Abstract
Prokaryotic type II CRISPR-Cas systems can be adapted to enable targeted genome modifications across a range of eukaryotes.1–7. Here we engineer this system to enable RNA-guided genome regulation in human cells by tethering transcriptional activation domains either directly to a nuclease-null Cas9 protein or to an aptamer-modified single guide RNA (sgRNA). Using this functionality we developed a novel transcriptional activation–based assay to determine the landscape of off-target binding of sgRNA:Cas9 complexes and compared it with the off-target activity of transcription activator–like (TAL) effector proteins8, 9. Our results reveal that specificity profiles are sgRNA dependent, and that sgRNA:Cas9 complexes and 18-mer TAL effector proteins can potentially tolerate 1–3 and 1–2 target mismatches, respectively. By engineering a requirement for cooperativity through offset nicking for genome editing or through multiple synergistic sgRNAs for robust transcriptional activation, we suggest methods to mitigate off-target phenomena. Our results expand the versatility of the sgRNA:Cas9 tool and highlight the critical need to engineer improved specificity.
Bacterial and archaeal CRISPR-Cas systems rely on short guide RNAs in complex with Cas proteins to direct degradation of complementary sequences present within invading foreign nucleic acids10–14. Recently the type II CRISPR-Cas system was engineered to effect robust RNA-guided genome modifications in multiple eukaryotic systems, significantly improving the ease of genome editing1–7. Here we expand the repertoire of sgRNA:Cas9-mediated control of eukaryotic genomes by developing sgRNA:Cas9 gene activators, thus enabling RNA-guided eukaryotic genome regulation. We use this expanded toolset to gain insights into the specificity of targeting by the S. pyogenes type II CRISPR-Cas system in human cells, compare the specificity profiles to those of TALE-based transcriptional activators and suggest the use of offset nicking to generate double strand breaks (DSBs) as a potential route to improving sgRNA:Cas9 genome editing specificity.
In S. pyogenes, Cas9 generates a blunt-ended double-stranded break 3bp upstream of the protospacer-adjacent motif (PAM) via a process mediated by two catalytic domains in the protein: an HNH domain that cleaves the complementary strand of the DNA and a RuvC-like domain that cleaves the non-complementary strand12. To enable RNA-guided genome regulation, it is essential to first eliminate Cas9 nuclease activity by ablating the natural activity of RuvC and HNH nucleases domains7. By searching for sequences with known structure that are homologous to Cas9 (refer Supplementary Note 1), we identified and mutated up to 4 amino acids likely involved in magnesium coordination (Supplementary Fig. 1a). The generated quadruple Cas9 mutant showed undetectable nuclease activity upon deep sequencing at the targeted loci (Supplementary Fig. 1b), implying that we had successfully reduced Cas9 nuclease activity to levels below the threshold of detection in our assay.
Nuclease-deficient Cas9 (hereafter referred to as Cas9N-) can in principle localize transcriptional regulatory domains to targeted loci by fusing these domains to either Cas9N- or to the sgRNA. We explored both approaches in parallel (Figs. 1a, 1b).
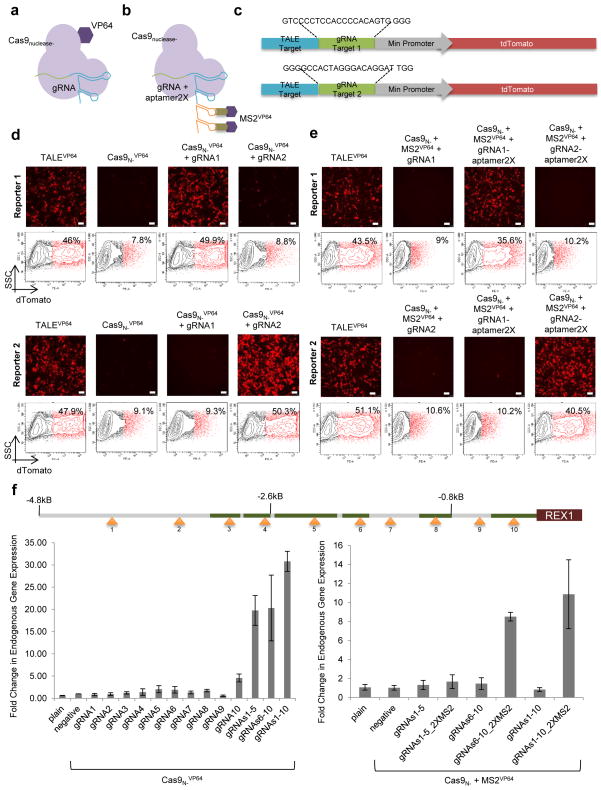
Fig. 1. RNA-guided transcriptional activation.
(a) To generate a Cas9N- fusion protein capable of transcriptional activation, we directly tethered the VP64 activation domain to the C terminus of Cas9N-. (b) To generate sgRNA tethers capable of recruiting activation domains, we appended two copies of the MS2 bacteriophage coat-protein binding RNA stem-loop to the 3′ end of the sgRNA and expressed these chimeric sgRNAs together with Cas9N- and MS2-VP64 fusion protein. (c) Design of reporter constructs used to assay transcriptional activation is shown. Note that the two reporters bear distinct sgRNA target sites, and share a control TALE-TF target site. (d) Cas9N-VP64 fusions display RNA-guided transcriptional activation as assayed by both fluorescence-activated cell sorting (FACS) and immunofluorescence assays (IF). Specifically, whereas the control TALE-TF activated both reporters, the Cas9N-VP64 fusion activates reporters in a sgRNA sequence specific manner. (e) As assayed by both FACS and IF we observed robust sgRNA sequence-specific transcriptional activation from reporter constructs only in the presence of all 3 components: Cas9N-, MS2-VP64 and sgRNA bearing the appropriate MS2 aptamer binding sites. The bar in the micrographs is 100μm. (f) For the REX1 gene we designed 10 sgRNAs (positions indicated in the figure) targeting a ~5kb stretch of DNA upstream of the transcription start site (DNase hypersensitive sites are highlighted in green), and assayed transcriptional activation using both the above approaches via qPCR of the endogenous genes. Although introduction of individual sgRNAs modestly stimulated transcription, multiple sgRNAs acted synergistically to stimulate robust multi-fold transcriptional induction. Note that in the absence of the 2X-MS2 aptamers on the sgRNA we do not observe transcriptional activation via the sgRNA-MS2-VP64 tethering approach. Data are means +/− SEM (N=3).
To generate a Cas9N- fusion protein capable of transcriptional activation, we directly fused the VP64 activation domain15 to the C terminus of Cas9N- (Fig. 1a). This Cas9N-VP64 protein robustly activated transcription of reporter constructs when combined with sgRNAs targeting sequences near the promoter, thereby displaying RNA-guided transcriptional activation (Figs. 1c, 1d, Supplementary Fig. 1c).
To generate sgRNA tethers capable of transcriptional regulation, we first determined which regions of the sgRNA will tolerate modifications by inserting random sequences into the sgRNA and assaying for sgRNA:Cas9 nuclease function. We found that sgRNAs bearing random sequence insertions at either the 5′ end of the crRNA portion or the 3′ end of the tracrRNA portion of a chimeric sgRNA retain functionality, whereas insertions into the tracrRNA scaffold portion of the chimeric sgRNA result in loss of function (Supplementary Fig. 2). To recruit VP64 to the sgRNA, we thus appended two copies of the MS2 bacteriophage coat-protein binding RNA stem-loop16 to the 3′ end of the sgRNA (Fig. 1b) and expressed these chimeric sgRNAs together with Cas9N- and a MS2-VP64 fusion protein. We observed robust sequence-specific transcriptional activation from reporter constructs only in the presence of all 3 components (Figs. 1c, 1e).
Having successfully activated reporter construct transcription, we next attempted to regulate endogenous genes. We initially chose to target ZFP42 (REX1) and POU5F1 (OCT4), both tightly regulated genes involved in maintenance of pluripotency. For each gene we designed multiple sgRNAs targeting a ~5kb stretch of DNA upstream of the transcription start site and assayed transcriptional activation using either a promoter-luciferase reporter construct17 or directly via qPCR of the endogenous genes. We observed that introduction of individual sgRNAs modestly stimulated transcription of both target genes, but multiple sgRNAs acted synergistically to stimulate robust multi-fold transcriptional induction (Fig. 1f, Supplementary Figs. 3, 4, and Supplementary Table 1) 18, 19. In these experiments, both the Cas9 and sgRNA tethering approaches were observed to be effective, with the former displaying ~1.5–3 fold higher potency (Fig. 1f, Supplementary Fig. 3). This difference is likely due to the requirement for 2-component as opposed to 3-component complex assembly. However, the sgRNA tethering approach, in principle, enables different effector domains to be recruited by distinct sgRNAs so long as each sgRNA uses a different RNA-protein interaction pair, enabling multiplex gene regulation using the same Cas9N- protein. We noted that a majority of the stimulation in the above experiments was by sgRNAs closer to the transcriptional start site, and thus correspondingly also attempted to regulate two additional genes, SOX2 and NANOG, via sgRNAs targeting within an upstream ~1kb stretch of promoter DNA (Supplementary Fig. 5). And indeed, this choice of sgRNAs proximal to the transcriptional start site resulted in robust gene activation.
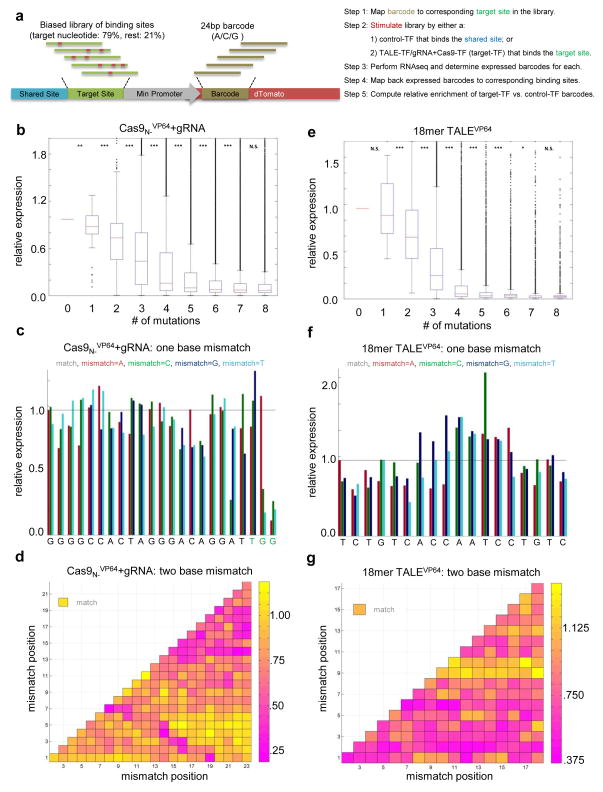
The ability to both edit and regulate genes using the above RNA-guided system opens the door to versatile multiplex genetic and epigenetic regulation of human cells. However, an increasingly recognized constraint on Cas9-mediated engineering is the apparently limited specificity of sgRNA:Cas9 targeting20. Resolution of this issue requires in-depth interrogation of Cas9 affinity for a very large space of target sequence variations. Towards this we adapted our RNA-guided transcriptional activation system to serve this purpose. Our approach provides a direct high-throughput readout of Cas9-targeting in human cells, avoids complications introduced by dsDNA cut toxicity and mutagenic repair incurred by specificity testing with native nuclease-active Cas9 and can be adapted to any programmable DNA binding system. To illustrate this latter point, we also applied this system to evaluate TALE specificity. The methodology of our approach is outlined in Fig. 2a (also see Supplementary Fig. 6): Briefly, we design a construct library in which each element of the library comprises a minimal promoter driving a dTomato fluorescent protein. Downstream of the transcription start site a 24bp (A/C/G) random transcript tag is inserted and two TF binding sites are placed upstream of the promoter: one is a constant DNA sequence shared by all library elements. The second is a variable feature that bears a ‘biased’ library of binding sites which are engineered to span a large collection of sequences that present many combinations of mutations of target sequence that the programmable DNA targeting complex was designed to bind. We achieved this using degenerate oligonucleotides engineered to have nucleotide frequencies at each position such that the target sequence nucleotide appears at a 79% frequency and each other nucleotide occurs at 7% frequency21. The reporter library is then sequenced to reveal the associations between the 24bp dTomato transcript tags and their corresponding ‘biased’ target site in the library element. The large diversity of the transcript tags assures that sharing of tags between different targets will be rare, whereas the biased construction of the target sequences means that sites with few mutations will be associated with more tags than sites with more mutations. Next we stimulate transcription of the dTomato reporter genes with either a control-TF engineered to bind the shared DNA site, or the target-TF that was engineered to bind the target site. As assayed by dTomato fluorescence, protein expression was observed to peak by ~48 hours. To prevent over-stimulation of the library total RNA was harvested within 24 hours. We then measure the abundance of each expressed transcript tag in each sample by conducting RNAseq on the stimulated cells, and then map these back to their corresponding binding sites using the association table established earlier. Note that one would expect the control-TF to excite all library members equally because its binding site is shared across all library elements, whereas the target-TF will skew the distribution of the expressed members to those that are preferentially targeted by it. This assumption is used to compute a final normalized expression level for each binding site by dividing the tag counts obtained for the target-TF by those obtained for the control-TF.
Fig. 2. Evaluating the landscape of targeting by sgRNA:Cas9 complexes and TALEs.
(a) The methodology of our approach is outlined (refer also Supplementary Fig. 6). (b) The targeting landscape of a sgRNA:Cas9 complex reveals that it is potentially tolerant to 1–3 mutations in its target sequences. (c) The sgRNA:Cas9 complex is also largely insensitive to point mutations, except those localized to the PAM sequence. Notably this data reveals that the predicted PAM for the S. pyogenes Cas9 is not just NGG but also NAG. (d) Introduction of 2 base mismatches significantly impairs the sgRNA:Cas9 complex activity, primarily when these are localized to the 8–10 bases nearer the 3′ end of the sgRNA target sequence (in the heat plot the target sequence positions are labeled from 1–23 starting from the 5′ end). (e) Similarly examining the TALE off-targeting data for an 18-mer TALE reveals that it can potentially tolerate 1–2 mutations in its target sequence, and fails to activate a large majority of 3 base mismatch variants in its targets. (f) The 18-mer TALE is, similar to the sgRNA:Cas9 complexes, largely insensitive to single base mismatched in its target. (g) Introduction of 2 base mismatches significantly impairs the 18-mer TALE activity. Notably we observe that TALE activity is more sensitive to mismatches nearer the 5′ end of its target sequence (in the heat plot the target sequence positions are labeled from 1–18 starting from the 5′ end). Statistical significance symbols are: *** for P<.0005/n, ** for P<.005/n, * for P<.05/n, and N.S. (Non-Significant) for P>= .05/n, where n is the number of comparisons (refer Supplementary Table 3).
We used the above approach to first analyze the targeting landscape of multiple sgRNA:Cas9 complexes. Our data reveals that these complexes can potentially tolerate 1–3 mutations in their target sequences (Fig. 2b). They are also largely insensitive to point mutations, except those localized to the PAM sequence (Fig. 2c). Introduction of 2 base mismatches significantly impairs activity, with highest sensitivity localized to the 8–10 bases nearest to the 3′ end of the sgRNA target sequence (Figs. 2d). These results are further reaffirmed by specificity data generated using two different sgRNA:Cas9 complexes (Supplementary Fig. 7 and Supplementary Tables 2, 3). Notably, we found that different sgRNAs can have vastly different specificity profiles (Supplementary Figs. 7a, 7d), specifically, sgRNA2 here tolerates up to 3 mismatches and sgRNA3 only up to 1. Again the greatest sensitivity to mismatches was localized to the 3′ end of the spacer, albeit mismatches at other positions were also observed to affect activity.
We next conducted additional experiments to validate these results. Specifically, we first confirmed the assay is specific for the sgRNA being evaluated, as a corresponding mutant sgRNA is unable to stimulate the reporter library (Supplementary Fig. 8). We also confirmed via targeted experiments that single-base mismatches within 12bp of the 3′ end of the spacer in the assayed sgRNAs indeed still result in detectable targeting, however 2bp mismatches in this region result in significant loss of activity (Supplementary Fig. 9). Furthermore, based on the observed insensitivity to mutations in the 5′ portion of the spacer, we conjectured that this region was not entirely required for sgRNA specificity and thus likely small truncations in this region would still result in retention of sgRNA activity. Supporting this hypothesis we observed that 1–3bp 5′ truncations are indeed well tolerated (Supplementary Fig. 10). Finally, an interesting revelation of the single-base mismatch data from both these experiments was that the predicted PAM for the S. pyogenes Cas9 is not just NGG but also NAG20. We confirmed this result with targeted experiments using the wild-type Cas9 in a nuclease assay (Supplementary Fig. 11).
Taken together, our data demonstrate that the sgRNA:Cas9 system can potentially tolerate multiple mismatches in its target sequence. Consequently, achieving high targeting specificity with current experimental formats will likely require judicious and potentially complicated bioinformatic choice of sgRNAs. Indeed, when we rescanned a previously generated set of ~190K Cas9 targets in human exons that had no alternate NGG targets sharing the last 13nt of the targeting sequence6 for the presence of alternate NAG sites or for NGG sites with a mismatch in the prior 13nt, only .04% were found to have no such alternate targets.
We note that our theoretical calculations suggest that there should be an exponential relationship between the cutting and mutation rates induced by a Cas9 nuclease and the expression level of a gene driven by a Cas9-TF (refer Supplementary Note 2), such that direct tests of specificity using Cas9 nucleases should be more sensitive and also more reflective of consequences of the underlying chromatin context. However, our TF assay offers a significant compensatory advantage in the form of convenient high-throughput multiplexing via RNAseq.
We next applied our transcriptional specificity assay to examine the mutational tolerance of another widely used genome engineering tool, TALE proteins. As a genome editing tool usually TALE-FokI dimers are used, and for genome regulation TALE-VP64 fusions have been shown to be highly effective. We used the latter as it was compatible with our transcriptional activation assay and this format also reveals the specificity profile of individual TALE proteins. Examining the TALE off-targeting data (Figs. 2e, 2f, 2g) reveals that 18-mer TALEs22 can potentially tolerate 1–2 mutations in their target sequences, but fail to activate a large majority of 3 base mismatch variants in their targets. They are also particularly sensitive to mismatches nearer the 5′ end of their target sequences23. Notably, certain mutations in the middle of the target lead to higher TALE activity, an aspect that needs further evaluation. We confirmed a subset of the above results via targeted experiments in a nuclease assay (Supplementary Fig. 12). We also observed that shorter TALEs (14-mer and 10-mer) are progressively less tolerant to mismatches but also reduced in activity by an order of magnitude (Supplementary Fig. 13) 24. To decouple the role of individual repeat-variable diresidues (RVDs), we confirmed that choice of RVDs25 does contribute to base specificity but TALE specificity is also a function of the binding energy of the protein as a whole (Supplementary Fig. 14). While a larger data-set would shed further light into the intricacies of TALE specificity profiles, our data imply that engineering shorter TALEs or TALEs bearing a judicious composition of high and low affinity monomers can potentially yield higher specificity in genome engineering applications and the requirement for FokI dimerization in nuclease applications enables a further dramatic reduction in off-target effects especially when using the shorter TALEs26.
Unlike TALEs where direct control of the size or monomer composition is a ready approach to modulating specificity, there are limited current avenues for engineering the sgRNA:Cas9 complex towards lower binding affinity (and hence higher specificity) for their targets27, 28. We therefore focused on exploiting cooperativity requirements to improve specificity, akin to the use of ZF/TALE fusions to the dimeric FokI endonuclease that creates the requirement for the simultaneous binding of two adjacent ZFs/TALEs. Because synergy between multiple complexes is critical to ensure robust target gene activation by Cas9N-VP64, transcriptional regulation applications of Cas9N- is naturally specific as individual off-target binding events should have minimal effect. Although it should be noted that since individual sgRNA:Cas9 complexes can result in measurable activation (Fig. 1f), potential off-target effects might be magnified when perturbations are highly multiplexed.
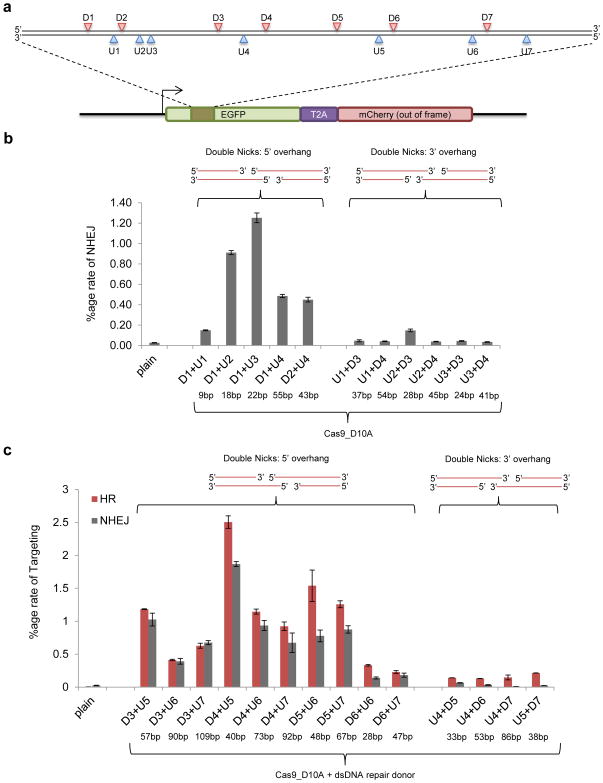
In the context of genome-editing, we chose to focus on creating off-set nicks to generate DSBs. Our motivation stems from the observation (Supplementary Fig. 15) that a large majority of nicks do not result in non-homologous end joining (NHEJ) mediated indels29., and thus when inducing off-set nicks, off-target single nick events will likely result in very low indel rates. Towards this we found that inducing off-set nicks to generate DSBs is highly effective at inducing gene disruption at both integrated reporter loci (Fig. 3) and at the native AAVS1 genomic locus (Supplementary Figs. 16, 17). Furthermore, we also noted that consistent with the standard model for homologous recombination (HR) mediated repair30 engineering of 5′ overhangs via off-set nicks generated more robust NHEJ events than 3′ overhangs (Fig. 3b). In addition to a stimulation of NHEJ, we also observed robust induction of HR when the 5′ overhangs were created. generation of 3′ overhangs did not result in improvement of HR rates (Figs. 3c). It remains to be determined if Cas9 biochemistry or chromatin state and nucleotide composition of the genomic loci also contributed to the observed results above. While we did not actually measure off-target activity of this methodology, we believe the use of cooperativity such as with off-set nicks for generating DSBs offers a promising route for mitigating the effects of off-target sgRNA:Cas9 activity.
Fig. 3. Off-set nicking.
(a) We employed the traffic light reporter29 to simultaneously assay for HR and NHEJ events upon introduction of targeted nicks or breaks: DNA cleavage events resolved through the HDR pathway restore the GFP sequence (via a donor template), whereas mutagenic NHEJ causes frame-shifts rendering the GFP out of frame and the downstream mCherry sequence in frame. For the assay, we designed 14 sgRNAs covering a 200bp stretch of DNA: 7 targeting the sense strand (U1–7) and 7 the antisense strand (D1–7). Using the Cas9D10A mutant, which nicks the complementary strand, we used different two-way combinations of the sgRNAs to induce a range of programmed 5′ or 3′ overhangs (the nicking sites for the 14 sgRNAs are indicated). (b) Inducing off-set nicks to generate DSBs is highly effective at inducing gene disruption. Notably off-set nicks leading to 5′ overhangs result in more NHEJ events as opposed to 3′ overhangs. (c) Again, off-set nicks leading to 5′ overhangs also result in more HR and NHEJ events as opposed to 3′ overhangs. In (b,c) the predicted overhang lengths are indicated below the corresponding x-axis legends. Data are means +/− SEM (N=3).
In summary, we have engineered the sgRNA:Cas9 system to enable RNA-guided genome regulation in human cells by tethering transcriptional activation domains to either a nuclease-null Cas9 or to guide RNAs. We expect the use of additional effector domains such as repressors, dimeric and monomeric nucleases, and epigenetic modulators to further expand this sgRNA:Cas9 toolset. As activation by individual sgRNA:Cas9 complexes was not observed to be strong and needed synergy among multiple complexes for robust transcription, exploring activity of Cas9-activators based on other Cas9 orthologs will be an important avenue for future studies.
Based on these RNA-guided regulators we additionally implemented a transcriptional activation based assay to determine the landscape of off-target binding by sgRNA:Cas9 complexes and compared them to TALE effectors. We observed that the sgRNA:Cas9 system can result in off-targeting events. We noted that there are large differences in specificity between evaluated sgRNAs (Supplementary Fig. 7). Based on this we speculate that sgRNA-DNA binding (and associated thermodynamic parameters) are a prominent determinant of specificity. Thus judicious choice of sgRNAs (such as avoidance of poly-G, poly-C rich spacers, and use of targets >3 mismatches away from the genome) will be a productive route to improved target specificity, albeit rules governing their precise design such as Tm, nucleotide composition, secondary structure of sgRNA spacer versus scaffold and role of the underlying chromatin structure of the target loci remain to be determined. Controlling the dose and duration of Cas9 and sgRNA expression will also be critical for engineering high specificity, and thus RNA based delivery will be an attractive genome editing route1. Although structure-guided design and directed evolution may eventually improve the specificity of individual Cas9 proteins, we have also shown here that engineering a requirement for cooperativity via off-set nicking to generate DSBs can potentially ameliorate off-target activity, and will perhaps be an useful approach for exploring therapeutic applications. Use of small molecule modulators of HR/NHEJ pathways and co-expression of associated end processing enzymes could further help refine this methodology. Overall, the ease and efficacy of editing and regulating genomes using the Cas9 RNA-guided genome engineering approach will have broad implications for our ability to tune and program complex biological systems.
Online Methods
Plasmid construction
The Cas9 mutants were generated using the Quikchange kit (Agilent technologies). The target sgRNA expression constructs were either directly ordered as individual gBlocks from IDT and cloned into the pCR-BluntII-TOPO vector (Invitrogen); or assembled using Gibson assembly of oligonucleotides into a sgRNA-cloning vector (plasmid #41824). Appending of MS2 binding RNA-stem loop domains16 to the 3′ end of sgRNAs was via pcr primers. The vectors for the HR reporter assay involving a broken GFP were constructed by fusion PCR assembly of the GFP sequence bearing the stop codon and appropriate fragment assembled into the EGIP lentivector from Addgene (plasmid #26777) 6. These lentivectors were then used to establish the GFP reporter stable lines. TALENs used in this study were constructed using standard protocols22. Cas9N-6 and MS216 (plasmid #27121) fusions to VP64 and NLS domains were performed using standard pcr fusion protocol procedures. Both C-terminus and N-terminus NLS fusion constructs were made for each. The Cas9m4 nuclease-null mutant and fusions thereof (described in Supplementary Fig. 1) were used for all experiments. The promoter luciferase constructs for OCT4 and REX1 were obtained from Addgene (plasmid #17221 and plasmid #17222). The choice of TALEs and sgRNAs 1, 2 and associated reagents was based on our earlier study targeting the AAVS1 locus6. sgRNA 3 also based on our earlier study targets the DNMT3a locus6. Reporter libraries were constructed as per the design in Fig. 2a, and involved Gibson assembly of PCR fragments generated using degenerate oligonucleotides from IDT. DNA reagents developed in this study will be made available via Addgene (http://www.addgene.org/crispr/church/).
Cell culture and transfections
HEK 293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) high glucose supplemented with 10% fetal bovine serum (FBS, Invitrogen), penicillin/streptomycin (pen/strep, Invitrogen), and non-essential amino acids (NEAA, Invitrogen). Cells were maintained at 37°C and 5% CO2 in a humidified incubator.
Transfections involving nuclease assays were as follows: 0.4×106 cells were transfected with 2μg Cas9 plasmid, 2μg sgRNA and/or 2μg DNA donor plasmid using Lipofectamine 2000 as per the manufacturer’s protocols. Cells were harvested 3 days after transfection and either analyzed by FACS, or for direct assay of genomic cuts the genomic DNA of ~1 × 106 cells was extracted using DNAeasy kit (Qiagen). For these PCR was conducted to amplify the targeting region with genomic DNA derived from the cells and amplicons were deep sequenced by MiSeq Personal Sequencer (Illumina) with coverage >200,000 reads. The sequencing data was analyzed to estimate NHEJ efficiencies.
For transfections involving transcriptional activation assays: 0.4×106 cells were transfected with (1) 2μg Cas9N-VP64 plasmid, 2μg sgRNA and/or 0.25μg of reporter construct; or (2) 2μg Cas9N- plasmid, 2μg MS2-VP64, 2μg sgRNA-2XMS2aptamer and/or 0.25μg of reporter construct. Cells were harvested 24–48hrs post transfection and assayed using FACS or immunofluorescence methods, or their total RNA was extracted and these were subsequently analyzed by RT-PCR. Here standard taqman probes from Invitrogen for REX1, OCT4, SOX2 and NANOG were used, with normalization for each sample performed against GAPDH.
For transfections involving transcriptional activation assays for specificity profile of sgRNA:Cas9 complexes and TALEs: 0.4×106 cells were transfected with (1) 2μg Cas9N-VP64 plasmid, 2μg sgRNA and 0.25μg of reporter library; or (2) 2μg TALE-TF plasmid and 0.25μg of reporter library; or (3) 2μg control-TF plasmid and 0.25μg of reporter library. Cells were harvested 24hrs post transfection (to avoid the stimulation of reporters being in saturation mode). Total RNA extraction was performed using RNAeasy-plus kit (Qiagen), and standard RT-pcr performed using Superscript-III (Invitrogen). Libraries for next-generation sequencing were generated by targeted pcr amplification of the transcript-tags.
Computational and sequence analysis for calculation of Cas9-TF and TALE-TF reporter expression levels
The high-level logic flow for this process is depicted in Supplementary Figure 6a, and additional details are given here. For details on construct library composition, see Supplementary Figures 6a (level 1) and 6b. Statistics are given in Supplementary Table 1.
Sequencing
For Cas9 experiments, construct library (Supplementary Figure 6a, level 3, left) and reporter gene cDNA sequences (Supplementary Figure 6a, level 3, right) were obtained as 150bp overlapping paired end reads on an Illumina MiSeq, whereas for TALE experiments, corresponding sequences were obtained as 51bp non-overlapping paired end reads on an Illumina HiSeq.
Construct library sequence processing
Alignment: For Cas9 experiments, novoalign V2.07.17 (http://www.novocraft.com/main/index.php) was used to align paired reads to a set of 250bp reference sequences that corresponded to 234bp of the constructs flanked by the pairs of 8bp library barcodes (see Supplementary Figure 6a, 3rd level, left). In the reference sequences supplied to novoalign, the 23bp degenerate Cas9 binding site regions and the 24bp degenerate transcript tag regions (see Supplementary Figure 6a, first level) were specified as Ns, wheras the construct library barcodes were explicitly provided. For TALE experiments, the same procedures were used except that the reference sequences were 203bp in length and the degenerate binding site regions were 18bp vs. 23bp in length. Validity checking: Novoalign output for comprised files in which left and right reads for each read pair were individually aligned to the reference sequences. Only read pairs that were both uniquely aligned to the reference sequence were subjected to additional validity conditions, and only read pairs that passed all of these conditions were retained. The validity conditions included: (i) Each of the two construct library barcodes must align in at least 4 positions to a reference sequence barcode, and the two barcodes must to the barcode pair for the same construct library. (ii) All bases aligning to the N regions of the reference sequence must be called by novoalign as As, Cs, Gs or Ts. Note that for neither Cas9 nor TALE experiments did left and right reads overlap in a reference N region, so that the possibility of ambiguous novoalign calls of these N bases did not arise. (iii) Likewise, no novoalign-called inserts or deletions must appear in these regions. (iv) No Ts must appear in the transcript tag region (as these random sequences were generated from As, Cs, and Gs only). Read pairs for which any one of these conditions were violated were collected in a rejected read pair file. These validity checks were implemented using custom perl scripts.
Induced sample reporter gene cDNA sequence processing
Alignment: SeqPrep (downloaded from https://github.com/jstjohn/SeqPrep on June 18, 2012) was first used to merge the overlapping read pairs to the 79bp common segment, after which novoalign (version above) was used to align these 79bp common segments as unpaired single reads to a set of reference sequences (see Supplementary Figure 6a, 3rd level, right) in which (as for the construct library sequencing) the 24bp degenerate transcript tag was specified as Ns whereas the sample barcodes were explicitly provided. Both TALE and Cas9 cDNA sequence regions corresponded to the same 63bp regions of cDNA flanked by pairs of 8bp sample barcode sequences. Validity checking: The same conditions were applied as for construct library sequencing (see above) except that: (a) Here, due prior SeqPrep merging of read pairs, validity processing did not have to filter for unique alignments of both reads in a read pair but only for unique alignments of the merged reads. (b) Only transcript tags appeared in the cDNA sequence reads, so that validity processing only applied these tag regions of the reference sequences and not also to a separate binding site region.
Assembly of table of binding sites vs. transcript tag associations
Custom perl was used to generate these tables from the validated construct library sequences (Supplementary Figure 6a, 4th level, left). Although the 24bp tag sequences composed of A, C, and G bases should be essentially unique across a construct library (probability of sharing = ~2.8e-11), early analysis of binding site vs. tag associations revealed that a non-negligible fraction of tag sequences were in fact shared by multiple binding sequences, likely mainly caused by a combination of sequence errors in the binding sequences, or oligo synthesis errors in the oligos used to generate the construct libraries. In addition to tag sharing, tags found associated with binding sites in validated read pairs might also be found in the construct library read pair reject file if it was not clear, due to barcode mismatches, which construct library they might be from. Finally, the tag sequences themselves might contain sequence errors. To deal with these sources of error, tags were categorized with three attributes: (i) safe vs. unsafe, where unsafe meant the tag could be found in the construct library rejected read pair file; shared vs. nonshared, where shared meant the tag was found associated with multiple binding site sequences, and 2+ vs. 1-only, where 2+ meant that the tag appeared at least twice among the validated construct library sequences and so presumed to be less likely to contain sequence errors. Combining these three criteria yielded 8 classes of tags associated with each binding site, the most secure (but least abundant) class comprising only safe, nonshared, 2+ tags; and the least secure (but most abundant) class comprising all tags regardless of safety, sharing, or number of occurrences.
Computation of normalized expression levels
Custom perl code was used to implement the steps indicated in Supplementary Figure 6a, levels 5–6. First, tag counts obtained for each induced sample were aggregated for each binding site, using the binding site vs. transcript tag table previously computed for the construct library (see Supplementary Figure 6c). For each sample, the aggregated tag counts for each binding site were then divided by the aggregated tag counts for the positive control sample to generate normalized expression levels. Additional considerations relevant to these calculations included:
For each sample, a subset of “novel” tags were found among the validity-checked cDNA gene sequences that could not be found in the binding site vs. transcript tag association table. These tags were ignored in the subsequent calculations.
The aggregations of tag counts described above were performed for each of the eight classes of tags described above in binding site vs. transcript tag association table. Because the binding sites in the construct libraries were biased to generate sequences similar to a central sequence frequently, but sequences with increasing numbers of mismatches increasingly rarely, binding sites with few mismatches generally aggregated to large numbers of tags, wheras binding sites with more mismatches aggregated to smaller numbers. Thus, although use of the most secure tag class was generally desirable, evaluation of binding sites with two or more mismatches might be based on small numbers of tags per binding site, making the secure counts and ratios less statistically reliable even if the tags themselves were more reliable. Some compensation for this consideration obtains from the fact that the number of separate aggregated tag counts for n mismatching positions grew with the number of combinations of mismatching positions (equal to ), and so dramatically increases with n; thus the averages of aggregated tag counts for different numbers n of mismatches (shown in Figs. 2b, 2e, and in Supplementary Figs. 7, 8, 13, 14) are based on a statistically very large set of aggregated tag counts for n ≥ 2. We note that for consistency in this study, however, all tags were used for all data sets.
Finally, the binding site built into the TALE construct libraries was 18bp and tag associations were assigned based on these 18bp sequences, but some experiments were conducted with TALEs programmed to bind central 14bp or 10bp regions within the 18bp construct binding site regions. In computing expression levels for these TALEs, tags were aggregated to binding sites based on the corresponding regions of the 18bp binding sites in the association table, so that binding site mismatches outside of this region were ignored.
Expression level boxplot P-values
For the expression level boxplots in Fig. 2, and Supplementary Figs. 7, 8, 13, P-values were computed comparing the mean expression levels between consecutive numbers of target sequence mismatches, so that for a boxplot showing expression level values associated with the 9 mismatch values 0, 1, 2, …, 8, there are 8 comparisons (0 vs. 1, 1 vs. 2, 2 vs. 3, …, 7 vs. 8). Because the 0 mismatch expression level data comprises a single value, the t-test performed for the 0 vs. 1 comparison is a single sample t-test comparing this single value against the distribution of 1 mismatch expression levels. For all other comparisons, two-sample, two-tailed, t-tests were performed assuming unequal variance. All P-values were calculated using MatLab (MathWorks, Waltham) version 2013b. Significance is portrayed using symbols * for P<.05, ** for P<.005, and *** for P<.0005, where all P values are Bonferroni-corrected for the number of comparisons presented in the boxplot. N.S. = Not Significant (P>=.05) (see Supplementary Table 3a).
Statistical characterization of seed region
The normalized expression data for Cas9N VP64+sgRNA for target sequences with two mutations (see Fig. 2d) was analyzed to identify the seed region at the 3′ end of the 20bp target region (excluding the PAM sequence in positions 21–23) by considering a range of candidate seed start positions. For each candidate start position, the normalized expression levels for position pairs, both of which were at or beyond the candidate start position, were accumulated in one set, and the expression values for position pairs, at least one of which was ahead of the candidate start position were accumulated in another set. The P-value of the separation of the central values of these two sets of normalized expression levels was then computed using the Wilcoxon rank sum test as calculated by the MatLab ranksum function. The start position associated with the lowest P-value in the range of positions tested was interpreted as the beginning of the seed region. An adjacent start position had virtually the same P-value as the minimum (see Supplementary Table 3b).
Supplementary Material
Acknowledgments
P.M. thanks Reza Kalhor for insightful discussions. This work was supported by NIH grant P50 HG005550 and Department of Energy grant DE-FG02-02ER63445.
Footnotes
Accession codes
All raw reads can be accessed at NCBI BioProject, accession number SRP028177. Files are described in Supplementary Table 2.
Author Contributions
P.M., J.A., G.M.C. conceived the study. P.M. designed and performed experiments. J.A. designed and performed bioinformatic analyses. M.M. performed experiments. P.B.S., K.M.E., S.K., L.Y. developed reagents and performed analyses. P.M. and J.A. wrote the manuscript with support from all authors.
References
- 1.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nature biotechnology. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 2.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dicarlo JE, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic acids research. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nature biotechnology. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jinek M, et al. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boch J, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 9.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 10.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host actor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2579–2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sapranauskas R, et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic acids research. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annual review of genetics. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 15.Zhang F, et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nature biotechnology. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fusco D, et al. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Current biology: CB. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 18.Maeder ML, et al. Robust, synergistic regulation of human gene expression using TALE activators. Nature methods. 2013;10:243–245. doi: 10.1038/nmeth.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez-Pinera P, et al. Synergistic and tunable human gene activation by combinations of synthetic transcription factors. Nature methods. 2013;10:239–242. doi: 10.1038/nmeth.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nature biotechnology. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patwardhan RP, et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nature biotechnology. 2012;30:265–270. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanjana NE, et al. A transcription activator-like effector toolbox for genome engineering. Nature protocols. 2012;7:171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meckler JF, et al. Quantitative analysis of TALE-DNA interactions suggests polarity effects. Nucleic acids research. 2013;41:4118–4128. doi: 10.1093/nar/gkt085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reyon D, et al. FLASH assembly of TALENs for high-throughput genome editing. Nature biotechnology. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Streubel J, Blucher C, Landgraf A, Boch J. TAL effector RVD specificities and efficiencies. Nature biotechnology. 2012;30:593–595. doi: 10.1038/nbt.2304. [DOI] [PubMed] [Google Scholar]
- 26.Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nature biotechnology. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 27.Pattanayak V, Ramirez CL, Joung JK, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nature methods. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gabriel R, et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nature biotechnology. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- 29.Certo MT, et al. Tracking genome engineering outcome at individual DNA breakpoints. Nature methods. 2011;8:671–676. doi: 10.1038/nmeth.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annual review of genetics. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.