. [DOI] [PubMed] [Google Scholar]
- 72.Nauta AJ, Trouw LA, Daha MR, Tijsma O, Nieuwland R, Schwaeble WJ, Gingras AR, Mantovani A, Hack EC, Roos A. Direct binding of C1q to apoptotic cells and cell blebs induces complement activation. Eur J Immunol. 2002;32:1726–36. doi: 10.1002/1521-4141(200206)32:6<1726::AID-IMMU1726>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 73.Bordin S, Whitfield D. Proliferating fibroblasts respond to collagenous C1q with phosphorylation of p38 mitogen-activated protein kinase and apoptotic features. J Immunol. 2003;170:667–71. doi: 10.4049/jimmunol.170.2.667. [DOI] [PubMed] [Google Scholar]
- 74.Sato T, van Dixhoorn MG, Heemskerk E, van Es LA, Daha MR. C1q, a subunit of the first component of complement, enhances antibody-mediated apoptosis of cultured rat glomerular mesangial cells. Clin Exp Immunol. 1997;109:510–7. doi: 10.1046/j.1365-2249.1997.4741372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schwaeble W, Schafer MK, Petry F, Fink T, Knebel D, Weihe E, Loos M. Follicular dendritic cells, interdigitating cells, and cells of the monocyte-macrophage lineage are the C1q-producing sources in the spleen. Identification of specific cell types by in situ hybridization and immunohistochemical analysis. J Immunol. 1995;155:4971–8. [PubMed] [Google Scholar]
- 76.Cao W, Bobryshev YV, Lord RS, Oakley RE, Lee SH, Lu J. Dendritic cells in the arterial wall express C1q: potential significance in atherogenesis. Cardiovasc Res. 2003;60:175–86. doi: 10.1016/s0008-6363(03)00345-6. [DOI] [PubMed] [Google Scholar]
- 77.Castellano G, Woltman AM, Nauta AJ, Roos A, Trouw LA, Seelen MA, Schena FP, Daha MR, Van Kooten C. Maturation of dendritic cells abrogates C1q production in vivo and in vitro. Blood. 2004;103:3813–20. doi: 10.1182/blood-2003-09-3046. [DOI] [PubMed] [Google Scholar]
- 78.Kuna P, Iyer M, Peerschke EI, Kaplan AP, Reid KBM, Ghebrehiwet B. Human C1q induces eosinophil migration. Clin Immunol Immunopathol. 1996;81:48–54. doi: 10.1006/clin.1996.0156. [DOI] [PubMed] [Google Scholar]
- 79.Leigh LE, Ghebrehiwet B, Perera TP, Bird IN, Strong P, Kishore U, Reid KBM, Eggleton P. C1q-mediated chemotaxis by human neutrophils: involvement of gClqR and G-protein signalling mechanisms. Biochem J. 1998;330:247–54. doi: 10.1042/bj3300247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Oiki S, Okada Y. C1q induces chemotaxis and K+ conductance activation coupled to increased cytosolic Ca2+ in mouse fibroblasts. J Immunol. 1988;141:3177–85. [PubMed] [Google Scholar]
- 81.Yokota T, Oritani K, Takahashi I, Ishikawa J, Matsuyama A, Ouchi N, Kihara S, Funahashi T, Tenner AJ, Tomiyama Y, Matsuzawa Y. Adiponectin, a new member of the family of soluble defense collagens, negatively regulates the growth of myelomonocytic progenitors and the functions of macrophages. Blood. 2000;96:1723–32. [PubMed] [Google Scholar]
- 82.Ghebrehiwet B, Kew RR, Gruber BL, Marchese MJ, Peerschke EI, Reid KBM. Murine mast cells express two types of C1q receptors that are involved in the induction of chemotaxis and chemokinesis. J Immunol. 1995;155:2614–9. [PubMed] [Google Scholar]
- 83.Ghebrehiwet B, Habicht GS, Beck G. Interaction of C1q with its receptor on cultured cell lines induces an anti-proliferative response. Clin Immunol Immunopathol. 1990;54:148–60. doi: 10.1016/0090-1229(90)90014-h. [DOI] [PubMed] [Google Scholar]
- 84.Peerschke EI, Ghebrehiwet B. Human blood platelet gC1qR/p33. Immunol Rev. 2001;180:56–64. doi: 10.1034/j.1600-065x.2001.1800105.x. [DOI] [PubMed] [Google Scholar]
- 85.Peerschke EI, Reid KBM, Ghebrehiwet B. Platelet activation by C1q results in the induction of αIIb/β3 integrins (GPIIb-IIIa) and the expression of P-selectin and procoagulant activity. J Exp Med. 1993;178:579–87. doi: 10.1084/jem.178.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cutler AJ, Botto M, van Essen D, Rivi R, Davies KA, Gray D, Walport MJ. T cell-dependent immune response in C1q-deficient mice: defective interferon-γ production by antigen-specific T cells. J Exp Med. 1998;187:1789–97. doi: 10.1084/jem.187.11.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Collins C, Tsui FW, Shulman MJ. Differential activation of human and guinea pig complement by pentameric and hexameric IgM. Eur J Immunol. 2002;32:1802–10. doi: 10.1002/1521-4141(200206)32:6<1802::AID-IMMU1802>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 88.Bickerstaff MC, Botto M, Hutchinson WL, Herbert J, Tennent GA, Bybee A, Mitchell DA, Cook HT, Butler PJ, Walport MJ, Pepys MB. Serum amyloid P component controls chromatin degradation and prevents antinuclear autoimmunity. Nat Med. 1999;5:694–7. doi: 10.1038/9544. [DOI] [PubMed] [Google Scholar]
- 89.Garlanda C, Hirsch E, Bozza S, Salustri A, De Acetis M, Nota R, Maccagno A, Riva F, Bottazzi B, Peri G, Doni A, Vago L, Botto M, De Santis R, Carminati P, Siracusa G, Altruda F, Vecchi A, Romani L, Mantovani A. Non-redundant role of the long pentraxin PTX3 in anti-fungal innate immune response. Nature. 2002;420:182–6. doi: 10.1038/nature01195. [DOI] [PubMed] [Google Scholar]
- 90.Nadesalingam J, Bernal AL, Dodds AW, Willis AC, Mahoney DJ, Day AJ, Reid KBM, Palaniyar N. Identification and characterization of a novel interaction between pulmonary surfactant protein D and decorin. J Biol Chem. 2003;278:25678–87. doi: 10.1074/jbc.M210186200. [DOI] [PubMed] [Google Scholar]
- 91.Ramamurthy P, Hocking AM, McQuillan DJ. Recombinant decorin glycoforms. Purification and structure. J Biol Chem. 1996;271:19578–84. doi: 10.1074/jbc.271.32.19578. [DOI] [PubMed] [Google Scholar]
- 92.Krumdieck R, Hook M, Rosenberg LC, Volanakis JE. The proteoglycan decorin binds C1q and inhibits the activity of the C1 complex. J Immunol. 1992;149:3695–701. [PubMed] [Google Scholar]
- 93.Ikeda F, Haraguchi Y, Jinno A, Iino Y, Morishita Y, Shiraki H, Hoshino H. Human complement component C1q inhibits the infectivity of cell-free HTLV-I. J Immunol. 1998;161:5712–9. [PubMed] [Google Scholar]
- 94.Nagy K, Clapham P, Cheingsong-Popov R, Weiss RA. Human T-cell leukemia virus type I: induction of syncytia and inhibition by patients’ sera. Int J Cancer. 1983;32:321–8. doi: 10.1002/ijc.2910320310. [DOI] [PubMed] [Google Scholar]
- 95.Sagara Y, Inoue Y, Shiraki H, Jinno A, Hoshino H, Maeda Y. Identification and mapping of functional domains on human T-cell lymphotropic virus type 1 envelope proteins by using synthetic peptides. J Virol. 1996;70:1564–9. doi: 10.1128/jvi.70.3.1564-1569.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Quinkal I, Hernandez JF, Chevallier S, Arlaud GJ, Vernet T. Mapping of the interaction between the immunodominant loop of the ectodomain of HIV-1 gp41 and human complement protein C1q. Eur J Biochem. 1999;265:656–63. doi: 10.1046/j.1432-1327.1999.00757.x. [DOI] [PubMed] [Google Scholar]
- 97.Thielens NM, Bally IM, Ebenbichler CF, Dierich MP, Arlaud GJ. Further characterization of the interaction between the C1q subcomponent of human C1 and the transmembrane envelope glyco-protein gp41 of HIV-1. J Immunol. 1993;151:6583–92. [PubMed] [Google Scholar]
- 98.Thielens NM, Bally IM, Ebenbichler CF, Dierich MP, Arlaud GJ. Interaction of C1 with HIV-1. Behring Inst Mitt. 1993:165–70. [PubMed] [Google Scholar]
- 99.Marschang P, Ebenbichler CF, Dierich MP. HIV and complement: role of the complement system in HIV infection. Int Arch Allergy Immunol. 1994;103:113–7. doi: 10.1159/000236616. [DOI] [PubMed] [Google Scholar]
- 100.Marschang P, Kruger U, Ochsenbauer C, Gurtler L, Hittmair A, Bosch V, Patsch JR, Dierich MP. Complement activation by HIV-1-infected cells: the role of transmembrane glycoprotein gp41. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;14:102–9. doi: 10.1097/00042560-199702010-00002. [DOI] [PubMed] [Google Scholar]
- 101.Tacnet-Delorme P, Boyer V, Thielens NM, Hernandez JF, Bally I, Sim RB, Desgranges C, Arlaud GJ. In vitro analysis of complement-dependent HIV-1 cell infection using a model system. J Immunol. 1999;162:4088–93. [PubMed] [Google Scholar]
- 102.Moir S, Malaspina A, Li Y, Chun TW, Lowe T, Adelsberger J, Baseler M, Ehler LA, Liu S, Davey RT, Jr, Mican JA, Fauci AS. B cells of HIV-1-infected patients bind virions through CD21-complement interactions and transmit infectious virus to activated T cells. J Exp Med. 2000;192:637–46. doi: 10.1084/jem.192.5.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Caffrey M. Model for the structure of the HIV gp41 ectodomain: insight into the intermolecular interactions of the gp41 loop. Biochim Biophys Acta. 2001;1536:116–22. doi: 10.1016/s0925-4439(01)00042-4. [DOI] [PubMed] [Google Scholar]
- 104.Kojouharova MS, Tsacheva IG, Tchorbadjieva MI, Reid KBM, Kishore U. Localization of ligand-binding sites on human C1q globular head region using recombinant globular head fragments and single-chain antibodies. Biochim Biophys Acta. 2003;1652:64–74. doi: 10.1016/j.bbapap.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 105.Gasque P, Dean YD, McGreal EP, VanBeek J, Morgan BP. Complement components of the innate immune system in health and disease in the CNS. Immunopharmacology. 2000;49:171–86. doi: 10.1016/s0162-3109(00)80302-1. [DOI] [PubMed] [Google Scholar]
- 106.Webster S, Glabe C, Rogers J. Multivalent binding of complement protein C1q to the amyloid β-peptide (Aβ) promotes the nucleation phase of Aβ aggregation. Biochem Biophys Res Commun. 1995;217:869–75. doi: 10.1006/bbrc.1995.2852. [DOI] [PubMed] [Google Scholar]
- 107.Tacnet-Delorme P, Chevallier S, Arlaud GJ. Beta-amyloid fibrils activate the C1 complex of complement under physiological conditions: evidence for a binding site for Aβ on the C1q globular regions. J Immunol. 2001;167:6374–81. doi: 10.4049/jimmunol.167.11.6374. [DOI] [PubMed] [Google Scholar]
- 108.Rostagno A, Revesz T, Lashley T, Tomidokoro Y, Magnotti L, Braendgaard H, Plant G, Bojsen-Moller M, Holton J, Fran-gione B, Ghiso J. Complement activation in chromosome 13 dementias. Similarities with Alzheimer's disease. J Biol Chem. 2002;277:49782–90. doi: 10.1074/jbc.M206448200. [DOI] [PubMed] [Google Scholar]
- 109.Ghiso J, Revesz T, Holton J, Rostagno A, Lashley T, Houlden H, Gibb G, Anderton B, Bek T, Bojsen-Moller M, Wood N, Vidal R, Braendgaard H, Plant G, Frangione B. Chromosome 13 dementia syndromes as models of neurodegeneration. Amyloid. 2001;8:277–84. doi: 10.3109/13506120108993826. [DOI] [PubMed] [Google Scholar]
- 110.Klein MA, Kaeser PS, Schwarz P, Weyd H, Xenarios I, Zinkernagel RM, Carroll MC, Verbeek JS, Botto M, Walport MJ, Molina H, Kalinke U, Acha-Orbea H, Aguzzi A. Complement facilitates early prion pathogenesis. Nat Med. 2001;7:488–92. doi: 10.1038/86567. [DOI] [PubMed] [Google Scholar]
- 111.Mabbott NA, Bruce ME, Botto M, Walport MJ, Pepys MB. Temporary depletion of complement component C3 or genetic deficiency of C1q significantly delays onset of scrapie. Nat Med. 2001;7:485–7. doi: 10.1038/86562. [DOI] [PubMed] [Google Scholar]
- 112.Casciola-Rosen LA, Anhalt G, Rosen A. Autoantigens targeted in systemic lupus erythematosus are clustered in two populations of surface structures on apoptotic keratinocytes. J Exp Med. 1994;179:1317–30. doi: 10.1084/jem.179.4.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Navratil JS, Watkins SC, Wisnieski JJ, Ahearn JM. The globular heads of C1q specifically recognize surface blebs of apoptotic vascular endothelial cells. J Immunol. 2001;166:3231–9. doi: 10.4049/jimmunol.166.5.3231. [DOI] [PubMed] [Google Scholar]
- 114.Tan LA, Kishore U, Yu BB, Sim RB. Complement activation by phospholipids and its regulation. Eur. J. Immunol. 2004 In Press. [Google Scholar]
- 115.Gorgani NN, Parish CR, Easterbrook Smith SB, Altin JG. Histidine-rich glycoprotein binds to human IgG and C1q and inhibits the formation of insoluble immune complexes. Biochemistry. 1997;36:6653–62. doi: 10.1021/bi962573n. [DOI] [PubMed] [Google Scholar]
- 116.Boackle RJ. The interaction of salivary secretions with the human complement system–a model for the study of host defense systems on inflamed mucosal surfaces. Crit Rev Oral Biol Med. 1991;2:355–67. doi: 10.1177/10454411910020030401. [DOI] [PubMed] [Google Scholar]
- 117.Boackle RJ, Connor MH, Vesely J. High molecular weight non-immunoglobulin salivary agglutinins (NIA) bind C1q globular heads and have the potential to activate the first complement component. Mol Immunol. 1993;30:309–19. doi: 10.1016/0161-5890(93)90059-k. [DOI] [PubMed] [Google Scholar]
- 118.Prakobphol A, Xu F, Hoang VM, Larsson T, Bergstrom J, Johansson I, Frangsmyr L, Holmskov U, Leffler H, Nilsson C, Boren T, Wright JR, Stromberg N, Fisher SJ. Salivary agglutinin, which binds Streptococcus mutans and Helicobacter pylori, is the lung scavenger receptor cysteine-rich protein gp-340. J Biol Chem. 2000;275:39860–6. doi: 10.1074/jbc.M006928200. [DOI] [PubMed] [Google Scholar]
- 119.Peerschke EI, Reid KBM, Ghebrehiwet B. Identification of a novel 33-kDa C1q-binding site on human blood platelets. J Immunol. 1994;152:5896–901. [PubMed] [Google Scholar]
- 120.Ghebrehiwet B, Lim BL, Peerschke EI, Willis AC, Reid KBM. Isolation, cDNA cloning, and overexpression of a 33-kD cell surface glycoprotein that binds to the globular heads of C1q. J Exp Med. 1994;179:1809–21. doi: 10.1084/jem.179.6.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jiang J, Zhang Y, Krainer AR, Xu RM. Crystal structure of human p32, a doughnut-shaped acidic mitochondrial matrix protein. Proc Natl Acad Sci USA. 1999;96:3572–7. doi: 10.1073/pnas.96.7.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ghebrehiwet B, Lim BL, Kumar R, Feng X, Peerschke EI. gC1q-R/p33, a member of a new class of multifunctional and multi-compartmental cellular proteins, is involved in inflammation and infection. Immunol Rev. 2001;180:65–77. doi: 10.1034/j.1600-065x.2001.1800106.x. [DOI] [PubMed] [Google Scholar]
- 123.Ghebrehiwet B, Jesty J, Peerschke EI. gC1q-R/p33: structure-function predictions from the crystal structure. Immunobiology. 2002;205:421–32. doi: 10.1078/0171-2985-00143. [DOI] [PubMed] [Google Scholar]
- 124.Braun L, Ghebrehiwet B, Cossart P. gC1q-R/p32, a C1q-binding protein, is a receptor for the InlB invasion protein of Listeria monocytogenes. EMBO J. 2000;19:1458–66. doi: 10.1093/emboj/19.7.1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pizarro-Cerda J, Sousa S, Cossart P. Exploitation of host cell cytoskeleton and signalling during Listeria monocytogenes entry into mammalian cells. CR Biol. 2004;327:115–23. doi: 10.1016/j.crvi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 126.Kirschfink M, Blase L, Engelmann S, Schwartz-Albiez R. Secreted chondroitin sulfate proteoglycan of human B cell lines binds to the complement protein C1q and inhibits complex formation of C1. J Immunol. 1997;158:1324–31. [PubMed] [Google Scholar]
- 127.Wuillemin WA, Te VH, Lubbers YT, de Ruig CP, Eldering E, Hack CE. Potentiation of C1 inhibitor by glycosaminoglycans: dextran sulfate species are effective inhibitors of in vitro complement activation in plasma. J Immunol. 1997;159:1953–60. [PubMed] [Google Scholar]
- 128.Sahu A, Pangburn MK. Identification of multiple sites of interaction between heparin and the complement system. Mol Immunol. 1993;30:679–84. doi: 10.1016/0161-5890(93)90079-q. [DOI] [PubMed] [Google Scholar]
- 129.Ghebrehiwet B, Galanakis DK. C1q inhibitor (chondroitin-4-sulfate proteoglycan): structure and function. Behring Inst Mitt. 1993:214–23. [PubMed] [Google Scholar]
- 130.Monell CR, Strand M. Structural and functional similarities between synthetic HIV gp41 peptides and defensins. Clin Immunol Immunopathol. 1994;71:315–24. doi: 10.1006/clin.1994.1092. [DOI] [PubMed] [Google Scholar]
- 131.Prohaszka Z, Nemet K, Csermely P, Hudecz F, Mezo G, Fust G. Defensins purified from human granulocytes bind C1q and activate the classical complement pathway like the trans-membrane glycoprotein gp41 of HIV-1. Mol Immunol. 1997;34:809–16. doi: 10.1016/s0161-5890(97)00097-7. [DOI] [PubMed] [Google Scholar]
- 132.van den Berg RH, Faber-Krol MC, van Wetering S, Hiemstra PS, Daha MR. Inhibition of activation of the classical pathway of complement by human neutrophil defensins. Blood. 1998;92:3898–903. [PubMed] [Google Scholar]
- 133.Reid KBM, Edmondson J. Location of the binding site in subcomponent C1q for plasma fibronectin. Acta Pathol Microbiol Immunol Scand Suppl. 1984;284:11–7. [PubMed] [Google Scholar]
- 134.Sorvillo J, Gigli I, Pearlstein E. Fibronectin binding to complement subcomponent C1q. Localization of their respective binding sites. Biochem J. 1985;226:207–15. doi: 10.1042/bj2260207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sorvillo JM, Pearlstein E. C1q, a subunit of the first component of complement, enhances binding of plasma fibronectin to bacteria. Infect Immun. 1985;49:664–9. doi: 10.1128/iai.49.3.664-669.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Entwistle RA, Furcht LT. C1q component of complement binds to fibrinogen and fibrin. Biochemistry. 1988;27:507–12. doi: 10.1021/bi00401a073. [DOI] [PubMed] [Google Scholar]
- 137.Kraulis PJ. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–50. [Google Scholar]
- 138.Kondo N, Kondo J. Identification of novel blood proteins specific for mammalian hibernation. J Biol Chem. 1992;267:473–8. [PubMed] [Google Scholar]
- 139.Murayama E, Takagi Y, Ohira T, Davis JG, Greene MI, Nagasawa H. Fish otolith contains a unique structural protein, otolin-1. Eur J Biochem. 2002;269:688–96. doi: 10.1046/j.0014-2956.2001.02701.x. [DOI] [PubMed] [Google Scholar]
- 140.Murayama E, Takagi Y, Nagasawa H. Immunohistochemical localization of two otolith matrix proteins in the otolith and inner ear of the rainbow trout. Oncorhynchus mykiss: comparative aspects between the adult inner ear and embryonic otocysts. Histochem Cell Biol. 2004;121:155–66. doi: 10.1007/s00418-003-0605-5. [DOI] [PubMed] [Google Scholar]
- 141.Davis JG, Oberholtzer JC, Burns FR, Greene MI. Molecular cloning and characterization of an inner ear-specific structural protein. Science. 1995;267:1031–4. doi: 10.1126/science.7863331. [DOI] [PubMed] [Google Scholar]
- 142.Urade Y, Oberdick J, Molinar-Rode R, Morgan JI. Precerebellin is a cerebellum-specific protein with similarity to the globular domain of complement C1q B chain. Proc Natl Acad Sci USA. 1991;88:1069–73. doi: 10.1073/pnas.88.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Greenhill NS, Ruger BM, Hasan Q, Davis PF. The α1(VIII) and α2(VIII) collagen chains form two distinct homotrimeric proteins in vivo. Matrix Biol. 2000;19:19–28. doi: 10.1016/s0945-053x(99)00053-0. [DOI] [PubMed] [Google Scholar]
- 144.Doliana R, Bot S, Mungiguerra G, Canton A, Cilli SP, Colombatti A. Isolation and characterization of EMILIN-2, a new component of the growing EMILINs family and a member of the EMI domain-containing superfamily. J Biol Chem. 2001;276:12003–11. doi: 10.1074/jbc.M011591200. [DOI] [PubMed] [Google Scholar]
- 145.Mayne R, Brewton RG. New members of the collagen superfamily. Curr Opin Cell Biol. 1993;5:883–90. doi: 10.1016/0955-0674(93)90039-s. [DOI] [PubMed] [Google Scholar]
- 146.Briles DE, Tart RC, Wu HY, Ralph BA, Russell MW, McDaniel LS. Systemic and mucosal protective immunity to pneumococcal surface protein A. Ann N Y Acad Sci. 1996;797:118–26. doi: 10.1111/j.1749-6632.1996.tb52954.x. [DOI] [PubMed] [Google Scholar]
- 147.Stenflo J, Stenberg Y, Muranyi A. Calcium-binding EGF-like modules in coagulation proteinases: function of the calcium ion in module interactions. Biochim Biophys Acta. 2000;1477:51–63. doi: 10.1016/s0167-4838(99)00262-9. [DOI] [PubMed] [Google Scholar]
- 148.Geer LY, Domrachev M, Lipman DJ, Bryant SH. CDART: protein homology by domain architecture. Genome Res. 2002;12:1619–23. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Maeda T, Abe M, Kurlsu K, Jikko A, Furukawa S. Molecular cloning and characterization of novel gene, CORS26, encoding a putative secretory protein and its possible involvement in skeletal development. J Biol Chem. 2001;276:3628–34. doi: 10.1074/jbc.M007898200. [DOI] [PubMed] [Google Scholar]
- 150.Schaffler A, Ehling A, Neumann E, Herfarth H, Paul G, Tarner I, Gay S, Schoimerich J, Muller-Ladner U. Genomic organization, promoter, amino acid sequence, chromosomal localization, and expression of the human gene for CORS-26 (collagenous repeat-containing sequence of 26-kDa protein). Biochim Biophys Acta. 2003;1630:123–9. doi: 10.1016/j.bbaexp.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 151.Haywards C, Shu X, Cideciyan AV, Lennon A, Barron P, Zareparsi S, Sawyer L, Hendry G, Dhillon B, Milam AH, et al. Mutation in a short-chain collages gene, CTRP5, results in extracellular deposit formation in late-onset retinal degeneration: a genetic model for age-related macular degeneration. Hum Mol Genet. 2003;12:2657–67. doi: 10.1093/hmg/ddg289. [DOI] [PubMed] [Google Scholar]