Abstract
The Pin1 prolyl isomerase regulates phosphorylation signaling by controlling protein conformation after phosphorylation and its upregulation promotes oncogenesis via acting on numerous oncogenic molecules. SUMOylation and deSUMOylation are dynamic mechanisms regulating a spectrum of protein activities. The SUMO proteases (SENPs) remove SUMO conjugate from proteins and their expression is deregulated in cancers. However, nothing is known about the role of SUMOylation in regulating Pin1 function. Here, we show that Pin1 is SUMOylated on Lys6 in the WW domain and on Lys63 in the PPIase domain. Pin1 SUMOylation inhibits its protein activity and oncogenic function. We further identify that SENP1 binds to and deSUMOylates Pin1. Importantly, either overexpression of SENP1 or disruption of Pin1 SUMOylation promotes the ability of Pin1 to induce centrosome amplification and cell transformation. Moreover, SENP1 also increases Pin1 protein stability in cell cultures and Pin1 levels are positively correlated with SENP1 levels in human breast cancer specimens. These results not only uncover Pin1 SUMOylation on Lys6/63 as a novel mechanism to inhibit its activity and function, but also identify a critical role for SENP1-mediated deSUMOylation in promoting Pin1 function during tumorigenesis.
Keywords: Pin1, SENP1, SUMOylation, phosphorylation signaling, Oncogenesis
Introduction
Proline-directed protein phosphorylation (pSer/Thr-Pro) is a central signaling mechanism in diverse cellular processes, notably cell proliferation and transformation. Certain pSer/Thr-Pro motifs in polypeptides exist in two completely distinct conformations, cis and trans, the conversion of which is markedly slowed down upon phosphorylation, but yet specifically catalyzed by the peptidyl-prolyl cis/trans isomerase Pin1 (1–3). This striking substrate specificity results from the unique N-terminal WW domain and C-terminal PPIase domain of Pin1 (1–3). The WW domain binds only to specific pSer/Thr-Pro-motifs and targets Pin1 close to its substrates, where the PPIase domain isomerizes specific pSer/Thr-Pro motifs and induces conformational changes in proteins (1–3). These cis and trans conformation-specific functions and their regulation by Pin1 have been directly demonstrated by the development of cis and trans conformation-specific antibodies (4).
Importantly, such Pin1-induced conformational changes following phosphorylation control various protein functions, including their catalytic activity, phosphorylation status, protein interaction, subcellular location, and/or protein stability (1–3). Functionally, Pin1 is important in many cellular processes involving Pro-directed phosphorylation, including the cell cycle, cell signaling, transcription and splicing, DNA damage responses, germ cell development and neuronal survival (1–3, 5, 6). Significantly, Pin1 deregulation contributes to certain pathological conditions, notably cancer and Alzheimer’s disease (1–3, 5, 6).
In human cancers, Pin1 is prevalently overexpressed and its overexpression level correlates with poor clinical outcome (2, 3). In contrast, the Pin1 genetic polymorphisms that reduce Pin1 expression are associated with reduced cancer risk in humans (2, 3). Significantly, Pin1 activates numerous oncogenes/growth enhancers, including β-catenin, cyclin D1, NF-κB, c-Jun, c-fos, Raf-1, Stat3, Neu/ErbB2, Notch, AKT, AIB1, Mcl-1, Hbx, and PKM2, and also inactivates a large number of tumor suppressors/growth inhibitors, including FOXOs, PML, SMRT, Smad, Pin2/TRF1, AMPK, Rb and Fbw7 (2, 3, 7–9). Furthermore, whereas Pin1 overexpression causes centrosome amplification, cell transformation and tumorigenesis (10, 11), Pin1 knockdown inhibits tumor growth in vitro and in vivo (1–3). Moreover, Pin1 knockout mice are fully resistant to tumorigenesis induced by oncogenes such as MMTV-Neu/ErbB2 or -Ras (12). In addition, Pin1 catalytic activity and oncogenic function are effectively suppressed by the tumor suppressor DAPK1 (13). These results demonstrate a major role for Pin1 in cancer development and make Pin1 as an attractive anticancer target (1, 2). However, how Pin1 function is upregulated during cancer development is still not fully understood.
Protein modification by a small ubiquitin-like modifier (SUMO) peptide on a lysine residue is important in controlling a spectrum of protein activities, including protein activity, stability and localization, (14–16). The conjugation and deconjugation of SUMO modification is a highly dynamic event, and only a small fraction of a substrate is SUMOylated at a given time (16). Among mammalian SUMO isoforms, the conjugation of SUMO1, SUMO2 and SUMO3 to protein substrates requires the E1-activating enzyme (SAE1/SAE2), the E2 conjugase (Ubc9) and, in some cases, the E3 ligases(15), while little is known about SUMO4, which has been linked to autoimmune diseases (17). DeSUMOylation mediated by the SUMO proteases (SENPs) has been shown to be also involved in many of the processes mentioned above (14). Six SENPs have been identified in humans, each with different cellular locations and substrate specificities (18). SENP1, a nuclear SUMO protease, has been shown to regulate androgen receptor transactivation by targeting histone deacetylase 1, to induce c-Jun activity through deSUMOylation of p300 and to increase expression of the cell cycle regulator Cyclin D1 (19). SENP1 deconjugates SUMO1 from hypoxia-inducible factor-1α (HIF1α) to control its stability and regulates hypoxic response (20). Recently, SENP1 has been shown to regulate STAT5 activation during early lymphoid development (21). Interestingly, SENP1 has been shown to overexpress in some human cancers including prostate and thyroid cancer (19, 22). However, so far only a handful of SENP1 substrates have been identified and its targets and molecular mechanisms during oncogenesis remain poorly understood.
In this study, we discover that SUMOylation inhibits Pin1 protein activity and cellular function, and also identify Lys6 in the WW domain of Pin1 as a major SUMOylation site and Lys63 in the PPIase domain as an additional site. Furthermore, either overexpression of SENP1 or disruption of Pin1 SUMOylation promotes the ability of Pin1 to induce centrosome amplification and cell transformation. Finally, Pin1 levels in human breast cancer tissue are positively correlated with SENP1 levels. Thus, SUMOylation of Pin1 on Lys6 and Lys63 is a novel mechanism inhibiting Pin1 activity and function, whereas SENP1-mediated deSUMOylation promotes Pin1 function during oncogenesis. Given that Pin1 activates and inactivates a large number of oncogenes and tumor suppressors, respectively, this new connection between SNEP1 and Pin1 may also provides additional attractive alternative targets for anticancer therapies.
Materials and Methods
Plasmids
The expression constructs for wild type and various mutants of SENP1 with N-terminal Flag- and HA-tag were described (20). The Pin1 wild type and deletion mutants with N-terminal GST, Flag or HA tag were described (13). The Pin1 mutants in which the Lys residue 6 and 63 were each replaced by an Arg or Ala residue, were generated by site-directed mutagenesis, and then subcloned to pLenti6/V5-GW/lacZ vector.
PPIase assay
The PPIase activity of Pin1 and SUMO1-modified Pin1 were determined using the protease-free PPIase activity assay with the substrate Suc-Ala-pSer-Pro-Phe-pNA, Suc-Ala-Glu-Pro-Phe-pNA or Suc-Ala-Ala-Pro-Phe-pNA (50 μM) in 35 mM HEPES pH 7.8 at 10°C, as described previously (23).
Analysis of centrosome duplication during S phase
Centrosome duplication assays in NIH3T3 cells were performed, as described previously (13). Briefly, cells were arrested in G1/S phase by adding aphidicolin at a final concentration of 10 μg/ml for 24 hr. Cells were then fixed with cold ethanol for 5 min and stained for centrosomes with anti-γ-tubulin antibodies and analyzed by fluorescent microscopy, as described previously (13).
Soft agar colony formation assay
Soft agar assays were done by seeding cells at a density of 103 in 60-mm tissue culture dishes containing 0.3% top low-melt agarose-0.5% bottom low-melt agarose as previously described (13). Cells were fed every 4 days and colonies were counted and measured after 3 weeks.
Additional Materials and Methods can be found in the Supplementary Data with this article online.
Results
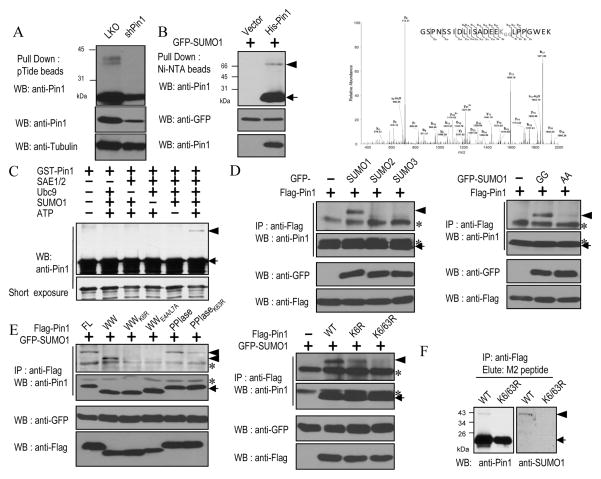
Pin1 is modified on Lys6 and Lys63 by SUMO1 in vitro and in vivo
Pin1 is an important regulator of many cellular processes involving Pro-directed phosphorylation (1, 2). Although recent studies have shown that Pin1 is regulated by protein phosphorylation, little is known whether Pin1 can be modified by any other post-translational modifications. While studying Pin1 post-translational modification (13, 24), we found that a modified Pin1 with a slower electrophoretic mobility was detected when we immunoprecipitated from normal breast cells (Fig. 1A). To examine whether Pin1 is modified by SUMOylation, we co-transfected His-Pin1 with GFP-tagged SUMO1 in 293T cells and found a modified Pin1 with a slower electrophoretic mobility (Fig. 1B, left). To confirm whether Pin1 is modified by SUMOylation and which residue is modified, we purified the SUMO1-modified GST-Pin1 in E. coli cells harboring a SUMO system plasmid (25), followed by identifying the Pin1 SUMOylation site(s) using mass spectrometric analysis. The major SUMOylation site was found on Lys6 in the WW domain (Fig. 1B, right). Interestingly, Lys6 in Pin1 is surrounded by EEKL, which resembles the inverted consensus SUMOylation site, ψKXE, where ψ is a hydrophobic residue, X is any amino acid, E is an acidic residue, and K is a lysine residue to which SUMO moiety is covalently bound (26).
Figure 1.
Pin1 is modified on Lys6 and Lys63 by SUMO1 in vitro and in vivo.
A, Pin1 was immunoprecipitated from normal MCF10A breast cells after stable expression Pin1 shRNA and lentiviral vector (LKO) virus, followed by immunoblotting. B, Pin1 SUMOylation in cells and identification of Pin1 SUMOylation on Lys6. Left, Cells expressing GFP-SUMO1 and His-Pin1 or vector control were extracted under denaturing conditions and GFP-SUMO1 conjugates were isolated with Ni-NTA agarose and immunoblotted. Right, SUMO1-modified GST-Pin1 was purified in E. coli cells and then subjected to Mass Spectrometry. C, In vitro SUMOylation reactions using recombinant Pin1 were carried out in the presence of the indicated components, followed by immunoblotting. D and E, Pin1 is modified on Lys6 and Lys63 in cells. 293T cell were transfected with GFP-SUMOs, together with Flag-Pin1 variants or a control vector. Cell extracts were immunoprecipitated and immunoblotted. F, Lys6/63 mutations ablate Pin1 SUMOylation in cells. MCF10A-Ras/Neu cells stably expressing Pin1 and Pin1K6/63R mutant were immunoprecipitated and immunoblotted. Arrowhead: SUMO1-modified Pin1, arrow: SUMO1-unmodified Pin1 and asterisk: IgG.
To further confirm that Pin1 is a SUMO substrate, we performed in vitro SUMOylation assays. GST-Pin1 was SUMOylated only when SAE1/2, Ubc9, ATP and SUMO-1 were present (Fig. 1C). To examine whether Pin1 is a SUMO1 specific substrate, we co-transfected Pin1 with GFP-tagged SUMO1, -SUMO2, or -SUMO3 into 293T cells. Only SUMO1 effectively promoted SUMOylation of Pin1 (Fig. 1D, left). Similarly, only wild-type SUMO1 (GFP-SUMO1 GG), but not its conjugation-defective mutant (GFP-SUMO1 AA), promoted Pin1 SUMOylation (Fig. 1D, right and Supplementary Fig. S1A). We further used Pin1 deletion and point mutation mutants to confirm the SUMOylation site(s) and found that the K6R mutation completely abolished SUMOylation of Pin1 WW domain (Fig. 1E, left). Surprisingly, the separate Pin1 PPIase was also found to be SUMOylated (Fig. 1E, left). To identify this new SUMOylation in the PPIase domain, we mutated all the seven Lys residues in the Pin1 PPIase domain together or in different combination to Ala in full length Pin1 and conducted in vivo SUMOylaion assay. Only the Lys63 point mutation significantly abrogated the formation of the characteristic shifted band corresponding to a single GFP-SUMO1 molecule conjugated to a specific Lys residue (Supplementary Fig. S1B). Moreover, the modification sites were also identified by site-directed mutagenesis and in vivo SUMOylation assay, which showed that the Pin1 SUMO1 modification was greatly impaired in WWK6R, PPIaseK63R, Pin1K6R, Pin1E4A and Pin1L7A mutants (Fig. 1E, left and Supplementary Fig. S1C). These results were also consistent with the data derived from in vitro SUMOylation assay (Supplementary Fig. S1D) and yeast two-hybrid assay (Supplementary Fig. S1E), as described previously (27), showing that the mutations at Lys6 and Lys63 or the surrounding inverted consensus SUMOylation sequence disrupted Pin1 SUMOylation. To further confirm that these two Lys residues are SUMO1 acceptor sites in Pin1, in vivo SUMOylation assay was conducted using full-length Pin1 as well as its K6R or K6/63R mutant (Fig. 1E, right). In contrast to Pin1, the SUMOylation-deficient Pin1K6/63R mutant failed to be SUMOylated in cells (Fig. 1F). The SUMOylation of Pin1 is also observed in different cell lines (Supplementary Fig. S1A, bottom). Taken together, these findings demonstrate that Pin1 is modified on Lys6 and Lys63 by SUMO1 in vitro and in vivo.
Pin1 SUMOylation inhibits its substrate binding, catalytic activity and cellular function
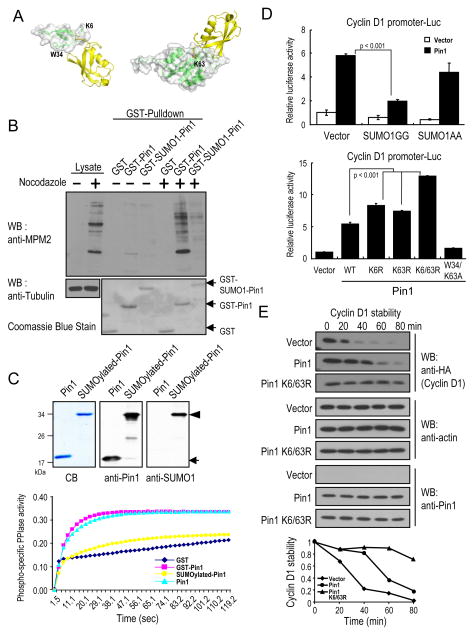
Given that Pin1 is SUMOylated on Lys6 in the WW domain and Lys63 in the PPIase domain, we asked whether these modifications affect Pin1 activity and function. Structural modeling analysis of SUMO1 (PDB code 1A5R) conjugated at Lys6 showed that the loop1 of SUMO1 was very close to the Trp34 residue of the WW domain (PDB code 1F8A) (Fig. 2A, left), the second invariant Trp residue that is essential for Pin1 to bind to its phosphorylated substrates (3). Similarly, SUMO1 conjugated at Lys63, a positively charged residue critical for anchoring the pSer/Thr-binding pocket in the PPIase domain (PDB code 3IK8) might prevent the substrates from entering the catalytic active site (Fig. 2A, right). These molecular modeling results suggest that Pin1 SUMOylation might affect its substrate binding and catalytic activity.
Figure 2.
Pin1 SUMOylation inhibits its substrate binding and catalytic activity.
A, Molecular modeling using Discovery studio software and graphically depicted using PyMol. B, Recombinant proteins were incubated with mitotic HeLa extracts followed by pulldown before subjecting to immunoblotting. C, Purified GST-fusion proteins were produced, as shown by Coomassie blue staining, immunoblotting. The Pin1 catalytic activity were determined using PPIase assay. D, HeLa Pin1 knockdown cells were transfected with SUMO1 or its inactive mutant and Pin1, or its Lys6 and/or Lys63 mutants or vector control, along with cyclin D1 promoter reporter construct, followed by assaying the luciferase activity and pRL-TK Renilla luciferase reporter activity as control. E, Pin1−/− MEFs were co-transfected with Pin1 or its Lys6 and Lys63 mutants and HA-cyclin D1 and then treated with cycloheximide.
To test whether Pin1 SUMOylation affects its substrate binding activity, we generated GST-SUMO1 fused to Lys6 of Pin1 protein because the first 5 amino acid of Pin1 is unstructured based on its X-ray structure (28). We examined the ability of GST-SUMO1-Pin1 to bind MPM-2 antigens, known Pin1 substrates present in mitotic cells, using a GST-Pin1 pulldown assay. Unlike GST-Pin1, GST-SUMO1-Pin1 greatly impaired its ability to bind the MPM-2 antigens (Fig. 2B), consistent with molecular modeling (Fig. 2A, left). To examine whether Pin1 SUMOylation also affects its catalytic activity, we purified both the unmodified and SUMO1-modified GST-Pin1 from E. coli cells harboring a SUMOylation system plasmid and a plasmid expressing GST-Pin1 (Fig. 2C, top) (25). In contrast to Pin1 or GST-Pin1, SUMOylated Pin1 displayed little phospho-specific PPIase activity (Fig. 2C, bottom). These results indicate that SUMOylation of Pin1 greatly inhibits its substrate binding and catalytic activity.
The above results suggest that SUMOylation of Pin1 might inhibit its function in the cell. One of the well-characterized Pin1 cellular functions in cell growth regulation is its ability to activate the promoter of cyclin D1 as well as to increase cyclin D1 protein stability (29, 30). Indeed, co-transfection of Pin1 and SUMO1, but not its inactive mutant inhibited the ability of Pin1 to activate the cyclin D1 promoter (Fig. 2D, top). Importantly, Pin1K6/63R, a mutant that is resistant to Pin1 SUMOylation or the mutants that are disrupted the inverted consensus SUMOylation site (Fig 1E, right and Supplementary Fig. S1C), were more active compared to wild-type Pin1 in activating the cyclin D1 promoter (Fig. 2D, bottom and Supplementary Fig. S2A and 2B). To examine whether SUMOylation of Pin1 affects turnover of cyclin D1, we co-transfected cyclin D1 with Pin1 and Pin1K6/63R mutants into Pin1−/− MEFs or MCF10A-Ras/Neu Pin1 knockdown cells, and then monitored cyclin D1 protein stability (13). Pin1 increased cyclin D1 protein stability, as shown previously (13) and importantly, Pin1K6/63R mutant was more potent than the wild-type protein in stabilizing cyclin D1 protein in Pin1 null cells (Fig. 2E, and Supplementary Fig. S2C). We further performed the immunostaining experiment to show that SUMOylation did not significantly disrupt the subcellular localization of Pin1 (Supplementary Fig. S3A). Together, these results show that Pin1 SUMOylation not only inhibits its substrate binding and catalytic activity in vitro, but also impairs its cellular function in cells.
SENP1 deSUMOylates Pin1
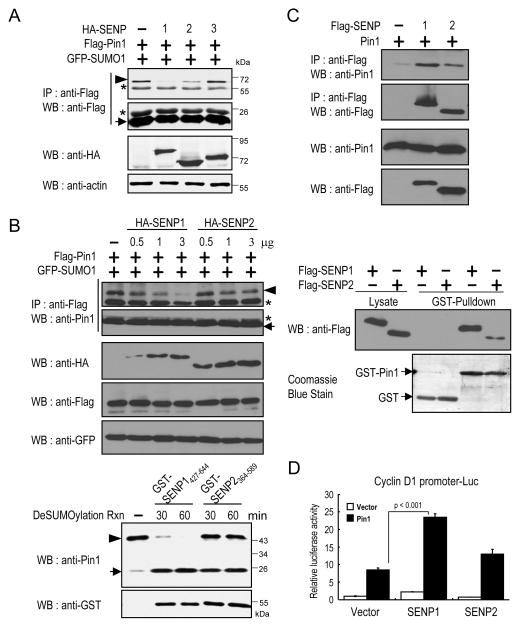
Given that Pin1 SUMOylation inhibits its protein activity and cellular function, the next question is whether Pin1 is subject to deSUMOylation. Since SENP1, SENP2 and SENP3 are primarily localized in the nucleus (31), we first investigated which SENPs might bind and deSUMOylate Pin1, a mainly nuclear protein (13). When SENP1, SENP2 or SENP3 were coexpressed with Pin1 in cells, we found that SENP1 was more efficient in deSUMOylating Pin1 than SENP2, while SENP3 was barely active (Fig. 3A). When different amounts of SENP1 and SENP2 expression plasmids were transfected to compare their abilities to deSUMOylate Pin1 in vivo, SENP1 was more efficient than SENP2 in promoting Pin1 deSUMOylation (Fig. 3B, top). When the same amount of recombinant SENPs catalytic domain proteins, GST-SENP1427–644 and -SENP2364–589, were analyzed to compare their abilities to deSUMOylate Pin1 in vitro, again SENP1 was found to be much more proficient than SENP2 (Fig. 3B, bottom). We further examined their interactions by coimmunoprecipitation (Co-IP) and GST pulldown assays. In both assays, SENP1 bound to Pin1 better than SENP2 (Fig. 3C). Accordingly, SENP1 was functionally more active than SENP2 in activating the cyclin D1 promoter activity (Fig. 3D).
Figure 3.
SENP1 binds to Pin1 and promotes Pin1-mediated cyclin D1 activation.
A,. 293T cells were co-transfected with Flag-Pin1, GFP-SUMO1 and HA-SENP1, HA-SENP2 or HA-SENP3 constructs and subjected to immunoprecipitation, followed by immunoblotting. B, Top, 293T cells were transfected with Pin1 with different amounts of HA-SENP1 or HA-SENP2 constructs, followed by Flag IP and immunoblotting. Bottom, The same amount of SENP1 or SENP2 with SUMOylated Pin1 were incubated in a deSUMOylation reaction buffer, followed by immunoblotting. C, Top, 293T cells were co-transfected with Flag-SENP1 or Flag-SENP2 and Pin1 constructs and then subjected to immunoprecipitation with anti-Flag, followed by immunoblotting with Pin1 Ab. Bottom, Glutathione-agarose beads containing GST or GST-Pin1 were incubated with cell extracts expressing Flag-SENP1 or Flag-SENP2. Proteins pulled down by GST beads were subjected to immunoblotting. D, HeLa cells were transfected with SENP1 or SENP2, and Pin1, or vector control, along with cyclin D1 promoter reporter construct, followed by assaying the luciferase and pRL-TK Renilla luciferase activity as control. Arrowhead: SUMO1-modified Pin1, arrow: SUMO1-unmodified Pin1 and asterisk: IgG.
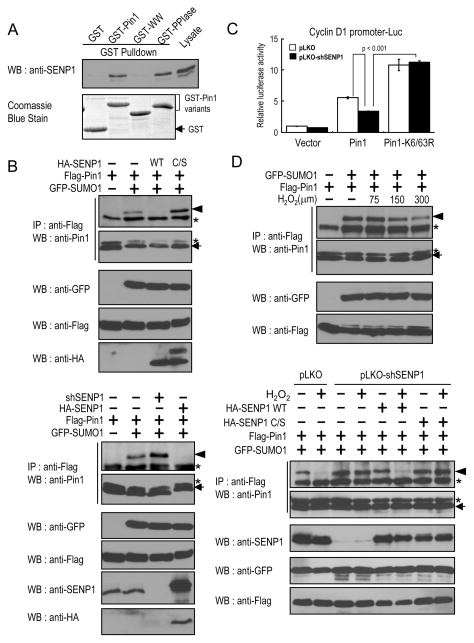
To map the Pin1 interaction domain within SENP1, we performed GST pulldown experiments using Pin1 and its deletion mutants and found that SENP1 bound only to the PPIase domain of Pin1 (Fig. 4A). Furthermore, we used purified SENP1 and Pin1 to show that Pin1 directly interacted with SENP1 and that the Pin1 PPIase domain is required for this interaction (Supplementary Fig. S3B). Given the SENP1 and Pin1 specific interaction, a central question is whether the enzymatic activity of SENP1 is essential for Pin1 deSUMOylation in vivo. By overexpressing a SENP1 catalytically inactive/dominant negative Cys-to-Ser SENP1 mutant (C/S) (32) or a SENP1 shRNA construct (33), we found inhibition of endogenous SENP1 via the SENP1 dominant negative mutant or shRNA induced Pin1 SUMOylation in vivo (Fig. 4B). Similarly, the inhibition of Pin1-mediated cyclin D1 activation by SUMO1 was recovered only by SENP1. Pin1K6/63R was more active than wild-type Pin1 in activating the cyclin D1 promoter, which was resistant to inhibition of SUMO1 or activation of SENP1 (Fig. 4C). Moreover, knockdown of endogenous SENP1 expression also reduced Pin1-mediated cyclin D1 activation and again the enhanced ability of Pin1K6/63R to activate the cyclin D1 promoter was not reduced by SENP1 (Supplementary Fig. S3C). Taken together, these data indicate that SENP1 is a Pin1 deSUMOylation protease that reverses Pin1 SUMOylation and inhibition.
Figure 4.
SENP1 binds and deSUMOylates Pin1 and thereby positively regulates cyclin D1 activation.
A, 293T cells expressing SNEP1 were incubated with glutathione-agarose beads containing GST, GST-Pin1 or GST-Pin1 truncated mutants, followed by GST-Pin1 pulldown assay. B, 293T cells were cotransfected with Flag-Pin1 and SUMO1 with Flag-SENP1, Flag-SENP1 C/S or shSENP1, followed by Flag IP and subjected to immunoblotting. C, SENP1 promotes Pin1-mediated cyclin D1 promoter activation. 293T cells were cotransfected with control or SENP1-shRNA followed by assaying the luciferase activity and pRL-TK Renilla luciferase activity as control, and relative luciferase activity was plotted (mean + SEM). *p <0.03; **p < 0.001. D, H2O2 triggers Pin1 deSUMOylation. Control cells or SENP1 knockdown cells were transfected with Flag-Pin1 and GFP-SUMO1 with control vector, HA-SENP1 or HA-SENP1C/S and then treated for 6 hr with increasing H2O2 concentration. Cells were subjected to immunoprecipitation with anti-Flag, followed by immunoblotting. Arrowhead: SUMO1-modified Pin1, arrow: SUMO1-unmodified Pin1 and asterisk: IgG.
Next we sought to search for signals that induce Pin1 de-SUMOylation. It has been shown that permissive and moderate concentrations of reactive oxygen species (ROS) are not only ubiquitous but also serve to regulate signaling events and to stimulate cell proliferation (34, 35). Furthermore, low concentrations of ROS results in the rapid disappearance of most SUMO conjugate and upregulation of SENP3 (36, 37). Interestingly, Pin1 has been shown to be involved in ROS production and ROS mediated cell proliferation (38, 39). These results suggest that ROS might regulate Pin1 deSUMOylation status. To examine this possibility, cells co-transfected with SENP1 and Pin1 were treated with H2O2. We found that increasing concentrations of H2O2 resulted in dose-dependent decrease in Pin1 SUMOylation (Fig. 4D, top). We further addressed whether the reduced SUMOylation of Pin1 is due to the SENP1-mediated deSUMOylation by generating stable SENP1 knockdown cells, followed by adding back SENP1 WT or its catalytic-defective(C/S) mutant. Upon the H2O2 treatment at 0.3 mM, SUMOylation of Pin1 was significantly reduced in SENP1 WT rescued cells, but not in the C/S mutant rescued cells (Fig. 4D, bottom). To investigate whether the stimulatory effect of H2O2 on Pin1 deSUMOylation is attributable to differential Pin1 and SENP1 interactions, Co-IP experiments were performed using cells expressing epitope-tagged versions of Pin1 and SENP1 following treatment with increasing concentrations of H2O2. Binding between Pin1 and SENP1 was increased with increasing concentrations of H2O2 (Supplementary Fig. S3D), showing the redox-regulated interaction between Pin1 and SENP1. These data suggest that oxidative stress induces Pin1 deSUMOylation through its interaction and deSUMOylation by SENP1.
SENP1 promotes Pin1-induced centrosome duplication, chromosome instability and cell transformation
The above results have shown that deSUMOylation of Pin1 by SENP1 reverses Pin1 inhibition by SUMOylation and that SENP1 also promotes the ability of Pin1 to activate its downstream targets such as cyclin D1, suggesting that SENP1 might promote Pin1-mediated cell proliferation and transformation. It has been shown that SENP1 is upregulated in some human cancers (18). Furthermore, tight regulation of Pin1 function during the cell cycle in normal cells is critical for the coordination of DNA synthesis and centrosome duplication (11, 13). By contrast, constitutive Pin1 overexpression disrupts this coordination, leading to centrosome amplification, abnormal spindle formation, chromosome instability and cell transformation (11, 13). Therefore, deSUMOylation of Pin1 by SENP1 might promote the ability of Pin1 to induce centrosome amplification and cell transformation.
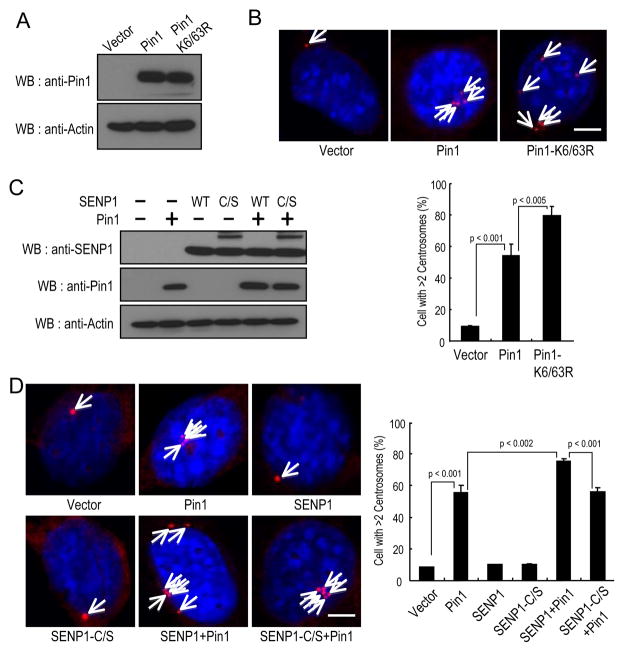
To examine this possibility, we first examined the effects of Pin1 deSUMOylation on centrosome duplication by transfecting non-transformed NIH3T3 cells with Pin1, Pin1K6/63R or a control vector (Fig. 5A), and then arrested them in G1/S phase by aphidicolin and counted centrosome number, as described previously (11, 13). It has been shown that these S phase arrested NIH3T3 cells permit centrosome duplication and are easily assayed to identify the role of a specific protein in centrosome duplication (11, 13). Indeed, more than 50% of Pin1-transfected cells contained >2 centrosomes (Fig. 5B), as described (13). Importantly, more than 80% of Pin1K6/63R mutant-transfected cells contained >2 centrosomes, indicating that Pin1K6/63R is more active than the wild-type protein in inducing centrosomes amplification. These results suggest that deSUMOylation of Pin1 promotes its ability to induce centrosome amplification.
Figure 5.
SENP1 promotes Pin1-induced centrosome amplification.
A and B, Pin1K6/63R mutant was more potent than wild-type Pin1 in inducing centrosome amplification. NIH3T3 cells were stably infected with lentiviruses expressing Pin1, its K6/63R mutants or control (A) and then arrested at the G1/S boundary for 24 hr by aphidicolin. Cells were stained with anti-γ-tubulin antibody (red) and DAPI (blue) (B, top). Bar, 10 μm (B, top). Cells containing >2 centrosomes were scored in 300 transfected cells (B, bottom). C and D, SENP1 promotes Pin1-induced centrosome amplification. NIH3T3H cells were transfected with SENP1 or its mutant and Pin1 (C). Cells were arrested at the G1/S boundary by aphidicolin. Cells were stained with anti-γ-tubulin antibody (red) and DAPI (D, top) (E). Bar, 10 μm (D, left). Cells containing >2 centrosomes were scored in 300 transfected cells (D, right).
To further determine whether Pin1-drived centrosome amplification is regulated by SENP1, we generated NIH3T3 cells overexpressing Pin1 and SENP1 or its C/S mutant (Fig. 5C). Co-expression of SENP1 significantly increased the ability of Pin1 to induce centrosome amplification (Fig. 5D). These results are consistent with the above findings showing that SENP1 binding increases Pin1 activity toward cyclin D1 (Fig. 3D). More importantly, the inhibitory effects of SENP1 on Pin1 appeared to be highly specific because they did not occur at all when the SENP1 C/S mutant was used (Fig. 5D). These findings indicate that deSUMOylation of Pin1 by SENP1 increases the ability of Pin1 overexpression to induce multiple rounds of centrosome duplication in S arrested NIH3T3 cells.
Given that Pin1 SUMOylation site mutations affect Pin1-derived centrosome amplification in non-transformed NIH3T3 cells, we next examined the functional consequences of Pin1 deSUMOylation on Pin1 function in cell proliferation. Specifically, we asked whether these SUMOylation-resistant Pin1 mutations also affect its ability to induce the transforming phenotypes. Firstly, we reconstituted MCF10A-Ras/Neu Pin1 knockdown cells with stably re-expressing Pin1 and Pin1K6/63R (Supplementary Fig. S1D). The MCF10A-Ras/Neu control, Pin1 knockdown cells and Pin1 knockdown cells stably re-expressing Pin1 or Pin1K6/63R were seeded on plastic plates, followed by MTT proliferation assay and three dimensional cell differentiation assay with exogenous basement membrane matrix (Matrigel), where Pin1 overexpression has been shown to induce the early transforming phenotype of mammary epithelial cells (10). Consistent with the previous findings (10), inhibition of Pin1 suppressed transforming phenotypes of mammary epithelial cells induced by Ras/Neu (Fig. 6A and B). More importantly, as compared with wild-type Pin1-expressing cells, Pin1K6/63R-expressing cells were more proficient in promoting cell proliferation both in MTT assay and three dimensional cell culture assay (Fig. 6A and B). Moreover, Pin1K6/63R-expressing cells formed a higher number of foci on plastic plates and colonies in soft agar than Pin1-expressing cells (Fig. 6C and D). These results together demonstrate that Pin1 deSUMOylation promotes its ability to induce centrosome amplification and cell transformation.
Figure 6.
Pin1 SUMOylation-deficient mutant Pin1K6/63R is more potent than wild-type Pin1 in inducing cell proliferation and cell transformation.
A and B, Pin1K6/63R mutant is more potent than wild-type Pin1 in inducing cell proliferation. Pin1 knockdown or rescued MCF10A Ras/Neu cells were seeded on plastic plates (A) and Matrigel gels (B), followed by MTT assay (A) or P-iodonitrotetrazolium violet staining (B). C, Pin1 knockdown or rescued MCF10A Ras/Neu cells were seeded on plastic plates followed by crystal violet staining (C, left). The number of colonies formed per 250 cells was seeded (C, right). Colony numbers are the mean ±SD of three independent experiments. D, Pin1K6/63R mutant is more potent than wild-type Pin1 in inducing cell transformation. Pin1 knockdown or rescued MCF10A Ras/Neu cells were seeded in soft agar (D, left) followed by P-iodonitrotetrazolium violet staining. The number of colonies formed per 1000 cells was seeded (D, right).
Pin1 levels correlate positively with SENP1 levels in human breast cancer
Pin1 plays a major role in the development of breast cancer. Intriguingly, it has been shown that SENP1 is overexpressed in human prostate cancer tissues (19) and that SENP1 deSUMOylates HIF1α and increases HIF1α protein stability, presumably contributing to tumorigenesis (20). Given that SENP1 deSUMOylates Pin1 and activates its function, we asked whether SENP1 might regulate Pin1 protein stability and correlate with Pin1 levels in human breast cancer tissues.
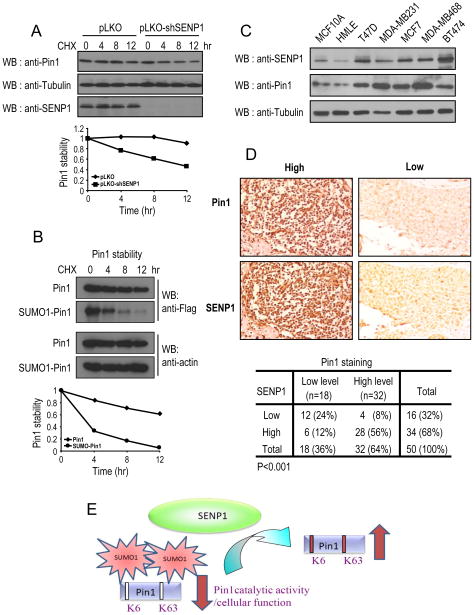
To investigate whether SENP1 regulates Pin1 stability, we stably knocked down SENP1 in breast cancer cells with shSENP1 and then examined Pin1 protein stability using the cycloheximide chase. SENP1 knockdown significantly reduced Pin1 stability (Fig. 7A). Moreover, we transfected Flag-Pin1, or Flag-SUMO-Pin1 in Pin1 knockdown cells and checked the protein stability. The protein half-life of SUMOylated-mimicking Pin1 is 4 hr, whereas Pin1 WT had a half-life longer than 12 hr (Fig. 7B). To further confirm these results, we have examined whether Pin1 protein levels are correlated with SENP1 in selected human normal and breast cancer cell lines, because we have previously shown that Pin1 is overexpressed in human breast cancer cell lines (30).. SENP1 was highly expressed and correlated with Pin1 overexpression in a number of breast cancer cell lines, including T47D, MDA-MB231, MCF7, MDA-MB468, and BT474 (Fig. 7C). We next investigated the relationship between SENP1 and Pin1 expression in human tissues by performing immunocytochemistry analysis on consecutive sections of tissue microarrays derived from human breast normal and cancer tissues using antibodies specifically against Pin1 and SENP1. Interestingly, most tumor specimens expressing low levels of SENP1 had marked reduced levels of Pin1, whereas most tumor specimens expressing high SENP1 had high level of Pin1 (Fig. 7B and C). There was a significant correlation between SENP1 and Pin1 levels in cancer specimens, as determined by the Spearman rank correlation test (P<0.001). These results on human breast cancer tissues are consistent with the findings that deSUMOylation of Pin1 on Lys6 and Lys63 by SENP1 promotes Pin1 protein activity, protein stability and oncogenic function (Fig. 7D).
Figure 7.
SENP1 levels positively correlate with Pin1 levels in human breast cancer tissues.
A, B, MCF10 Ras/Neu cells expressing control and SENP1 shRNA or MCF10 Ras/Neu cells expressing Pin1shRNA and SENP1 shRNA were transfected with Flag-Pin1, or Flag-SUMO-Pin1 were treated with cycloheximide (100 μg/ml) for indicated times, followed by immunoblotting. C, The same amounts of total lysates prepared from spontaneously immortalized normal human mammary epithelial cell lines and human breast carcinoma-derived cell lines were subjected to immunoblotting. D, Serial sections of tissue arrays of 50 breast cancer tissue specimen were subjected to immunohistochemistry using anti-Pin1 antibodies (upper panel) or anti-SENP1 antibodies (low panel), and visualized by the DAB staining (B). In each sample, SENP1 expression and Pin1 levels were semi-quantified in a double-blind manner as high or low according to the standards presented in (B) and summarized in (C) Their correlation was analyzed by Spearman rank correlation test (P<0.001). E, A scheme depicts that SENP1 binds and deSUMOylates Pin1 and promotes its Pin1 activity and function.
Discussion
Pin1 regulates over 30 oncogenes and tumor suppressors (1, 3). Emerging evidence suggests that Pin1 function may be regulated at multiple levels (2). Pin1 expression is generally correlated with cell proliferation in normal human tissues, but further upregulated in many human cancer tissues(2, 3). Indeed, Pin1 expression is subject to E2F-mediated transcriptional regulation in response to growth factors (2, 3). In addition, Pin1 is one of the genes suppressed by up-regulation of Brca1(2, 3). Pin1 belongs to a very small number of genes whose SNPs in the germ line are linked to somatic gene expression in breast tumors (2, 3). Indeed, the Pin1 genetic polymorphisms that reduce Pin1 expression are associated with reduced risk for multiple cancers in humans (2, 3, 40–42). Increasing evidence suggests that Pin1 is also subject to post-translational modifications. Pin1 Ser16 phosphorylation is regulated in a cell cycle-dependent manner, which abolishes the ability of Pin1 to interact with its substrates (2, 3). Pin1 is also phosphorylated on Ser65 by Polo-like kinase, which appears to increase Pin1 protein stability (2, 3). Our recent studies show that DAPK1 phosphorylates Pin1 on Ser71 and inhibits its prolyl isomerase activity and cellular function, providing the first explanation for the observed link between DAPK1 and Pin1 mediated malignancy in caner progression (13). Finally, Pin1 is also modified by oxidization, which impairs its PPIase activity (43). Nevertheless, little is known about whether Pin1 is regulated by other post-translational modifications.
SUMOylation is a highly dynamic process and an important regulator of the functional properties of many proteins (14). The SUMO conjugation enzymes (E1 and E2) are also frequently up-regulated in breast cancers and closely associated with patient survival (44, 45). The current studies indicate that deregulation of either SUMO conjugation or deconjugation can contribute to cancer progression (46–48). Most of the reports have focused on the effect of SUMO modification through the action of the conjugation enzyme Ubc9 or E3 ligases (14). However, the function of the SUMO deconjugating systems is still emerging (49). In mammalian cells, there are at least six different SUMO-specific proteases that have been identified. Notably, SENP1 was the first identified SUMO-specific protease. Data from SENP1 knockout mice indicate that the loss of SENP1 potentiates HIF1α degradation and consequently lowers VEGF levels (20). Reduction of VEGF hinders development of new vasculature and contributes to the lethality of SENP1 knockout. These studies suggest that induction of SENP1 in cancers might facilitate angiogenesis via enhanced stability of HIF1α. Elevated SENP1 levels are observed in thyroid oncocytic adenocarcinoma and prostate cancer (19, 22), and the upregulation of SENP1 is a relatively early event in the carcinogenesis of the prostate (19). While it is intriguing to speculate that SENP1 may play an important role in the growth regulation and cancer progression, only a handful of its substrates have been identified so far, and its targets and molecular mechanisms during tumorigenesis remain poorly understood.
Our studies of Pin1 SUMOylation have not only identified the inhibitory role of SUMOylation in controlling Pin1 activity and function, but also discovered a potent role of SENP1 in promoting Pin1 activity and function during cell proliferation and transformation. First, Pin1 is SUMOylated on Lys6 in the WW domain and Lys63 in the catalytic domain in vitro and in vivo. Second, the Pin1 SUMOylation fully inactivates Pin1 substrate binding and phosphorylation-specific PPIase activity. Third, SUMOylation-resistant mutation (K6/63R) is more active than the wild-type protein in cells, as assayed by activating transcription factors, stabilizing proteins as well as inducing centrosome amplification and cell transformation. More importantly, SENP1 over-expression promotes the ability of Pin1 to activate transcription factors, stabilize proteins and induce centrosome amplification and cell transformation. The significance of these findings is further substantiated by the demonstrations that Pin1 levels correlated positively with SENP1 levels in breast cancer tissues. Thus, Pin1 catalytic activity and cellular function is activated by SENP1 mediated Pin1 deSUMOylation. Future studies will be required to determine the specific relationship between overexpression of SENP1 and pathogenesis of breast cancer by using transgenic mice model.
The findings that Pin1 is deSUMOylated and activated by SENP1 suggest an exciting novel mechanism to control Pin1 catalytic activity in human disease. The most notable example is cancer, where Pin1 and SENP1 have been shown to have many correlated functions in tumorigenesis, although they have not been studied together before. Both Pin1 and SENP1 are upregulated in many human cancers (19, 22). Moreover, reducing Pin1 or SENP1 upregulation in cancer cells effectively suppresses tumorigenic phenotypes (12, 33, 50).
Supplementary Material
Acknowledgments
Grant Support
We thank Fu-An Li for mass spectrometry analysis. C-H.C. is the recipient DOD Breast Cancer Postdoctoral Award (W81XWH09-1-0481) and Ruth L. Kirschstein National Research Service Award, S. W. is the recipient of Susan G. Komen Postdoctoral Fellowship (KG111233) and T.H.L is the recipient of NIH Pathway to Independence (PI) Award (R00AG033104). The work was supported by Taipei Medical University Grant TMU101-AE1-B41 to C-C.C., NIH grants R01CA167677 to K.P.L., and Komen Investigator-initiated Research Grant to X. Z. Z.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Lee TH, Pastorino L, Lu KP. Peptidyl-prolyl cis-trans isomerase Pin1 in ageing, cancer and Alzheimer disease. Expert Rev Mol Med. 2011;13:e21. doi: 10.1017/S1462399411001906. [DOI] [PubMed] [Google Scholar]
- 2.Liou YC, Zhou XZ, Lu KP. Prolyl isomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends Biochem Sci. 2011;36:501–14. doi: 10.1016/j.tibs.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu KP, Zhou XZ. The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat Rev Mol Cell Biol. 2007;8:904–16. doi: 10.1038/nrm2261. [DOI] [PubMed] [Google Scholar]
- 4.Nakamura K, Greenwood A, Binder L, Bigio EH, Denial S, Nicholson L, et al. Proline isomer-specific antibodies reveal the early pathogenic tau conformation in Alzheimer’s disease. Cell. 2012;149:232–44. doi: 10.1016/j.cell.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan WC, Lee YR, Huang SF, Lin YM, Chen TY, Chung HC, et al. A Cullin3-KLHL20 Ubiquitin ligase-dependent pathway targets PML to potentiate HIF-1 signaling and prostate cancer progression. Cancer Cell. 2011;20:214–28. doi: 10.1016/j.ccr.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 6.Girardini JE, Napoli M, Piazza S, Rustighi A, Marotta C, Radaelli E, et al. A Pin1/mutant p53 axis promotes aggressiveness in breast cancer. Cancer Cell. 2011;20:79–91. doi: 10.1016/j.ccr.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Min SH, Lau AW, Lee TH, Inuzuka H, Wei S, Huang P, et al. Negative regulation of the stability and tumor suppressor function of Fbw7 by the Pin1 prolyl isomerase. Mol Cell. 2012;46:771–83. doi: 10.1016/j.molcel.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khanal P, Kim G, Yun HJ, Cho HG, Choi HS. The prolyl isomerase Pin1 interacts with and downregulates the activity of AMPK leading to induction of tumorigenicity of hepatocarcinoma cells. Mol Carcinog. 2012 doi: 10.1002/mc.21920. [DOI] [PubMed] [Google Scholar]
- 9.Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F, et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012;14:1295–304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryo A, Liou YC, Wulf G, Nakamura M, Lee SW, Lu KP. PIN1 is an E2F target gene essential for Neu/Ras-induced transformation of mammary epithelial cells. Mol Cell Biol. 2002;22:5281–95. doi: 10.1128/MCB.22.15.5281-5295.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suizu F, Ryo A, Wulf G, Lim J, Lu KP. Pin1 regulates centrosome duplication and its overexpression induces centrosome amplification, chromosome instability and oncogenesis. Mol Cell Biol. 2006;26:1463–79. doi: 10.1128/MCB.26.4.1463-1479.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wulf G, Garg P, Liou YC, Iglehart D, Lu KP. Modeling breast cancer in vivo and ex vivo reveals an essential role of Pin1 in tumorigenesis. EMBO J. 2004;23:3397–407. doi: 10.1038/sj.emboj.7600323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee TH, Chen CH, Suizu F, Huang P, Schiene-Fischer C, Daum S, et al. Death-associated protein kinase 1 phosphorylates Pin1 and inhibits its prolyl isomerase activity and cellular function. Mol Cell. 2011;42:147–59. doi: 10.1016/j.molcel.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol. 2010;11:861–71. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–56. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 16.Hannoun Z, Greenhough S, Jaffray E, Hay RT, Hay DC. Post-translational modification by SUMO. Toxicology. 2010;278:288–93. doi: 10.1016/j.tox.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 17.Wang CY, Podolsky R, She JX. Genetic and functional evidence supporting SUMO4 as a type 1 diabetes susceptibility gene. Ann N Y Acad Sci. 2006;1079:257–67. doi: 10.1196/annals.1375.039. [DOI] [PubMed] [Google Scholar]
- 18.Bawa-Khalfe T, Yeh ET. SUMO Losing Balance: SUMO Proteases Disrupt SUMO Homeostasis to Facilitate Cancer Development and Progression. Genes Cancer. 2010;1:748–52. doi: 10.1177/1947601910382555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng J, Bawa T, Lee P, Gong L, Yeh ET. Role of desumoylation in the development of prostate cancer. Neoplasia. 2006;8:667–76. doi: 10.1593/neo.06445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng J, Kang X, Zhang S, Yeh ET. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell. 2007;131:584–95. doi: 10.1016/j.cell.2007.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Nguyen T, Angkasekwinai P, Dou H, Lin FM, Lu LS, Cheng J, et al. SUMO-specific protease 1 is critical for early lymphoid development through regulation of STAT5 activation. Mol Cell. 2012;45:210–21. doi: 10.1016/j.molcel.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacques C, Baris O, Prunier-Mirebeau D, Savagner F, Rodien P, Rohmer V, et al. Two-step differential expression analysis reveals a new set of genes involved in thyroid oncocytic tumors. J Clin Endocrinol Metab. 2005;90:2314–20. doi: 10.1210/jc.2004-1337. [DOI] [PubMed] [Google Scholar]
- 23.Yaffe MB, Schutkowski M, Shen M, Zhou XZ, Stukenberg PT, Rahfeld J, et al. Sequence-specific and phosphorylation-dependent proline isomerization: A potential mitotic regulatory mechanism. Science. 1997;278:1957–60. doi: 10.1126/science.278.5345.1957. [DOI] [PubMed] [Google Scholar]
- 24.Rangasamy V, Mishra R, Sondarva G, Das S, Lee TH, Bakowska JC, et al. Mixed-lineage kinase 3 phosphorylates prolyl-isomerase Pin1 to regulate its nuclear translocation and cellular function. Proc Natl Acad Sci U S A. 2012;109:8149–54. doi: 10.1073/pnas.1200804109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huh YH, Kim YE, Kim ET, Park JJ, Song MJ, Zhu H, et al. Binding STAT2 by the acidic domain of human cytomegalovirus IE1 promotes viral growth and is negatively regulated by SUMO. J Virol. 2008;82:10444–54. doi: 10.1128/JVI.00833-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matic I, Schimmel J, Hendriks IA, van Santen MA, van de Rijke F, van Dam H, et al. Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Molecular cell. 2010;39:641–52. doi: 10.1016/j.molcel.2010.07.026. [DOI] [PubMed] [Google Scholar]
- 27.Chang CC, Lin DY, Fang HI, Chen RH, Shih HM. Daxx mediates the small ubiquitin-like modifier-dependent transcriptional repression of Smad4. J Biol Chem. 2005;280:10164–73. doi: 10.1074/jbc.M409161200. [DOI] [PubMed] [Google Scholar]
- 28.Ranganathan R, Lu KP, Hunter T, Noel JP. Structural and functional analysis of the mitotic peptidyl-prolyl isomerase Pin1 suggests that substrate recognition is phosphorylation dependent. Cell. 1997;89:875–86. doi: 10.1016/s0092-8674(00)80273-1. [DOI] [PubMed] [Google Scholar]
- 29.Ryo A, Suizu F, Yoshida Y, Perrem K, Liou YC, Wulf G, et al. Regulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol Cell. 2003;12:1413–26. doi: 10.1016/s1097-2765(03)00490-8. [DOI] [PubMed] [Google Scholar]
- 30.Wulf GM, Ryo A, Wulf GG, Lee SW, Niu T, Petkova V, et al. Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. The EMBO journal. 2001;20:3459–72. doi: 10.1093/emboj/20.13.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hang J, Dasso M. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J Biol Chem. 2002;277:19961–6. doi: 10.1074/jbc.M201799200. [DOI] [PubMed] [Google Scholar]
- 32.Bailey D, O’Hare P. Characterization of the localization and proteolytic activity of the SUMO-specific protease, SENP1. J Biol Chem. 2004;279:692–703. doi: 10.1074/jbc.M306195200. [DOI] [PubMed] [Google Scholar]
- 33.Wang Q, Xia N, Li T, Xu Y, Zou Y, Zuo Y, et al. SUMO-specific protease 1 promotes prostate cancer progression and metastasis. Oncogene. 2012 doi: 10.1038/onc.2012.250. [DOI] [PubMed] [Google Scholar]
- 34.de la Vega L, Grishina I, Moreno R, Kruger M, Braun T, Schmitz ML. A redox-regulated SUMO/acetylation switch of HIPK2 controls the survival threshold to oxidative stress. Mol Cell. 2012;46:472–83. doi: 10.1016/j.molcel.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 35.Hamanaka RB, Chandel NS. Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes. Trends Biochem Sci. 2010;35:505–13. doi: 10.1016/j.tibs.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bossis G, Melchior F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell. 2006;21:349–57. doi: 10.1016/j.molcel.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 37.Han Y, Huang C, Sun X, Xiang B, Wang M, Yeh ET, et al. SENP3-mediated de-conjugation of SUMO2/3 from promyelocytic leukemia is correlated with accelerated cell proliferation under mild oxidative stress. J Biol Chem. 2010;285:12906–15. doi: 10.1074/jbc.M109.071431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boussetta T, Gougerot-Pocidalo MA, Hayem G, Ciappelloni S, Raad H, Arabi Derkawi R, et al. The prolyl isomerase Pin1 acts as a novel molecular switch for TNF-alpha-induced priming of the NADPH oxidase in human neutrophils. Blood. 2010;116:5795–802. doi: 10.1182/blood-2010-03-273094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim SE, Lee MY, Lim SC, Hien TT, Kim JW, Ahn SG, et al. Role of Pin1 in neointima formation: down-regulation of Nrf2-dependent heme oxygenase-1 expression by Pin1. Free Radic Biol Med. 2010;48:1644–53. doi: 10.1016/j.freeradbiomed.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 40.Han CH, Lu J, Wei Q, Bondy ML, Brewster AM, Yu TK, et al. The functional promoter polymorphism (−842G>C) in the PIN1 gene is associated with decreased risk of breast cancer in non-Hispanic white women 55 years and younger. Breast cancer research and treatment. 2010;122:243–9. doi: 10.1007/s10549-009-0682-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu J, Hu Z, Wei S, Wang LE, Liu Z, El-Naggar AK, et al. A novel functional variant (−842G>C) in the PIN1 promoter contributes to decreased risk of squamous cell carcinoma of the head and neck by diminishing the promoter activity. Carcinogenesis. 2009;30:1717–21. doi: 10.1093/carcin/bgp171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu J, Yang L, Zhao H, Liu B, Li Y, Wu H, et al. The polymorphism and haplotypes of PIN1 gene are associated with the risk of lung cancer in Southern and Eastern Chinese populations. Hum Mutat. 2011;32:1299–308. doi: 10.1002/humu.21574. [DOI] [PubMed] [Google Scholar]
- 43.Butterfield DA, Abdul HM, Opii W, Newman SF, Joshi G, Ansari MA, et al. Pin1 in Alzheimer’s disease. J Neurochem. 2006;98:1697–706. doi: 10.1111/j.1471-4159.2006.03995.x. [DOI] [PubMed] [Google Scholar]
- 44.Mo YY, Yu Y, Theodosiou E, Ee PL, Beck WT. A role for Ubc9 in tumorigenesis. Oncogene. 2005;24:2677–83. doi: 10.1038/sj.onc.1208210. [DOI] [PubMed] [Google Scholar]
- 45.Kessler JD, Kahle KT, Sun T, Meerbrey KL, Schlabach MR, Schmitt EM, et al. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science. 2012;335:348–53. doi: 10.1126/science.1212728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mooney SM, Grande JP, Salisbury JL, Janknecht R. Sumoylation of p68 and p72 RNA helicases affects protein stability and transactivation potential. Biochemistry. 2010;49:1–10. doi: 10.1021/bi901263m. [DOI] [PubMed] [Google Scholar]
- 47.Driscoll JJ, Pelluru D, Lefkimmiatis K, Fulciniti M, Prabhala RH, Greipp PR, et al. The sumoylation pathway is dysregulated in multiple myeloma and is associated with adverse patient outcome. Blood. 2010;115:2827–34. doi: 10.1182/blood-2009-03-211045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matunis MJ, Guzzo CM. Pigment cell & melanoma research. 2012. SUMO, PTEN and Tumor Suppression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu B, Shuai K. Regulation of the sumoylation system in gene expression. Curr Opin Cell Biol. 2008;20:288–93. doi: 10.1016/j.ceb.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ryo A, Uemura H, Ishiguro H, Saitoh T, Yamaguchi A, Perrem K, et al. Stable suppression of tumorigenicity by Pin1-targeted RNA interference in prostate cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11:7523–31. doi: 10.1158/1078-0432.CCR-05-0457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.