Abstract
We report a method for fabricating permeable polymer microstructure barriers in polydimethylsiloxane (PDMS) microfluidic devices and the use of the devices to capture and transport DNA and cells. The polymer microstructure in a desired location in a fluidic channel is formed in situ by the polymerization of acrylamide and polyethylene diacrylate cross-linker (PEG-DA) monomer in a solution which is trapped in the location using a pair of PDMS valves. The porous polymer microstructure provides a mechanical barrier to convective fluid flow in the channel or between two microfluidic chambers while it still conducts ions or small charged species under an electric field, allowing for the rapid capture and transport of biomolecules and cells by electrophoresis. We have demonstrated the application of the devices for the rapid capture and efficient release of bacteriophage λ genomic DNA, solution exchange and for the transport and capture of HeLa cells. Our devices will enable the multi-step processing of biomolecules and cells or individual cells within a single microfluidic chamber.
Introduction
Permeable polymer gels or microplugs fabricated in microfluidic devices allow for the active manipulations of ions and charged biomolecular species using electric fields for many applications. These include, for example, the transport and concentration of biomolecules by electrophoresis1–7, DNA hybridization and sequencing8–11, protein separation and detection12, 13, and formation of chemical gradients.14 The polymer gels or microplugs in the desired locations of the device are usually formed by photo-initiated radical polymerization of monomers in a solution using photomasks, focused beams produced by a laser or shaping optics1, 8, 15, or digital micro-mirrors (DMM).16, 17 In general, the patterns of the polymer structures are defined by the layout on the photomasks or the positioning of the illuminating beams. The sizes of the polymer structures are controlled by the careful timing of the polymerization process followed by the subsequent removal of the unpolymerized monomer solution in other parts of the devices. The surfaces of the polymer structures produced using these methods are usually not very smooth and affected by many parameters.18–20 For certain applications where smooth polymer surfaces are desirable to minimize potential sample loss, the photopatterning method may not be the ideal approach. In addition, the technique requires photomasks, lasers, DMMs and mechanisms for their accurate alignment to the microfluidic devices.
We present a method for fabricating precisely defined permeable polymer microstructures in desired locations in the fluidic channels of PDMS microfluidic devices. The feature of each polymer structure is defined by the channel and a pair of valves designed as an integral part of the fluidic devices. A solution containing the monomers and a photo-activatable initiator is trapped at the location and the device is flood exposed to light to initiate the polymerization of the monomers to form the structure. Upon polymerization, the PDMS valves are released to open the connections to the channel and adjacent chambers. Our method allows for the rapid fabrication of permeable polymer microstructures with well-defined dimensions and very smooth surfaces using valves and structures designed in the microfluidic devices. The properties of the polymer can be tuned by varying the concentration of the monomer and the cross-linker. The polymer microstructure barriers prevent the convective fluid flow and diffusion of biomolecules and cells through the channels or between adjacent microfluidic chambers but are permeable to small ions and other charged species, allowing for the active manipulations of biomolecules and cells using electric fields. To demonstrate the potential applications of our devices, we have shown that genomic DNA can be rapidly captured from the solution in the channels or chambers onto the surface of the polymer structures using an electric field and the DNA can be efficiently released by reversing the electric field. Using HeLa cells, we have also demonstrated that mammalians cells can be captured and transported between microfluidic chambers
Materials and Methods
Design and operation of microfluidic devices with permeable polymer microstructures
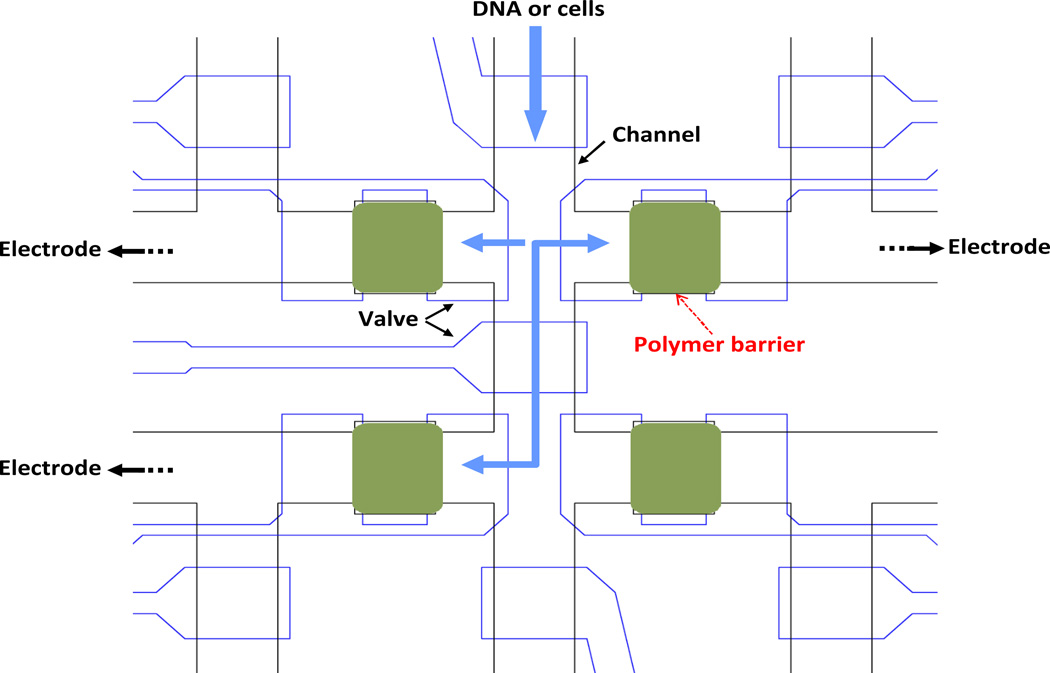
Fig. 1 illustrates an example device with four porous polymer microstructures. The microfluidic device consists of two layers, a bottom layer for flow channels and a top layer for control valves. The cross section of the flow channels is arch-shaped, 20 µm high in the center and 200 µm wide. The cross section of valve control channels is rectangular, 25 µm high and 200 µm wide. The PDMS microstructures and the valves for fabricating the porous polymer barriers are designed into the microfluidic devices. The PDMS structures for holding the polymers are designed to be slightly larger than the channels to enhance the physical resistance of the polymer structures against dislodging by fluid flow. The barriers allow for the active electrophoretic manipulations of biomolecules and cells. The biomolecules such as DNA and cells in the flow channel can be transported, captured and released by selectively applying an electric potential across the desired polymer microstructure barriers.
Fig. 1.
Design and operation of a microfluidic device with permeable polymer barriers. The flow channels in the bottom layer are defined by the black lines. The valves and control channels in the top layer are defined by the blue lines. The polymer barriers are located in the green areas. The barriers allow for the active electrophoretic transport, capture and release of biomolecules such as DNA and cells using electric fields. The biomolecules or cells injected into the channels and chambers are captured and/or transported by applying a voltage to a pair of electrodes submerged in the inlets of the selected channels.
Fabrication of PDMS devices
The PDMS microfluidic device was fabricated using soft lithography according to the procedures of Unger et al.21 To fabricate the mold for the fluidic channel layer, a silicon wafer was primed with hexamethyldisilazane (HMDS) by spin-coating at 4000 rpm for 40 s and then coated with a positive photoresist (Shipley Microposit SPR 220-7.0, Rohm & Haas Electronic Materials, LLC) by spin-coating at 1250 rpm for 45 s. After soft-baking at 115 °C for 5 minutes, the photoresist was exposed using a transparency photomask (FineLine Imaging) on a Karl Suss MA6 aligner for 60 s at 11 mW/cm2 in hard contact mode. After 40 minutes of holding time, the photoresist was developed in MF-24A (Microposit) for 5 minutes, rinsed with de-ionized H2O, and dried with nitrogen gas. The patterned photoresist on the mold was reflowed on a hotplate at 200 °C for 120 minutes to produce round PDMS channels. To fabricate the mold for the valve control layer, a negative photoresist SU 8–2025 (MicroChem Corp.) was coated onto a silicon wafer by spin-coating at 3000 rpm for 35 s. After soft-baking at 65 °C for 1 minute and 95 °C for 5 minutes, the photoresist was exposed with a transparency photomask for 13.7 s at 11 mW/cm2 in hard contact mode. The mold was baked at 65 °C for 1 minute and 95 °C for 5 minutes and then developed in SU-8 developer (MicroChem Corp.) for 4 minutes, followed by rinsing with isopropyl alcohol, and drying with nitrogen gas. The photoresist was hard-baked at 150 °C for 10 minutes to enhance its strength and durability.
The molds were passivated with tridecafluoro-1, 1, 2, 2-tetrahydrooctyl-1-trichlorosilane (Pfaltz and Bauer, Inc.) by vapor deposition for 1 hr in a vacuum chamber prior to use. The PDMS layers were fabricated with molds using Sylgard 184 (Dow Corning). A 20:1 mixture (part A:part B = 20:1) was used for the fluidic layer while a 5:1 mixture was used for the valve control layer. The mixture was de-gassed in a vacuum chamber for 30 minutes. The 5:1 mixture was poured onto the mold for the control valve layer in a custom polycarbonate carrier. The 20:1 mixture was spin-coated onto the mold for the fluidic channel layer at 1500 rpm for 1 minute. After both PDMS layers were cured on the molds in an oven at 75 °C for 25 minutes, the valve control layer was peeled off from the mold and holes were created using a 0.75 mm diameter punch (Ted Pella, Inc.). It was then aligned and laid onto the thin fluidic channel layer. The two PDMS layers were bonded together by incubating in an oven at 80 °C for 2 hours. The bonded layers were peeled off from the fluidic channel mold and inlet and outlet holes for the fluidic channels were punched. The surface of the fluidic channel layer and a cover glass (50 × 50 mm, #1.5 thickness) were treated with oxygen plasma in a UV-ozone cleaner (Jelight Company, Inc.) for 3 minutes, and then bonded together by heating at 80 °C in an oven for at least 10 hours.
Fabrication of polymer microstructures in PDMS microfluidic devices
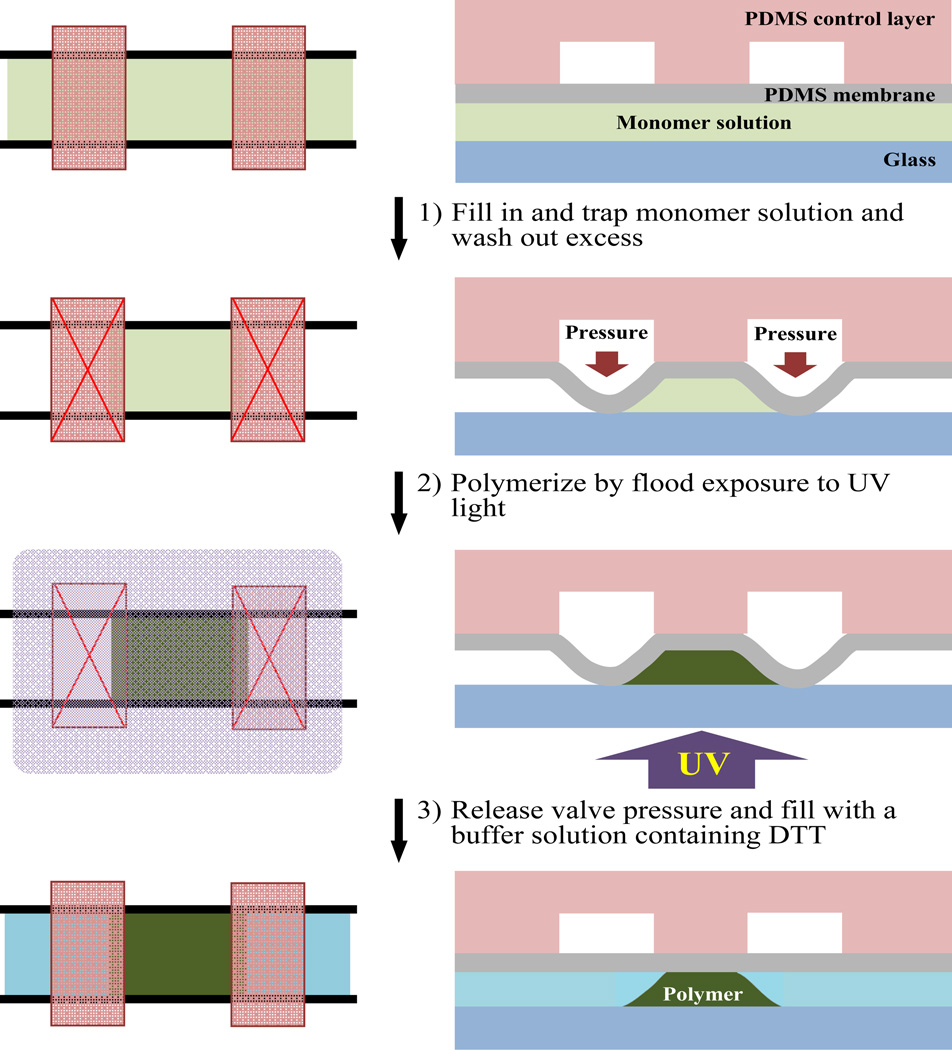
The permeable microstructures were formed in situ by photo-activated polymerization of monomer and a cross-linker. Acrylamide was used as the monomer. Both N,N'-mthylenebisacrylamide (bisacrylamide) and polyethylene diacrylate cross-linker (PEG-DA) (MW of 575, Sigma Aldrich) were used as cross-linkers. The water-soluble 2,2’-azobis[2-methyl-N-(2-hydroxyethyl)propionamide] (AMHP) (Wako Chemicals) was used as the photo-activatable initiator. To promote strong anchoring of the polymer to the walls, the surface of the PDMS channels was pre-treated with a photo-activatable initiator in an organic solvent22. The channels were filled with 10% 2-2-dimethoxy-2-phenylacetophenone (DMPA) in acetone and incubated for 10 minutes, and dried by nitrogen gas. The general procedure for fabricating the permeable polymer microstructures is illustrated in Fig. 2. First, the channels pre-treated with DMPA were filled with a solution containing the monomer, cross-likers and a photo-activatable initiator. The solution was trapped in the desired location for the polymer microstructure using the pair of valves designed for the structure. The valves were closed by deflecting down the PDMS membrane using 10 psi of pressure. The solution in the rest of the device was thoroughly washed away with a buffer solution. For most of the work described here, 1X phosphate-buffered saline (PBS, 137 mM NaCl, 3 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) was used as the buffer solution, and the monomer solution contains 19 % acrylamide, 1% bisacrylamide, 10% PEG-DA and 0.5% AMHP in 1X PBS. Second, the device was flood exposed to UV light to initiate the polymerization of the monomer and cross-linkers. The 365 nm UV light from the tip of a liquid light guide of the light source (Omni Cure S2000, EXFO) was projected to the bottom glass substrate of the device for 30 s. The density of the light at the substrate was about 3.36W/cm2. Third, the PDMS valves were opened by releasing the pressure applied to the valves and driving a solution of 10 mM dithiothreitol (DTT) in PBS into the channels using 2 psi of pressure. Soaking the polymer in DTT may help annihilate any residual radicals from the polymerization reaction.
Fig. 2.
Fabrication of permeable polymer microstructures in PDMS microfluidic devices. The microstructures are fabricated in three steps: (1) The monomer solution is filled into the channels and trapped at each site where the polymer microstructure is to be formed using a pair of valves. Excess solution is rinsed out. (2) The polymer microstructures are formed by flood exposure of the device to UV light to initiate the polymerization of the monomers. (3) The PDMS valves are opened and the polymer microstructures are soaked and rinsed with a solution of PBS buffer containing DTT. The left and right panels show the aerial view and the cross-sectional view of the device, respectively.
Capturing and release of DNA
Active manipulations of DNA and cells are carried out by applying electric fields across the selected polymer microstructure barriers via the solution in the fluid channels. To generate an electric field (E-field), a DC voltage is applied using a pair of platinum wires submersed into the inlet and outlet of the selected channels using a DC power supply (Power PAC 1000, BioRad). Since the electrical resistance of the PBS solution in the channel is very high (a few MΩ), which resulted in a current smaller than that of the minimum limit required by the power supply, a 50 kΩ resistor is connected in parallel to the pair of platinum wires in order to produce a proper amount of current flow through the barriers and channels. The direction and speed of the capture and transportation of biomolecules and cells are controlled by the polarity and magnitude of the applied electric fields respectively. The fluid flow is driven by purified compressed air though the system with multiple solenoid valves. The PDMS microfluidic valves and fluid flow are controlled using a customized electronic control system.
To demonstrate the active manipulation of DNA, the polymer barrier microstructures were fabricated with 20% polyacrylamide solution (acrylamide:bisacrylamide =19:1) plus 10% of PEG-DA. Genomic DNA from bacteriophage lambda (λ DNA, 48502 base pairs) was used. Stock λ DNA (New England Biolabs) was diluted to 2.5 ng/uL in PBS with 1X SYBR Gold (Invitrogen), a fluorescent DNA stain. The DNA solution was injected into the channel to fill a chamber with polymer barriers on both the left and right sides. The chamber was then closed off by activating a pair of PDMS valves on the entrance and exit side of the chamber. A pair of platinum wires was submerged into the inlets of the channels connected to the left and right sides of the polymer microstructures of the chamber holding the DNA solution. The total length of the fluid channel between the two inlets is about 15 mm. The DNA was captured by applying a 20V DC for 10 seconds. The DNA was released by reversing the polarity of the electrodes. The processes were monitored by fluorescence microscopy using an epifluorescence microscope (Zeiss Axiovert 200M) and a filter set for SYBRGold imaging (SYBRGold/LP-A, Semrock Inc. Excitation: 495 nm enter wavelength, 16 nm bandwidth; Emission: 519 nm edge, 534–653 nm bandwidth; Dichroic: longpass with 516 nm edge).. The images and movies were acquired using a 20X objective (0.8 NA, Zeiss Plan-Apochromat) and an EM-CCD camera (Andor iXon Plus) with excitation from X-Cite 120 (EXO).
To assess the efficiency of DNA release from the polymer barriers after capture, λ DNA was filled into the chamber and captured from the solution by applying the voltage for 10 s, and then the DNA was released by reversing the polarity of the applied voltage for 30 s and flushed out of the chamber. This process was repeated five times and each time a series of fluorescence images were acquired. The fluorescent images were analyzed using ImageJ and Matlab (Mathworks). First, the fluorescence intensity profile along the center of channel in each image was plotted using ImageJ. Second, background fluorescence was subtracted. Third, the efficiency of DNA release was estimated by taking the ratio of the fluorescence intensity of the residual DNA remaining on the polymer to that of the captured DNA. An algorithm was used to align the images by creating a mask from the first image prior to DNA release by recording the location of the pixels with signal greater than certain threshold. The subsequent images were processed using the mask to ensure the intensity from the same location was used.
Capturing and transportation of cells
HeLa cells were used to demonstrate the ability to capture and transport mammalian cells. The cells were cultured under standard conditions (37 °C and 5% CO2) in Dulbecco’s Modified Eagle’s Medium (DMEM; Mediatech) supplemented with 10% fetal bovine serum (Mediatech) and 1% penicillin/streptomycin (MP Biomedicals). The cells were detached from the surface of the culture flask using 2 mL of 0.25% trypsin-EDTA (Mediatech), re-suspended in 8 mL DMEM. The cells were then centrifuged down at 1,000 rpm for 6 minutes and suspended in PBS to a density of 105 cells per mL. The cell suspension was injected from the upper inlet of the fluidic channel, and one or two cells were enclosed in the 2nd chamber by closing the valves. A platinum wire submerged into the upper right inlet and another wire into the lower left inlet were used as a pair of electrodes to apply the potential. A 150 V DC voltage over 18.5 mm distance between the inlets was applied to capture and transport the cells. The cells are moved back and forth by applying a DC voltage and reversing the polarity of the voltage.
Solution exchange and delivery
One of the motivations for our current work is to develop the capability for active manipulations and multi-step processing of biomolecules and cells within a single microfluidic chamber. A device as illustrated in Fig. 1 but with only one pair of polymer microstructure barriers (lower pair) was used. The upper part of the device was used for DNA loading, buffer exchange and delivery. λ DNA (4.5 ng/µL) stained with 1X SYBR Gold in water was loaded into the lower chamber defined by two valves and a pair of polymer microstructures. 20 VDC was applied across the polymer barriers to capture the DNA onto the surface of one of the polymer barrier. The DNA outside the chamber was rinsed out using a PBS solution driven by 2 psi pressure. While the voltage was maintained, the valves of the chamber were opened and a PBS solution was flowed through the chamber. After a few seconds, the voltage was turned off while the fluid flow was maintained. Finally, the polarity of the voltage was reversed. The entire process was monitored by fluorescence imaging.
Results and discussion
Fabrication and characterization of permeable polymer microstructure barriers
Using our method, permeable polymer microstructures can be fabricated in situ reproducibly at desired locations in microfluidic devices. The shape, size and location of each polymer microstructures are defined by the channel and a pair of valves which are designed as integral parts of the PDMS-based microfluidic devices. The trapping of the monomer solution is guaranteed by the use of a pair of PDMS valves. As long as the concentrations of the monomer and cross-linker(s) are high enough to produce a polymer with sufficient mechanical strength, the polymer microstructures can be formed with certainty. Two notches are also designed in the channel to hold the polymer microstructure in position to increase its resistance to dislodging by force impinged to the polymer by fluidic flow. An example device is shown in Fig. 3. Since the polymer structure assumes the shape of the space surrounded by the channel walls and the valve membranes, the physical dimension of the structure is predictable and could be tailored to some degree by design. In the work described here, we used push-down PDMS valves. Since the PDMS membrane is deflected down by applying a pressure, the polymer structure assumes a trapezoid shape as illustrated in Fig. 2. Structures with other geometries can be designed and fabricated perhaps by using other types of valves, such as push-up valves.23 We characterized the mechanical sealing property of the permeable polymer structures using a device with two polymer microstructures. As shown in Fig. 3e, a PBS solution containing a blue dye solution was flowed through the vertical channel which is saperated by two polymer microstructures from the left and right horizontal channels. The polymer barriers effectively blocked the flow of the bulk solution from the vertical channel into the horizontal side channels. Since the polymer is porous with pore sizes from a few nanometers to tens of nanometers or larger depending on the concentration of the monomer and crosslinker used24, which are permeable to small molecules and ions, the small dye molecules eventually diffuse through the barriers in about 10 minutes in our test devices.
Fig. 3.
Characteristics and sealing property of permeable polymer microstructures fabricated in microfluidic channels. The polymer structures were formed in the channels by the polymerization of the monomers in the solution trapped by a set of PDMS valves. (a) and (b): Brightfield images of the device filled with air (a) and PBS solution (b). The polymer microstructure is inside the box outlined by the dashed line. (c) An enlarged portion of the phase-contrast image in (b) showing the boundary between the polymer structure and the PBS solution in the channel. (d) Fluorescence image showing the boundaries of the polymer structure which was fabricated with a monomer solution containing a fluorescence dye. The boundaries on both sides of the structure in the fluidic channel are visible, represented by the brighter curved lines. (e) Sealing property of polymer microstructure barriers. The polymer barriers block the convective fluid flow of the dye solution into the side channels while the solution was flowed through the vertical channel by applying a 2 psi of pressure at the fluid inlet.
The mechanical sealing property of the polymer microstructures is influenced by certain factors, including the strength of the adhesion of the polymer surfaces to the glass substrate and the PDMS channel walls, the mechanical strength of the polymer gel, and the geometry of the polymer microstructures. The polymer usually adheres very well to the glass substrate but not very strong to un-treated PDMS surfaces. We found that the pre-treatment of the microfluidic channels with a hydrophobic photo-activatable initiator such as AMHP significantly enhanced the sealing property of the polymer microstructures, probably by promoting the covalent attachment of the polymer to the PDMS walls as reported by Hu et al.22 Without the pre-treatment the polymer tended to separate from the PDMS walls even under a moderate fluid flow pressure of 2 psi. We also found that the mechanical sealing property of the polymer barrier is enhanced by using a high concentration of PEG-DA. As an added benefit, the PEG moiety also helps reduce non-specific binding of the polymer surface. The notched microstructures are designed to be slightly larger than the channels to accommodate the polymer barriers to increase its mechanical resistance to potential dislodging by fluid flow. Very robust polymer microstructures with good sealing property and very low non-specific binging can be fabricated using 19% acrylamide plus 1% bisacrylamide and 10% PEG-DA. We also experimented with various concentrations of acrylamide and PEG-DA. The sealing property of some of the polymer microstructures fabricated is summarized in Table S1 in ESI.
We also measured the electrical conductivity of electrolyte solutions through the polymer barriers in our devices. The conductivity of the fluidic channel filled with 1x PBS without polymer microstructures was measured to be 17.91 mS/cm, whereas the channel with two polymer microstructures formed with a total 30% (w/v) of monomer and cross-linker as shown in Fig. 4 had a conductivity of 13.17 mS/cm. Therefore, the polymer microstructure reduces the conductivity by about 26%. This is as expected considering the microstructures are slightly larger than the channel and the polymer takes up ~30% of the volume in the channel.
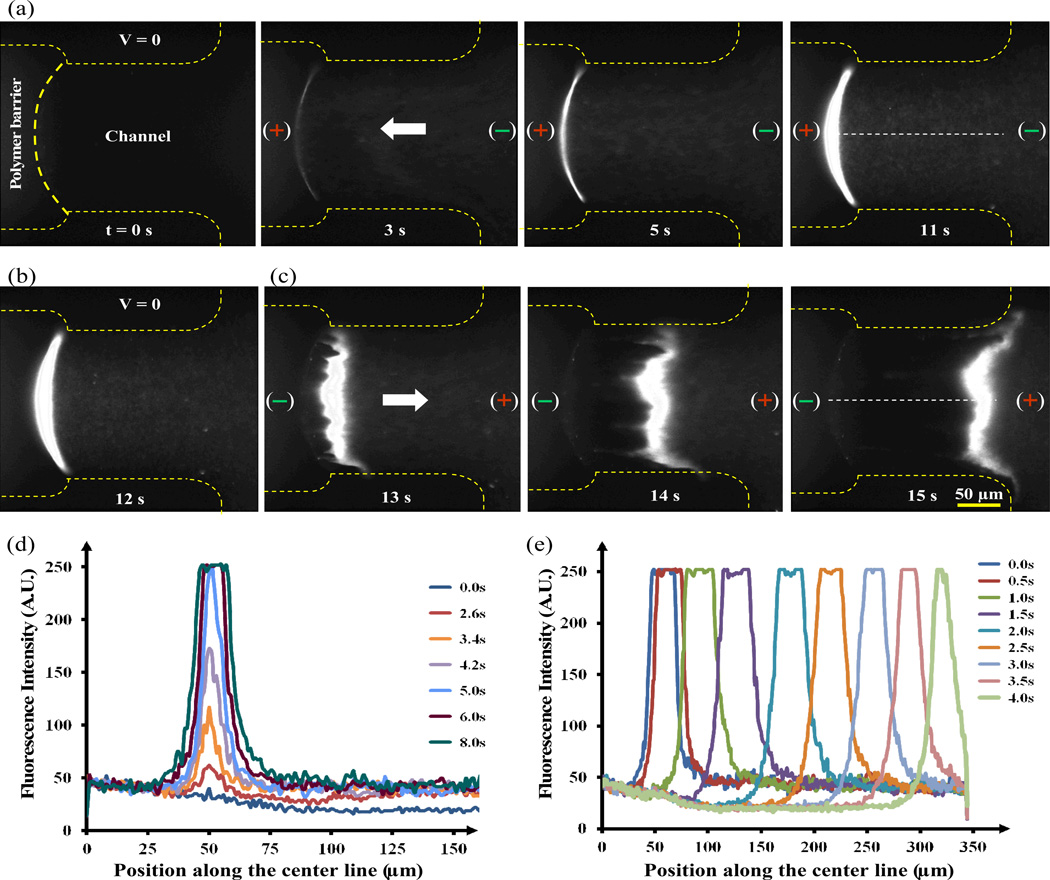
Fig. 4.
Capture and release of genomic DNA using electric fields. (a) Selected frames of the time-lapse images of DNA capturing onto the surface of the polymer barriers using an electric field. (b) Fluorescence image after the field is turned off. (c) Selected frames of the time-lapse images of DNA released from the surface of the barrier by reversing the electric field. The channel is outlined with narrower yellow dashed lines while the surface of the polymer barrier is outlined with a thicker dashed line. Genomic λ DNA in a solution containing SYBR gold was used. The images were captures using an epifluorescence microscope. (d) and (e) Fluorescence intensity profiles of the DNA being captured on the surface the polymer barrier (d) and the release of the captured DNA from the polymer surface by reversing the electric field (e). The fluorescence intensity profiles were acquired along a line in the center of the channel as indicated by a white dashed straight line in (a) and (c) with the origin to the left of the DNA band.
Capture, release and transport of DNA
Microfluidic devices with built-in permeable polymer microstructures allow for the active on-chip manipulations of biomolecules and cells using electric fields. First we demonstrated the ability to capture and release DNA using the polymer microstructure barriers fabricated with 20% polyacrylamide plus 10% PEG-DA. Since λ genomic DNA is ~48.5kbp long, it should not get into the polymer barrier which has pore size of only a few nanometers. A series of time-lapse fluorescence images in Fig. 4 and the movie (Movie M1 in ESI) show the rapid capture and release of the genomic DNA molecules. As shown in Fig. 4a, the DNA molecules were immediately pulled and collected at the surface of polymer barrier connected via the fluid channel to the positive electrode as soon as a DC voltage (20 VDC across a 13 mm distance) was applied.
All the DNA molecules were captured within seconds under the experimental condition. When the electric field was turned off, the DNA was observed to diffuse slightly into the solution by Brownian motion (Fig. 4b). The captured DNA was also rapidly released by reversing the electric field. As soon as the polarity of electric field was reversed, the molecules were pulled away from the surface of the polymer (Fig. 4c). Interestingly, in contrast to the capture process where the DNA molecules in free solution are pulled toward and accumulate on the surface of the polymer barrier, the DNA molecules are released together into the free solution with a group velocity as indicated by the bright moving bands. The shape of the band becomes only slightly more dispersed over time perhaps due to diffusion and slight spatial variation of the electric field. The overall process can be described more quantitatively by the fluorescence intensity profiles along a line in the center of the channel in the time-serial images. Perhaps these characteristics can be utilized to transport the captured DNA to other location for downstream processes.
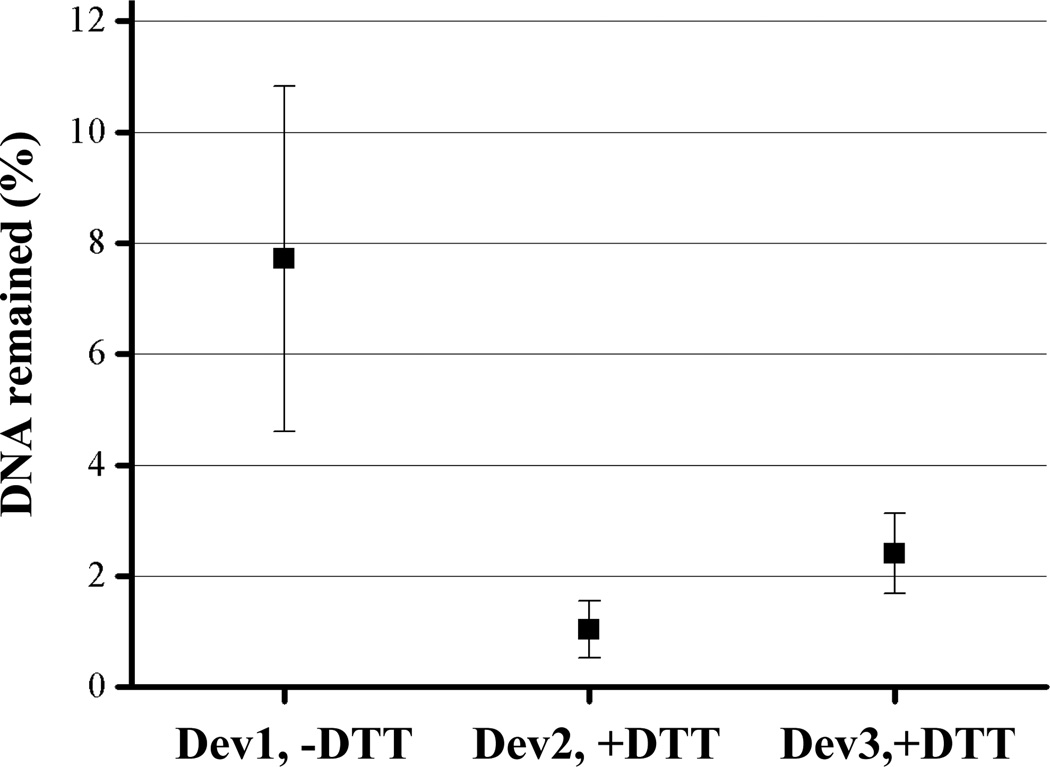
To investigate the property of the polymer surface and the efficiency of releasing the captured DNA, we performed repeated capture and release of DNA in the same device. λ DNA in PBS was captured with a constant 20 VDC for 30 s and the collected DNA was released by reversing the polarity of the potential for 60 s. After each cycle, the solution was rinsed out and replaced with a fresh solution. The percentage of the residual DNA remained on the surface was calculated from the fluorescence images. As shown in Fig. 5, the captured DNA can be released with very high efficiency (>97%) for polymer microstructures fabricated with a high concentration of PEG-DA (10% in this case) and treated with 10 mM DTT.
Fig. 5.
Effective release of captured DNA from the surface of the polymer barrier. DNA capture and release were repeated five times with several devices. The captured DNA was released by reversing the electric field for 30 s. The effect of DTT treatment on release efficiency was evaluated. Each error bar represents the standard deviation calculated from five independent measurements.
The captured DNA may not be released effectively from the surface due to some factors, including the covalent binding of the DNA to potential reactive species on the polymer, strong non-specific binding of DNA to the surface, and penetration and entanglement of the DNA with the polymer mesh. We found that 7–8% of DNA remained on the surface if the polymer was not pre-treated with a DTT solution. The pre-treatment of the polymer with DTT, which is known to be a very effective radical quencher, may help eliminate any potential residual radicals on the polymer, preventing covalent binding of DNA to the surface. PEG is commonly used to reduce non-specific binding of biomolecules to surfaces and as an anti-fouling reagent. Therefore the use of PEG-DA is also beneficial in reducing non-specific binding. Considering the small pore sizes of the polymer formed with a high concentration of polyacrylamide, it his very unlikely that the large λ genomic DNA would migrate appreciably into the polymer. However, if the surface of the polymer is not very smooth, some entanglement may still occur, preventing the rapid release of DNA from the surface. The polymer microstructures produced by our method have smooth surfaces defined by the PDMS membranes. Even though the release of the PDMS valves may cause some tearing of the polymer surfaces as sometimes we observed (The jagged boundary line in Fig. 3c), which could potentially decrease the smoothness of the surfaces. our results demonstrated that the captured DNA molecules can be rapidly and efficiently released from the polymer surfaces by electrophoresis using an electric field. Further work will be requied to characterize the smoothness of the surfaces of the polymer microstructures. Smoother polymer surfaces can be produced by pre-treating the PDMS surfaces with a photo-initiator which are not hydrogen-abstracting but promote the non-covalent interwining anchoring of the polymer to the PDMS surface.25 This may eliminate the potential the tearing of the polymer surface. In addition, certain structures can be designed in the channels and valves to physically confine the polymer barriers to increase the sealing strenghth between the polymer barriers and PDMS surfaces in the channels to eliminate the pre-treatment of the PDMS channels with photoinitiator. Work is in progress in our lab to investigate polymer barriers designed and fabricated with such structures.
Capture and transport of cells
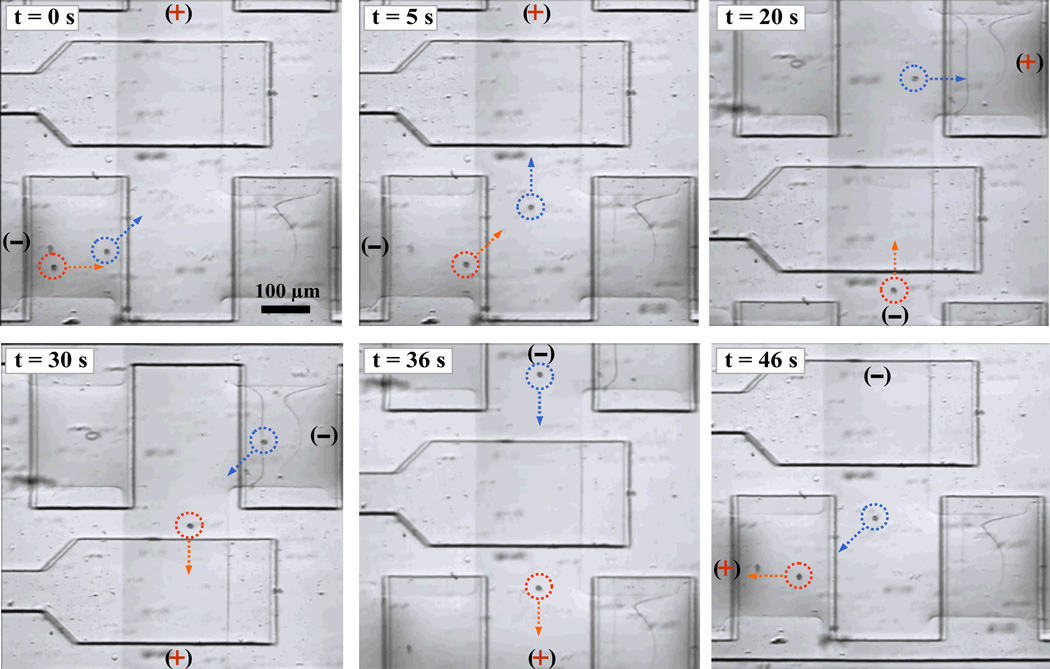
We have also demonstrated the ability to capture and transport mammalian cells using our devices. Fig. 6 shows the capture and transport of HeLa cells. The cells in suspension were flowed into the device and two cells were enclosed in a chamber. The cells were transported into another chamber and captured onto the polymer by applying a 150 VDC to a pair of electrodes immersed in the channel outlets about 18.5 mm apart. The cells are transported toward the chamber connected to the electrode with a positive potential, indicating that they carry a net negative charge. In this case the HeLa cells were transported with a velocity of about 100 µm/s. The cells eventually stopped at the surface of the polymer barrier inside the chamber. Even though a very high DC voltage was applied, no perceivable bubbles were observed at the electrodes. This is because the current flow is very small, about 37 µA. Since the transport velocity depends on the net charge, shape and size, and dielectric property of the cells, different cells may be separated as well26. We found that the cells may be pulled into the narrow space between the polymer and the upper PDMS membrane if they are pulled with a very strong electric field. In principle, the cells can be transported to any chamber in the microfluidic device by applying electric potential to the selected channels connected to the polymer barriers on the sides of the chamber. This capability may allow for the rapid sorting and downstream analysis of cells or single cells in microfluidic devices.
Fig. 6.
Capture, transport and separation of cells. Cells were transported across two chambers by electric fields using a microfluidic device with a layout similar to the one illustrated in Fig. 1. The time-serial brightfield images show the transportation of two HeLa cells initially in the lower chamber. The cells were moved toward the upper chamber by applying an electric field. One cell was moved into the upper chamber and captured against the polymer barrier. By reversing the electric field, the cells were moved back toward the barrier of the lower chamber. It is possible to separate the cells by closing the valve separating the two chambers, in this case, at the 36 s time point. The video clip of cell capture and transport is available online (Movie M2 in ESI).
Solution exchange and delivery
Furthermore, we have also demonstrated the application of our devices for the rapid capture of biomolecules, washing and exchange of the solution in the microfluidic chambers. After the λ genomic DNA was captured on to the surface using an electric field, the solution in the chamber was flushed out and replaced with another solution while the electric field was maintained. We have shown that captured DNA remained on the surface of the polymer microstructure when a buffer solution was flowed through the chamber. If the electric field was turned off, the DNA was released slowly from the surface to the flow stream. The captured DNA was rapidly released into the flow stream by reversing the electric field. A movie of the processes are available online (Movie M3 in ESI). The ability to capture and hold the molecules or cells to allow for solution exchange in the chamber will enable the multi-step processing such as performing enzymatic reactions on biomolecules and cell lysis in a single microfluidic chamber without transferring the molecules or cells to downstream chambers, which would be difficult or not possible with microfluidic devices using conventional PDMS valves and chambers.
Conclusions
We have demonstrated a new method of fabricating permeable polymer microstructure in microfluidic devices. The polymer microstructures are formed in situ by photo-initiated polymerization of monomers in a solution trapped at location using a pair of PDMS valves. Our method allows for the facile fabrication of the polymer microstructures with well defined dimensions without using photomasks, lasers and beam-shaping optics or digital micromirrors. We have demonstrated that the polymer forms a very tight seal with the PDMS walls in the channel, preventing convective fluid flow through the polymer barrier. The polymer microstructures are permeable to small ions and molecules. This enables the active manipulation of biomolecules and cells in microfluidic devices using electric fields. We have demonstrated the rapid capture and almost quantitative release of λ genomic DNA. This capability may enable the rapid concentration, transport, and biochemical processing and analysis of biomolecules. We have also shown the ability to capture, transport and separate mammalian cells between different microfluidic chambers using electric fields instead of convective fluid flow. Our method will enable the fabrication of multiple permeable polymer microstructures, perhaps even with different characteristics, at designed locations in large-scale microfluidic devices with ease. The capability to capture and transport cells between different chambers in the microfluidic devices by applying electric potentials to selected channel inlets/outlets will be useful for transport, sorting and further downstream analysis of cells or single cells. The electrodes could be integrated into the devices using microfabrication techniques so that much lower voltages can be used for the capture and transport process. Work is in progress in our lab in the design, fabrication and operation of such devices.
Supplementary Material
Acknowledgements
This work was supported in part by grants from the NIH (Grants R01HG004804 and R21HG005096 to X.H and R01GM097253 to K. Z.).
Footnotes
Electronic Supplementary Information (ESI) available: Video clips of capture and release of DNA and cells, and a supporting table. See DOI: 10.1039/b000000x/
References
- 1.Song S, Singh AK, Kirby BJ. Anal. Chem. 2004;76:4589–4592. doi: 10.1021/ac0497151. [DOI] [PubMed] [Google Scholar]
- 2.Dhopeshwarkar R, Li SA, Crooks RM. Lab Chip. 2005;5:1148–1154. doi: 10.1039/b509063f. [DOI] [PubMed] [Google Scholar]
- 3.Hatch AV, Herr AE, Throckmorton DJ, Brennan JS, Singh AK. Anal. Chem. 2006;78:4976–4984. doi: 10.1021/ac0600454. [DOI] [PubMed] [Google Scholar]
- 4.Sommer GJ, Singh AK, Hatch AV. Anal. Chem. 2008;80:3327–3333. doi: 10.1021/ac702523g. [DOI] [PubMed] [Google Scholar]
- 5.Li ZM, He QH, Ma D, Chen HW, Soper SA. Anal. Chem. 2010;82:10030–10036. doi: 10.1021/ac101768j. [DOI] [PubMed] [Google Scholar]
- 6.Sommer GJ, Mai J, Singh AK, Hatch AV. Anal. Chem. 2011;83:3120–3125. doi: 10.1021/ac200073p. [DOI] [PubMed] [Google Scholar]
- 7.Mai J, Sommer GJ, Hatch AV. Anal. Chem. 2012;84:3538–3545. doi: 10.1021/ac203076p. [DOI] [PubMed] [Google Scholar]
- 8.Seong GH, Zhan W, Crooks RM. Anal. Chem. 2002;74:3372–3377. doi: 10.1021/ac020069k. [DOI] [PubMed] [Google Scholar]
- 9.Paegel BM, Yeung SHI, Mathies RA. Anal. Chem. 2002;74:5092–5098. doi: 10.1021/ac0203645. [DOI] [PubMed] [Google Scholar]
- 10.Bromberg A, Jensen EC, Kim J, Jung YK, Mathies RA. Anal. Chem. 2011;84:963–970. doi: 10.1021/ac202303f. [DOI] [PubMed] [Google Scholar]
- 11.Thaitrong N, Toriello NM, Del Bueno N, Mathies RA. Anal. Chem. 2009;81:1371–1377. doi: 10.1021/ac802057f. [DOI] [PubMed] [Google Scholar]
- 12.Lo CT, Throckmorton DJ, Singh AK, Herr AE. Lab Chip. 2008;8:1273–1279. doi: 10.1039/b804485f. [DOI] [PubMed] [Google Scholar]
- 13.He M, Herr AE. Nature Protocols. 2010;5:1844–1856. doi: 10.1038/nprot.2010.142. [DOI] [PubMed] [Google Scholar]
- 14.Choi E, Jun I, Chang HK, Park KM, Shin H, Park KD, Park J. Lab Chip. 2012;12:302–308. doi: 10.1039/c1lc20777f. [DOI] [PubMed] [Google Scholar]
- 15.Olsen KG, Ross DJ, Tarlov MJ. Anal. Chem. 2002;74:1436–1441. doi: 10.1021/ac0156969. [DOI] [PubMed] [Google Scholar]
- 16.Sun C, Fang N, Wu DM, Zhang X. Sens. and Actuators A: Phys. 2005;121:113–120. [Google Scholar]
- 17.Chung SE, Park W, Park H, Yu K, Park N, Kwon S. Appl. Phys. Lett. 2007;91:041106–041103. [Google Scholar]
- 18.Itoga K, Yamato M, Kobayashi J, Kikuchi A, Okano T. J Biomed Mater Res A. 2004;69:391–397. doi: 10.1002/jbm.a.30010. [DOI] [PubMed] [Google Scholar]
- 19.Mair DA, Schwei TR, Dinio TS, Svec F, Frechet JM. Lab Chip. 2009;9:877–883. doi: 10.1039/b816521a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He M, Bao JB, Zeng Y, Harrison DJ. ELECTROPHORESIS. 31:2422–2428. doi: 10.1002/elps.200900774. [DOI] [PubMed] [Google Scholar]
- 21.Unger MA, Chou H-P, Thorsen T, Scherer A, Quake SR. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 22.Hu SW, Ren XQ, Bachman M, Sims CE, Li GP, Allbritton NL. Anal. Chem. 2004;76:1865–1870. doi: 10.1021/ac049937z. [DOI] [PubMed] [Google Scholar]
- 23.Studer V, Hang G, Pandolfi A, Ortiz M, Anderson WF, Quake SR. J. Appl. Phys. 2004;95:393–398. [Google Scholar]
- 24.Chen Z, Graham R, Burns MA, Larson RG. ELECTROPHORESIS. 2007;28:2783–2800. doi: 10.1002/elps.200600684. [DOI] [PubMed] [Google Scholar]
- 25.Burke JM, Smela E. Biomicrofluidics. 2012;6:16506–1650610. doi: 10.1063/1.3693589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vahey MD, Voldman J. Methods Mol. Biol. 2012;853:53–63. doi: 10.1007/978-1-61779-567-1_6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.