Summary
T cells are key players of the mammalian adaptive immune system. They experience different mechanical microenvironments during their life cycles, from the thymus, secondary lymph organs, and peripheral tissues that are free of externally applied force but display variable substrate rigidities, to the blood and lymphatic circulation systems where complicated hydrodynamic forces are present. Regardless of whether T cells are subject to external forces or generate their own internal forces, they response and adapt to different biomechanical cues to modulate their adhesion, migration, trafficking, and triggering of immune functions through mechanical regulation of various molecules that bear force. These include adhesive receptors, immunoreceptors, motor proteins, cytoskeletal proteins, and their associated molecules. Here we discuss the forces acting on various surface and cytoplasmic proteins of a T cell in different mechanical milieus. We review existing data on how force regulates protein conformational changes and interactions with counter molecules, including integrins, actin, and the T-cell receptor, and how each relates to T-cell functions.
Keywords: T cell, mechanochemistry, mechanosensing, mechanotransduction, catch bonds, conformational change, cytoskeleton, actin, integrin, T-cell receptors
Introduction
T cells are key players of the mammalian adaptive immune system. As a sensory organ, interactions of the dispersed and circulating T cells with other cells differ from tissue cells of other solid organs in a number of ways. Some of these differences have important biomechanical implications. To appreciate their changing mechanical environments, let us consider the variable milieus in which T cells function at different developmental stages. After being derived from hematopoietic stem cells at the bone marrow, lymphoid precursors travel to the thymus where they migrate from the medulla to the cortex and then back to medulla again. During their migration through the thymus, these hematopoietic precursor cells differentiate into CD4+CD8+ double positive thymocytes that display surface αβ T-cell receptors (TCR). Those cells whose TCRs interact with self-peptides presented by major histocompatibility complex (pMHC) molecules expressed on thymic epithelial cells survive and differentiate into CD4+CD8− or CD4−CD8+ single positive immature thymocytes. After purging those that express strongly self-reactive TCRs from the repertoire by apoptosis, the selected cells mature into functional naive T cells and exit the thymus (1–3). Exported naive T cells migrate across high endothelial venules (HEV) and home to secondary lymphatic organs, e.g. lymph node and spleen. Upon encountering antigen-presenting cells (APCs), e.g. dendritic cells (DCs), antigen-reactive lymphocytes form immunological synapses (IS) or kinapses and differentiate into activated T cells. Activated T cells undergo clonal expansion and exit lymph nodes into the circulation to patrol peripheral tissues to provide immunological surveillance. During inflammation, circulating T cells are guided by biochemical cues (e.g. chemokines) to inflamed tissues where they adhere to vascular surface and transmigrate across the blood vessel wall to search for and form IS and kinapses with infected cells to carry out immune effector functions (4–7).
Substantial changes in mechanical microenvironments of T cells are clearly evident during their life cycle. For example, fluid flow is minimal in the thymus, lymph nodes, spleen, and peripheral tissues, whereas complex hemodynamic forces exert on T cells in the blood and lymphatic circulations. On the other hand, flowing T cells usually take a spherical shape, indicating their resting state with minimal cytoskeletal contractile activity. By comparison, adhered T cells spread and migrate on substrates of different rigidities and form IS and kinapses with APCs. Cell motility and shape changes require dynamic rearrangement of the cytoskeleton and generation of molecular motor-based intracellular forces. Thus, an important aspect of the changing mechanical milieus is the variable physical forces externally applied to and internally generated by the T cell. These forces must be borne by cellular structures, e.g. membrane and cytoplasmic proteins, which may regulate their activities, and by so doing, impact T-cell functions depending on the biological processes. There has been an increasing recognition that T cells respond and adapt to changing mechanical microenvironments (8, 9) just like other tissue cells (10–13). However, detail mechanisms at the molecular level are still missing. In this review, we discuss the forces acting on various surface and cytoplasmic proteins of a T cell in different mechanical milieus and review existing data on how force regulates protein conformational changes and interactions with counter molecules. Efforts are made in every step to relate these to T-cell functions.
T-cell trafficking in the circulation system
T-cell trafficking is a critical step for T-cell development, surveillance, and immune response (4, 6, 14). It is a multiple-step adhesion, migration, and signaling process. Steps that involve mechanical aspects include tethering, rolling (Fig. 1A), slow rolling, arrest, spreading and intraluminal crawling (Fig. 1B), and paracellular and intercellular transmigration across the vessel wall (6). Initial tethering and rolling are mediated by selectins and α 4 integrins (6, 15). For example, L-selectin on leukocytes binds to peripheral node addressin (PNAd) mucins on HEV during lymphocyte homing to lymph nodes. The leukocyte mucin P-selectin glycoprotein ligand 1 (PSGL-1) binds to P- and E-selectins on activated endothelial cells during inflammatory response. These interactions occur under dynamic flow conditions of the circulation, providing several transport mechanisms to enhance T-cell tethering to the vascular surface (16). Blood flow also applies external forces on the transient selectin–ligand bonds that form and dissociate alternatively to enable rolling adhesion (Fig. 1A). Force strengthens these interactions by forming catch bonds to prolong bond lifetimes (17–19). Together, these biophysical mechanisms give rise to a counter-intuitive phenomenon called flow-enhanced adhesion (20). Catch bonds and the structural basis of force-prolongation of bond lifetime will be discussed in later sections.
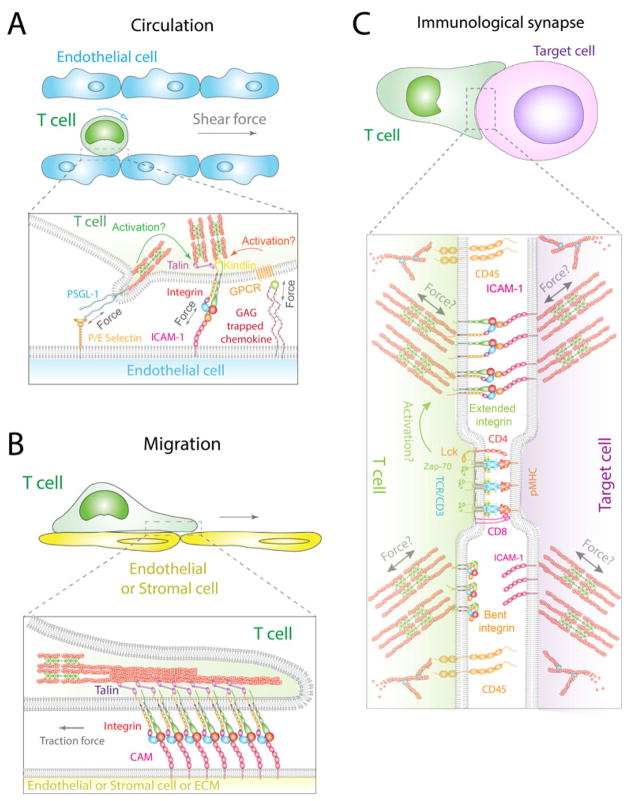
Fig. 1. Schematics of T-cell functions and molecular interactions regulated by external and/or internal mechanical forces in circulation (A), migration (B), and immunological synapse (C).
A circulating T cell adheres to the vessel wall lined by endothelial cells through selectin and integrin interactions with their respective ligands (e.g. PSGL-1 and ICAM-1) under shear force. Integrin conformations and ligand binding are modulated by inside-out signaling from PSGL-1 interacting with P/E selectin and/or GPCR interacting with GAG-trapped chemokine (A). When a T cell migrates on the endothelium or stromal cells or ECM, protruding and contracting actomyosins can exert traction forces on clustered integrins and regulate their interactions with ligands [cell adhesion molecules (CAMs)] on the leading edge of migrating T cells (B). Once a T cell recognizes antigens on a target cell by TCRs, an IS is formed on the contact zone between the T-cell and the APC (C). In the IS, large molecules, such as LFA-1, are squeezed out of the contact zone center where TCRs, coreceptors (CD4/CD8) and their associated cytoplasmic proteins (e.g. Lck and Zap-70) cluster. Larger molecules, such as CD45, are further segregated away to distal zones of the IS. TCRs may induce inside-out signaling to activate LFA-1. Actin retrograde flow, dynamic actomyosins and bending of cell membrane may generate mechanical forces on and regulate the functions of adhesion molecules, immunoreceptors or their associated cytoplasmic proteins (C). The molecular organization of the lamellipodium depicted in the enlarged box in B is similar to the architecture shown in the enlarged box in C, but with additional details and shown as a symmetric radially arranged version in C.
Not only does engagement of PSGL-1 and CD44 by P- and/or E-selectin mediate leukocyte tethering and rolling, but it also initiates signaling to prime β2 integrins (21–25) (Fig. 1A). Such priming extends the integrin ectodomain to induce a state that binds ligands [e.g. intercellular adhesion molecule-1 (ICAM-1)] with an intermediate affinity, resulting in leukocyte slow rolling on the vessel wall (21, 24, 25). This signaling cascade involves the Src family tyrosine kinase (e.g. Fgr) and spleen tyrosine kinase (Syk) but is independent of the anchorage of PSGL-1 to cytoskeleton (21–24). Details of integrin conformational changes and their regulation of ligand binding are discussed in a later section.
Rolling and slow rolling mediated by respective ligand binding of selectins and integrins in the intermediate state reduce the relative motion between the T cell and the endothelial cell, allowing the T cell to encounter more endothelial chemokines and integrin ligands (26). Binding of endothelial glycosaminoglycans (GAG)-trapped chemokines to G-protein-coupled receptors (GPCRs) rapidly activates β integrins LFA-1 (lymphocyte function-associated antigen-1)(αLβ2) and Mac-1 (macrophage-1 antigen) (αMβ2) to a high affinity state to arrest the T cell. The high affinity binding of β2 integrins mediates firm adhesion to endothelial cells and the subsequent steps that require transmitting traction forces from the cell to the extracellular matrix (ECM) (Fig. 1B). The GPCR-stimulated integrin activation is dependent on mechanical forces on integrin–ligand bonds and potentially also on GPCR-chemokine bond (6, 14, 26). As discussed in detail in a later section, mechanical forces can induce conformational changes in the integrin. GPCR-induced signaling extends the ectodomain of integrin through unclasping its cytoplasmic tails by binding of key integrin cytoplasmic adapters, talin and kindlin, to the cytoplasmic domain of the β subunit (25, 26). Talin binds to actin directly or indirectly through other cytoplasmic proteins (e.g. vinculin), thereby anchoring integrins to newly generated actin bundles with the help of Rho (26). This anchoring provides a physical linkage for transmitting mechanical forces bi-directionally across cell membrane between the integrin ligand-binding site and the interior of the cell. Recruitment of talin-1 to the β subunit cytoplasmic region for integrin activation requires local elevation of PI(4,5)P2 (PIP2), GPCR-stimulated RhoA, Rac1 and Rap-1 near talin head (27, 28). Kindlin-3 is the exclusive kindlin family member expressed on T cells based on the knowledge to date. It does not directly associate with actin or talin and may need to cooperate with other molecules [e.g. migfilin or integrin-linked kinase (ILK)] to activate integrins. The regulation of kindlin-3 on α Lβ 2 integrin-mediated T-cell functions is through its pleckstrin homology (PH) domain (29). As these cytoplasmic molecules and membrane receptors form a complex interaction network, mechanical forces should affect the functions of all members. Force regulation on some of these members are discussed in the following sections.
T-cell migration
Once transmigrated across the vessel wall, the T cell moves into a lymph node, the spleen, or injured tissue to look for APCs, where the mechanical microenvironment switches to a flow-free condition. Responding to the changing mechanical cues and the presence of abundant biochemical cues (e.g. chemokines), the T cell changes to a motile mode by adjusting its adhesiveness via modulating integrin functional states and remolding the actin cytoskeleton (Fig. 1B). In many biophysical aspects, migration of matured T cells in peripheral tissues is similar to migration of immature thymocytes in the thymus.
T-cell migration is rapid. Typically, its migration speed is 10–40 μm/min, 100 times faster than many tissue cells (e.g. fibroblast) (27). To reach such fast migrating speed, integrins are distributed in at least three different zones of activity in the migrating T cell (30): (i) the protruding lamellae at the leading edge where intermediate affinity LFA-1 functions, (ii) the mid-cell zone where LFA-1 is activated to high affinity by talin, and (iii) the uropod at the trailing edge with LFA-1 of unknown activity status. Active integrin–ligand engagement can direct integrin clustering and trigger F-actin reorganization that supports cell adhesion and spreading. The dynamics of the leading edge of the migrating T cell is mainly regulated by actomyosin contraction with assistance from myosin-light chain kinase (MLCK), whereas the relatively less dynamic uropod is enriched with Rho-associated protein kinase (ROCK) that has much slower kinetics on phosphorylation of myosin-light chain (MLC) than MLCK (30–32). Thus, not only the magnitude of the traction force generated by actomyosin can regulate integrin–ligand binding but also its frequency can alter integrin functions. Such precise regulation of integrin conformations and ligand-binding affinity with cooperation from cytoskeleton rearrangement determines T-cell migration in lymph nodes and injured tissues.
Immunological synapse and kinapse
Once encountered a rare APC, the motile T cell quickly slows down from a migrating speed of >10 μm/min to <2 μm/min to form a stable contact with the APC (27, 33). This deceleration is triggered by the TCR interaction with antigen pMHC with helps from coreceptor, costimulatory [e.g. cytotoxic T-lymphocyte antigen 4 (CTLA-4)], and adhesion (e.g. LFA-1) molecules. After intimate contact, the T cell forms an asymmetric kinapse and symmetric IS (Fig. 1C) with the APC on their contact zone (34).
The IS has a unique ‘bulls-eye’ pattern (34–37). TCRs and its associated molecules, e.g. lymphocyte-specific protein tyrosine kinase (Lck), and ζ-chain-associated protein kinase-70 (Zap-70), are actively transported into the IS center by dynamic actin retrograde flow, forming the center region of the supramolecular activation clusters (cSMACs) (Fig. 1C), which is lacking actomyosin filaments (38, 39). Adhesion molecules, such as LFA-1, and their associated cytoplasmic linker (e.g. talin), form a peripheral ring zone surrounding the cSMAC, called pSMAC (Fig. 1C). Acto-myosin II arcs are also found to locate at the pSMAC (39, 40). Large and bulky molecules such as phosphatase CD45 are squeezed out to a distal region (dSMAC) outside the pSMAC. Compared to the pSMAC, the dSMAC is relatively more dynamic and composed of branched actin network created by Arp2/3-depedent nucleation (7, 27, 41). It has been proposed that the pSMAC and the dSMAC respectively mimic lamella and lamellipodium on migrating cells (e.g. fibroblast) (34, 42), spreading at the leading edge and stretching at the trailing edge, with the cSMAC in the middle. When a T cell is about to migrate, the IS breaks its circular symmetry and turns into a asymmetric kinapse (34, 42) (Fig. 1B).
Due to dynamic nature of the IS and kinapse, mechanical forces are inevitably applied on membrane receptors as well as on their physically associated cytoplasmic adapter proteins (43, 44). Mechanical forces can be generated from various sources. First of all, the active transport process driven by the actin retrograde flow can produce drag forces on membrane receptors engaged with ligands on the APC, as most of them anchor to actin cytoskeleton through adapter molecules (27, 39, 40, 44, 45). For example, LFA-1 anchors to cytoskeleton via talin, and the TCR/CD3 complex via molecular complexes that involve coreceptor CD4/CD8, Lck, Zap-70 and others (46). Secondly, segregation of small and large molecules into cSMAC and pSMAC respectively can produce membrane bending on both the T-cell and APC surfaces (47–49), which may result in pulling force on short molecules, such as the TCR/CD3 complex. Furthermore, the dSMAC undergoes cycles of actin polymerization-dependent protrusion and myosin II-mediated contraction (39, 43, 45). Such cyclic protrusion and contraction may exert force on and reinforce receptor–ligand interactions, which may provide a biophysical mechanism for the T cell to regulate IS and kinapse formation and to sense the mechanical properties of the APC.
T-cell triggering
A T cell integrates a wide array of external and internal signals to precisely control its differentiation, adhesion, migration, IS/kinapse formation, and various immune responses. These biological processes and their changes thereof are triggered and modulated by the biochemical and biomechanical cues sensed by the T cell. T-cell triggering is usually initiated by antigen recognition by the TCR, but may also involve ligand binding of the coreceptor, costimulatory/co-inhibitory molecules, and adhesion molecules. The molecular details of how binding to the membrane distal end of the extracellular portion of any of these receptors communicates the information encoded in the ligand across the plasma membrane to initiate the first biochemical signal (e.g. phosphorylation of the CD3 cytoplasmic tails) is still unclear. However, physical forces have been suggested to play a driving or regulatory role in light of the rich and variable mechanical milieus experienced by the T cell. This view is natural for adhesive receptors such as integrins, which have long been proposed as mechanosensors (50–60). Even for molecules that are known for their roles in receiving biochemical signals, such as the TCR and coreceptors, increasing evidence suggests that they can also sense biomechanical signals (44, 61). This is because the ligands of these immunoreceptors are immobilized on the surface of the APC rather than being soluble in a fluid phase. Mechanical force may act on the TCR–pMHC bonds when the T cell membrane moves relative to the APC membrane as a result of cell motility (Fig. 1B) or in the case of stable T-cell–APC conjugates as TCR microclusters form and stream along the actin cytoskeleton to the IS cSMAC (Fig. 1C). Recently published data suggest that the TCR can mediate sensing of mechanical force (44, 61) and substrate rigidity (8, 9) via engaged pMHCs or antibodies.
Biochemical sensing is usually localized at the surface receptor and requires a cascade of chemical reactions to relay the signal from the plasma membrane inwards, which takes time. By comparison, biomechanical sensing can act instantaneously and transmit long distances through intracellular structures. As such, mechanosensing may occur not only at the cell surface but also inside the cell, not only by cell surface molecules but also by cytoplasmic molecules. Candidate sensory molecules may include those that have a load-bearing role in supporting and/or regulating the forms and shapes of cellular and subcellular structures, such as structural, scaffolding, and connective proteins.
In the preceding sections, we have described various mechanical settings where T-cell functions. In the following sections, we review existing data on mechanochemistry of proteins with focuses on how mechanical force regulates molecular interactions and conformational changes of proteins.
Slip bonds, catch bonds, and ideal bonds
The description of molecular interaction usually adopts that of chemical reaction based on mass action laws. The association on-rate kon, dissociation off-rate koff, and binding affinity Ka follow the Arrhenius equation to depend on temperature explicitly and also on pressure implicitly through the ideal gas law. It is therefore natural to conceptualize that koff may depend on tensile force applied on a molecular bond, as proposed by Bell (62) and Dembo et al. (63). Bell’s original model postulates that koff increase exponentially with force (62), which is termed ‘slip bond’ (63). The opposite behavior is ‘catch bond’ where koff decreases with force. The case in which koff is independent of force is called ‘ideal bond’. The classification of three types of bonds provides simple and useful definitions of how force may regulate molecular interactions (63).
Slip bonds have been found in most molecular interactions analyzed to date, e.g. antigen–antibody interactions (17, 18, 54, 55) that play a prominent role in immunology. The first measurements of force regulation of TCR–pMHC interaction also observed slip bonds for most of the peptides studied (64). However, since their first experimental demonstration a decade ago, the once thought unusual and counterintuitive catch bonds have been observed in more than a dozen of molecular interactions. These include both cell surface receptors (e.g. adhesion molecules) and cytoplasmic proteins (e.g. structural and motor molecules). Cell adhesion molecules shown to form catch bonds include the following: three (P-, L- and E-) selectins interacting with their common and distinct carbohydrate ligands (17–19); two integrins, α5β1 and αLβ2, interacting with their respective ligands, fibronectin and ICAM-1 (54, 55); platelet glycoprotein Ibα (GPIbα ) interacting with von Willebrand factor (VWF) (65); E. coli fimbrial adhesin FimH interacting with mannose ligand (66), and homotypical interaction between E-cadherins (67). Published catch bonds between intracellular structural and motor proteins include that of actomyosin (68), kinetochore proteins (69), and actin (70). In addition, catch bonds have also been found in force-dependent intramolecular interactions (55, 71) and enzymatic reaction (71, 72). More recently, ideal bonds have also been observed (67).
Most of these molecular interactions mediate T-cell functions, and they have one thing in common: one of their functional roles is to bear or transmit force. As such, catch bonds may regulate T-cell functions where mechanical loads have to be supported or overcome to carry out such functions. Indeed, it has been suggested that catch bond may be related to TCR triggering (73). One of the pMHCs in the aforementioned study of force-dependent TCR–pMHC dissociation exhibited catch bond behavior, although this is based on a single data point (64). The catch mechanism allows force to prolong bond lifetime, one of the TCR–pMHC interaction parameters that correlates well with T-cell response to antigen (74–76). Interestingly, all catch bonds observed to date only exist in a finite force regime beyond which they transition to slip bonds. The force where catch-slip transition occurs defines an optimal force. Under such force, the molecular interaction becomes most stable in a range of forces. It may also provide a mechanism for the cell to select for or adapt to a mechanical microenvironment most suitable for its survival, proliferation, differentiation, and carrying out its functions. Further, catch bonds are usually formed by force-induced formation of new noncovalent contacts (e.g. hydrogen bonds and salt bridges) at the complex interface of the two interacting molecules that are not observed in the structures co-crystallized in the absence of force (65, 70, 77). Furthermore, point mutations that prevent such new atomic-level interactions from forming under tensile force could suppress or even eliminate catch bonds, notwithstanding that such mutations are predicted not to impact the complex interface at zero force (65, 70, 78). Conversely, single-residue replacements that enhance these new noncovalent contacts could produce more pronounced catch bonds, despite that these residues are far away from the complex interface (65, 79, 80). Moreover, some of these mutations that alter catch bond behaviors correlate with human diseases, e.g. von Willebrand diseases (65) and nemaline myopathy (70), supporting the physiological importance of catch bonds. More studies are required to identify catch bonds in key molecular interactions in T cells and more definitive evidence is needed to elucidate their precise roles. Nevertheless, available data suggest several possible links between catch bond and the mechanical regulation of various T-cell functions. In later sections, we use integrin–ligand and actin–actin interactions to exemplify various features of catch bonds.
Integrin structural-functional states
Integrins are essential to T-cell functions, as their interactions with ligands mediate T-cell trafficking in the circulation systems, migration inside the thymus, secondary lymphoid organs and infected tissues, formation of the IS/kinapse, and execution of immune responses. In mammals, the integrin family consists of 18 α and 8 β subunits that combine to form 24 αβ heterodimeric membrane receptors. At least 12 of them are expressed on T cells (4, 81, 82), including four leukocyte-specific β2 integrins with αL, αM, αX, and αD subunits that bind ICAMs, two β7 integrins with α4 and αE subunits that bind mucosal addressin cell adhesion molecule 1 (MAdCAM-1), and six β1 integrins with α1–α6 subunits that bind ECM proteins. Each subunit has a large ectodomain, a transmembrane domain, and a short cytoplasmic tail. The ectodomains form a head supported by two long legs (Fig. 2). The ligand-binding head comprises the β-propeller domain of the α subunit and the VWF type A domain (called βA or βI) inserted into the hybrid domain of the β subunit. Half of the α subunits have an additional αA (or αI) domain inserted into the β-propeller domain, which contains the ligand binding-site for these integrins (83) (Fig. 2).
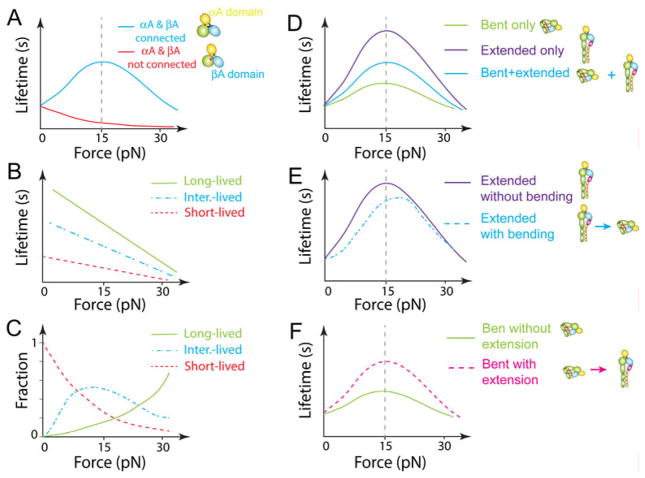
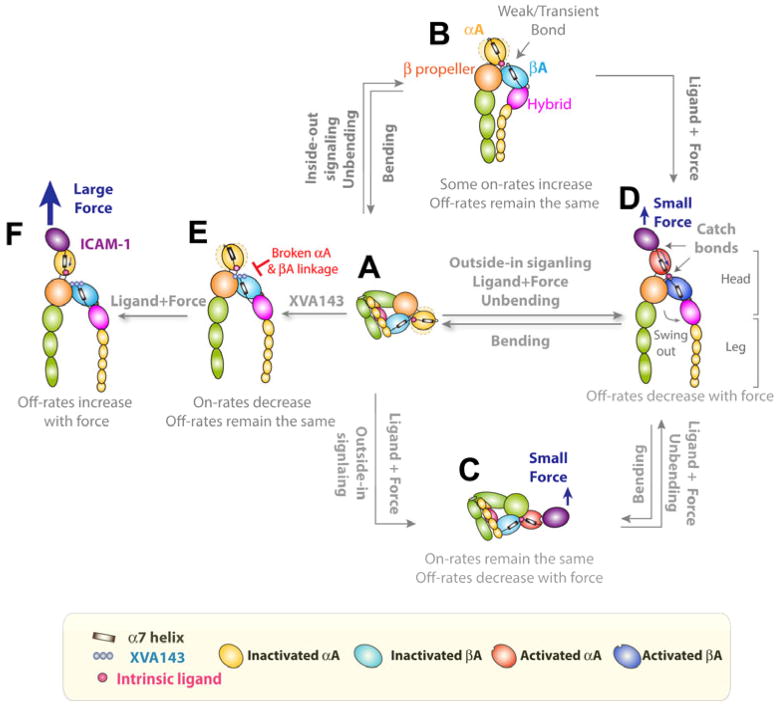
Fig. 2. Force-regulated integrin conformational changes.
(A) Inactive, bent LFA-1 with a closed headpiece and a flexible αA domain depicted by the dotted light yellow ellipsoid moving around the solid yellow αA domain. The headpiece is closed, and the αA domain is in the inactive conformation with the α7-helix in the up position and the MIDAS in the closed conformation. (B) Priming by inside-out signaling or metal ions extends LFA-1, increasing on-rate of LFA-1 for ICAM-1 but unchanging zero-force off-rate. The intrinsic ligand (Glu310, small pink circle) on the αA domain may form weak or transient bond with the βA MIDAS. (C) Small mechanical force applied to the bent LFA-1 activates the αA domain by pulling down the αA α7 helix. This activation decreases the off-rate of bent LFA-1 for ICAM-1. (D) Force fully activates LFA-1 with activated αA and βA domains, open headpiece and extended ectodomains. Two catch bonds in series are formed under pulling force. One is the intermolecular catch bond between ICAM-1 and the αA domain; the other is the intramolecular catch bond between the αA domain and the βA domain. Force can accelerate unbending from the bent to extended state [from (A) to (B) or from (C) to (D)] but impede bending. (E) Regardless of priming, the binding of XVA143 blocks the αA and βA ligation, makes the αA domain more flexible, and pushes the βA domain α7-helix further downward, inducing hybrid domain swing-out and ectodomain extension. On-rate for ICAM-1 is reduced, but zero-force off-rate remains the same. (F) Blocking the internal ligation impairs the force transmission along the pathway that favors the αA and/or βA α7 helixes downward movements. A much larger external force applied by ICAM-1 is therefore needed to generate a similar internal force to pull down the αA α7 helix to induce the intermediate- and long-lived states, but the mechanism of forcing acceleration of dissociation dominates at such high forces, converting the LFA-1–ICAM-1 catch-slip bonds to that of slip-only. Adopted from Ref. (55).
Integrins can adopt multiple conformations that exhibit different ligand-binding properties (Fig. 2). Crystallography (84–88), electronic microscopy (EM) (89–91), monoclonal antibody (mAb) mapping (87, 90, 92), Förster resonance energy transfer (FRET) (93–95), nuclear magnetic resonance (96), and force probes (97) have revealed distinct conformations for different regions of integrins, including bent and extended ectodomains, clasped and separated legs, closed and open headpieces, and closed, intermediate, and open αA domain (83) (Fig. 2). Under physiological conditions without stimulations, integrins are in the resting state with a bent ectodomain. The headpiece is closed, and its ligand-binding site is only <5 nm from their membrane anchor (90, 91, 98) (Fig. 2A). Upon exposure of the T cell to various biochemical cues, integrins rapidly extend their ectodomains and displace their ligand-binding site 15–20 nm away from the cell membrane (89–91, 97) as a result of inside-out signaling (Fig. 2B). The upstream events depend on which membrane receptor is triggered, e.g. PSGL-1, GPCRs, or TCR, but the final steps involve recruitment of talin or kindlin to associate respectively with the integrin β subunit via a membrane-proximal NPxY motif or a membrane-distal NxxY motif (99). These interactions can unclasp integrin cytoplasmic tail and transmembrane domain through a basic ‘snorkeling’ amino acid (100), leading to the leg separation and switchblade-like ectodomain extension.
The conformational changes of ectodomain extension and/or leg separation may propagate to other integrin domains (83) by 8-step transitions of headpieces to fully swing out the hybrid domain (101). This may pull the α7 helix of the βA domain at the bottom, which activates its metal ion dependent adhesion site (MIDAS) on the top, thereby upregulating the ligand binding affinity for αA domain-lacking integrins (Fig. 2D). For αA domain-containing integrins, the activated βA domain may bind an intrinsic ligand on the C-terminus of the α7 helix of the αA domain, providing a physical connection to transmit conformational changes to the αA domain or vice versa (Fig. 2C, D). This may pull the α7 helix on αA domain from the up to intermediate and down conformations, which opens up the MIDAS on the top of the αA domain (83), resulting in transitions from low- to intermediate- and high-affinity for ligand.
Cations are artificial stimulatory agents that can regulate integrin conformations and binding affinities. Usually in physiological condition of Ca2+ and Mg2+ (Ca2+Mg2+), integrins adopt a bent conformation with a closed headpiece and low affinity for ligand (Fig. 2A). Changing the cation compositions to Mg2+ plus EGTA to chelate Ca2+ (Mg2+) or to Mn2+ (Mn2+) extends integrins and induces headpiece opening, resulting in a higher affinity state by enhancing ligand association on rate (55, 102). In the absence of force, however, the off-rates of LFA-1–ICAM-1 dissociation are the same in both Ca2+Mg2+ and Mn2+, suggesting that the αA domain MIDAS remains in the short-lived state (55) (Fig. 2B). This is consistent with the cation-independent staining by the HI-111 mAb that reports the closed conformation of the αA domain MIDAS (55, 103). Mn2+ was also found to increase the on-rate but not to change the off-rate of αIIbβ3–fibrinogen bonds in the absence of force (104). Even under highly stimulating (e.g. by chemokine CXCL12) conditions that readily induce strong integrin-dependent adhesion (105, 106), the zero-force off-rate of LFA-1–ICAM-1 dissociation remains unchanged (55) (Fig. 2B). But the situation is completely changed under force because of catch bonds. When a 10 pN force is applied, the bond lifetime could increase as much as two orders of magnitude (55) (Fig. 2C,D), revealing an LFA-1 integrin catch bond with ICAM-1.
Integrin catch bonds
Integrin catch bonds were experimentally demonstrated by force-clamp experiments with an atomic force microscopy (AFM) and a biomembrane force probe (BFP) using purified α5β1 constructs interacting with fibronectin (54) and LFA-1 expressing cells interacting with ICAM-1 (55) (Fig. 3A). Both catch bonds exist at low forces (<20 pN) (54, 55). A recent study by optical tweezers also found that force reduced the off-rate of and increased the population of the high affinity state of platelet integrin αIIbβ3 bond with fibrinogen in Ca2+Mn2+ (104).
Fig. 3. Effects of force-regulated conformational change of integrins on their ligand dissociation.
(A) LFA-1 and ICAM-1 form catch-slip bond (blue) under force but pure slip bond (red) when the internal ligation between the αA and βA domains is blocked. (B-C) Biophysical model for LFA-1–ICAM-1 catch bond based on force-induced transition among three states. Force can switch LFA-1 from a short-lived (red dashed line) to intermediate-lived (blue dotted-dashed line) and long-lived (green solid line) states. Each state follows the Bell-model, i.e. its lifetime exponentially decreases with force. Fractions of the states also change with force (C). (D) Effect of initial conformations of LFA-1 on ligand dissociation under force. Initially extended LFA-1 forms a more pronounced catch bond with ICAM-1 than initially bent LFA-1 in the absence of subsequent bending or unbending. Catch-slip bonds curves for initially bent, extended LFA-1 and mixture of these two conformers without subsequent ectodomain unbending and bending during lifetime measurements are respectively indicated by green, purple and blue. (E and F) Effects of bending (E) and unbending (F) on LFA-1–ICAM-1 catch-slip bonds. (E) Force-dependent lifetime of extended LFA-1 without bending (solid purple curve) is compared to that with bending (dotted dash blue curve). (F) Force-dependent lifetime of bent LFA-1 without unbending (solid green curve) is compared to that with unbending (dashed pink curve). Bending of extended LFA-1 shortens LFA-1–ICAM-1 bond lifetimes at forces <20 pN (E), while unbending of bent LFA-1 prolongs LFA-1–ICAM-1 bond lifetimes (F).
Integrin catch bonds may be explained by allosteric mechanisms via force-induced conformational changes. Many lines of evidence indicate that the LFA-1–ICAM-1 catch bond is induced allosterically by forcing movement of the αA domain α7 helix from the up to intermediate and down positions. Recombinant proteins locking this α7 helix by disulfide bonds at these three conformations have been crystalized and they display a wide range of affinities as measured by surface plasma resonance (SPR) (107) and an adhesion frequency assay (102). Steered molecular dynamics (SMD) simulations predict that force facilitate shifting the α7 helix transition (108, 109) and open up the αA domain MIDAS. Cells expressing C- but not N-terminally anchored isolated recombinant LFA-1 αA domain rolled stably under shear flow (53), suggesting that the C-terminal link may guide the force transmission along a pathway to pull the αA domain α7 helix to induce a high affinity MIDAS conformation. The authors also showed that locking the αA domain in the open conformation with disulfide bonds support firm adhesion of cells on ICAM-1 coated surfaces (53). Shear flow has been shown to further activate chemokine-primed extended LFA-1 to mediate firm adhesion of T-cells with intact cytoskeleton (110).
Using force-clamp assay by a BFP, force has been shown to shift the αA domain α7 helix from up to intermediate and down conformations that respectively correspond to short-, intermediate-, and long-lived states (Fig. 3B). Although force exponentially decreases the lifetimes of all three states (Fig. 3B) as predicted by the Bell model (62) and hence all are slip bonds (63), force also progressively shifts the respective fractions associated with the short- and intermediate-lived states to those of the intermediate- and long-lived states (Fig. 3C) to prolong the overall bond lifetime averaged over all three states (Fig. 3A), thereby producing a phenomenological catch bond at low forces (<20 pN). Further increase in force (>20 pN) completely switches short- and intermediate-lived states to the long-lived state, whose lifetime exponentially decreases with force, thereby converting the LFA-1–ICAM-1 catch bond to slip bond (55) (Fig. 3A).
The LFA-1–ICAM-1 catch bond requires binding of the C-terminal intrinsic ligand (Glu310) of the αA domain α7 helix to the βA domain MIDAS (83) (Figs 2E, F and 3A). Blocking this binding by a small molecule antagonist XVA143 keeps the αA MIDAS in the closed and low affinity state and abolishes the LFA-1–ICAM-1 catch bond (55, 90, 111). Importantly, to prolong the engagement of ICAM-1 to the αA domain MIDAS requires an internal catch bond formed between the aforementioned intrinsic ligand and the βA domain MIDAS. This is because the dissociation of the intramolecular catch bond would release the αA domain α7 helix, which relieves the intermolecular catch bond (55). Thus, two catch bonds work in series to maintain the durable force transmission from ICAM-1 through the αA domain from the MIDAS down the α7 helix to the βA domain and other downstream domains (Fig. 2). This observation may be extended to other scenarios because intracellular forces are usually borne by multiple proteins interacting with each other in series along the pathway of force transmission. The formation of catch bond in one linkage may imply that other connecting points also form catch bonds in order to avoid failure at the weakest link. An example of this is the actin catch bond, which is discussed later.
Integrin catch bonds do not require the ectodomain and headpiece to be at a particular conformation (bent or extend and opening or closed, respectively). Both LFA-1 and α5β1 form catch bonds with their respective ligands but have similar off-rate at zero-force in both Ca2+Mg2+ and Mn2+ (54, 55). Even a leg-less α5β1 construct formed catch bond with fibronectin. Furthermore, LFA-1 extension stably induced by the small molecule antagonist, XVA143 (55, 90, 112), changes the LFA-1–ICAM-1 catch bond to a pure slip bond (55). Chemokine-extended LFA-1 cannot mediate T-cell firm adhesion in the absence of shear force (110). These data indicate that unclasping of the cytoplasmic tails, separation of the αβ legs, swing-out of the hybrid domain, and extension of the ectodomain of an integrin induced by divalent-cations, allosteric small molecule, or inside-out signaling from GPCRs are not required for integrin catch bond. Note that extension makes integrins ready to associate with their ligands by significantly increasing on-rates (54, 55, 102, 104) (Fig. 2B). It also favors force-induced conformational changes on the ligand binding domain as well as the intrinsic ligand docking of the αA domain α7 helix to the βA domain MIDAS to stabilize the flexible αA domain (87), giving rise to stronger catch bonds with longer peak lifetimes than that of the bent integrins in the absence force-induced unbending (97).
Force-induced integrin conformational changes
Like other proteins, integrins can also be deformed by force, as measured by stretching or thermal fluctuation using a biomembrane force probe (97). But deformation is different from conformational change. Deformation occurs when an external force is applied to the integrin. The atomic coordinates of the elastically deformed integrin displace from their original positions, but return to their original positions upon force removal. By comparison, conformational change occurs among multiple conformations that are stable even in the absence of externally applied force, as revealed by crystallographic studies (88, 101, 113, 114). Such change may occur spontaneously in the absence of external force, giving rise to the coexistence of multiple conformers in equilibrium, as revealed by EM studies (89–91, 98). However, force may tilt such equilibrium, alter the fractions of different conformers by regulating their stability, and accelerate or decelerate the rate of conformational transition by shortening or prolonging the dwell times before conformational change occurs, as directly observed by BFP experiments (below) (97). Indeed, the multi-domain quaternary structures of integrins resemble protein machines with moving parts connected by ratchets, ropes, and hinges. It would seem reasonable to hypothesize that not only would force transmit across such structures but it would also perturb their stability and alter the rates of their conformational changes.
In addition to inducing the integrin αA or βA α7 helix downward movement as discussed in the preceding section, mechanical force may also induce other integrin conformational changes. SMD simulations have suggested that force can activate the headpiece (57), swing out the hybrid domain (59), separate the αβ legs (59), and extend the ectodomain (115), leading to propagation of conformational changes to the ligand binding site (Fig. 2A–D).
The first real-time observation of force-regulated dynamic bending and unbending conformational changes of single LFA-1 on living cells was recently made by a mechanical method using a BFP (97). The study demonstrated that force accelerates unbending, which occurs in <0.1 s, but decelerates bending, which takes >1 s. Unbending facilitates force to prolong LFA-1–ICAM-1 bond lifetime, whereas bending slightly shorten LFA-1–ICAM-1 bond lifetime in the catch bond regime (<20 pN) but does not change the bond lifetime at higher forces (97) (Fig. 3D–F).
Mechanical force can regulate integrin conformations and ligand–binding in two ways. Firstly, force facilitates integrin unbending but impedes bending, increasing ligand-association rate to enhance ligand binding. Secondly, force induces unclasp of integrin cytoplasmic and transmembrane domains, separation of αβ legs, opening the headpiece by fully swing-out of the hybrid domain, and pulling down of the α7 helix in both the αA and βA domains to activate their respective MIDASs to prolong bond lifetimes. These conformational changes can also be induced by chemical cues except the last one. Pulling down of the α7 helix of the αA domain has to be induced by external force, thereby producing catch bonds (55).
Mechanical regulation of actin cytoskeleton dynamics
Regulation of actin cytoskeleton organization is important for T-cell immune functions, as these functions are related to T-cell morphology and mobility, which critically depend on actin polymerization and depolymerization dynamics. For example, circulating T cells in the blood stream are rounded, which are maintained by actin network in the cortical layer with actin bundles filling the microvilli on the T-cell surface. Different adhesion molecules are segregated to concentrate at microvilli (e.g., PSGL-1, L-selectin, α4β1 integrin) or the cell body (e.g. LFA-1) to facilitate T-cell adhesion to the vessel wall during an inflammatory response. After transmigrating across the vessel wall, T cells change to a ‘hand mirror’ morphology (116). On the leading edge, branched actin filaments form pseudopodia and lamellipodia and work with Rho GTPase to push the T-cell forward. On the trailing edge, concentrated actin filaments work with RhoA-activated myosin to generate contractile force to detach the trailing edge from the ECM (32). Once a T cell recognizes appropriate antigenic pMHCs on an APC, it pulls the trailing edge forward and extends its large pseudopodia and lamellipodia toward the APC to form an IS (116). During this dynamic process, actin retrograde flow transports TCRs inward to the cSMAC, while large adhesion molecules (e.g. LFA-1) and phosphatase CD45 are sent outward to the pSMAC and dSMAC, respectively. By the end of IS formation, actin-rich filament and myosin II form a peripheral ring in the dSMAC within which concentric circular waves mediated by cyclic actin polymerization and myosin contraction propagate inward towards the IS center (27, 39, 45). In the above processes, external and internal forces exerted on and generated by the T cell are mainly supported by membrane receptors and the actin cytoskeleton. Thus, mechanical forces may regulate actin dynamics, especially actin depolymerization.
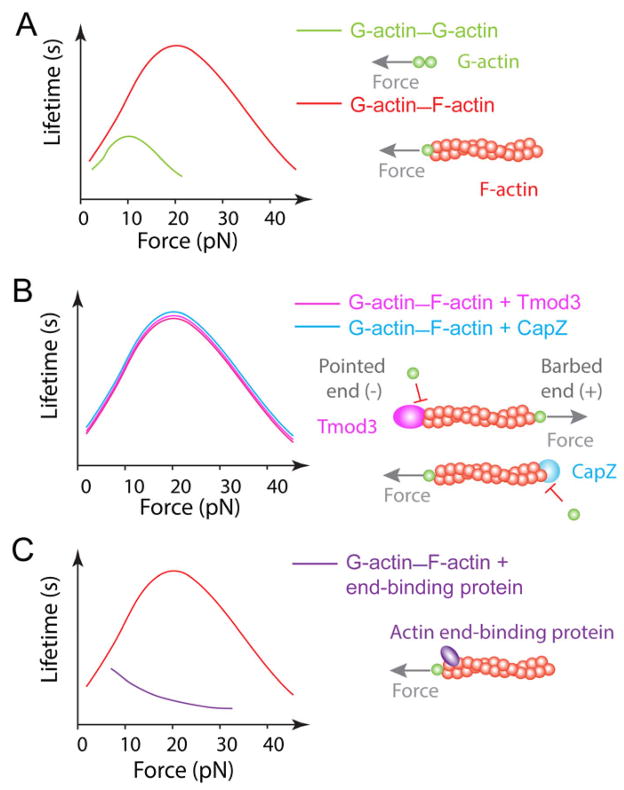
A single filament can bear a force as high as 100 pN without rupture in the middle, as measured by a glass microneedle (117). However, actin depolymerization, which occurs at the ends, can be regulated by much smaller forces. This has recently been shown by AFM single-bond measurements of G-actin–G-actin and G-actin–F-actin dissociation under constant tensile forces (70). Remarkably, low forces prolonged bond lifetimes of these two interactions, resulting in catch bonds, whereas higher forces shortened bond lifetimes, generating slip bonds. The optimal force and the corresponding peak lifetime of the G-actin–F-actin bond are about twice the respective values of the G-actin–G-actin bond, suggesting that the G-actin–G-actin bond at the G-actin–F-actin interface sustains half of the force applied to stretch the actin filament (Fig. 4A). This is reasonable because depolymerization of the terminal actin subunit from the filament tip involves the dissociation of two G-actin–G-actin bonds, an intrastrand long-pitch bond and interstrand short-pitch bond arranged in parallel (118). Since these interactions should also be present periodically between neighboring actin subunits of an F-actin, they can also be considered as catch bonds arranged in series within the actin filament. Thus, F-actin should be strengthened by force not only at the ends but also along the entire filament.
Fig. 4. Mechanical regulation of actin dynamics.
(A) G-actin and F-actin form more pronounced catch-slip bonds (red) than that between two G-actins (green). (B) Catch bonds of G-actin–F-actin remain the same in the presence of actin end-binding proteins, Tmod3 (magenta) or CapZ (blue). (C) Hypothesized transition of G-actin–F-actin catch-slip bond (red) to pure slip bond (purple) in the presence of an actin end-binding protein.
The existence of an optimal force where the actin bond lifetime becomes the longest provides a mechanism for actin microfilaments to orient their organization depending on the anisotropic force field within the cell. Thus, the actin catch bonds may explain tension-induced assembly and stabilization of actin cytoskeleton in microvilli of trafficking T-cells, in protruding lamella of migrating T-cells, and in cyclically waving lamellipodia of the IS between T-cell and APC. Interestingly, catch-slip bonds at the barbed and pointed ends of actin filaments are qualitatively similar (Fig. 4B), suggesting that common structural mechanism underlies the catch-slip bonds at both ends. Lee et al. (70) used SMD simulations to identify a pair of salt bridges between residues K113 and E195 that would be enhanced by force. The contributions of these force-induced interactions to catch bond were verified by mutagenesis studies, showing that eliminating these salt bridges by residue replacements K113S and/or E195S suppressed both the G-actin–G-actin and G-actin–F-actin catch bonds (70). The K113 residue is related to nemalin myopathy mutations in the human actin gene ACTA1. Taken together, these data support the importance of actin catch-slip bonds and suggest that mechanical regulation of actin dynamics may be essential to T-cell functions.
Loading history of force application can also regulate actin dynamics and actin network mechanical properties. Before reaching a threshold force that ceases growth, the growth velocity of the actin network was observed to be force-independent, but rather unexpectedly, loading-history dependent (119). The Bausch group (120) showed that actin network bundled by α-actinin can be hardened by cyclic shear. Given the recently observed cyclic mechanical reinforcement of α5β1-FN interactions (121), it would seem reasonable to predict that cyclic force may also reinforce G-actin–G-actin and G-actin–F-actin interactions. Combined with structural analysis, more insights may be revealed to explain the molecular mechanism of how cyclic mechanical loading affects actin dynamics.
Mechanical force may also affect the functions of actin-associated molecules as related to the assembly of actin filaments under force. Jegou et al. (122) and Courtemanche et al. (123) independently demonstrated that piconewton forces from hydrodynamic flow exerted on a single F-actin filament and a formin [mDia1 from mouse (122) or Bni1p from yeast (123)] could increase F-actin elongation at the barbed end mediated by formin and profilin. Jegou et al. (122) also showed that such small force could slow down depolymerization of F-actin filaments at the barbed end in the presence of formin and profilin. These data lead to a new model to explain formin-mediated F-actin polymerization at the barbed end. The model proposes that tension applied to membrane tethered formin dimers could induce conformational change on FH2 dimer to the open state, which favors the binding of actin monomers to the barbed end of growing actin filaments in the presence of profilin (122). These studies expanded previous work on mechanical regulation of cytoskeleton and its associated regulatory molecules, and developed advanced tension-based single-molecule imaging assays to study other actin-associated molecules under force. It will be of great interest to further investigate force regulation on the functions of other actin-regulating molecules (Fig. 4C), such as Arp2/3 complex, Wiskott-Aldrich syndrome family protein (WASp), as these molecules are critical to actin dynamics and assembly during T-cell trafficking, migration, and IS formation.
Besides actin filament and its binding/regulatory molecules, many adapter, scaffolding, or signaling molecules that link actin filament to membrane receptors are also under tension or even cyclic tension. These molecules consist of direct and indirect binders to actin filaments. For example, direct binders include talin, vinculin, and paxilin. Indirect binders include Lck, Zap-70, Csk (C-terminal Src kinase), LAT (linker for activation of T cells), SLP-76 [Src homology 2 (SH2)-domain-containing leukocyte protein of 76 kDa], SHP-1 (SH2-domain-containing protein tyrosine phosphatase 1), and Itk (interleukin-2-inducible T-cell kinase) (46). Many of these molecules have multiple conformational states, which correspond to different functional activities (e.g. different enzymatic activities). Using single-molecule approaches, Sheetz and colleagues (124) demonstrated that physiologically relevant force (~12 pN) applied on a single talin rod can expose cryptic sites for the binding of multiple vinculins, which could lead to actin cytoskeleton reorganization, exemplifying how mechanical force may be translated to chemical signal. This force-induced conformational change and cryptic site exposure could be a general mechanism operative in other actin-associated signaling molecules. For example, Lck, the first kinase immediately downstream of the TCR triggering, is known to have resting, primed and activated states. In the resting state, Lck adopts a closed conformation in which its kinase domain binds the SH2 and SH3 domains and its terminal domain binds to the SH2 domain through a phosphorylated tyrosine at position 505. Once Tyr505 is dephosphorylated, the terminal domain is released from the SH2 domain, leaving the Lck in the primed sate. Further phosphorylation on Tyr394 induces dissociation of the kinase domain from the SH2 and SH3 domains, resulting in the open conformation and activated state of Lck. This activated Lck may further phosphorylate immunoreceptor tyrosine-based activation motifs (ITAMs) on the CD3 cytoplasmic tails (46). Mechanical force may induce Lck conformational changes and expose tyrosine sites to favor other kinases and/or phosphatases (e.g. Csk and/or CD45) to bind Lck and change its phosphorylation states to fully activate Lck (46). Activated Lck can cluster to regulate T-cell early signaling (125). Such force-regulated protein conformational changes to favor enzymatic activity have been reported in VWF, in which force-induced exposure of the cryptic cleavage site on the A2 domain facilitates enzymatic cleavage by a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13 (ADTAMS-13) (71, 126).
Mechanical regulation of immunoreceptors
T-cell functions depend not only on adhesive and cytoskeletal molecules but also on immunoreceptors. The TCR is of particular interest, because it is arguably the most important immunoreceptor of the adaptive immunity. Binding of the TCR to different pMHCs exhibits different interaction characteristics, which are believed to be the basis for distinctive decisions that lead to different T-cell fates or functions, e.g. T-cell development, thymic selection, lineage commitment and differentiation into effector T cells, or memory T-cell response to foreign antigen (46). The αβ TCR itself does not contain any signaling motif but noncovalently associates with the homo- or hetero-dimeric CD3 subunits, ζζ, εδ, and ε. These signaling subunits contain a total of 10 ITAMs that can be phosphorylated by Src family tyrosine kinases (e.g. Lck). Phosphorylated CD3 ITAMs recruit Zap-70 to transduce signals further downstream (127, 128) (Fig. 1C). However, the mechanism of TCR triggering remains unclear, i.e., how the information embedded in the characteristics of interactions with distinct pMHCs communicates from the binding interface on the top of the αβ TCR along the molecular structure across the cell membrane to the CD3 ITAMs and transduces into different biochemical signals.
Much efforts has been devoted to understand TCR triggering and T cell activation. Several models have been proposed, including kinetic proofreading (129–133), serial triggering (76, 134), kinetic segregation (135–137), TCR dimerization or oligomerization (138, 139), conformational changes (44, 140–143), two competing feedback pathways (144), digital triggering (145) and receptor deformation (146, 147) models. Since T cells experience a wide range of mechanical environments under which the TCR and other load-bearing membrane receptors (e.g. integrins) engage their respective ligands concurrently on the one hand, and connect to their respective cytoskeletal linkages simultaneously on the other hand, we assume that TCR–pMHC bonds are also subjected to forces either externally applied to or internally generated by the T cell (Fig. 1C). Mossman et al. (148) found that blocking the free transport of TCR microclusters in planar bilayers with chrome barriers enhanced the levels of early TCR-associated phosphorylated tyrosine and elevated cytoplasmic Ca2+. These data suggest that chrome barriers may introduce mechanical effect on TCR triggering (148). During IS formation, a T cell generates cyclic protrusion-contraction in the periphery of the IS. This cyclic contraction may allow the T cell and the APC to exert force on TCR–pMHC bonds to mediate TCR triggering (43). Similarly, actively transporting TCR microcluster from the dSMAC and pSMAC to cSMAC by actin retrograde flow may also induce drag force on TCR–pMHC bonds, thereby modulating TCR triggering (39, 40, 44). This is supported by the finding that the disrupting actin cytoskeleton by pharmacological agents abolished TCR downstream signaling (149). Based on crystal structure and NMR studies (150), Reinhertz and colleagues (44) suggested that the relatively rigid CD3ε and the protruding FG loops on Cβ domain of the TCR may help transmitting mechanical force from the membrane distal site to induce a ‘piston-like’ movement on transmembrane domains of CD3s (100, 128). Tolar and colleagues (151) demonstrated that B cells use mechanical energy to discriminate antigen. Although a recent AFM study shows unbinding forces of single TCR–pMHC bonds are not dependent on altered peptides or the presence or absence of co-receptors (152), it seems reasonable to hypothesize that force may serve as a key concept to integrate these models of T-cell triggering.
One aspect of this concept is the ability of the TCR to sense mechanical signals by converting them into chemical signals. Reinherz and colleagues (44) observed that tangential, but not normal, force applied to the TCR via an antibody or pMHC can induce Ca2+ signals. These authors proposed that force might induce conformational changes of the β constant domain F-G loop that might propagate to other parts of the TCR/CD3 complex to initiate T-cell signaling (73). This proposal integrates force into the conformational change model. Li et al. (61) also showed that both a mild shear force from micropipette suction and pulling the TCR/CD3 complex with an elongated CD3 ligand could induce Ca2+. As this elongated CD3 ligand could not induce T-cell activation in the absence of externally applied forces, their data imply that force may enhance segregation of large-size phosphatases (e.g. CD45) from the small-sized TCR to shift their local balance with kinases (e.g. Lck) in favor of phosphorylation. This may integrate force into the kinetic segregation model. Lim et al. (153) found that adhesion strengths (as assessed by AFM pulling) between a T cell and a DC loaded with different pMHCs correlate with T-cell responsiveness, suggesting that mechanically stable DC–T cell contacts are crucial for driving T-cell activation. Lam and coworkers (8) showed that T cells can respond to changing substrate rigidity in a TCR-dependent manner, supporting the mechanosensing ability of the TCR. These studies support the hypothesis that mechanical force could activate T cells by regulating TCR triggering.
Another aspect of mechanical force as an integrating concept is that force may regulate TCR–pMHC dissociation kinetics and antigen discrimination. A major recent development in the analysis of TCR–pMHC interaction has been in situ measurements of the binding kinetics by two-dimensional (2D) methods. These include a single-molecule FRET (smFRET) assay (154), two single-molecule mechanical assays (76, 155–158), a single-molecule diffusion assay (159) and a single-molecule tracking assay (160). Both the smFRET and mechanical studies found much faster 2D TCR–pMHC off-rates than their three dimensional (3D) counterparts measured using soluble protein constructs by SPR (76, 154). These results imply that surface anchoring of both the TCR and the pMHC and association of the αβ TCR with CD3 in the lipid environment may regulate TCR–pMHC interaction. Interestingly, Huppa et al. (154) found that disrupting the actin polymers decreased the 2D off-rates of TCR–pMHC dissociation, suggesting that cytoskeleton dynamics destabilizes this interaction. A 2D study using a flow chamber with recombinant constructs of TCR and pMHC also observed force-dependent off-rates (64). Theoretical studies suggest that segregation of long surface molecules (e.g. CD45) away from TCRs may introduce time-dependent tension stretching the TCR–pMHC bonds leading to an increase in the 2D off-rates (49) and that force may amplify the dynamic range of antigen discrimination (161). These works have provided preliminary evidence for the concept of force regulation of TCR–pMHC dissociation.
The ability for force to regulate TCR–pMHC dissociation may greatly broaden the spectrum of interaction parameters that may potentially correlate with T-cell functions. In particular, TCR may form catch bonds and slip bonds with different pMHCs. A puzzling result of the study by Huang et al. (76) is that the 2D off-rates negatively correlate with the 3D off-rates and with the peptide potency. This is counter-intuitive and opposite to the generally accepted assertion as expressed in the kinetic proofreading model (129). However, these measurements were made in the absence of force. Ligand-specific force-regulated dissociation would allow force to differentially decelerate the off-rate of catch bonds while accelerate the off-rate of slip bonds. This would provide the possibility for agonist-specific catch bonds to invert, in a force-dependent manner, the aforementioned negative correlation found with zero-force off-rates. The TCR is known to be triggered by pMHCs that differ by as little as a single amino acid. Conversion of catch bonds to slip bonds by single-residue replacements has been demonstrated in other molecular systems, including L-selectin–PSGL-1 (78) and GPIbα –VWF (65) interactions. Since the T cell can generate forces on TCR–pMHC bonds, their force-regulated dissociation may provide a feedback mechanism for the T cell to control how it is activated by distinct pMHCs, e.g. to generate different force levels to amplify different triggering signals to be differentially activated.
Force on the TCR–pMHC bond has to be transmitted from the αβ TCR through its interactions with the CD3, the proximal lipid membrane, coreceptors, kinases, phosphatases, and/or adapter/scaffolding proteins to the cytoskeleton. This would provide many possibilities for molecules along the force transmission pathways to be mechanically regulated for their interactions, conformations, or both, which, in turn, regulate their functions and activities. Although at present very little is known, it seems that the possible roles for force to play in regulating TCR triggering and T-cell functions are so great, so broad, and so important, that they can no longer be ignored; rather, considerations of mechanical forces have to be integrated into mainstream immunology. It is our hope that the data and arguments presented in this article would raise awareness to this emerging area of fruitful research in T-cell biology.
Acknowledgments
We thank Janis Burkhardt and Hai-tao He for helpful comments on the manuscript. We thank former and current Zhu lab members and collaborators who contributed to the results reviewed in this article. This work was supported by NIH grants AI38282 and GM096187.
Footnotes
The authors declare no competing financial interests.
References
- 1.Petrie HT, Zúñiga-Pflücker JC. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu Rev Immunol. 2007;25:649–679. doi: 10.1146/annurev.immunol.23.021704.115715. [DOI] [PubMed] [Google Scholar]
- 2.Koch U, Radtke F. Mechanisms of T cell development and transformation. Annu Rev Cell Dev Biol. 2011;27:539–562. doi: 10.1146/annurev-cellbio-092910-154008. [DOI] [PubMed] [Google Scholar]
- 3.Love PE, Bhandoola A. Signal integration and crosstalk during thymocyte migration and emigration. Nat Rev Immunol. 2011;11:469–477. doi: 10.1038/nri2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.von Andrian UH, Mackay CR. T-cell function and migration. Two sides of the same coin. N Engl J Med. 2000;343:1020–1034. doi: 10.1056/NEJM200010053431407. [DOI] [PubMed] [Google Scholar]
- 5.Huppa JB, Davis MM. T-cell-antigen recognition and the immunological synapse. Nat Rev Immunol. 2003;3:973–983. doi: 10.1038/nri1245. [DOI] [PubMed] [Google Scholar]
- 6.Ley K, Laudanna C, Cybulsky MI, Nourshargh S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat Rev Immunol. 2007;7:678–689. doi: 10.1038/nri2156. [DOI] [PubMed] [Google Scholar]
- 7.Dustin ML. Modular design of immunological synapses and kinapses. Cold Spring Harb Perspect Biol. 2009;1:a002873. doi: 10.1101/cshperspect.a002873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Judokusumo E, Tabdanov E, Kumari S, Dustin ML, Kam LC. Mechanosensing in T lymphocyte activation. Biophys J. 2012;102:L5–7. doi: 10.1016/j.bpj.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Connor RS, Hao X, Shen K, Bashour K, Akimova T, Hancock WW, Kam LC, et al. Substrate rigidity regulates human T cell activation and proliferation. J Immunol. 2012;189:1330–1339. doi: 10.4049/jimmunol.1102757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 11.Fu J, Wang Y-K, Yang MT, Desai RA, Yu X, Liu Z, Chen CS. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat Meth. 2010;7:733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saha K, Keung AJ, Irwin EF, Li Y, Little L, Schaffer DV, Healy KE. Substrate modulus directs neural stem cell behavior. Biophys J. 2008;95:4426–4438. doi: 10.1529/biophysj.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin J-W, Swift J, Ivanovska I, Spinler KR, Buxboim A, Discher DE. Mechanobiology of bone marrow stem cells: From myosin-II forces to compliance of matrix and nucleus in cell forms and fates. Differentiation. 2013 doi: 10.1016/j.diff.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luster AD, Alon R, von Andrian UH. Immune cell migration in inflammation: present and future therapeutic targets. Nat Immunol. 2005;6:1182–1190. doi: 10.1038/ni1275. [DOI] [PubMed] [Google Scholar]
- 15.McEver RP, Zhu C. Rolling cell adhesion. Annu Rev Cell Dev Biol. 2010;26:363– 396. doi: 10.1146/annurev.cellbio.042308.113238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yago T, Zarnitsyna VI, Klopocki AG, McEver RP, Zhu C. Transport governs flow- enhanced cell tethering through L-selectin at threshold shear. Biophys J. 2007;92:330–342. doi: 10.1529/biophysj.106.090969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marshall BT, Long M, Piper JW, Yago T, McEver RP, Zhu C. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 2003;423:190–193. doi: 10.1038/nature01605. [DOI] [PubMed] [Google Scholar]
- 18.Sarangapani KK, Yago T, Klopocki AG, Lawrence MB, Fieger CB, Rosen SD, McEver RP, et al. Low force decelerates L-selectin dissociation from P-selectin glycoprotein ligand-1 and endoglycan. J Biol Chem. 2004;279:2291–2298. doi: 10.1074/jbc.M310396200. [DOI] [PubMed] [Google Scholar]
- 19.Wayman AM, Chen W, McEver RP, Zhu C. Triphasic force dependence of E- selectin/ligand dissociation governs cell rolling under flow. Biophys J. 2010;99:1166–1174. doi: 10.1016/j.bpj.2010.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu C, Yago T, Lou J, Zarnitsyna VI, McEver RP. Mechanisms for flow-enhanced cell adhesion. Annals of Biomedical Engineering. 2008;36:604–621. doi: 10.1007/s10439-008-9464-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miner JJ, Xia L, Yago T, Kappelmayer J, Liu Z, Klopocki AG, Shao B, et al. Separable requirements for cytoplasmic domain of PSGL-1 in leukocyte rolling and signaling under flow. Blood. 2008;112:2035–2045. doi: 10.1182/blood-2008-04-149468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zarbock A, Abram CL, Hundt M, Altman A, Lowell CA, Ley K. PSGL-1 engagement by E-selectin signals through Src kinase Fgr and ITAM adapters DAP12 and FcR gamma to induce slow leukocyte rolling. J Exp Med. 2008;205:2339–2347. doi: 10.1084/jem.20072660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuwano Y, Spelten O, Zhang H, Ley K, Zarbock A. Rolling on E- or P-selectin induces the extended but not high-affinity conformation of LFA-1 in neutrophils. Blood. 2010;116:617–624. doi: 10.1182/blood-2010-01-266122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shao B, Yago T, Coghill PA, Klopocki AG, Mehta-D’souza P, Schmidtke DW, Rodgers W, et al. Signal-dependent slow leukocyte rolling does not require cytoskeletal anchorage of P-selectin glycoprotein ligand-1 (PSGL-1) or integrin αLβ2. J Biol Chem. 2012;287:19585–19598. doi: 10.1074/jbc.M112.361519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lefort CT, Rossaint J, Moser M, Petrich BG, Zarbock A, Monkley SJ, Critchley DR, et al. Distinct roles for talin-1 and kindlin-3 in LFA-1 extension and affinity regulation. Blood. 2012;119:4275–4282. doi: 10.1182/blood-2011-08-373118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alon R, Feigelson SW. Chemokine-triggered leukocyte arrest: force-regulated bidirectional integrin activation in quantal adhesive contacts. Curr Opin Cell Biol. 2012;24:670–676. doi: 10.1016/j.ceb.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Alon R, Dustin ML. Force as a facilitator of integrin conformational changes during leukocyte arrest on blood vessels and antigen-presenting cells. Immunity. 2007;26:17–27. doi: 10.1016/j.immuni.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 28.Bolomini-Vittori M, Montresor A, Giagulli C, Staunton D, Rossi B, Martinello M, Constantin G, et al. Regulation of conformer-specific activation of the integrin LFA-1 by a chemokine-triggered Rho signaling module. Nat Immunol. 2009;10:185–194. doi: 10.1038/ni.1691. [DOI] [PubMed] [Google Scholar]
- 29.Hart R, Stanley P, Chakravarty P, Hogg N. The kindlin 3 pleckstrin homology domain has an essential role in lymphocyte function-associated antigen 1 (LFA-1) integrin-mediated B cell adhesion and migration. J Biol Chem. 2013;288:14852–14862. doi: 10.1074/jbc.M112.434621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stanley P, Smith A, McDowall A, Nicol A, Zicha D, Hogg N. Intermediate-affinity LFA-1 binds alpha-actinin-1 to control migration at the leading edge of the T cell. EMBO J. 2008;27:62–75. doi: 10.1038/sj.emboj.7601959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith A, Carrasco YR, Stanley P, Kieffer N, Batista FD, Hogg N. A talin-dependent LFA-1 focal zone is formed by rapidly migrating T lymphocytes. J Cell Biol. 2005;170:141–151. doi: 10.1083/jcb.200412032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith A, Stanley P, Jones K, Svensson L, McDowall A, Hogg N. The role of the integrin LFA-1 in T-lymphocyte migration. Immunol Rev. 2007;218:135–146. doi: 10.1111/j.1600-065X.2007.00537.x. [DOI] [PubMed] [Google Scholar]
- 33.Shakhar G, Lindquist RL, Skokos D, Dudziak D, Huang JH, Nussenzweig MC, Dustin ML. Stable T cell-dendritic cell interactions precede the development of both tolerance and immunity in vivo. Nat Immunol. 2005;6:707–714. doi: 10.1038/ni1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dustin ML. T-cell activation through immunological synapses and kinapses. Immunol Rev. 2008;221:77–89. doi: 10.1111/j.1600-065X.2008.00589.x. [DOI] [PubMed] [Google Scholar]
- 35.Monks CR, Freiberg BA, Kupfer H, Sciaky N, Kupfer A. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature. 1998;395:82–86. doi: 10.1038/25764. [DOI] [PubMed] [Google Scholar]
- 36.Grakoui A, Bromley SK, Sumen C, Davis MM, Shaw AS, Allen PM, Dustin ML. The immunological synapse: a molecular machine controlling T cell activation. Science. 1999;285:221–227. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- 37.Dustin ML, Groves JT. Receptor signaling clusters in the immune synapse. Annu Rev Biophys. 2012;41:543–556. doi: 10.1146/annurev-biophys-042910-155238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Babich A, Burkhardt JK. Lymphocyte Signaling Converges on Microtubules. Immunity. 2011;34:825–827. doi: 10.1016/j.immuni.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 39.Yi J, Wu XS, Crites T, Hammer JA. Actin retrograde flow and actomyosin II arc contraction drive receptor cluster dynamics at the immunological synapse in Jurkat T cells. Mol Biol Cell. 2012;23:834–852. doi: 10.1091/mbc.E11-08-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Babich A, Li S, O’Connor RS, Milone MC, Freedman BD, Burkhardt JK. F-actin polymerization and retrograde flow drive sustained PLCγ1 signaling during T cell activation. J Cell Biol. 2012;197:775–787. doi: 10.1083/jcb.201201018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hammer JA, Burkhardt JK. Controversy and consensus regarding myosin II function at the immunological synapse. Current Opinion in Immunology. 2013;25:300–306. doi: 10.1016/j.coi.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dustin ML. The cellular context of T cell signaling. Immunity. 2009;30:482–492. doi: 10.1016/j.immuni.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sims TN, Soos TJ, Xenias HS, Dubin-Thaler B, Hofman JM, Waite JC, Cameron TO, et al. Opposing effects of PKCtheta and WASp on symmetry breaking and relocation of the immunological synapse. Cell. 2007;129:773–785. doi: 10.1016/j.cell.2007.03.037. [DOI] [PubMed] [Google Scholar]
- 44.Kim ST, Takeuchi K, Sun Z-YJ, Touma M, Castro CE, Fahmy A, Lang MJ, et al. The alphabeta T cell receptor is an anisotropic mechanosensor. J Biol Chem. 2009;284:31028–31037. doi: 10.1074/jbc.M109.052712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ilani T, Vasiliver-Shamis G, Vardhana S, Bretscher A, Dustin ML. T cell antigen receptor signaling and immunological synapse stability require myosin IIA. Nat Immunol. 2009;10:531–539. doi: 10.1038/ni.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Acuto O, Di Bartolo V, Michel F. Tailoring T-cell receptor signals by proximal negative feedback mechanisms. Nat Rev Immunol. 2008;8:699–712. doi: 10.1038/nri2397. [DOI] [PubMed] [Google Scholar]
- 47.Stachowiak JC, Schmid EM, Ryan CJ, Ann HS, Sasaki DY, Sherman MB, Geissler PL, et al. Membrane bending by protein–protein crowding. Nat Cell Biol. 2012;14:944–949. doi: 10.1038/ncb2561. [DOI] [PubMed] [Google Scholar]
- 48.James JR, Vale RD. Biophysical mechanism of T-cell receptor triggering in a reconstituted system. Nature. 2012;487:64–69. doi: 10.1038/nature11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allard JF, Dushek O, Coombs D, van der Merwe PA. Mechanical modulation of receptor-ligand interactions at cell-cell interfaces. Biophys J. 2012;102:1265–1273. doi: 10.1016/j.bpj.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shyy JY-J, Chien S. Role of integrins in endothelial mechanosensing of shear stress. Circ Res. 2002;91:769–775. doi: 10.1161/01.res.0000038487.19924.18. [DOI] [PubMed] [Google Scholar]
- 51.Moore SW, Roca-Cusachs P, Sheetz MP. Stretchy proteins on stretchy substrates: the important elements of integrin-mediated rigidity sensing. Dev Cell. 2010;19:194–206. doi: 10.1016/j.devcel.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen KD, Li YS, Kim M, Li S, Yuan S, Chien S, Shyy JY. Mechanotransduction in response to shear stress. Roles of receptor tyrosine kinases, integrins, and Shc. J Biol Chem. 1999;274:18393–18400. doi: 10.1074/jbc.274.26.18393. [DOI] [PubMed] [Google Scholar]
- 53.Astrof NS, Salas A, Shimaoka M, Chen J, Springer TA. Importance of force linkage in mechanochemistry of adhesion receptors. Biochemistry. 2006;45:15020–15028. doi: 10.1021/bi061566o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kong F, García AJ, Mould AP, Humphries MJ, Zhu C. Demonstration of catch bonds between an integrin and its ligand. J Cell Biol. 2009;185:1275–1284. doi: 10.1083/jcb.200810002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen W, Lou J, Zhu C. Forcing switch from short- to intermediate- and long-lived states of the alphaA domain generates LFA-1/ICAM-1 catch bonds. J Biol Chem. 2010;285:35967–35978. doi: 10.1074/jbc.M110.155770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Friedland JC, Lee MH, Boettiger D. Mechanically activated integrin switch controls alpha5beta1 function_sup. Science. 2009;323:642–644. doi: 10.1126/science.1168441. [DOI] [PubMed] [Google Scholar]
- 57.Puklin-Faucher E, Vogel V. Integrin activation dynamics between the RGD-binding site and the headpiece hinge. J Biol Chem. 2009;284:36557–36568. doi: 10.1074/jbc.M109.041194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Puklin-Faucher E, Gao M, Schulten K, Vogel V. How the headpiece hinge angle is opened: New insights into the dynamics of integrin activation. J Cell Biol. 2006;175:349–360. doi: 10.1083/jcb.200602071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu J, Luo B-H, Xiao T, Zhang C, Nishida N, Springer TA. Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol Cell. 2008;32:849–861. doi: 10.1016/j.molcel.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schwartz MA. Integrins and extracellular matrix in mechanotransduction. Cold Spring Harb Perspect Biol. 2010;2:a005066. doi: 10.1101/cshperspect.a005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Y-C, Chen B-M, Wu P-C, Cheng T-L, Kao L-S, Tao M-H, Lieber A, et al. Cutting Edge: mechanical forces acting on T cells immobilized via the TCR complex can trigger TCR signaling. J Immunol. 2010;184:5959–5963. doi: 10.4049/jimmunol.0900775. [DOI] [PubMed] [Google Scholar]
- 62.Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 63.Dembo M, Torney DC, Saxman K, Hammer D. The reaction-limited kinetics of membrane-to-surface adhesion and detachment. Proc R Soc Lond, B, Biol Sci. 1988;234:55–83. doi: 10.1098/rspb.1988.0038. [DOI] [PubMed] [Google Scholar]
- 64.Robert P, Aleksic M, Dushek O, Cerundolo V, Bongrand P, van der Merwe PA. Kinetics and mechanics of two-dimensional interactions between T cell receptors and different activating ligands. Biophys J. 2012;102:248–257. doi: 10.1016/j.bpj.2011.11.4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yago T, Lou J, Wu T, Yang J, Miner JJ, Coburn L, López JA, et al. Platelet glycoprotein Ibalpha forms catch bonds with human WT vWF but not with type 2B von Willebrand disease vWF. J Clin Invest. 2008;118:3195–3207. doi: 10.1172/JCI35754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yakovenko O, Sharma S, Forero M, Tchesnokova V, Aprikian P, Kidd B, Mach A, et al. FimH forms catch bonds that are enhanced by mechanical force due to allosteric regulation. J Biol Chem. 2008;283:11596–11605. doi: 10.1074/jbc.M707815200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rakshit S, Zhang Y, Manibog K, Shafraz O, Sivasankar S. Ideal, catch, and slip bonds in cadherin adhesion. Proc Natl Acad Sci USA. 2012;109:18815–18820. doi: 10.1073/pnas.1208349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guo B, Guilford WH. Mechanics of actomyosin bonds in different nucleotide states are tuned to muscle contraction. Proc Natl Acad Sci USA. 2006;103:9844–9849. doi: 10.1073/pnas.0601255103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Akiyoshi B, Sarangapani KK, Powers AF, Nelson CR, Reichow SL, Arellano-Santoyo H, Gonen T, et al. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 2010;468:576–579. doi: 10.1038/nature09594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee C-Y, Lou J, Wen K-K, McKane M, Eskin SG, Ono S, Chien S, et al. Actin depolymerization under force is governed by lysine 113:glutamic acid 195-mediated catch-slip bonds. Proc Natl Acad Sci USA. 2013;110:5022–5027. doi: 10.1073/pnas.1218407110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu T, Lin J, Cruz MA, Dong J-F, Zhu C. Force-induced cleavage of single VWFA1A2A3 tridomains by ADAMTS-13. Blood. 2010;115:370–378. doi: 10.1182/blood-2009-03-210369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wiita AP, Perez-Jimenez R, Walther KA, Gräter F, Berne BJ, Holmgren A, Sanchez-Ruiz JM, et al. Probing the chemistry of thioredoxin catalysis with force. Nature. 2007;450:124–127. doi: 10.1038/nature06231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim ST, Shin Y, Brazin K, Mallis RJ, Sun Z-YJ, Wagner G, Lang MJ, et al. TCR Mechanobiology: Torques and Tunable Structures Linked to Early T Cell Signaling. Front Immunol. 2012;3:76. doi: 10.3389/fimmu.2012.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Davis MM, Boniface JJ, Reich Z, Lyons D, Hampl J, Arden B, Chien Y. Ligand recognition by alpha beta T cell receptors. Annu Rev Immunol. 1998;16:523–544. doi: 10.1146/annurev.immunol.16.1.523. [DOI] [PubMed] [Google Scholar]
- 75.Stone JD, Chervin AS, Kranz DM. T-cell receptor binding affinities and kinetics: impact on T-cell activity and specificity. Immunology. 2009;126:165–176. doi: 10.1111/j.1365-2567.2008.03015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang J, Zarnitsyna VI, Liu B, Edwards LJ, Jiang N, Evavold BD, Zhu C. The kinetics of two-dimensional TCR and pMHC interactions determine T-cell responsiveness. Nature. 2010;464:932–936. doi: 10.1038/nature08944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lou J, Zhu C. A structure-based sliding-rebinding mechanism for catch bonds. Biophys J. 2007;92:1471–1485. doi: 10.1529/biophysj.106.097048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Klopocki AG, Yago T, Mehta P, Yang J, Wu T, Leppänen A, Bovin NV, et al. Replacing a lectin domain residue in L-selectin enhances binding to P-selectin glycoprotein ligand-1 but not to 6-sulfo-sialyl Lewis x. J Biol Chem. 2008;283:11493–11500. doi: 10.1074/jbc.M709785200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thomas WE, Trintchina E, Forero M, Vogel V, Sokurenko EV. Bacterial adhesion to target cells enhanced by shear force. Cell. 2002;109:913–923. doi: 10.1016/s0092-8674(02)00796-1. [DOI] [PubMed] [Google Scholar]
- 80.Lou J, Yago T, Klopocki AG, Mehta P, Chen W, Zarnitsyna VI, Bovin NV, et al. Flow-enhanced adhesion regulated by a selectin interdomain hinge. J Cell Biol. 2006;174:1107–1117. doi: 10.1083/jcb.200606056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hogg N, Laschinger M, Giles K, McDowall A. T-cell integrins: more than just sticking points. J Cell Sci. 2003;116:4695–4705. doi: 10.1242/jcs.00876. [DOI] [PubMed] [Google Scholar]
- 82.Pribila JT, Quale AC, Mueller KL, Shimizu Y. Integrins and T cell-mediated immunity. Annu Rev Immunol. 2004;22:157–180. doi: 10.1146/annurev.immunol.22.012703.104649. [DOI] [PubMed] [Google Scholar]
- 83.Luo B-H, Carman CV, Springer TA. Structural basis of integrin regulation and signaling. Annu Rev Immunol. 2007;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xiong J-P, Stehle T, Diefenbach B, Zhang R, Dunker R, Scott DL, Joachimiak A, et al. Crystal structure of the extracellular segment of integrin alphaVbeta3. Science. 2001;294:339–345. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xiong J-P, Stehle T, Zhang R, Joachimiak A, Frech M, Goodman SL, Arnaout MA. Crystal structure of the extracellular segment of integrin alphaVbeta3 in complex with an Arg-Gly-Asp ligand. Science. 2002;296:151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- 86.Xiong J-P, Mahalingham B, Alonso JL, Borrelli LA, Rui X, Anand S, Hyman BT, et al. Crystal structure of the complete integrin alphaVbeta3 ectodomain plus an alpha/beta transmembrane fragment. J Cell Biol. 2009;186:589–600. doi: 10.1083/jcb.200905085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xie C, Zhu J, Chen X, Mi L, Nishida N, Springer TA. Structure of an integrin with an alphaI domain, complement receptor type 4. EMBO J. 2010;29:666–679. doi: 10.1038/emboj.2009.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yu Y, Zhu J, Mi L-Z, Walz T, Sun H, Chen J, Springer TA. Structural specializations of α(4)β(7), an integrin that mediates rolling adhesion. J Cell Biol. 2012;196:131–146. doi: 10.1083/jcb.201110023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Takagi J, Petre BM, Walz T, Springer TA. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 2002;110:599–511. doi: 10.1016/s0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- 90.Nishida N, Xie C, Shimaoka M, Cheng Y, Walz T, Springer TA. Activation of leukocyte beta2 integrins by conversion from bent to extended conformations. Immunity. 2006;25:583–594. doi: 10.1016/j.immuni.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 91.Ye F, Hu G, Taylor D, Ratnikov B, Bobkov AA, McLean MA, Sligar SG, et al. Recreation of the terminal events in physiological integrin activation. J Cell Biol. 2010;188:157–173. doi: 10.1083/jcb.200908045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Beglova N, Blacklow SC, Takagi J, Springer TA. Cysteine-rich module structure reveals a fulcrum for integrin rearrangement upon activation. Nat Struct Biol. 2002;9:282–287. doi: 10.1038/nsb779. [DOI] [PubMed] [Google Scholar]
- 93.Kim M, Carman CV, Springer TA. Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science. 2003;301:1720–1725. doi: 10.1126/science.1084174. [DOI] [PubMed] [Google Scholar]
- 94.Chigaev A, Buranda T, Dwyer DC, Prossnitz ER, Sklar LA. FRET detection of cellular alpha4-integrin conformational activation. Biophys J. 2003;85:3951–3962. doi: 10.1016/S0006-3495(03)74809-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chigaev A, Zwartz GJ, Buranda T, Edwards BS, Prossnitz ER, Sklar LA. Conformational regulation of alpha 4 beta 1-integrin affinity by reducing agents. “Inside-out” signaling is independent of and additive to reduction-regulated integrin activation. J Biol Chem. 2004;279:32435–32443. doi: 10.1074/jbc.M404387200. [DOI] [PubMed] [Google Scholar]
- 96.Kim C, Schmidt T, Cho E-G, Ye F, Ulmer TS, Ginsberg MH. Basic amino-acid side chains regulate transmembrane integrin signalling. Nature. 2012;481:209–213. doi: 10.1038/nature10697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen W, Lou J, Evans EA, Zhu C. Observing force-regulated conformational changes and ligand dissociation from a single integrin on cells. J Cell Biol. 2012;199:497–512. doi: 10.1083/jcb.201201091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen X, Yu Y, Mi L-Z, Walz T, Springer TA. Molecular basis for complement recognition by integrin αXβ2. Proc Natl Acad Sci USA. 2012;109:4586–4591. doi: 10.1073/pnas.1202051109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Moser M, Legate KR, Zent R, Fässler R. The tail of integrins, talin, and kindlins. Science. 2009;324:895–899. doi: 10.1126/science.1163865. [DOI] [PubMed] [Google Scholar]
- 100.Kim C, Ye F, Hu X, Ginsberg MH. Talin activates integrins by altering the topology of the β transmembrane domain. J Cell Biol. 2012;197:605–611. doi: 10.1083/jcb.201112141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhu J, Zhu J, Springer TA. Complete integrin headpiece opening in eight steps. J Cell Biol. 2013;201:1053–1068. doi: 10.1083/jcb.201212037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang F, Marcus WD, Goyal NH, Selvaraj P, Springer TA, Zhu C. Two-dimensional kinetics regulation of alphaLbeta2-ICAM-1 interaction by conformational changes of the alphaL-inserted domain. J Biol Chem. 2005;280:42207–42218. doi: 10.1074/jbc.M510407200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ma Q, Shimaoka M, Lu C, Jing H, Carman CV, Springer TA. Activation-induced conformational changes in the I domain region of lymphocyte function-associated antigen 1. J Biol Chem. 2002;277:10638–10641. doi: 10.1074/jbc.M112417200. [DOI] [PubMed] [Google Scholar]
- 104.Litvinov RI, Mekler A, Shuman H, Bennett JS, Barsegov V, Weisel JW. Resolving two-dimensional kinetics of the integrin αIIbβ3-fibrinogen interactions using binding- unbinding correlation spectroscopy. J Biol Chem. 2012;287:35275–35285. doi: 10.1074/jbc.M112.404848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Horn J, Wang X, Reichardt P, Stradal TE, Warnecke N, Simeoni L, Gunzer M, et al. Src homology 2-domain containing leukocyte-specific phosphoprotein of 76 kDa is mandatory for TCR-mediated inside-out signaling, but dispensable for CXCR4-mediated LFA-1 activation, adhesion, and migration of T cells. J Immunol. 2009;183:5756–5767. doi: 10.4049/jimmunol.0900649. [DOI] [PubMed] [Google Scholar]
- 106.Lee D, Kim J, Baker RG, Koretzky GA, Hammer DA. SLP-76 is required for optimal CXCR4-stimulated T lymphocyte firm arrest to ICAM-1 under shear flow. Eur J Immunol. 2012;42:2736–2743. doi: 10.1002/eji.201142303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shimaoka M, Lu C, Salas A, Xiao T, Takagi J, Springer TA. Stabilizing the integrin alpha M inserted domain in alternative conformations with a range of engineered disulfide bonds. Proc Natl Acad Sci USA. 2002;99:16737–16741. doi: 10.1073/pnas.252633099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jin M, Andricioaei I, Springer TA. Conversion between three conformational states of integrin I domains with a C-terminal pull spring studied with molecular dynamics. Structure. 2004;12:2137–2147. doi: 10.1016/j.str.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 109.Xiang X, Lee C-Y, Li T, Chen W, Lou J, Zhu C. Structural Basis and Kinetics of Force-Induced Conformational Changes of an αA Domain-Containing Integrin. PLoS ONE. 2011;6:e27946. doi: 10.1371/journal.pone.0027946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Woolf E, Grigorova I, Sagiv A, Grabovsky V, Feigelson SW, Shulman Z, Hartmann T, et al. Lymph node chemokines promote sustained T lymphocyte motility without triggering stable integrin adhesiveness in the absence of shear forces. Nat Immunol. 2007;8:1076–1085. doi: 10.1038/ni1499. [DOI] [PubMed] [Google Scholar]
- 111.Shimaoka M, Salas A, Yang W, Weitz-Schmidt G, Springer TA. Small molecule integrin antagonists that bind to the beta2 subunit I-like domain and activate signals in one direction and block them in the other. Immunity. 2003;19:391–402. doi: 10.1016/s1074-7613(03)00238-3. [DOI] [PubMed] [Google Scholar]
- 112.Salas A, Shimaoka M, Kogan AN, Harwood C, von Andrian UH, Springer TA. Rolling adhesion through an extended conformation of integrin alphaLbeta2 and relation to alpha I and beta I-like domain interaction. Immunity. 2004;20:393–406. doi: 10.1016/s1074-7613(04)00082-2. [DOI] [PubMed] [Google Scholar]
- 113.Shimaoka M, Takagi J, Springer TA. Conformational regulation of integrin structure and function. Annu Rev Biophys Biomol Struct. 2002;31:485–516. doi: 10.1146/annurev.biophys.31.101101.140922. [DOI] [PubMed] [Google Scholar]
- 114.Xiao T, Takagi J, Coller BS, Wang J-H, Springer TA. Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature. 2004;432:59–67. doi: 10.1038/nature02976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chen W, Lou J, Hsin J, Schulten K, Harvey SC, Zhu C. Molecular dynamics simulations of forced unbending of integrin α(v)β. PLoS Comput Biol. 2011;7:e1001086. doi: 10.1371/journal.pcbi.1001086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Burkhardt JK, Carrizosa E, Shaffer MH. The actin cytoskeleton in T cell activation. Annu Rev Immunol. 2008;26:233–259. doi: 10.1146/annurev.immunol.26.021607.090347. [DOI] [PubMed] [Google Scholar]
- 117.Kishino A, Yanagida T. Force measurements by micromanipulation of a single actin filament by glass needles. Nature. 1988;334:74–76. doi: 10.1038/334074a0. [DOI] [PubMed] [Google Scholar]
- 118.Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 119.Parekh SH, Chaudhuri O, Theriot JA, Fletcher DA. Loading history determines the velocity of actin-network growth. Nat Cell Biol. 2005;7:1219–1223. doi: 10.1038/ncb1336. [DOI] [PubMed] [Google Scholar]
- 120.Schmoller KM, Fernández P, Arevalo RC, Blair DL, Bausch AR. Cyclic hardening in bundled actin networks. Nat Commun. 2010;1:134. doi: 10.1038/ncomms1134. [DOI] [PubMed] [Google Scholar]
- 121.Kong F, Li Z, Parks WM, Dumbauld DW, García AJ, Mould AP, Humphries MJ, et al. Cyclic mechanical reinforcement of integrin-ligand interactions. Mol Cell. 2013;49:1060–1068. doi: 10.1016/j.molcel.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jégou A, Carlier M-F, Romet-Lemonne G. Formin mDia1 senses and generates mechanical forces on actin filaments. Nat Commun. 2013;4:1883. doi: 10.1038/ncomms2888. [DOI] [PubMed] [Google Scholar]
- 123.Courtemanche N, Lee JY, Pollard TD, Greene EC. Tension modulates actin filament polymerization mediated by formin and profilin. Proc Natl Acad Sci USA. 2013;110:9752–9757. doi: 10.1073/pnas.1308257110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rio AD, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rossy J, Owen DM, Williamson DJ, Yang Z, Gaus K. Conformational states of the kinase Lck regulate clustering in early T cell signaling. Nat Immunol. 2013;14:82–89. doi: 10.1038/ni.2488. [DOI] [PubMed] [Google Scholar]
- 126.Zhang X, Halvorsen K, Zhang C-Z, Wong WP, Springer TA. Mechanoenzymatic cleavage of the ultralarge vascular protein von Willebrand factor_supplemental. Science. 2009;324:1330–1334. doi: 10.1126/science.1170905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Reinherz EL, Acuto O. Molecular T cell biology -- basic and translational challenges in the twenty-first century. Front Immunol. 2011;2:3. doi: 10.3389/fimmu.2011.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.van der Merwe PA, Dushek O. Mechanisms for T cell receptor triggering. Nat Rev Immunol. 2011;11:47–55. doi: 10.1038/nri2887. [DOI] [PubMed] [Google Scholar]
- 129.McKeithan TW. Kinetic proofreading in T-cell receptor signal transduction. Proc Natl Acad Sci USA. 1995;92:5042–5046. doi: 10.1073/pnas.92.11.5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rabinowitz JD, Beeson C, Lyons DS, Davis MM, McConnell HM. Kinetic discrimination in T-cell activation. Proc Natl Acad Sci USA. 1996;93:1401–1405. doi: 10.1073/pnas.93.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kersh GJ, Kersh EN, Fremont DH, Allen PM. High- and low-potency ligands with similar affinities for the TCR: the importance of kinetics in TCR signaling. Immunity. 1998;9:817–826. doi: 10.1016/s1074-7613(00)80647-0. [DOI] [PubMed] [Google Scholar]
- 132.Rosette C, Werlen G, Daniels MA, Holman PO, Alam SM, Travers PJ, Gascoigne NR, et al. The impact of duration versus extent of TCR occupancy on T cell activation: a revision of the kinetic proofreading model. Immunity. 2001;15:59–70. doi: 10.1016/s1074-7613(01)00173-x. [DOI] [PubMed] [Google Scholar]
- 133.Dushek O, Das R, Coombs D. A role for rebinding in rapid and reliable T cell responses to antigen. PLoS Comput Biol. 2009;5:e1000578. doi: 10.1371/journal.pcbi.1000578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Valitutti S, Müller S, Cella M, Padovan E, Lanzavecchia A. Serial triggering of many T-cell receptors by a few peptide-MHC complexes. Nature. 1995;375:148–151. doi: 10.1038/375148a0. [DOI] [PubMed] [Google Scholar]
- 135.Davis SJ, van der Merwe PA. The structure and ligand interactions of CD2: implications for T-cell function. Immunol Today. 1996;17:177–187. doi: 10.1016/0167-5699(96)80617-7. [DOI] [PubMed] [Google Scholar]
- 136.Anton van der Merwe P, Davis SJ, Shaw AS, Dustin ML. Cytoskeletal polarization and redistribution of cell-surface molecules during T cell antigen recognition. Semin Immunol. 2000;12:5–21. doi: 10.1006/smim.2000.0203. [DOI] [PubMed] [Google Scholar]
- 137.Davis SJ, van der Merwe PA. The kinetic-segregation model: TCR triggering and beyond. Nat Immunol. 2006;7:803–809. doi: 10.1038/ni1369. [DOI] [PubMed] [Google Scholar]
- 138.Irvine DJ, Purbhoo MA, Krogsgaard M, Davis MM. Direct observation of ligand recognition by T cells. Nature. 2002;419:845–849. doi: 10.1038/nature01076. [DOI] [PubMed] [Google Scholar]
- 139.Krogsgaard M, Li Q-J, Sumen C, Huppa JB, Huse M, Davis MM. Agonist/endogenous peptide-MHC heterodimers drive T cell activation and sensitivity. Nature. 2005;434:238–243. doi: 10.1038/nature03391. [DOI] [PubMed] [Google Scholar]
- 140.Aivazian D, Stern LJ. Phosphorylation of T cell receptor zeta is regulated by a lipid dependent folding transition. Nat Struct Biol. 2000;7:1023–1026. doi: 10.1038/80930. [DOI] [PubMed] [Google Scholar]
- 141.Xu C, Gagnon E, Call ME, Schnell JR, Schwieters CD, Carman CV, Chou JJ, et al. Regulation of T cell receptor activation by dynamic membrane binding of the CD3epsilon cytoplasmic tyrosine-based motif. Cell. 2008;135:702–713. doi: 10.1016/j.cell.2008.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kuhns MS, Girvin AT, Klein LO, Chen R, Jensen KDC, Newell EW, Huppa JB, et al. Evidence for a functional sidedness to the alphabetaTCR. Proc Natl Acad Sci USA. 2010;107:5094–5099. doi: 10.1073/pnas.1000925107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang H, Cordoba S-P, Dushek O, van der Merwe PA. Basic residues in the T-cell receptor ζ cytoplasmic domain mediate membrane association and modulate signaling. Proc Natl Acad Sci USA. 2011;108:19323–19328. doi: 10.1073/pnas.1108052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Stefanová I, Hemmer B, Vergelli M, Martin R, Biddison WE, Germain RN. TCR ligand discrimination is enforced by competing ERK positive and SHP-1 negative feedback pathways. Nat Immunol. 2003;4:248–254. doi: 10.1038/ni895. [DOI] [PubMed] [Google Scholar]
- 145.Altan-Bonnet G, Germain RN. Modeling T cell antigen discrimination based on feedback control of digital ERK responses. PLoS Biol. 2005;3:e356. doi: 10.1371/journal.pbio.0030356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ma Z, Sharp KA, Janmey PA, Finkel TH. Surface-anchored monomeric agonist pMHCs alone trigger TCR with high sensitivity. PLoS Biol. 2008;6:e43. doi: 10.1371/journal.pbio.0060043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ma Z, Janmey PA, Finkel TH. The receptor deformation model of TCR triggering. FASEB J. 2008;22:1002–1008. doi: 10.1096/fj.07-9331hyp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mossman KD, Campi G, Groves JT, Dustin ML. Altered TCR signaling from geometrically repatterned immunological synapses. Science. 2005;310:1191–1193. doi: 10.1126/science.1119238. [DOI] [PubMed] [Google Scholar]
- 149.Valitutti S, Dessing M, Aktories K, Gallati H, Lanzavecchia A. Sustained signaling leading to T cell activation results from prolonged T cell receptor occupancy. Role of T cell actin cytoskeleton. J Exp Med. 1995;181:577–584. doi: 10.1084/jem.181.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sun ZJ, Kim KS, Wagner G, Reinherz EL. Mechanisms contributing to T cell receptor signaling and assembly revealed by the solution structure of an ectodomain fragment of the CD3 epsilon gamma heterodimer. Cell. 2001;105:913–923. doi: 10.1016/s0092-8674(01)00395-6. [DOI] [PubMed] [Google Scholar]
- 151.Natkanski E, Lee W-Y, Mistry B, Casal A, Molloy JE, Tolar P. B cells use mechanical energy to discriminate antigen affinities. Science. 2013;340:1587–1590. doi: 10.1126/science.1237572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Puech P-H, Nevoltris D, Robert P, Limozin L, Boyer C, Bongrand P. Force measurements of TCR/pMHC recognition at T cell surface. PLoS ONE. 2011;6:e22344. doi: 10.1371/journal.pone.0022344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lim TS, Mortellaro A, Lim CT, Hämmerling GJ, Ricciardi-Castagnoli P. Mechanical interactions between dendritic cells and T cells correlate with T cell responsiveness. J Immunol. 2011;187:258–265. doi: 10.4049/jimmunol.1100267. [DOI] [PubMed] [Google Scholar]
- 154.Huppa JB, Axmann M, Mörtelmaier MA, Lillemeier BF, Newell EW, Brameshuber M, Klein LO, et al. TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature. 2010;463:963–967. doi: 10.1038/nature08746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sabatino JJ, Huang J, Zhu C, Evavold BD. High prevalence of low affinity peptide-MHC II tetramer-negative effectors during polyclonal CD4+ T cell responses. J Exp Med. 2011;208:81–90. doi: 10.1084/jem.20101574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jiang N, Huang J, Edwards LJ, Liu B, Zhang Y, Beal CD, Evavold BD, et al. Two-stage cooperative T cell receptor-peptide major histocompatibility complex-CD8 trimolecular interactions amplify antigen discrimination. Immunity. 2011;34:13–23. doi: 10.1016/j.immuni.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Adams JJ, Narayanan S, Liu B, Birnbaum ME, Kruse AC, Bowerman NA, Chen W, et al. T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity. 2011;35:681–693. doi: 10.1016/j.immuni.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rosenthal KM, Edwards LJ, Sabatino JJ, Hood JD, Wasserman HA, Zhu C, Evavold BD. Low 2-dimensional CD4 T cell receptor affinity for myelin sets in motion delayed response kinetics. PLoS ONE. 2012;7:e32562. doi: 10.1371/journal.pone.0032562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Axmann M, Huppa JB, Davis MM, Schütz GJ. Determination of interaction kinetics between the T cell receptor and peptide-loaded MHC class II via single-molecule diffusion measurements. Biophys J. 2012;103:L17–9. doi: 10.1016/j.bpj.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.O’Donoghue GP, Pielak RM, Smoligovets AA, Lin JJ, Groves JT. Direct single molecule measurement of TCR triggering by agonist pMHC in living primary T cells. eLife. 2013;2013:e00778. doi: 10.7554/eLife.00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Klotzsch E, Schütz GJ. Improved ligand discrimination by force-induced unbinding of the T cell receptor from peptide-MHC. Biophys J. 2013;104:1670–1675. doi: 10.1016/j.bpj.2013.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]