Abstract
Bypassing estrogen receptor (ER) signaling during development of endocrine resistance remains the most common cause of disease progression and mortality in breast cancer patients. To date, the majority of molecular research on ER action in breast cancer has occurred in cell line models derived from late stage disease. Here we describe patient-derived ER + luminal breast tumor models for the study of intratumoral hormone and receptor action. Human breast tumor samples obtained from patients post surgery were immediately transplanted into NOD/SCID or NOD/SCID/ILIIrg−/− mice under estrogen supplementation. Five transplantable patient-derived ER + breast cancer xenografts were established, derived from both primary and metastatic cases. These were assessed for estrogen dependency, steroid receptor expression, cancer stem cell content, and endocrine therapy response. Gene expression patterns were determined in select tumors ±estrogen and ±endocrine therapy. Xenografts morphologically resembled the patient tumors of origin, and expressed similar levels of ER (5–99 %), and progesterone and androgen receptors, over multiple passages. Four of the tumor xenografts were estrogen dependent, and tamoxifen or estrogen withdrawal (EWD) treatment abrogated estrogen-dependent growth and/or tumor morphology. Analysis of the ER transcriptome in select tumors revealed notable differences in ER mechanism of action, and downstream activated signaling networks, in addition to identifying a small set of common estrogen-regulated genes. Treatment of a na¨ıve tumor with tamoxifen or EWD showed similar phenotypic responses, but relatively few similarities in estrogen-dependent transcription, and affected signaling pathways. Several core estrogen centric genes were shared with traditional cell line models. However, novel tumor-specific estrogen-regulated potential target genes, such as cancer/testis antigen 45, were uncovered. These results evoke the importance of mapping both conserved and tumor-unique ER programs in breast cancers. Furthermore, they underscore the importance of primary xenografts for improved understanding of ER+ breast cancer heterogeneity and development of personalized therapies.
Keywords: Breast cancer, Estrogen receptors, Progesterone receptors, Xenografts, Tamoxifen, Gene regulation, Cancer/testis antigens
Introduction
Steroid hormone receptors play important prognostic and biological roles in breast cancer. Despite many advances in our understanding breast malignancies during the last several decades, estrogen receptor (ER) still remains the single most important prognostic and predictive factor determining treatment outcomes. Thus, endocrine therapies targeting ER and estrogen production remain the cornerstone of breast cancer treatment for most patients [1]. Receptors for progesterone (PR) and androgens (AR) also play significant roles in breast cancer biology. PR, an estrogen-regulated gene, is often co-expressed with ER, independently predicts outcome in breast cancer [2], and has been implicated in regulating stem-like cancer cells [reviewed in 3]. AR is expressed in ~70–80 % of breast cancers [4, 5], is generally considered to be growth inhibitory in ER+ tumors [6], and potentially mitogenic in some ER− tumors [7]. In a recent breast cancer classification study, six subgroups with distinct clinical features could be distinguished by gene clusters centered on ER, AR, and cell cycle regulators [8]. Thus, dissecting the roles of steroid receptors, especially ER, in breast cancer biology remains essential for understanding and overcoming treatment resistance.
Breast cancers are divided into three main entities that inform treatment decisions: ER+ PR(+/−), HER2 amplified, and triple negative (TN). Molecular subtyping based on gene expression signatures has further divided breast tumors into multiple groups with distinct outcomes in anticipation of developing new treatment targets and strategies [9]. Luminal breast cancers are defined by ER positivity but otherwise represent a diverse group of tumors in regard to their origin (ductal or lobular), histological features, and expression of nuclear (PR or AR) and cell surface receptors (HER2). Although luminal tumors have an overall better prognosis compared to ER–/HER2 amplified and TN tumors, up to 40 % eventually escape endocrine therapies, recur in a more untreatable state, often many years later, and still account for the majority of overall breast cancer mortality [10]. ER expression in luminal tumors can range from 1 to 99 % positive cells; more highly ER+ tumors are generally slower growing, low-mid grade, and have a more “pure” luminal phenotype. Lower ER scores usually correlate with higher proliferation indices, higher grade, and sometimes “mixed” luminal/basal features [11]. Nonetheless, even patients with 1 % of strongly ER+ tumor cells clinically benefit from endocrine therapies [12]. Thus, the determinants that define responsiveness lie beyond ER levels and perhaps with the ER transcriptional program defining each tumor.
The majority of refractory luminal tumors retain ER, but transition from estrogen dependency to utilize alternate signaling pathways for growth and survival, including PI3K/AKT/mTOR, EGFR/HER2, AR, and cell cycle regulators such as Cyclin D1 [13–15]. To date, targeting alternative molecules in resistant tumors has improved outcomes but has failed to replicate the clinical success of targeting HER2. Thus, there remains a pressing and unmet clinical need for understanding the process of inherent and acquired endocrine resistance. A second but not mutually exclusive theory of endocrine resistance posits that breast tumors contain populations of cancer stem cells that are less susceptible to eradication with conventional therapies [16]. Most ER+ luminal tumors, however, contain low (<1 %) levels of cancer stem cells as defined by the two predominant signatures CD44+CD24(−/low) and aldehyde dehydrogenase (ALDH)+ [17–19]. Many luminal tumors contain cell subpopulations that express cytokeratin 5 (CK5), a marker of stem and progenitor cells in the normal breast [20, 21]. These luminal tumor CK56+ cells are ER− PR− and have increased resistance to endocrine and chemotherapies [22]. The near absence of currently defined cancer stem cells in many advanced luminal tumors suggests the existence of as yet undefined luminal-specific cancer stem cells, or that tumor progression is undertaken stochastically by the bulk ER+ cells themselves.
The molecular mechanisms of ER action have historically been defined in a cohort of ER+ breast cancer cell lines, predominantly ER-rich MCF7 cells, derived from late stage metastatic disease. These include, among many things, the global ER transcriptome and cistrome [23–25], and anti-estrogen actions in ER+ solid tumor xenografts [26, 27]. Fresh tumor samples offer insight into mechanisms of endocrine resistance including changes in gene expression pre- and post-neoadjuvant endocrine therapy in responsive and non-responsive tumors [28, 29], and alterations in the ER cistrome in drug resistant ER+ breast cancers [30]. Individual patient centric models are now needed to address heterogeneity of response and the development of more personalized therapies. Transplantable in vivo models of ER+ breast cancers have been historically difficult to develop, and only a few successes have been described [31, 32]. Loss of ER expression, low take rate, and preferential engraftment of high grade, rapidly proliferating tumors have contributed to this difficulty. A recent study describes highly improved establishment of breast cancer grafts, both TN and ER+, with phenotypic similarities to the original samples [33]. Overall, luminal tumors remain underrepresented and engraftment still tends to favor the faster growing TN breast cancers. Herein we describe the development and characterization of five transplantable luminal ER+ human breast cancers, derived from both primary untreated tumors and late stage metastases. These contain various levels of ER (5–99 %), PR, and AR, and continue to express these steroid receptors for multiple passages at similar levels to the original tumors. The in vivo estrogen dependency of these tumors is marked and without change over passages. Profiling of select ER+ tumor grafts with or without estrogen or endocrine treatments disclosed signaling networks underlying each individual tumor, as well as a common base ER transcriptome that is also retained in historic ER+ breast cancer cell line models. These studies highlight the importance of modeling widely diverse patient-derived ER+ breast cancers in vivo to advance our understanding and treatment of this disease.
Materials and methods
Tumor transplantation and propagation
Initial transplantation
Tumors or metastases were collected at surgical resection at University of Colorado Hospital (Aurora, CO). These studies were performed with institutional review board approval and informed consent of all patients. A trained pathologist identified normal versus tumor tissue from the surgical specimens and placed fresh procured tumor tissue into sterile vials on ice for transport. Female NOD/SCID or NOD/SCID/ILIIrg−/− mice age 4–7 weeks were purchased from Jackson Laboratories. All animal studies were performed under an IACUC approved protocol. For primary tumors, 5–10 mm3 pieces were dipped in Matrigel (BD Biosciences) and inserted into the #4 mammary fat pads of anesthetized recipient mice using a 10 gauge trochar. For pleural effusions or ascites, cells were collected by centrifugation, incubated for 2–3 min on ice with hemolysis buffer (BD Biosciences), collected by centrifugation, suspended in Matrigel, and injected into the #4 mammary fat pads with 27-gauge insulin syringes. A single silastic pellet containing 17β-estradiol was implanted into animals receiving ER+ tumor specimens, as determined by clinical pathology. Tumors were palpated weekly for assessment of tumor take and thereafter measured weekly, where possible, with a digital caliper. Tumor volume was estimated by the formula lw 2/2.
Tumor propagation
When tumors reached >1 cm, or after 5–6 months, these were removed from euthanized animals, partitioned into 10-mm3 pieces, and placed into new recipient animals under various hormone conditions. This was facilitated by insertion of single silastic pellets containing placebo, 17β-estra-diol, progesterone, medroxyprogesterone acetate (MPA), or combinations (all from Sigma), at the time of surgery sub-cutaneously in the upper lumbar region. Pellet preparation was as described previously [34]. For tamoxifen treatment, animals were given 1-mg tamoxifen dissolved in peanut oil IP 3× weekly for 3 weeks, or vehicle only. For estrogen withdrawal (EWD), estrogen pellets were surgically removed from animals for the last 3 weeks of the experiment. Tumor volumes were measured as described above.
Immunohistochemistry and microscopy
Immunohistochemistry (IHC) was as described previously [35]. Antibodies used were as follows: AR (AR441, 1:500, DAKO), CK5 (XM26, 1:100, Leica), ER (SP1, 1:100, Thermo-Fisher), HER2/ErbB2 (29D8, 1:2,000, Cell Signaling), Ki67 (MIB-1, 1:1,000, DAKO), and PR (1294, 1:500, DAKO). Images were captured using an Olympus BH2 microscope equipped with a Microfire camera and Picture-frame software. Adobe Photoshop CS5 was used to perform minimal linear adjustments to brightness/contrast and to assemble pictures into multipanel figures. All IHC staining was evaluated and scored by a pathologist (J.W.). For ER, PR, and AR, these were calculated as intensity and percent according to clinical standards. HER2 and Ki67 scores were calculated via Vias Imaging system software (Ventana).
Gene expression profiling
Freshly cut pieces (50–100 mg) of each tumor were placed into 0.5 mL Qiazol (Qiagen) immediately on necropsy and stored at −80 °C. RNA was prepared using Fast-Prep Lysing Matrix A (MP Biomedicals) followed by miRNeasy (Qiagen) isolation. RNA concentration was measured using a Nanodrop 2000 (Thermo Scientific), and integrity analyzed using an Agilent 2100 Bioanalyzer, and the RNA 6000 Nano kit. Expression profiling was performed using the Gene Atlas System (Fisher). Briefly, amplified and biotinylated sense strand DNA targets were hybridized to Affymetrix Human Gene 1.1 ST Array Strips representing 28,875 genes. Microarray data were analyzed using Partek Genomics Suite® v6.5 and Ingenuity Pathways Analysis®. Array data have been deposited in the Gene Expression Omnibus (GEO) database under accession GSE36075. Additional details on microarray analyses, including comparison to MCF7 datasets GSE848 and GSE3834 and molecular subtype analysis, are described in supplementary methods.
Flow cytometry
Processing of tumors into a single-cell suspension and flow cytometry was performed as described previously [36]. For these studies, cells were incubated for 30 min with antibodies CD44-Alexafluor647 (Serotec), CD24-Alexafluor488 (Biolegend), and Pacific Blue conjugated anti-mouse CD45, CD31, and H-2Kd (Biolegend). Dead cells (propidium iodide (PI)+) and mouse host cells (Pacific Blue+) were excluded from the analysis.
Quantitative PCR (qPCR)
Total RNA was isolated from frozen pieces of PE4 tumors (placebo, estrogen, or estrogen plus MPA treated) or breast cancer cell lines BT474, MCF7, T47D, and ZR75 using the Trizol protocol (Invitrogen). cDNA was prepared using the Versa cDNA kit (Fisher Scientific) following manufacturer protocols. qPCR was performed using primers specific to total CT45 (5′:CTCTGCCATGTCCAAAGCAA, 3′:AAG TCATCAATCTGAGAATCCAATTG), PR (5′:TCGAGCT CACAGCGTTTCTA, 3′:CCCGGGACTGGATAAATGT), and β-actin (5′:GATCATTGCTCCTCCTGAGC, 3′:ACT-CCTGCTTGCTGATCCAC) (Fisher Scientific) using AbsoluteBlue SYBR Green with Fluorescein Mix (Fisher Scientific), 70 nM primers, and 25 ng of the cDNA in 96-well plates. Standard curves were generated from 2 to 50 ng of standard cDNA (mixed known cDNAs) run alongside the samples on a Bio-Rad iCycler with MyIQ Single Color Real-Time PCR Detection System (Bio-Rad). Values were calculated by subtracting the experimental Ct scores from the untreated Ct scores and controlling for the efficiency of the reaction, then dividing by the β-actin scores.
Results
Retention of hormone receptor expression in transplantable patient-derived luminal breast tumor xenografts
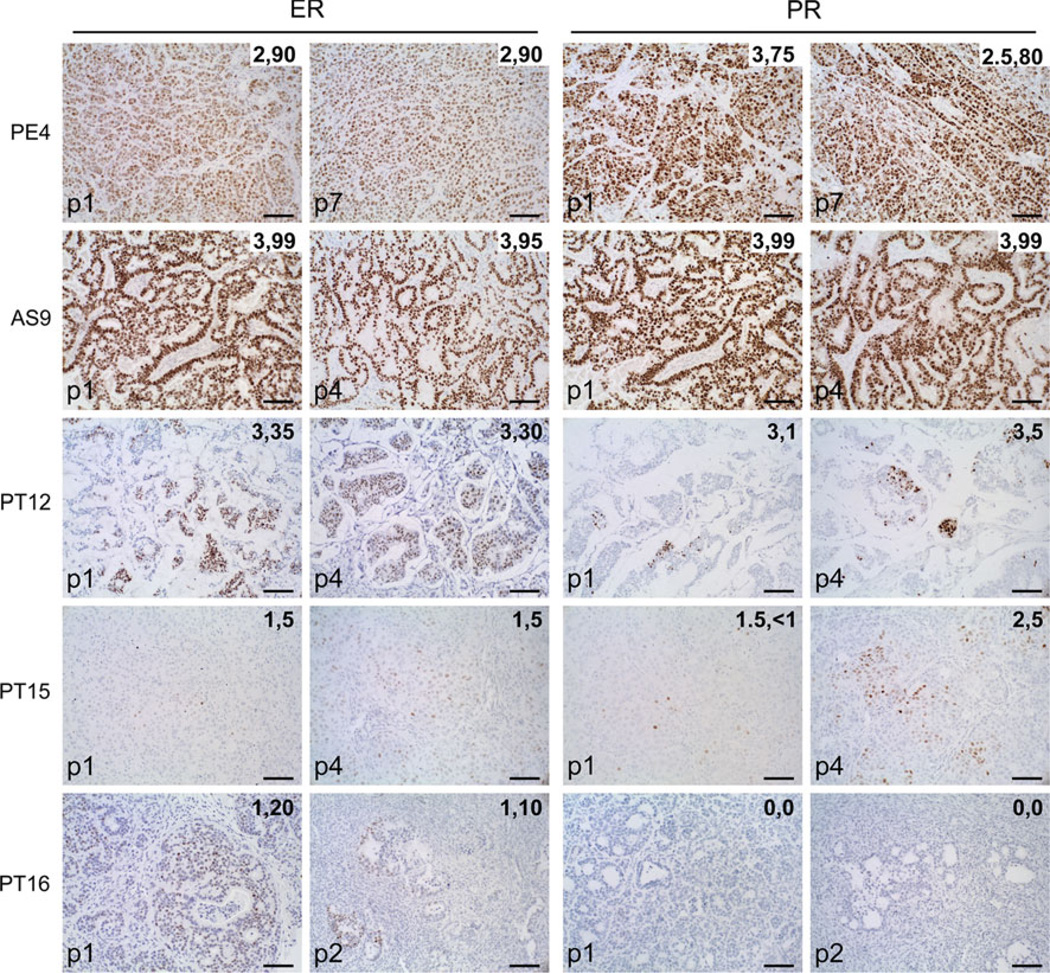
Twenty-four samples were implanted into either NOD/ SCID or NOD/SCID/ILIIrg−/− mice as described. A total of 8/18 ER+ (four metastatic and four primary), and 2/6 TN (both primary) tumor samples were successfully grafted and propagated beyond passage 1. The median latency to achieve transplantable tumors (>300 mm3 volume) was 119 days (range 73–228 days). For the purposes of this study, we characterized five ER+ and one TN tumor sample. Table 1 summarizes the characteristics of the six breast cancers and their corresponding xenografts. A hemotoxylin and eosin (H&E) stain of each xenograft is supplied in Supplementary Fig. 1. Tumors were named according to their origin as pleural effusion (PE), ascites (AS), or primary tumor (PT). Overall, these morphologically resembled the patient tumor of origin, including grade, ER and PR expression levels, HER2 status, and histological features such as extracellular matrix (mucin) secretion (Table 1; Supplementary Fig. 2). To determine if hormone receptors would be retained at similar levels over multiple passages, sections of each tumor at passage 1 and passages 2–7 were stained by IHC for ER and PR (Fig. 1). Serially transplanted tumors retained consistent levels and intensities of ER and PR expression, with some remaining >90 % ER+ PR+ at passage 7 (PE4). TN PT18 remained ER− PR− for four passages (not shown). HER2 scores for xenografts (Table 1; Supplementary Fig. 3) matched the clinical diagnosis. AS9 was HER2 amplified (3+) similar to the cancer of origin and remained 3+ consistently through passage 3 (Supplementary Fig. 3). AR expression levels were measured by IHC in all six xenograft tumor lines (Supplementary Fig. 4). Two tumors were 30 % (PT15) and 80 % (PE4) AR positive, three were AR negative/low (<1 %) (PT12, PT16, PT18), and one had diffuse cytoplasmic AR staining (AS9). Overall ER+ luminal tumor xenografts resembled the patient tumor of origin, especially with regard to hormone receptor status, and serve as a faithful representation of the range of ER levels observed clinically, from low ER (5 %) to highly ER+ PR+ (>90 %) tumors.
Table 1.
Characteristics and hormone receptor status of patient breast tumors and their corresponding xenografts
| Patient tumor |
Xenografta |
|||||||
|---|---|---|---|---|---|---|---|---|
| Sample | Origin | Clinical description | Description | ER (intensity, %) |
PR (intensity, %) |
AR (%) |
HER2 intensity |
CK5 (%) |
| PE4 | Pleural effusion | Invasive mammary carcinoma ER+ PR+ HER2− | Poorly differentiated with necrosis | 2, 90 | 3, 75 | 80 | 1+ | <0.1 |
| AS9 | Ascites | Invasive mammary carcinoma ER+ PR+ HER2+ | Moderately differentiated with necrosis | 3, 99 | 3, 99 | 5 | 3+ | 0 |
| PT12 | Primary tumor | IDCb with mucinous features, grade 3, ER+ (93 %), PR+ (15 %), HER2− | IDC with mucinous features | 3, 35 | 3, 1 | 0 | 1+ | <0.1 |
| PT15 | Primary tumor | IDC grade 3, ER+ (8 %), PR (0 %), HER2− | IDC, poorly differentiated | 1, 5 | 1.5,<1 | 30 | 1+ | 30 |
| PT16 | Primary tumor | IDC grade 3, ER (74 %), PR (0 %), HER2− (FISH ratio 1.46) | IDC, moderately differentiated | 1, 20 | 1, <1 | 1 | 2+ | 0 |
| PT18 | Primary tumor | Metaplastic carcinoma, grade 3, ER− PR− HER2− | Metaplastic carcinoma, poorly differentiated | 0 | 0 | 0 | 1+ | 30 |
At first passage, estrogen-treated except for PT18
Invasive ductal carcinoma
Fig 1.
Retention of ER and PR expression in patient-derived luminal breast cancer xenografts over multiple passages. Patient-derived xenografts were established in NOD/SCID or NOD/SCID/ILIIrg−/− mice, and subsequently propagated via direct transplantation of solid tumor pieces into new recipient mice. Tumors were grown under continuous estrogen supplementation. Sections of five xenograft tumor lines were stained by IHC for ER (left panels) and PR (right panels) at passage 1, and at subsequent passages 2–7 as indicated. The intensity (1–3) and percent of ER+ and PR+ cells are indicated on each panel. Scale bars, 100 µM
To determine the intrinsic molecular subtype of the patient-derived xenografts, RNA from first passage tumors (estrogen treated) was profiled on custom Agilent gene expression microarrays. Each tumor was intrinsically subtyped with PAM50 and claudin-low predictors to identify luminal A, luminal B, HER2-enriched, basal-like, and claudin-low tumors, according to previously described methods [37, 38]. Four of the tumors were classified as luminal B (PE4, AS9, PT12, and PT16), one was HER2-enriched (PT15), and the TN tumor was basal-like (PT18) (Supplementary Fig. 5). Although PT15 did not overexpress HER2 protein (Table 1; Supplementary Fig. 3), it did express other HER2-associated genes.
Estrogen dependency of ER+ luminal tumor xenografts
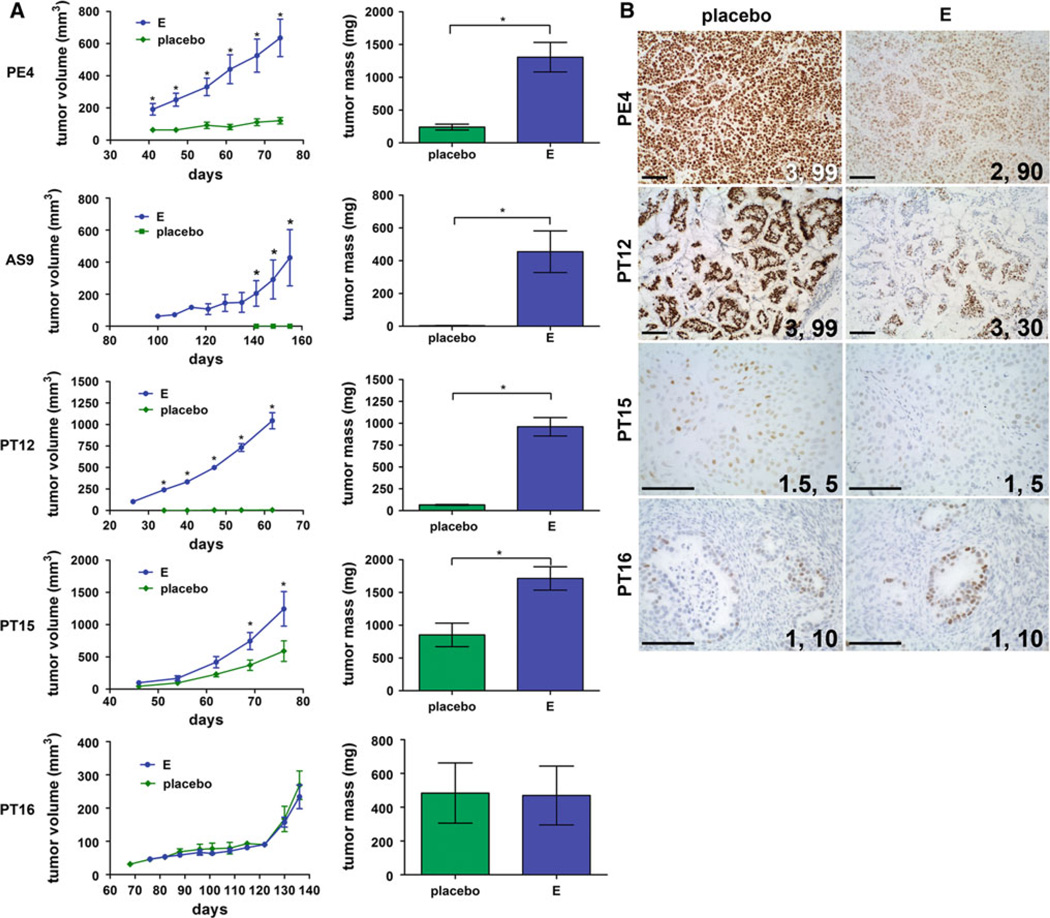
To determine hormone responsiveness, tumors were resected from euthanized animals, partitioned, and placed into new recipient animals supplemented with pellets containing placebo or estrogen. Tumor growth was measured over time and is plotted in Fig. 2a. Four/five tumors depended on continuous levels of estrogen for tumor growth, with tumors growing ~2–20-fold larger in estrogen compared with placebo supplemented animals. In the case of AS9, no residual tumors could be found in the mammary fat pads of placebo animals. PT16 was the exception and grew equally well in placebo and estrogen supplemented animals. This tumor originated from a male patient and thus may be accustomed to growing under lower estrogen conditions. Note that placebo-treated animals had intact ovaries and low circulating estrogens [34] but that this was not sufficient for significant tumor growth in most ER+ samples, that seemingly require continuously sustained high estrogen levels. TN PT18 tumor xenografts grew robustly without estrogen supplementation (>1 g tumors by 70 days, not shown). In all cases except for PT16 (p2), tumors were grown with/without estrogen in subsequent passages with similar results. The proliferation rates of early passages of each tumor were compared by immunostaining for Ki67 followed by computerized analysis (Supplementary Table 1). Average Ki67 indices for luminal ER+ tumors ranged from 36 to 50. TN PT18 had a higher Ki67 score of 68, comparable to the original tumor score of 65, suggesting the relative rate of tumor cell proliferation is preserved in these xenograft models.
Fig 2.
Growth kinetics of ER+ breast cancer xenografts in the absence or presence of estrogens. Tumors were transplanted into recipient mice either in the absence of exogenously added hormones (placebo), or in the presence of estrogen (17β-estradiol, E). a Tumor volumes were measured weekly (once attainable) and plotted versus the number of days of incubation ±SEM. Final tumor mass at necropsy ±SEM was plotted for each hormone treatment. Sample numbers (tumors): PE4 placebo (6), E (8); AS9 placebo (2), E (5); PT12 placebo (6), E (8); PT15, placebo (4), E (10); PT16 placebo (2), E (4). Tumor volumes at each time point, and final tumor masses, were compared using Student’ s t test. For tumor volume, *p < 0.05 E compared to placebo. For final tumor mass, *p < 0.05. b Sections from tumors grown in mice containing placebo versus estrogen (E) pellets were stained by IHC for ER. The staining intensity and percent of ER positive cells are indicated on each panel. Scale bars, 100 µM
Because circulating ligand is known to affect receptor turnover, or “downregulation” [39], we assayed ER expression in tumors without (placebo) or with continuous estrogen supplementation (Fig. 2b). ER expression was markedly dependent on the hormonal background of the host animal. In PE4, PT12, and PT15, ER staining (percentages and/or intensities) was higher in placebo compared with estrogen tumors. PT12 transitioned from 30 to 99 % positive cells in estrogen versus placebo animals. Even PT15 with low ER (1, 5 %) also had a small but consistent and measurable increase in ER (1.5, 8 %) in placebo-treated animals. PT16 was the exception and had no difference in ER levels without/with estrogens, correlating to the observed similarity in tumor growth between the two conditions (Fig. 2a). There was insignificant material in placebo-treated AS9 animals for analysis, although ER was already at a maximum (3, 99 %) in estrogen-treated tumors (Fig. 1). These data suggest that, under continuous estrogens necessary for tumor propagation, ER is often actively downregulated, and thus the true “resting” ER scores may often be underestimated in these models.
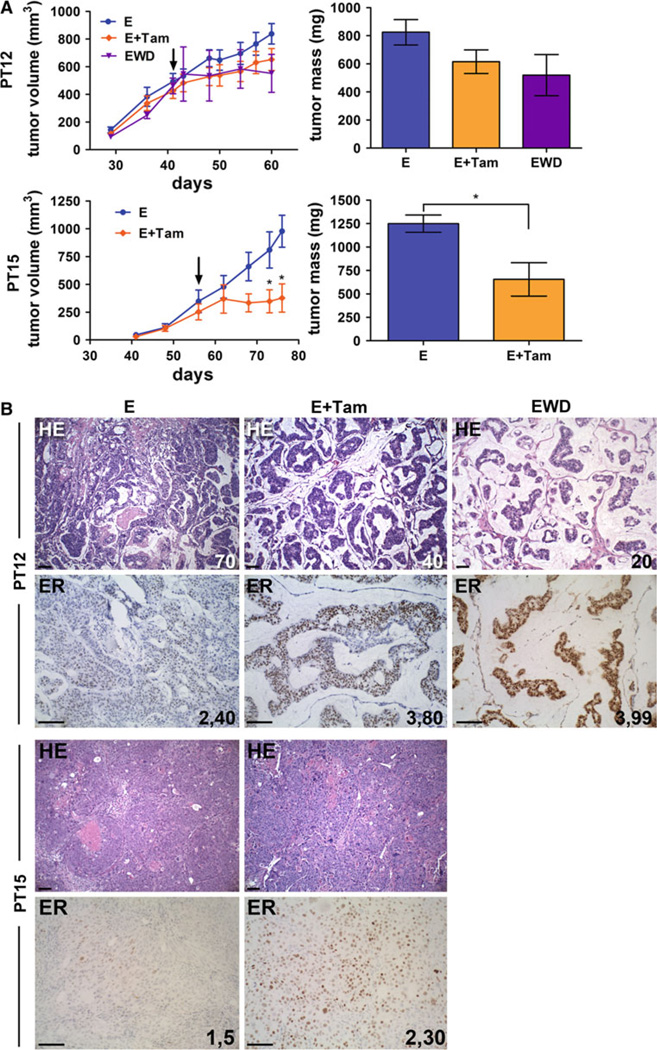
Endocrine therapy of drug untested ER+ tumors
To determine if primary tumor xenografts obtained from previously untreated patients would respond to ER-targeted therapies, naive tumors were treated with tamoxifen (PT12 and PT15) or EWD (PT12) (Fig. 3). For these experiments, tumors were established with estrogen pellets until they reached 300-mm3 volume, and then randomized into control, tamoxifen (estrogen pellet retained), and EWD (estrogen pellet removed) groups (PT12 only) for an additional 3 weeks. EWD is surrogate for aromatase inhibitor (AI) therapy, depleting high circulating estrogens. PT12 tumor volume and final tumor mass did not change significantly following tamoxifen treatment or EWD, although these trended toward a decrease, whereas PT15 tumor volume and mass significantly decreased in tamoxifen compared with estrogen animals (Fig. 3a). The cellularity of PT12 decreased from 70 % in estrogen-treated tumors to 40 and 20 % in tamoxifen or EWD tumors, respectively (Fig. 3b), indicating a positive response to treatment not measurable by size alone. ER staining increased in tamoxifen (80 %) and EWD (99 %) PT12 tumors compared with estrogen alone (40 %), and in tamoxifen PT15 (30 %) compared with estrogen alone (5 %) (Fig. 3b). This parallels the reversal of ER downregulation observed in tumor slices transplanted to placebo animals (Fig. 2b). Ki67 proliferation scores decreased in endocrine-treated PT12 (44 estrogen, 17 estrogen plus tamoxifen, 10.5 EWD), but remained similar in tamoxifen-treated PT15 tumors (50.7 estrogen, 47.3 estrogen plus tamoxifen) (Supplementary Table 1). Collectively, these studies demonstrate endocrine therapy of patient grafted tumors show features consistent with clinical responses, and therefore will provide unique opportunities to study the early response of untreated ER+ breast cancers to endocrine treatments and dissect mechanisms of acquired resistance.
Fig 3.
Endocrine therapy response of previously untreated primary tumor xenografts. a Tumors PT12 and PT15 were grown under estrogen supplementation (E) until they reached 300 mm3 (40–50 days), then treated for 3 weeks with tamoxifen, 0.6 mg/kg IP 3×/ week (E + Tam), or removal of the estrogen pellet (PT12, estrogen withdrawal, EWD). Arrows indicate the start of tamoxifen injections and EWD. Tumor volumes are plotted versus number of days of incubation. Final tumor masses are shown on the right panel. *p < 0.05 PT15 tumor volume and tumor mass (E + Tam compared with E). b Sections of PT12 and PT15 tumors from untreated (E), tamoxifen treated (E + Tam), or EWD animals (PT12 only) were stained by H&E or IHC for ER. For PT12 H&E, the percent of tumor cellularity is indicated; the intensity and percent of ER staining is indicated for PT12 and PT15. Scale bars, 100 µM
Cancer stem cell populations in ER+ tumor xenografts
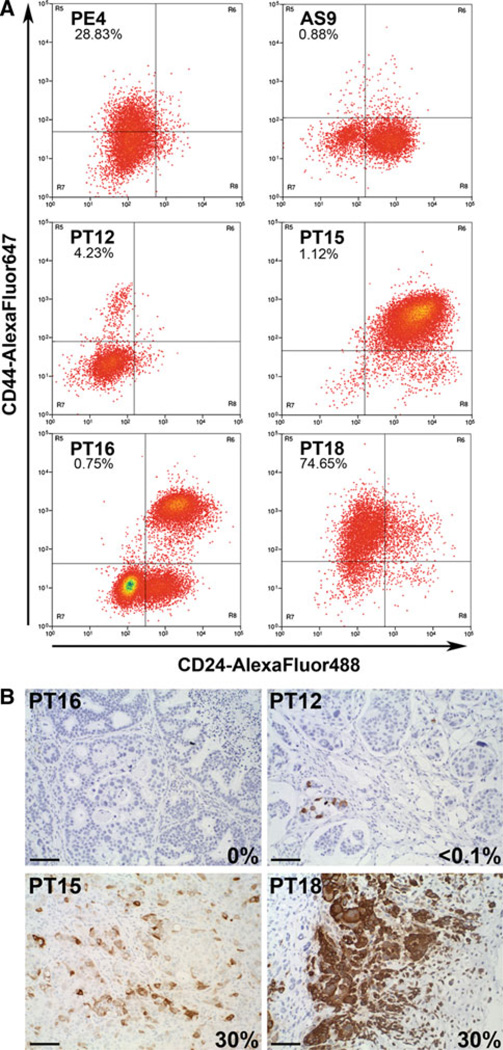
To determine if xenografted tumors contain currently defined cancer stem cells, we measured the CD44+CD24 (−/low) population in the five ER+ luminal tumors (under estrogen supplementation), and TN PT18. Among the luminal tumors, only PE4 (metastatic disease) contained a significant population of CD44+CD24(−/low) cells (28 %). PT12 and PT15 had low CD44+CD24(−/low) cells (4 and ~1 %, respectively) and moderately differentiated tumors AS9 and PT16 xenografts had a virtual absence of CD44+CD24(−/low) cells (<0.1 %). In contrast, PT18 had a high percent (~75 %) of CD44+CD24(−/low) cells, typical for TN tumors. Flow analyses were repeated two to three times for each tumor with consistent results. Where measured, we found no statistically significant differences in the CD44+CD24(−/low) populations in placebo- versus estrogen-treated animals (PT12 and PT16) (not shown). Thus, ER-positive tumor xenografts generally contained low CD44+CD24(−/low), with the exception of PE4, consistent with that described for ER+ primary patient tumors.
Within luminal tumors, CK5 marks populations of ER− PR− cells that express several stem-like genes including CD44 [36]. We, therefore, determined the relative expression of CK5 by IHC in the six xenograft tumors, all under estrogen conditions except PT18 (Table 1; Fig. 4b). AS9 and PT16 (shown for PT16) were absent for CK5, and PE4 and PT12 (shown for PT12) had <1 % CK5+ cells. PT15 and TN PT18 both had 30 % CK5+ cells, albeit in different patterns. PT15 had somewhat evenly dispersed CK5+ cells throughout the tumor, whereas PT18 had focal areas of high CK5 expression adjacent to areas with CK5− chondrocyte-like differentiation, typical for metaplastic tumors. Note that the presence of CK5 and CD44+ CD24(−/low) populations did not necessarily correlate; PE4 and PT12 contained CD44+CD24(−/low) populations (4 and 26 %) but were virtually absent for CK5 (<0.1 %), whereas PT15 had a significant CK5+ population (30 %) but low CD44+CD24(−/low) cells (1 %). Thus, under estrogen conditions, some luminal tumors contain cells expressing cancer stem cell markers, while others are void of either, underscoring a need to better understand the factors that drive tumor propagation, drug resistance, and long-term recurrence in ER+ tumors. We speculate that under conditions where estrogens and progestins are both present, these levels may fluctuate.
Fig 4.
Relative abundance of cancer stem cell markers in ER+ tumor xenografts. a Fresh pieces of tumors (all from estrogen-treated animals except for PT18, no treatment) were processed into single cell suspensions, and stained with a cocktail of antibodies to exclude murine host cells, and antibodies to CD44 (AlexaFLuor647) and CD24 (AlexaFLuor488). Dead cells were excluded by PI. Unstained cells from each tumor were used to set gates. The percent of cells with the cancer stem cell signature CD449+CD24(−/low) for each tumor is indicated in the upper left quadrant. b Sections of tumors were stained by IHC for CK5. The percent of cells stained is indicated on each panel. Scale bars, 100 µM
ER-dependent signaling networks in patient-derived tumor xenografts
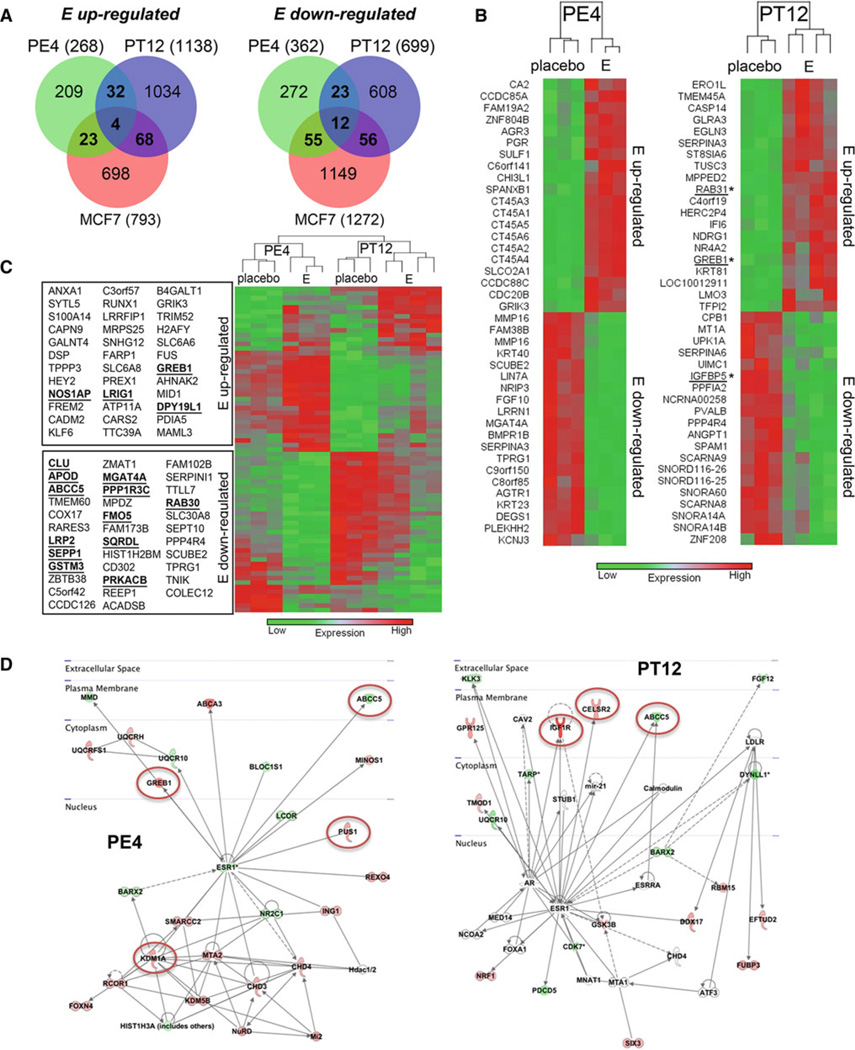
To determine the functional aspect of the transcription factor ER in luminal xenografts, the gene expression profiles of PE4 (90 % ER+, +/−estrogen) and PT12 (30 % ER+, +/−estrogen, tamoxifen or EWD) were assessed using Affymetrix human gene ST 1.1 arrays. Subsequent analyses were performed with Partek software. We also sought to establish if ER gene regulation in these tumor models has any cross relevance to ER-regulated genes mapped in traditional ER+ breast cancer cell line models. To do this, we compared the gene expression profiles in PE4 and PT12 to select publically available datasets previously reported for the standard ER+ MCF7 human breast cancer cell line, without or with estrogen, tamoxifen, and EWD treatment [24, 25, 40]. Bioinformatic details are found in supplementary methods. The majority of estrogen-regulated genes in PE4 and PT12 were distinct, whereas 36 up- and 35 down-regulated genes were shared between PE4/PT12 (Fig. 5a). Among these, 4 up- and 12 down-regulated genes were shared with MCF7 cells. Each individual tumor shared as many estrogen-regulated genes with MCF7 cells as they did with each other, suggesting the ER transcriptome is cell-specific more so than model-specific. The top 20 estrogen up- and down-regulated genes in PE4 or PT12 are shown by dendogram in Fig. 5b. Among the highest estrogen-regulated genes in PE4 are six cancer/ testis antigens, not previously reported to be estrogen regulated in breast cancers. A supervised hierarchical cluster using the 71 common gene set between PE4/PT12 segregates the expression patterns by tumor, followed by hormone (placebo vs. estrogen) (Fig. 5c). Genes that were estrogen regulated in the MCF7 data sets are indicated, and include the highly estrogen-sensitive GREB1. Many known estrogen-regulated genes were uniquely regulated in either PE4 (CXCL12) or PT12 (AREG, IGF1R), but not both tumors. Shared estrogen-regulated genes between PE4/PT12, PE4/MCF7, and PT12/MCF7 are supplied in (Supplementary Tables 2–4). The signaling networks activated or deactivated by ER in each tumor were determined by Ingenuity Pathway Analyses (Fig. 5d). Commonalities include downregulation of the transporter ABCC5 (PE4/PT12 and MCF7), and activation of GREB1 (PE4/MCF7) and IGF1R (PT12/MCF7). However, most affected networks are unique to each tumor, including activation of enzymes in the mitochondrial respiratory chain in PE4 (UQCRH and UQCRFS1), and activation of additional plasma membrane signaling molecules in PT12 (GPR125 and CELSR2). The molecule/pathway with the most significant difference between placebo- and estrogentreated tumors centered on MYC in PE4, and YWHAG (14-3-3 gamma) in PT12 (Supplementary Fig. 6). These data highlight the complexity of the ER transcriptome in breast cancers, and that disease-unique, as well as common factors, may play critical roles in disease progression.
Fig 5.
Estrogen-regulated genes and signaling networks in luminal tumor xenografts PE4 and PT12. The gene expression profiles of tumors PE4 and PT12 grown without (placebo) or with continuous estrogen (E) were determined using Affymetrix human gene 1.1 ST arrays. An n of 3 tumors were profiled for all groups except for PT12 E, n of 4. A list of estrogen-regulated genes in MCF7 cells was assembled by performing a union of significantly changed genes in data sets GSE848 or GSE3834 (supplementary methods). a Venn diagrams depicting E up- and down-regulated genes in xenografts PE4, PT12, and MCF7 cells (p < 0.05, > 1.5-fold). b The top 20 E up-and down-regulated genes in PE4 or PT12 are depicted by hierarchical cluster. Genes that also changed in MCF7 cells in the same direction are indicated by underlines/asterisks. c Common E up- (36) and down- (35) regulated genes in PE4 and PT12 graphed by hierarchical cluster. Underlines/asterisks indicate genes that were also regulated in the same direction in MCF7 cells. d Schematic map of ER-regulated pathways, based on cellular location, which are activated (red) or deactivated (green) in PE4 or PT12 tumors. Pathways also significantly affected in MCF7 cells are circled
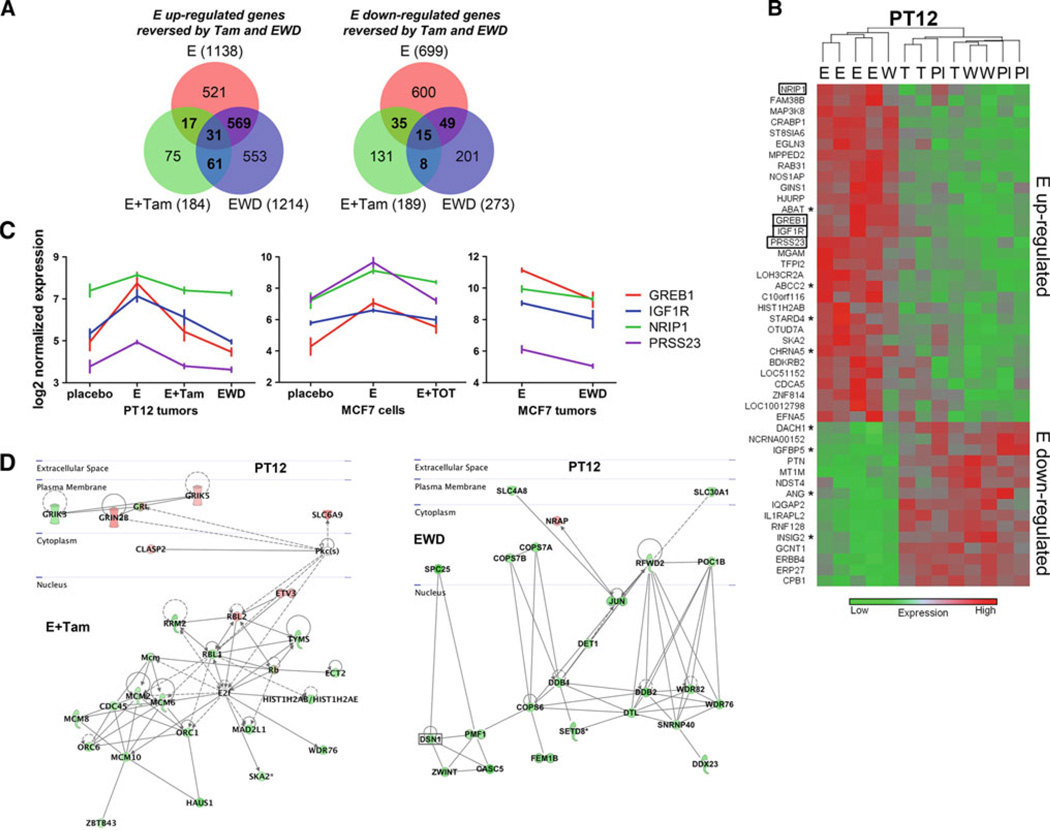
To map changes in estrogen signaling affected by endocrine therapy in PT12, we cross-compared estrogen-regulated genes that were reversed by both tamoxifen and EWD (Fig. 6a, b). Overall, EWD abrogated estrogen signaling more robustly than tamoxifen (1,487 vs. 373 total affected genes) (Fig. 6a). The observed difference is possibly because of more complete blockade of estrogen signaling with EWD compared with direct ligand competition for ER with tamoxifen. In addition, tamoxifen acts as a partial agonist on ER responsive genes [41], contributing to the overall differences in gene expression profiles. Hierarchical cluster analysis was used to map the genes (46 total) that were estrogen upregulated and decreased by Tam and EWD (35 genes), or estrogen downregulated and increased by Tam and EWD (15 genes) (Fig. 6b). Overall, these clustered by estrogen versus non-estrogen conditions (placebo, tamoxifen, EWD). One tumor was partially resistant to EWD, and still clustered with the estrogen samples. Twelve/46 common tamoxifen/EWD sensitive genes were also affected in short term MCF7 tamoxifen and/or EWD models determined from datasets GSE848 and GSE3834 (marked by asterisks). Four estrogen upregulated genes (GREB1, IGFR1, NRIP, and PRSS; none downregulated) were abrogated by both tamoxifen and EWD in PT12 tumors, and in the tamoxifen and EWD MCF7 datasets (Fig. 6d). Indeed, GREB1 and IFG1R are estrogen-regulated across multiple breast cancer models [42–44]. Ingenuity analysis determined the major networks affected in tamoxifen- versus EWD-treated PT12. Tamoxifen affected Rb-associated genes (Rb, RBL1, RBL2) and replication proteins in the mini chromosome maintenance family (MCM2, MCM6, MCM8, andMCM10), whereas EWD most highly affected molecules involved in solute transport (SLC4A8 and SLC30A1), the ubiquitin protein ligase complex (COPS7A, COPS7B, COPS6, and DET1), and DNA damage response (DDB1, DDB2, and DTL) (Fig. 6d). While these molecules were affected in responsive tumors, longer treatment and resistance studies in these models will determine which pathways eventually drive recurrent tumors. These data put forward that while some estrogen-regulated genes and signaling networks (i.e., GREB1 and IGF1R) are apparently broadly conserved among ER+ tumors, each tumor also has its own unique estrogen-dependent targetable pathways that will need to be considered for more individualized treatments.
Fig 6.
Gene expression networks in endocrine therapy treated PT12. The gene expression profiles of tumor PT12 grown with continuous estrogen for 8 weeks (E), plus tamoxifen for the last 3 weeks (E + Tam), or EWD for the last 3 weeks, were determined using Affymetrix human gene 1.1 ST arrays. An n of 3 tumors were profiled for all groups except for PT12 E, n of 4. a Venn diagrams comparing estrogen (E) up-regulated genes (compared with placebo) and tamoxifen (E + Tam) and EWD down-regulated genes (compared with E), and E down-regulated genes (compared with placebo) and E + Tam and EWD up-regulated genes (compared with E). b Hierarchical cluster of E up- and down-regulated genes reversed by both endocrine therapies (Pl placebo, T E+ tamoxifen, W EWD). Genes changed in the same direction in MCF7 cells with both tamoxifen and EWD are indicated by boxes and either tamoxifen or EWD are indicated by asterisks. c Normalized expression (log2) plots of four genes that significantly changed in both tamoxifen and EWD PT12 tumors, and tamoxifen-treated MCF7 cells (24 h E vs. 24 h E + 4-hydroxy-tamoxifen [GSE848]), and EWD MCF7 tumors (E vs. 48 h EWD [GSE3834]). d Schematic map based on cellular location of the top significant gene networks altered in tamoxifen (E + Tam) or EWD PT12 tumors
Unique estrogen-regulated genes in tumors
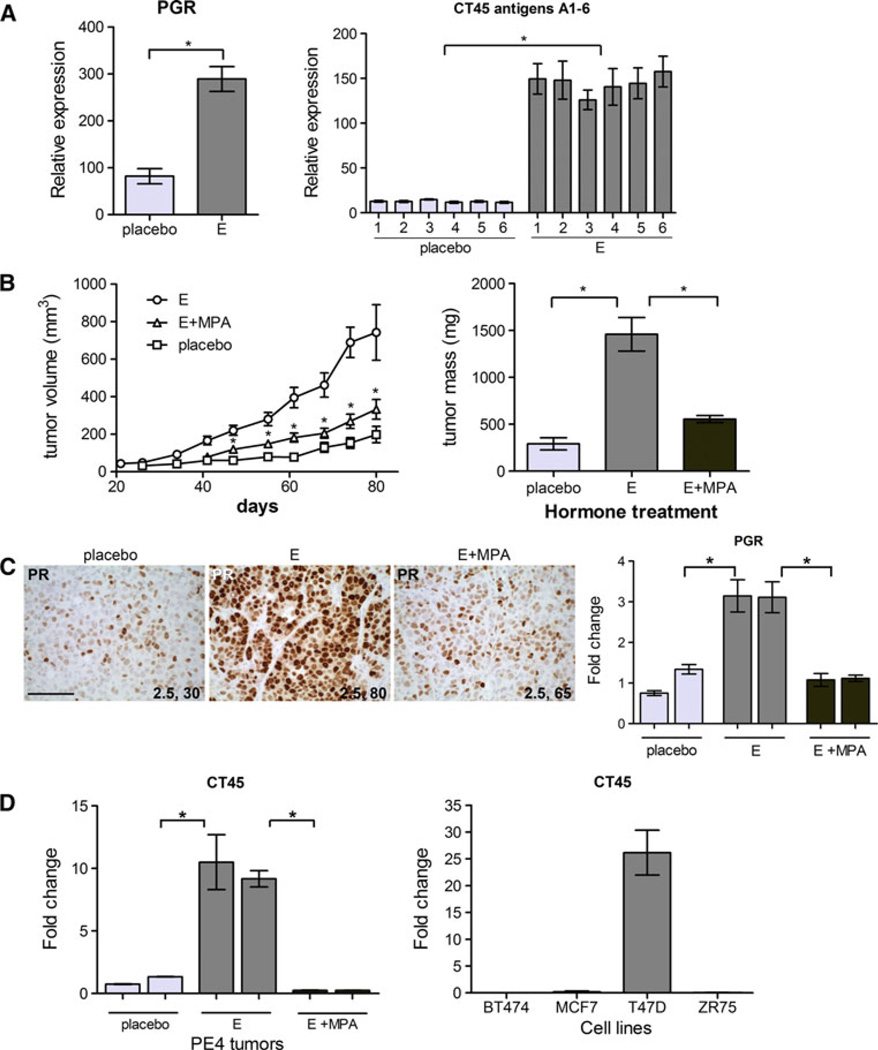
To confirm the estrogen regulation of select genes in tumors, we chose a well-known estrogen-responsive gene and a group of previously undescribed estrogen-regulated genes for analysis. The gene for PR, PGR, is well described as estrogen/ER regulated in many breast tumors, and is used regularly as a marker of functional ER. Cancer/testis antigen family 45 members A1–6 belong to a group of genes normally expressed only in germ cells, but are aberrantly expressed in many tumor types, including about 10 % of breast cancers [45, 46]. By microarray analysis, both PGR and all six CT45 genes (A1–6) were significantly regulated by estrogen compared with placebo in PE4 (Fig. 7a). PE4 is indicative of a subset of advanced luminal tumors that respond to treatment with high-dose synthetic progestins [47]. Estrogen plus progestin (MPA) combination significantly inhibited the estrogen alone induced growth of PE4 to near placebo controls as measured by tumor volume and mass (Fig. 7b). Of note is the MPA progestin levels we used can be extrapolated to be near 20 ng/mL based on measuring serum progesterone give at the same dose [34], whereas the high doses used in MPA progestin therapy are >150 ng/mL [48]. The natural hormone progesterone is also effective at blocking estrogen-induced growth of PE4 (not shown), and we therefore conclude that blockade of estrogen signaling occurs at physiological levels. IHC for PR confirmed its upregulation by estrogen, compared with placebo, and that addition of MPA abrogates that increase (Fig. 7c). Progestin treatment may be contributing to PR loss through receptor downregulation, similar to that observed for ER (Fig. 2). However, PGR mRNA was also significantly decreased in estrogen plus MPA-treated tumors as measured by qPCR (Fig. 7c). Using a probe that measures all CT45 family members, qPCR demonstrated that MPA treatment significantly decreased expression of CT45 in PE4 tumors (Fig. 7d). Additionally, only select luminal breast cancer cell lines express CT45 (Fig. 7d). T47D cells express the CT45 genes whereas BT474, MCF7, and ZR75 were absent for CT45 expression. At present, there are no commercially available CT45 antibodies that are competent for IHC analysis. Cancer/testis antigens are being explored as therapeutic targets, because of their antigenicity and virtual absence in normal adult tissues, and ability to elicit cytotoxic T cell response in cancer vaccines [46]. The finding that some of these genes are potently estrogen regulated and that their expression is decreased in regressing tumors suggests they may provide a tumor-unique target in combination with endocrine therapies in a select set of ER+ breast tumors.
Fig 7.
Estrogen-regulated target genes in PE4. a Relative expression levels of the genes for PR (PGR) and cancer/testis antigen family 45 (CT45) members A1-6 in placebo versus estrogen (E) treated PE4 tumors. Expression derived from Affymetrix microarray data analysis, n of 3 each, *p < 0.05 E compared with placebo. b Tumor volume (left) and tumor mass (right) ±SEM of PE4 grown under placebo, estrogen (E), or estrogen plus MPA (E + MPA) conditions. *p < 0.05, E + MPA tumor volume compared with E, tumor mass E relative to placebo or E + MPA. c Sections of PE4 tumors treated with placebo, E, or E + MPA were stained by IHC for PR (left). Intensity and percent of cells stained are indicated. Scale bars, 100 µM. qPCR for PGR was performed on cDNA prepared from duplicate samples of PE4 tumors treated with placebo, E, or E + MPA. *p < 0.05. d qPCR for CT45 (probes cover all members) on cDNA prepared from duplicate placebo, E, or E + MPA-treated PE4 tumors (left) or four luminal breast cancer cell lines (right). Values were normalized to β-actin and plotted as fold change relative to placebo. *p < 0.05 E compared with placebo or E + MPA
Discussion
Despite many advances in our knowledge of ER+ breast cancer, we are still relatively naive in our mechanistic understanding of refractory endocrine-resistant disease. One reason is that this largest group of breast cancers has considerable intra-subtype diversity with differences in histopathologic features, and expression of other growth factors and receptors, among genetic and epigenetic factors. It is clear that ER dynamics play a central role in tumor behavior [49], and it is likely that the ER cistrome/ transcriptomes are divergent in individual patient samples. While a battery of breast cancer cell lines representing different subtypes are available for study, we believe that the presented model provides a complementary approach that has distinct advantages. First, intra-tumoral heterogeneity in ER expression clearly plays an important role in treatment response—patient-derived tumor models are more likely to maintain such cellular diversity. Second, translating pre-clinical findings into clinical successes is likely to be more efficacious from tumor models. The described tumors maintain stable ER expression levels over multiple passages and will allow for seamless manipulation and specific study of both ER+ and ER− tumor cell populations. In some cases, tumors have not been previously drug treated, affording a unique opportunity to map the nascent ER transcriptome, as well as changes incurred under endocrine therapy pressure.
Although endocrine therapies remain fundamental to treating ER+ breast cancers, several alternate signaling molecules have been targeted in these tumors alone or in combination therapies. HER2-directed therapies have vastly improved treatment outcomes, providing reason to believe other molecules exist with similar potential [50]. However, HER1/EGFR-targeted agents, mainly gefitinib, have offered only modest improvement in clinical trials [51]. Everolimus, an mTOR inhibitor, is the first in a long list of combination therapies to prove clinically beneficial in a phase III trial [52]. However, even in mTOR responsive tumors, we inevitably need to narrow down the ideal target patient population. Non-responsive tumors to targeted therapies likely have different pathways driving tumor survival. Here, we demonstrate that two ER+ estrogen-dependent breast cancers isolated from patients and directly propagated in vivo, PE4 and PT12, have predominantly different ER transcriptomes, although they share a small set of common estrogen-regulated genes. The most significant pathway active in estrogen-treated PE4 centers on MYC, whereas in PT12 it centers on YWHAG (14-3-3 gamma) (Supplementary Fig. 6). PE4 originates from a previously endocrine-treated cancer, that metastasized, and it is therefore likely that its progression was partially MYC-driven [53]. PT12, a previously untreated, but relatively rapid growing tumor, seemingly utilizes the highly conserved 14-3-3 gamma protein, a member of a family of signal transduction modulating molecules potentially involved in tumor cell survival [54]. EWD effectively reversed 14-3-3 gamma expression while tamoxifen did not, highlighting important mechanistic differences between the two predominant endocrine therapies. The partial agonist activity of tamoxifen through ER compared with the effective absence of liganded ER in EWD presumably led to differentially affected estrogen responsive pathways. By contrast, four highly estrogen-dependent genes (GREB1, IGF1R, NRIP1, and PRSS23) were significantly reduced across cell line (MCF7) and tumor (PT12) models with both therapies (tamoxifen and EWD). Development of treatment-resistant sublines of these models analogous to those described for MCF7 tumor xenografts [26, 27] will ultimately determine which escape pathways become activated in each tumor. While the number of samples analyzed in these models is likely to be moderate, they will provide more in depth mechanistic data on each individual tumor, including the potential to test combination therapies in pre-clinical models. For example, we show here that a group of X-linked aberrantly expressed cancer/testis antigens is estrogen-regulated and treatment-sensitive, and therefore could serve as a possible additional target or marker of response in a subset of luminal breast tumors.
Cancer stem cells offer an intriguing theory for endocrine resistance in ER+ tumors, especially because they are routinely described as ER− [55, 56], and would thus naturally avoid anti-ER-targeted therapies. ER+ cell lines and their corresponding xenograft tumors contain low cancer stem cell populations characterized by either CD44+CD24(−/low) or CK5+ER−PR− [19, 36], while both of these markers are in much higher abundance in TN cell lines and tumors [17, 18]. Treatment of ER+ tumors with the aromatase inhibitor letrozole increased the CD44+CD24(−/low) population [28], and ER+ cells treated with tamoxifen or EWD have an accumulation of CK5+ cells [22, 57]. Here we confirm that ER+ luminal tumor xenografts generally contain low numbers of CD44+CD24(−/low) and CK5+ cells. PT12 had a small (4 %) CD44+CD24(−/low) population and negligible (<0.1 %) CK5+ cells, under estrogen conditions. PT15 by contrast contained low (~1 %) CD44+CD24(−/low) cells (albeit nearly uniformly CD44+CD24 +), but had a relatively high (~30 %) CK5+ cell population. It is likely that CSC populations will fluctuate under different hormone conditions as both circulating estradiol and progesterone affect their numbers [36, 58]. Although we have not fully explored these scenarios in this report, tamoxifen treatment of PT15 did not lead to a significant increase in the CK5+ cell population (not shown), although the ER+ cell population notably intensified. Because PR levels and progesterone are important for the regulation of stem cells in the normal breast and perhaps some cancers [3], it is likely these will fluctuate under different hormone conditions. Therefore, while cancer stem cell populations likely contribute to drug resistance in some ER+ breast cancers, a combination of stem-like cells and a general adaptation of the bulk ER+ tumor cells may culminate to produce resistant luminal disease. Of note is that tumors such as AS9 and PT16, a metastatic and grade 3 lesion, have near absent populations of both CD44+CD24(−/low) and CK5+ cells (<0.1 % each). We speculate that some ER+ tumors have luminal-specific cancer stem cells that are as yet undiscovered, and some moderate to well-differentiated breast cancers may not fit the current cancer stem cell model [59].
It is intriguing that molecular subtyping categorized four of the five ER+ tumors as luminal B and one as having the HER2-enriched gene signature, while none fell into the luminal A category. Likewise, ER+ tumor grafts were all characterized as luminal B in the DeRose et al. study [33]. The five luminal xenografts described here displayed a wide variety in ER expression levels (5–99 %) and moderate to high proliferation rates (Ki67 scores 36–50). It is possible that the amount of proliferation necessary to produce transplantable tumors in these mouse models under the experimental time constraint may necessarily lead to a luminal B phenotype on output. This seems likely as near half of transplanted ER+ tumors would fall into the luminal A category if subtyped from the original biopsy without further manipulation [37]. The successful grafting of moderately differentiated and slower growing tumors (AS9 and PT16) lends optimism for the eventual grafting and maintenance of luminal A subtype tumors. Nonetheless, the demonstration of functional ER in diverse individual breast cancers supports the use of this model to dissect ER action and response to therapy regardless of subtype category.
Patient-derived xenografts will also provide unique opportunities to dissect the contributions of other steroid hormones and their receptors in breast cancer biology, particularly AR and PR. Because AR is a tumor suppressor in most ER+ tumors, but a tumor promoter in some ER− AR+ tumors [5, 6, 60], grafted tumors with different ER, PR, and AR status will allow the exploration of AR-specific tumor effects, alone or in the context of each other, under tightly controlled ligand conditions (estrogens, progestins, and androgens). While PR is a superb prognostic indicator, and progestins clearly are detrimental in the inception of breast disease [2, 61], the influence of progestins on established tumors has remained elusive [47]. These models will allow the dissection of PR action in tumors with a wide heterogeneity of PR expression. The PR transcriptome has been described in relatively few human models including 3D organoid models of normal breast tissue and PR-rich T47D breast cancer cells [62, 63]. In addition, the PR cistrome was recently reported for the first time in an immortalized breast cancer cell line (MCF10A stably expressing PR) and T47D cells [64]. Of note, there was little overlap between normal and cancer models in regard to both PR-binding sites and PR-regulated genes [63, 64]. We now have the prospect of mapping PR gene regulation patterns in diverse breast tumor samples with various PR levels.
In summary, patient-derived ER+ tumor xenografts display estrogen-dependent growth and have unique ER transcriptomes. While multiple other factors such as genetic and epigenetic signatures, and co-regulator expression patterns, collectively influence tumor behavior, ER remains the common denominator driving tumor growth and survival, at least initially. Because ER is retained in most drug-resistant tumors, determining the “switch” in activated/deactivated genes and signaling networks under treatment pressure in individual tumors is cornerstone to overcoming persistent ER+ breast disease. This approach will advance the study of ER and other steroid receptors in individual breast tumors, by allowing relatively rapid analysis of transcriptome dynamics and therapeutic responses. Clinically, there is a push toward more individualized medicine. It is conceivable that the pathways involved in estrogen promotion, and on estrogen deprivation could be discerned in a matter of months—yielding patient centric tumor data that may ultimately inform treatment options.
Supplementary Material
Acknowledgments
The authors thank Robert Tsay for providing tissue from pathological specimens, Lisa Litzenberger for graphics help, Audrie Van Bokhoven for the AR control slides, Britta Jacobsen and Elizabeth Wellberg for critical review of the manuscript, and the University of Colorado Cancer Center Breast Cancer Tissue Bank, Tissue Procurement, and Flow Cytometry Cores for their excellent services. This work was supported, in part, by grants from the Grohne Cancer Research Fund (P. Kabos), ASCO Young Investigator Award (P. Kabos), the National Institutes of Health R01 CA140985 (C. Sartorius), the Wendy Will Case Foundation (C. Sartorius), the University of Colorado Cancer Center (P. Kabos and C. Sartorius), and the Martha Cannon Dear Professorship (A. Elias).
Abbreviations
- AI
Aromatase inhibitors
- ALDH
Aldehyde dehydrogenase
- AR
Androgen receptor
- AS
Ascites
- CK5
Cytokeratin 5
- CT45
Cancer/testis antigen 45
- ER
Estrogen receptor
- EWD
Estrogen withdrawal
- GEO
Gene expression omnibus
- IHC
Immunohistochemisty
- MPA
Medroxyprogesterone acetate
- PE
Pleural effusion
- PR
Progesterone receptor
- PT
Primary tumor
- qPCR
Quantitative PCR
- TN
Triple negative
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10549-012-2164-8) contains supplementary material, which is available to authorized users.
Conflict of interest The authors have no conflicts of interest to disclose.
Contributor Information
Peter Kabos, Department of Medicine, Division of Medical Oncology, University of Colorado Denver Anschutz Medical Campus, Aurora 80045, CO, USA.
Jessica Finlay-Schultz, Department of Pathology, University of Colorado Denver, Anschutz Medical Campus Center, 12801 E 17th Ave MS8104, Aurora 80045, CO, USA.
Chunling Li, Department of Medicine, Division of Medical Oncology, University of Colorado Denver Anschutz Medical Campus, Aurora 80045, CO, USA.
Enos Kline, Department of Medicine, Division of Medical Oncology, University of Colorado Denver Anschutz Medical Campus, Aurora 80045, CO, USA.
Christina Finlayson, Department of Surgery, University of Colorado Denver Anschutz Medical Campus, Aurora 80045, CO, USA.
Joshua Wisell, Department of Pathology, University of Colorado Denver Anschutz Medical Campus Center, 12801 E 17th Ave MS8104, Aurora 80045, CO, USA.
Christopher A. Manuel, Department of Pathology, University of Colorado Denver Anschutz Medical Campus Center, 12801 E 17th Ave MS8104, Aurora 80045, CO, USA
Susan M. Edgerton, Department of Pathology, University of Colorado Denver Anschutz Medical Campus Center, 12801 E 17th Ave MS8104, Aurora 80045, CO, USA
J. Chuck Harrell, Lineberger Comprehensive Cancer Center, University of North Carolina, Chapel Hill 27599, NC, USA.
Anthony Elias, Department of Medicine, Division of Medical Oncology, University of Colorado Denver Anschutz Medical Campus, Aurora 80045, CO, USA.
Carol A. Sartorius, Department of Pathology, University of Colorado Denver Anschutz Medical Campus Center, 12801 E 17th Ave MS8104, Aurora 80045, CO, USA Carol.Sartorius@ucdenver.edu
References
- 1.Obiorah I, Jordan VC. Progress in endocrine approaches to the treatment and prevention of breast cancer. Maturitas. 2011;70(4):315–321. doi: 10.1016/j.maturitas.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bardou VJ, Arpino G, Elledge RM, Osborne CK, Clark GM. Progesterone receptor status significantly improves outcome prediction over estrogen receptor status alone for adjuvant endocrine therapy in two large breast cancer databases. J Clin Oncol. 2003;21(10):1973–1979. doi: 10.1200/JCO.2003.09.099. [DOI] [PubMed] [Google Scholar]
- 3.Axlund SD, Sartorius CA. Progesterone regulation of stem and progenitor cells in normal and malignant breast. Mol Cell Endocrinol. 2012;357(1–2):71–79. doi: 10.1016/j.mce.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moinfar F, Okcu M, Tsybrovskyy O, Regitnig P, Lax SF, Weybora W, Ratschek M, Tavassoli FA, Denk H. Androgen receptors frequently are expressed in breast carcinomas: potential relevance to new therapeutic strategies. Cancer. 2003;98(4):703–711. doi: 10.1002/cncr.11532. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez LO, Corte MD, Vazquez J, Junquera S, Sanchez R, Alvarez AC, Rodriguez JC, Lamelas ML, Vizoso FJ. Androgen receptor expression in breast cancer: relationship with clinicopathological characteristics of the tumors, prognosis, and expression of metalloproteases and their inhibitors. BMC Cancer. 2008;8:149. doi: 10.1186/1471-2407-8-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peters AA, Buchanan G, Ricciardelli C, Bianco-Miotto T, Centenera MM, Harris JM, Jindal S, Segara D, Jia L, Moore NL, Henshall SM, Birrell SN, Coetzee GA, Sutherland RL, Butler LM, Tilley WD. Androgen receptor inhibits estrogen receptor-alpha activity and is prognostic in breast cancer. Cancer Res. 2009;69(15):6131–6140. doi: 10.1158/0008-5472.CAN-09-0452. [DOI] [PubMed] [Google Scholar]
- 7.Farmer P, Bonnefoi H, Becette V, Tubiana-Hulin M, Fumoleau P, Larsimont D, Macgrogan G, Bergh J, Cameron D, Goldstein D, Duss S, Nicoulaz AL, Brisken C, Fiche M, Delorenzi M, Iggo R. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene. 2005;24(29):4660–4671. doi: 10.1038/sj.onc.1208561. [DOI] [PubMed] [Google Scholar]
- 8.Guedj M, Marisa L, de Reynies A, Orsetti B, Schiappa R, Bibeau F, Macgrogan G, Lerebours F, Finetti P, Longy M, Bertheau P, Bertrand F, Bonnet F, Martin AL, Feugeas JP, Bieche I, Lehmann-Che J, Lidereau R, Birnbaum D, Bertucci F, de The H, Theillet C. A refined molecular taxonomy of breast cancer. Oncogene. 2011 doi: 10.1038/onc.2011.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5(1):5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osborne CK, Schiff R. Mechanisms of endocrine resistance in breast cancer. Annu Rev Med. 2011;62:233–247. doi: 10.1146/annurev-med-070909-182917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abd El-Rehim DM, Pinder SE, Paish CE, Bell J, Blamey RW, Robertson JF, Nicholson RI, Ellis IO. Expression of luminal and basal cytokeratins in human breast carcinoma. J Pathol. 2004;203(2):661–671. doi: 10.1002/path.1559. [DOI] [PubMed] [Google Scholar]
- 12.Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS, Hayes M, Hicks DG, Lester S, Love R, Mangu PB, McShane L, Miller K, Osborne CK, Paik S, Perlmutter J, Rhodes A, Sasano H, Schwartz JN, Sweep FC, Taube S, Torlakovic EE, Valenstein P, Viale G, Visscher D, Wheeler T, Williams RB, Wittliff JL, Wolff AC. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28(16):2784–2795. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Creighton CJ, Fu X, Hennessy BT, Casa AJ, Zhang Y, Gonzalez-Angulo AM, Lluch A, Gray JW, Brown PH, Hilsenbeck SG, Osborne CK, Mills GB, Lee AV, Schiff R. Proteomic and transcriptomic profiling reveals a link between the PI3K pathway and lower estrogen-receptor (ER) levels and activity in ER+ breast cancer. Breast Cancer Res. 2010;12(3):R40. doi: 10.1186/bcr2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Amicis F, Thirugnansampanthan J, Cui Y, Selever J, Beyer A, Parra I, Weigel NL, Herynk MH, Tsimelzon A, Lewis MT, Chamness GC, Hilsenbeck SG, Ando S, Fuqua SA. Androgen receptor overexpression induces tamoxifen resistance in human breast cancer cells. Breast Cancer Res Treat. 2010;121(1):1–11. doi: 10.1007/s10549-009-0436-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lange CA, Yee D. Killing the second messenger: targeting loss of cell cycle control in endocrine-resistant breast cancer. Endocr Relat Cancer. 2011;18(4):C19–C24. doi: 10.1530/ERC-11-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Brien CS, Howell SJ, Farnie G, Clarke RB. Resistance to endocrine therapy: are breast cancer stem cells the culprits. J Mammary Gland Biol Neoplasia. 2009;14(1):45–54. doi: 10.1007/s10911-009-9115-y. [DOI] [PubMed] [Google Scholar]
- 17.Honeth G, Bendahl PO, Ringner M, Saal LH, Gruvberger-Saal SK, Lovgren K, Grabau D, Ferno M, Borg A, Hegardt C. The CD44+/CD24− phenotype is enriched in basal-like breast tumors. Breast Cancer Res. 2008;10(3):R53. doi: 10.1186/bcr2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, Hur MH, Diebel ME, Monville F, Dutcher J, Brown M, Viens P, Xerri L, Bertucci F, Stassi G, Dontu G, Birnbaum D, Wicha MS. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69(4):1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to pheno-typically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10(2):R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boecker W, Moll R, Poremba C, Holland R, Van Diest PJ, Dervan P, Burger H, Wai D, Ina Diallo R, Brandt B, Herbst H, Schmidt A, Lerch MM, Buchwallow IB. Common adult stem cells in the human breast give rise to glandular and myo-epithelial cell lineages: a new cell biological concept. Lab Invest. 2002;82(6):737–746. doi: 10.1097/01.lab.0000017371.72714.c5. [DOI] [PubMed] [Google Scholar]
- 21.Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A, Feleppa F, Huschtscha LI, Thorne HJ, Fox SB, Yan M, French JD, Brown MA, Smyth GK, Visvader JE, Lindeman GJ. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15(8):907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 22.Kabos P, Haughian JM, Wang X, Dye WW, Finlayson C, Elias A, Horwitz KB, Sartorius CA. Cytokeratin 5 positive cells represent a steroid receptor negative and therapy resistant subpopulation in luminal breast cancers. Breast Cancer Res Treat. 2011;128(1):45–55. doi: 10.1007/s10549-010-1078-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38(11):1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 24.Creighton CJ, Cordero KE, Larios JM, Miller RS, Johnson MD, Chinnaiyan AM, Lippman ME, Rae JM. Genes regulated by estrogen in breast tumor cells in vitro are similarly regulated in vivo in tumor xenografts and human breast tumors. Genome Biol. 2006;7(4):R28. doi: 10.1186/gb-2006-7-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, Katzenellenbogen BS. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology. 2003;144(10):4562–4574. doi: 10.1210/en.2003-0567. [DOI] [PubMed] [Google Scholar]
- 26.Creighton CJ, Massarweh S, Huang S, Tsimelzon A, Hilsenbeck SG, Osborne CK, Shou J, Malorni L, Schiff R. Development of resistance to targeted therapies transforms the clinically associated molecular profile subtype of breast tumor xenografts. Cancer Res. 2008;68(18):7493–7501. doi: 10.1158/0008-5472.CAN-08-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Massarweh S, Osborne CK, Creighton CJ, Qin L, Tsimelzon A, Huang S, Weiss H, Rimawi M, Schiff R. Tamoxifen resistance in breast tumors is driven by growth factor receptor signaling with repression of classic estrogen receptor genomic function. Cancer Res. 2008;68(3):826–833. doi: 10.1158/0008-5472.CAN-07-2707. [DOI] [PubMed] [Google Scholar]
- 28.Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, Rimm DL, Wong H, Rodriguez A, Herschkowitz JI, Fan C, Zhang X, He X, Pavlick A, Gutierrez MC, Renshaw L, Larionov AA, Faratian D, Hilsenbeck SG, Perou CM, Lewis MT, Rosen JM, Chang JC. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA. 2009;106(33):13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harvell DM, Spoelstra NS, Singh M, McManaman JL, Finlayson C, Phang T, Trapp S, Hunter L, Dye WW, Borges VF, Elias A, Horwitz KB, Richer JK. Molecular signatures of neoadjuvant endocrine therapy for breast cancer: characteristics of response or intrinsic resistance. Breast Cancer Res Treat. 2008;112(3):475–488. doi: 10.1007/s10549-008-9897-4. [DOI] [PubMed] [Google Scholar]
- 30.Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, Ali S, Chin SF, Palmieri C, Caldas C, Carroll JS. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481(7381):389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bergamaschi A, Hjortland GO, Triulzi T, Sorlie T, Johnsen H, Ree AH, Russnes HG, Tronnes S, Maelandsmo GM, Fodstad O, Borresen-Dale AL, Engebraaten O. Molecular profiling and characterization of luminal-like and basal-like in vivo breast cancer xenograft models. Mol Oncol. 2009;3(5–6):469–482. doi: 10.1016/j.molonc.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marangoni E, Vincent-Salomon A, Auger N, Degeorges A, Assayag F, de Cremoux P, de Plater L, Guyader C, De Pinieux G, Judde JG, Rebucci M, Tran-Perennou C, Sastre-Garau X, Sigal-Zafrani B, Delattre O, Dieras V, Poupon MF. A new model of patient tumor-derived breast cancer xenografts for preclinical assays. Clin Cancer Res. 2007;13(13):3989–3998. doi: 10.1158/1078-0432.CCR-07-0078. [DOI] [PubMed] [Google Scholar]
- 33.DeRose YS, Wang G, Lin YC, Bernard PS, Buys SS, Ebbert MT, Factor R, Matsen C, Milash BA, Nelson E, Neumayer L, Randall RL, Stijleman IJ, Welm BE, Welm AL. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17(11):1514–1520. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sartorius CA, Shen T, Horwitz KB. Progesterone receptors A and B differentially affect the growth of estrogen-dependent human breast tumor xenografts. Breast Cancer Res Treat. 2003;79(3):287–299. doi: 10.1023/a:1024031731269. [DOI] [PubMed] [Google Scholar]
- 35.Sartorius CA, Harvell DM, Shen T, Horwitz KB. Progestins initiate a luminal to myoepithelial switch in estrogen-dependent human breast tumors without altering growth. Cancer Res. 2005;65(21):9779–9788. doi: 10.1158/0008-5472.CAN-05-0505. [DOI] [PubMed] [Google Scholar]
- 36.Horwitz KB, Dye WW, Harrell JC, Kabos P, Sartorius CA. Rare steroid receptor-negative basal-like tumorigenic cells in luminal subtype human breast cancer xenografts. Proc Natl Acad Sci USA. 2008;105(15):5774–5779. doi: 10.1073/pnas.0706216105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, Marron JS, Nobel AB, Mardis E, Nielsen TO, Ellis MJ, Perou CM, Bernard PS. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harrell JC, Prat A, Parker JS, Fan C, He X, Carey L, Anders C, Ewend M, Perou CM. Genomic analysis identifies unique signatures predictive of brain, lung, and liver relapse. Breast Cancer Res Treat. 2012;132(2):523–535. doi: 10.1007/s10549-011-1619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pink JJ, Jordan VC. Models of estrogen receptor regulation by estrogens and antiestrogens in breast cancer cell lines. Cancer Res. 1996;56(10):2321–2330. [PubMed] [Google Scholar]
- 40.Frasor J, Stossi F, Danes JM, Komm B, Lyttle CR, Katzenellenbogen BS. Selective estrogen receptor modulators: discrimination of agonistic versus antagonistic activities by gene expression profiling in breast cancer cells. Cancer Res. 2004;64(4):1522–1533. doi: 10.1158/0008-5472.can-03-3326. [DOI] [PubMed] [Google Scholar]
- 41.Santen RJ. Long-term tamoxifen therapy: can an antagonist become an agonist. J Clin Endocrinol Metab. 1996;81(6):2027–2029. doi: 10.1210/jcem.81.6.8964823. [DOI] [PubMed] [Google Scholar]
- 42.Ghosh MG, Thompson DA, Weigel RJ. PDZK1 and GREB1 are estrogen-regulated genes expressed in hormone-responsive breast cancer. Cancer Res. 2000;60(22):6367–6375. [PubMed] [Google Scholar]
- 43.Rae JM, Johnson MD, Scheys JO, Cordero KE, Larios JM, Lippman ME. GREB 1 is a critical regulator of hormone dependent breast cancer growth. Breast Cancer Res Treat. 2005;92(2):141–149. doi: 10.1007/s10549-005-1483-4. [DOI] [PubMed] [Google Scholar]
- 44.Fagan DH, Yee D. Crosstalk between IGF1R and estrogen receptor signaling in breast cancer. J Mammary Gland Biol Neoplasia. 2008;13(4):423–429. doi: 10.1007/s10911-008-9098-0. [DOI] [PubMed] [Google Scholar]
- 45.Chen YT, Ross DS, Chiu R, Zhou XK, Chen YY, Lee P, Hoda SA, Simpson AJ, Old LJ, Caballero O, Neville AM. Multiple cancer/testis antigens are preferentially expressed in hormone-receptor negative and high-grade breast cancers. PLoS One. 2011;6(3):e17876. doi: 10.1371/journal.pone.0017876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5(8):615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 47.Muss HB, Cruz JM. High-dose progestin therapy for metastatic breast cancer. Ann Oncol. 1992;3(Suppl 3):15–20. doi: 10.1093/annonc/3.suppl_3.s15. [DOI] [PubMed] [Google Scholar]
- 48.Mahlke M, Grill HJ, Knapstein P, Wiegand U, Pollow K. Oral high-dose medroxyprogesterone acetate (MPA) treatment: cortisol/MPA serum profiles in relation to breast cancer regression. Oncology. 1985;42(3):144–149. doi: 10.1159/000226021. [DOI] [PubMed] [Google Scholar]
- 49.Tyson JJ, Baumann WT, Chen C, Verdugo A, Tavassoly I, Wang Y, Weiner LM, Clarke R. Dynamic modelling of oestrogen signalling and cell fate in breast cancer cells. Nat Rev Cancer. 2011;11(7):523–532. doi: 10.1038/nrc3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krop I, Winer EP. Further progress in HER2-directed therapy. Lancet Oncol. 2012;13(1):2–3. doi: 10.1016/S1470-2045(11)70388-6. [DOI] [PubMed] [Google Scholar]
- 51.Osborne CK, Neven P, Dirix LY, Mackey JR, Robert J, Underhill C, Schiff R, Gutierrez C, Migliaccio I, Anagnostou VK, Rimm DL, Magill P, Sellers M. Gefitinib or placebo in combination with tamoxifen in patients with hormone receptor-positive metastatic breast cancer: a randomized phase II study. Clin Cancer Res. 2011;17(5):1147–1159. doi: 10.1158/1078-0432.CCR-10-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baselga J, Campone M, Piccart M, Burris HA, III, Rugo HS, Sahmoud T, Noguchi S, Gnant M, Pritchard KI, Lebrun F, Beck JT, Ito Y, Yardley D, Deleu I, Perez A, Bachelot T, Vittori L, Xu Z, Mukhopadhyay P, Lebwohl D, Hortobagyi GN. Ever-olimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med. 2012;366(6):520–529. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolfer A, Wittner BS, Irimia D, Flavin RJ, Lupien M, Gunawardane RN, Meyer CA, Lightcap ES, Tamayo P, Mesirov JP, Liu XS, Shioda T, Toner M, Loda M, Brown M, Brugge JS, Ramaswamy S. MYC regulation of a “poor-prognosis” metastatic cancer cell state. Proc Natl Acad Sci USA. 2010;107(8):3698–3703. doi: 10.1073/pnas.0914203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tzivion G, Gupta VS, Kaplun L, Balan V. 14-3-3 proteins as potential oncogenes. Semin Cancer Biol. 2006;16(3):203–213. doi: 10.1016/j.semcancer.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 55.Asselin-Labat ML, Shackleton M, Stingl J, Vaillant F, Forrest NC, Eaves CJ, Visvader JE, Lindeman GJ. Steroid hormone receptor status of mouse mammary stem cells. J Natl Cancer Inst. 2006;98(14):1011–1014. doi: 10.1093/jnci/djj267. [DOI] [PubMed] [Google Scholar]
- 56.Sleeman KE, Kendrick H, Robertson D, Isacke CM, Ashworth A, Smalley MJ. Dissociation of estrogen receptor expression and in vivo stem cell activity in the mammary gland. J Cell Biol. 2007;176(1):19–26. doi: 10.1083/jcb.200604065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haughian JM, Pinto MP, Harrell JC, Bliesner BS, Joensuu KM, Dye WW, Sartorius CA, Tan AC, Heikkila P, Perou CM, Horwitz KB. Maintenance of hormone responsiveness in luminal breast cancers by suppression of Notch. Proc Natl Acad Sci USA. 2012;109(8):2742–2747. doi: 10.1073/pnas.1106509108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fillmore CM, Gupta PB, Rudnick JA, Caballero S, Keller PJ, Lander ES, Kuperwasser C. Estrogen expands breast cancer stem-like cells through paracrine FGF/Tbx3 signaling. Proc Natl Acad Sci USA. 2010;107(50):21737–21742. doi: 10.1073/pnas.1007863107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell. 2012;21(3):283–296. doi: 10.1016/j.ccr.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Farmer P, Bonnefoi H, Becette V, Tubiana-Hulin M, Fumoleau P, Larsimont D, MacGrogan G, Bergh J, Cameron D, Goldstein D, Duss S, Delorenzi M, Iggo R. Identification of molecular apocrine breast tumours by microarray analysis. Cancer Treat Rev. 2005;31:S15. doi: 10.1038/sj.onc.1208561. [DOI] [PubMed] [Google Scholar]
- 61.Chlebowski RT, Anderson GL, Gass M, Lane DS, Aragaki AK, Kuller LH, Manson JE, Stefanick ML, Ockene J, Sarto GE, Johnson KC, Wactawski-Wende J, Ravdin PM, Schenken R, Hendrix SL, Rajkovic A, Rohan TE, Yasmeen S, Prentice RL. Estrogen plus progestin and breast cancer incidence and mortality in postmenopausal women. JAMA. 2010;304(15):1684–1692. doi: 10.1001/jama.2010.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jacobsen BM, Richer JK, Sartorius CA, Horwitz KB. Expression profiling of human breast cancers and gene regulation by progesterone receptors. J Mammary Gland Biol Neoplasia. 2003;8(3):257–268. doi: 10.1023/b:jomg.0000010028.48159.84. [DOI] [PubMed] [Google Scholar]
- 63.Graham JD, Mote PA, Salagame U, van Dijk JH, Balleine RL, Huschtscha LI, Reddel RR, Clarke CL. DNA replication licensing and progenitor numbers are increased by progesterone in normal human breast. Endocrinology. 2009;150(7):3318–3326. doi: 10.1210/en.2008-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clarke CL, Graham JD. Non-overlapping progesterone receptor cistromes contribute to cell-specific transcriptional outcomes. PLoS One. 2012;7(4):e35859. doi: 10.1371/journal.pone.0035859. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.