Abstract
Traditional read-across approaches typically rely on the chemical similarity principle to predict chemical toxicity; however, the accuracy of such predictions is often inadequate due to the underlying complex mechanisms of toxicity. Here we report on the development of a hazard classification and visualization method that draws upon both chemical structural similarity and comparisons of biological responses to chemicals measured in multiple short-term assays (”biological” similarity). The Chemical-Biological Read-Across (CBRA) approach infers each compound's toxicity from those of both chemical and biological analogs whose similarities are determined by the Tanimoto coefficient. Classification accuracy of CBRA was compared to that of classical RA and other methods using chemical descriptors alone, or in combination with biological data. Different types of adverse effects (hepatotoxicity, hepatocarcinogenicity, mutagenicity, and acute lethality) were classified using several biological data types (gene expression profiling and cytotoxicity screening). CBRA-based hazard classification exhibited consistently high external classification accuracy and applicability to diverse chemicals. Transparency of the CBRA approach is aided by the use of radial plots that show the relative contribution of analogous chemical and biological neighbors. Identification of both chemical and biological features that give rise to the high accuracy of CBRA-based toxicity prediction facilitates mechanistic interpretation of the models.
INTRODUCTION
The chemical similarity principle1, 2 posits that chemically similar compounds are likely to exhibit similar effects. Consequently, a variety of chemical similarity-based methods have been developed to predict chemical-induced responses from chemical structure alone. The chemical similarity principle provides the basis for both straightforward read-across analysis,3-7 and more complex machine learning-based approaches used in Quantitative Structure Activity-Relationship (QSAR) modeling.8-10
Chemical structure-based prediction methods face limitations, especially when the challenge is to accurately predict complex in vivo outcomes.8, 11 Data from in vitro screening of thousands of chemicals in hundreds of experimental systems provide additional biological activity information at molecular and cellular levels potentially useful for predictive toxicology modeling.12-14 Indeed, integration of chemical structural features and biological screening data provides important advantages over traditional QSAR modeling, such as improved prediction accuracy,15, 16 greater coverage of chemical space and a better interpretation of chemical and biological features.17
While QSAR modeling approaches have grown in popularity and complexity, end-users often show preference for simple and more transparent methods such as read-across conducted, e.g., using the OECD QSAR Toolbox (http://www.qsartoolbox.org/). The read-across methodology requires chemical (i.e., structure-based) similarity as a starting point. The objective of this method is to predict the toxicity behavior of a compound (i.e., produce an equivalent of a test result) by inferring from structurally similar chemicals with available toxicity data. Grouping and read-across of chemicals in a hazard and/or risk assessment context is well established and can be used to satisfy information requirements under Registration, Evaluation, Authorization and Restriction of Chemicals (REACH) regulation in the European Union. For example, more than 20% of high production volume chemicals submitted for the first REACH deadline relied on read-across for hazard information on a number of toxicity endpoints necessary for registration (http://echa.europa.eu/documents/10162/13639/alternatives_test_animals_2011_en.pdf). Under REACH, flexibility exists for how the analogues are selected; however, the read-across argument needs to be convincingly substantiated with scientifically credible justification. Thus, even though chemical structure-based read-across represents an alternative to standard animal-based tests, the inherited uncertainty of the prediction, combined with a lack of a standardized framework for its application by the decision-makers, creates a need to increase confidence in prediction and utilize visual aids for presenting evidence in a transparent manner.
Although chemical and biological factors are sometimes considered using a weight-of-evidence framework,4, 6, 7 these approaches are largely qualitative, not completely transparent, and may be prone to bias. Hence, there is a need to automate a read-across process that would combine both chemical and biological factors and yet, keep the process transparent for expert interrogation. To that end, we introduce a quantitative toxicity prediction approach combined with a visualization methodology, termed chemical-biological read-across (CBRA), that relies not only on inherent chemical properties (chemical descriptors), but also on biological profiles measured by short-term experimental assays (biological descriptors). A graphical display of the compound's classification, along with the identity of the neighbors and weights applied, is employed to increase transparency and interpretability. Using several data sets with short-term bioassay profiles, we demonstrate the advantages of CBRA over other methods that rely on biological and/or chemical descriptors alone.
MATERIALS AND METHODS
Data sets
Four data sets were used in this study (descriptor matrices and prediction endpoints of compounds are available as part of the Supporting Information). The first data set contained 127 compounds from the Toxicogenomics Project-Genomics Assisted Toxicity Evaluation system (TG-GATES18). The target property for prediction is sub-chronic hepatotoxicity previously modeled in Low et al17 based on liver histopathology and clinical chemistry findings over 28 days of repeat dosing.18 Gene expression data (biological features) and chemical descriptors (inherent chemical features) were processed as explained in Low et al.17 Briefly, of the 31,042 probes on the arrays, we removed those that were consistently not expressed or did not change their expression value across all compounds between treated vs. vehicle control groups. Next, 2,991 transcripts were selected that varied in their expression across all the compounds based on the following criteria: the largest change of any transcript over its untreated equivalent was over 1.5 fold and the smallest false discovery rate (Welch t-test) was less than 0.05. Then, transcripts with low variance (all, or all but one value is constant) were excluded and further, one of each pair of transcripts with high pairwise correlation (r2>0.9) chosen randomly, was removed; this left 2,923 transcript variables which were range scaled and used for model building. Models were built using supervised variable selection methods and five-fold external validation procedure each of the five subgroups (20% of the dataset) was systematically treated as an external set while the model was developed with the remaining four groups used as training set (80% of the dataset). In the end, we selected 85 unique transcripts (5 sets of 30 top-ranked transcripts per fold) to avoid selection bias introduced by a supervised selection process. Similar filtering procedures were applied prior to model building to chemical descriptors, i.e., prior to model building low variance (SD<10−6) and highly correlated descriptors (one of a pair of descriptors with pair-wise r2>0.9) were removed).
The second data set contained 132 compounds (DrugMatrix®, https://ntp.niehs.nih.gov/drugmatrix). The biological descriptors were the expression of 200 genes in the rat liver (5 day repeat dosing), that were selected as detailed in Natsoulis et al.20 The data for the prediction target, hepatocarcinogenicity, was compiled in a related study by Fielden et al19 using literature sources and the Carcinogenicity Potency Database (CPDB, http://www.epa.gov/ncct/dsstox/sdf_cpdbas.html).
The third and fourth data sets were from Lock et al21 in which 240 compounds were tested for cytotoxicity (intracellular ATP and caspase-3/7 apoptosis) in 84 lymphoblastoid cell lines with different genotypes. The 148 biological descriptors used here were the consolidated cytotoxicity profiles derived as detailed in Sedykh et al.15 Of the original 240 compounds, 185 compounds were assigned mutagenicity labels according to CCRIS (http://toxnet.nlm.nih.gov/cgi bin/sis/htmlgen?CCRIS) and 122 compounds were assigned rat oral LD50 according to ChemIDplus (http://chem.sis.nlm.nih.gov/chemidplus/). Because CCRIS provides detailed-level mutagenicity test results (including test strains, concentrations, and type of metabolic activation), the final mutagenicity assignments were determined using the protocol described in Mortelmans et al:22 mutagens must test positively in any one of the five standard Ames Salmonella test strains; non-mutagens, in contrast, must be consistently negative (in at least 4 out of 5 test strains). Additionally, when Ames Salmonella strains beyond the standard five were available, we defined mutagens as those tested positively in at least 20% of the strains and non-mutagens as those tested negatively in at least 80% of the strains regardless of metabolic activation. For the other classification endpoint, continuous rat oral LD50 values were split into two classes based on a threshold of 300 mg/kg, consistent with the threshold separating categories 1-3 and 4-6 in Globally Harmonized System of Classification and Labeling of Chemicals.23
Chemical descriptors and data processing
Chemicals utilized in all data sets underwent structural curation according to the procedures described in Fourches et al.24 This involved standardizing the molecular structures and removing salts, duplicates and problematic structures (e.g., metal-containing, molecular weight > 2000). Next, Dragon (v.5.5, Talete SRL, Milan, Italy) descriptors were computed for all chemicals.
After additional treatment of gene expression data (data sets 1 and 2, see above), all chemical and biological descriptors were range scaled to fall between 0 and 1. Furthermore, descriptors with low-variance (standard deviation <10−6) or one of any pair of descriptors with high intercorrelation (pairwise r2 >0.9) were removed.
Quantitative read-across methodology
For read-across, the predicted activity of a compound (Apred) was calculated using the following equation (Equation 1) from the similarity (Si) weighted aggregate of the activities Ai of k nearest neighbors.
| [1] |
The pairwise Tanimoto similarity, Si, between the molecule of interest (A) and its ith neighbor (B), was calculated from the Jaccard distance dJac [Equation 2, (Willett et al., 1998)] across descriptors x1,..., xp. For a set of range-scaled continuous descriptors, Tanimoto similarity is normalized between 0 and 1 with 1 corresponding to identical pairs.
| [2] |
The similarity-weighted aggregate in Equation 1 ensures that the activities of more similar neighbors are given higher weights when calculating the predicted activity.
For CBRA, compound activity was estimated from sets of neighbors in both the biological (bio) and chemical (chem) data (Equation 3).
| [3] |
Two sets of nearest neighbors (kbio biological neighbors in the biological space characterized by bioassay profiles and kchem neighbors in the chemical space characterized by Dragon descriptors) were used for the estimation of the toxicity for each test compound. Activities of nontoxic compounds were assigned “−1” while those of toxic compounds were assigned “+1”. The predicted classification threshold was set at zero such that compounds with negative predicted activity were considered as nontoxic and toxic otherwise.
Read-across was performed in two ways depending on how the maximum number of neighbors was determined: 1) by a similarity threshold (RA-sim), or 2) by a set value of k (RA-kNN). RA-sim included all neighbors with similarity greater than or equal to a similarity threshold set at 0, 0.6, 0.7, 0.8, or 0.9; RA-kNN included only k nearest neighbors (possible k values: integers from 1 to 5). Limiting the number of nearest neighbors by 5 is arbitrary; generally it should be understood that the selection of a large number of nearest neighbors would undermine the nearest neighbor selection principle so “5” is a threshold number we have been using typically in our implementation of the kNN QSAR method.25
Other models using both biological and chemical descriptors
In addition to CBRA, other approaches combining biological and chemical data for toxicity prediction were examined. First, biological and chemical descriptors were pooled together constituting a “hybrid” space which generated a single set of k nearest neighbors for each molecule (see Equation 1 where k=khybrid). Second, compounds’ activities were predicted by developing independent biological read-across and chemical read-across models and pooling the resulting predictions, essentially forming an ensemble model (Equation 4).
| [4] |
Model evaluation
External 5-fold cross validation
All models were evaluated using an external 5-fold cross validation. Briefly, each data set was randomly divided into five equal parts with the same toxic/nontoxic ratio before modeling. Each of the five parts was left out in turn to form an external set for validating the model developed on the remaining four parts (modeling set). For each target compound in the external set, neighbors were selected from the modeling set and not from the external set.
Internal 10-fold cross validation in read across kNN method
In RA-kNN, optimal kbio and kchem were selected from values between 1 and 5 by additional internal 10-fold cross-validation. Briefly, each modeling set was further divided according to a 10-fold cross-validation scheme, forming ten pairs of training and test sets. For each training set, we performed a grid search across all 25 possible pairs of kbio and kchem values and validated the models using the corresponding test set. For each pair of kbio and kchem values, the balanced accuracies across the ten test sets were averaged. The pair yielding the highest mean balanced accuracy was considered to be optimal and its kbio and kchem values were subsequently applied to the corresponding external validation set. Therefore, the five modeling sets resulted in five optimal models with various kbio and kchem values.
Y-randomization
The y-randomization test (randomization of the response) was performed to ensure that models were robust and not due to chance correlations. After random permutation of the activity labels in the modeling sets, models were rebuilt following the same workflow as described above. This protocol was repeated 30 times. Performance of the models generated from the permuted labels was compared to that of the models derived from the original data sets. Statistical significance of the difference in classification accuracy was determined with one-tailed one-sample t-test.
Prediction performance metrics
All metrics characterizing model performance [i.e., specificity, sensitivity, accuracy, balanced accuracy, and area under curve (AUC)] were obtained from external 5-fold cross-validation. Metric values close to 1 indicate high classification accuracy while 0.5 serves as the random baseline for binary classification. Specificity is the fraction of compounds predicted correctly within the nontoxic class; conversely, sensitivity is the fraction of compounds predicted correctly within the toxic class. Accuracy is the fraction of compounds predicted correctly in total. Balanced accuracy is the average of the rates correctly predicted within each class ((specificity + sensitivity)/2). AUC is the area under the receiver operating characteristic curve of sensitivity against (1-specificity). Thus, AUC is a function of sensitivity and specificity, providing an overall accuracy metric independent of a predefined activity threshold unlike the other prediction metrics which were calculated using a predefined activity threshold of zero (for activity values ranging between −1 to +1).
Coverage of the models is reported as the fraction of compounds in the external set that are within the applicability domain (AD) for which reliable predictions are expected to be obtained. In RA-sim, a target compound is within the AD if there exists at least one neighbor in the modeling set whose similarity is above the similarity threshold; in RA-kNN, a compound is within the AD if there exists at least one neighbor with a minimum similarity of 0.3. Standard errors were calculated by the bootstrap method26 using 1000 sampling trials.
Model interpretation
Identification of informative descriptors
Adapted from the local importance score used in the random forest method,27 we use a local importance score based on x-randomization to rank descriptors by their contribution to a target compound's predicted activity. X-randomization involves the random permutation of a descriptor x across the modeling set such that the descriptor's effect on the model before and after permutation is compared. This difference is expected to be more pronounced for important descriptors. Specifically, after permutation, the similarity between the target compound and its k neighbors (previously used for RA-kNN) will change. This resultant change in similarity is averaged over 99 random permutations to obtain the local importance score I(x,compound) which measures the descriptor x's contribution towards the target compound's predicted activity. This procedure was repeated for each descriptor per target compound. A high local importance score indicates that the descriptor is highly contributory to the target compound's prediction.
Visualization of nearest neighbors using radial plots
To visualize the information used to generate the predicted activity of a compound (Apred), the radial plot is used (see Figures 1-3 for examples). The central node marks the target compound. Surrounding it are nodes representing biological neighbors (left hand side) and chemical neighbors (right hand side), all colored according to their known toxicity assignments (red=toxic, black=nontoxic). The relative position of each neighbor from the central node (i.e., edge length) reflects the Jaccard distance (Equation 2) from the target compound. The nearest neighbors (shortest edges) are placed closest to the 12 o'clock position. Each radial plot displays all the neighbors relevant to a compound's prediction (i.e., kchem chemical neighbors and kbio biological neighbors above 0.3 Tanimoto similarity, consistent with the AD similarity threshold for RA-kNN). The algorithm for generating the radial plots was written in R Statistical Software (version 2.14; R Foundation for Statistical Computing, Vienna, Austria) and is available as part of the Supporting Information.
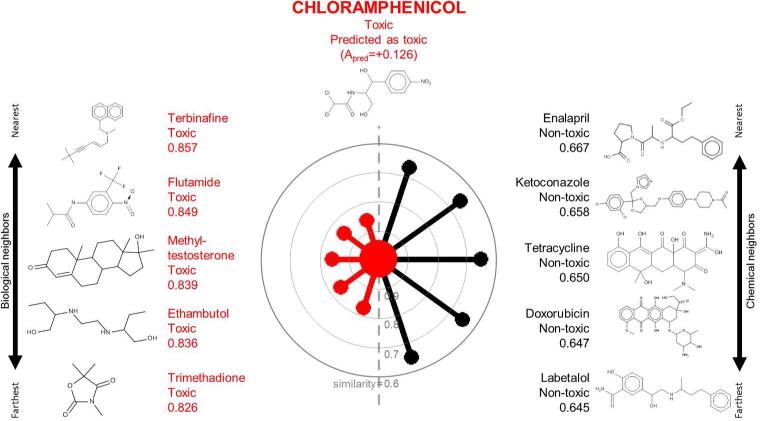
Figure 1.
A radial plot for chloramphenicol in the TG-GATES data set. The central node representing the target compound chloramphenicol is surrounded by biological neighbors (left hand side) and chemical neighbors (right hand side). Nearest (i.e., most similar) neighbors are placed at the top. Neighbors are positioned from the target compound at a distance proportional to the Jaccard distance. Edges and nodes are colored according to the known activity classification (black: nontoxic; red: toxic).
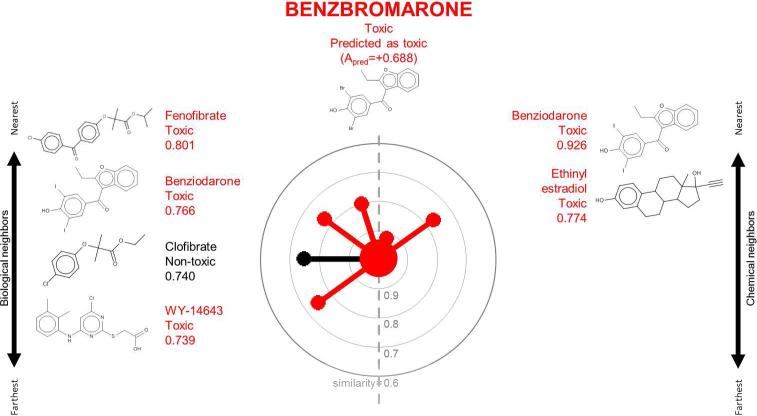
Figure 3.
A radial plot for benzbromarone in the TG-GATES data set. See legend to Figure 1 for details.
RESULTS
Visualisation of the chemical-biological read across classification
The premise of this study was to establish a transparent methodology for inferring a compound's potential toxicity from its biological and chemical analogs. Here, we use graphical means to illustrate how CBRA integrates information from both biological and chemical analogs of a compound to predict its toxicity. Because the relevant information (the analogs, their similarities and known toxicity assignments) can be communicated using the radial plot, CBRA offers a highly transparent and interpretive method for hazard classification.
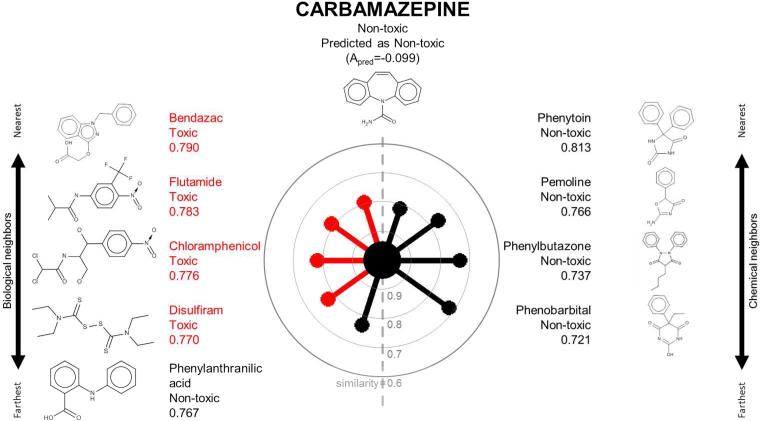
Figures 1-3 show the radial plots of three case study compounds, classifying them as hepatotoxic or not (see Methods for description of hepatotoxicity class designation) using both their biological (similar toxicogenomic profiles) and chemical (similar structures) analogs in the TG-GATES data set. Figure 1 depicts the basis for classifying chloramphenicol as “toxic”. The central node was colored red to denote chloramphenicol's known toxicity. On the left hand side, all five biological neighbors were labeled as toxic (red) and they are highly similar to chloramphenicol (similarities: 0.826-0.857). On the right hand side, the five closest chemical neighbors are nontoxic (black, similarities: 0.645-0.667). All neighbors’ activities are aggregated according to their similarity weights by CBRA (Equation 3), yielding Apred=+0.126, i.e., a “toxic” prediction concordant with the known “toxic” assignment. Figure 2 shows the opposite case in which the correct classification of carbamazepine (known to be nontoxic; Apred=−0.099) was due to its greater similarity with its chemical neighbors (similarities: 0.721-0.813). Figure 3 shows that benzbromarone's biological and chemical neighbors were mostly toxic (red), yielding concordant predictions (Apred=+0.688), in agreement with its known toxicity.
Figure 2.
A radial plot for carbamazepine in the TG-GATES data set. See legend to Figure 1 for details.
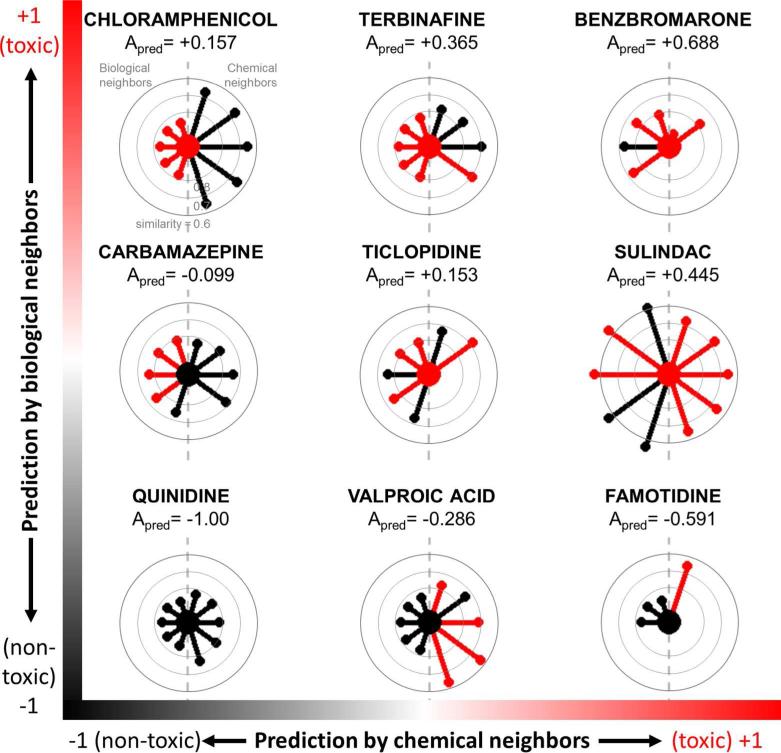
Figure 4 provides a visual comparison of radial plots for selected compounds from the TG-GATES data set that may facilitate expert judgment of each predicted classification. As with previous radial plots, each central node represents the compound of interest and is colored according to its experiment-derived toxicity (black=nontoxic, red=toxic). Radial plots were organized by the predicted activities using only chemical neighbors (horizontal axis) and those using only biological neighbors (vertical axis). As such, the compounds can be assessed by whether the chemical or biological neighbors had higher contribution to the final classification. The lower left corner (e.g., quinidine) is populated by radial plots with mostly nontoxic (black) neighbors while the upper right corner (e.g., benzbromarone) is filled by those with mostly toxic (red) neighbors. Conversely, other radial plots involve discordant predictions. Such radial plots are surrounded by neighbors of various toxicities (i.e., edges of various colors). Despite the discordance, the target compounds were still correctly predicted by CBRA because the activities of more similar neighbors (shorter edges) were given higher weight than those of less similar neighbors (longer edges). This simple visualization identifying neighbors based on an objective and standardized similarity metric such as the Tanimoto similarity allows users to assess the relevance of the neighbors and their contribution to the final prediction for every compound of interest.
Figure 4.
Radial plots of compounds in TG-GATES data set ordered by predictions based on chemical neighbors (horizontal axis) and biological neighbors (vertical axis). Along the horizontal axis, nontoxic predictions (black) by chemical neighbors are shown on the left and toxic predictions (red) are shown on the right. Along the vertical axis, nontoxic predictions (black) by biological neighbors are shown at the bottom while toxic predictions (red) predominate at the top. Compounds predicted with high confidence by near neighbors of similar toxicities are indicated by radial plots with short edges of the same color.
Model performance
RA-kNN vs RA-sim
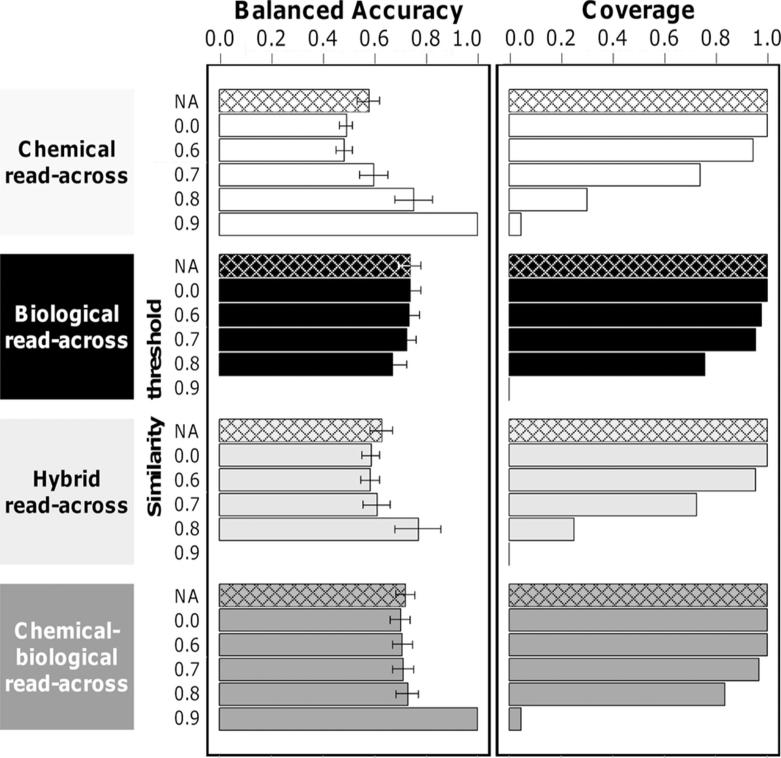
As read-across can be performed in two ways using either a similarity threshold (RA-sim) or a set value of k (RA-kNN), we first compared these two approaches on the TG-GATES data set (Figure 5 and Supporting Information Table 1). The first approach (RA-sim, solid filled bars in Figure 5) utilizing chemical descriptors only (“chemical read-across”, white solid bars), showed that higher balanced accuracy may be achieved by restricting chemical similarity thresholds; however, the cost of such improved accuracy is much reduced coverage. RA-sim using gene expression data only (“biological read-across”, black solid bars) had a higher balanced accuracy as compared to chemical read-across when all compounds were considered (i.e., 100% coverage). However, the accuracy of biological read-across did not increase markedly when more stringent similarity thresholds were applied. Finally, CBRA (dark gray solid fill) showed the highest balanced accuracy while being the least affected by the increasing similarity threshold. RA-sim utilizing hybrid descriptors resulting from pooling both chemical and biological descriptors (light gray solid bars) together, exhibited intermediate accuracy. The second read-across approach (RA-kNN, patterned fill) showed comparable or higher balanced accuracy across all four types of methods integrating chemical and biological descriptors when compared to RA-sim (solid fill). For this reason, RA-kNN was selected as the preferred algorithm for read-across.
Figure 5.
Performance of RA-sim (solid fill) and RA-kNN (patterned fill) models for the TG-GATES data set. RA-sim models were varied using various similarity thresholds for neighbor selection. Models based on various spaces are denoted by colors: chemical space (white), biological space (black), hybrid space (light gray), and chemical-biological spaces (dark gray).
Comparison of read-across in biological and/or chemical spaces
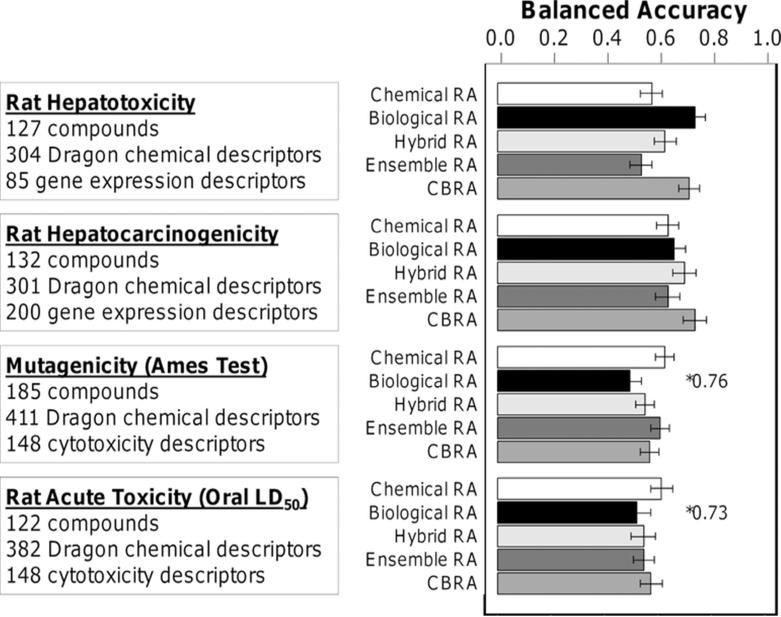
Next, we tested the performance of various read-across methods for different toxicity endpoints (liver toxicity and carcinogenicity, mutagenicity and acute lethality) and for different “biological descriptor” types (e.g. gene expression data from two different studies and in vitro cytotoxicity screening data). For this, we used both chemical and/or biological descriptors and the RA-kNN algorithm. In all four data sets, we applied: 1) chemical read-across (white bars); 2) biological read-across (black bars); 3) hybrid read-across by pooling chemical and biological descriptors (Equation 1, light gray bars); 4) ensemble read-across from pooling predictions from (1) and (2) (Equation 4, dark gray bars); and 5) CBRA (Equation 3, medium gray bars). Figure 6 and Supporting Information Tables 1 and 2 show a comparison of the performance of the various read-across methods in each data set.
Figure 6.
Performance of various RA-kNN models for four data sets. The models are colored as follows: chemical RA (white), biological RA (black), hybrid RA (from pooling chemical and biological descriptors, light gray), ensemble RA (consensus of chemical RA and biological RA, dark gray) and CBRA (gray). *Coverage values <0.95% are indicated on the chart.
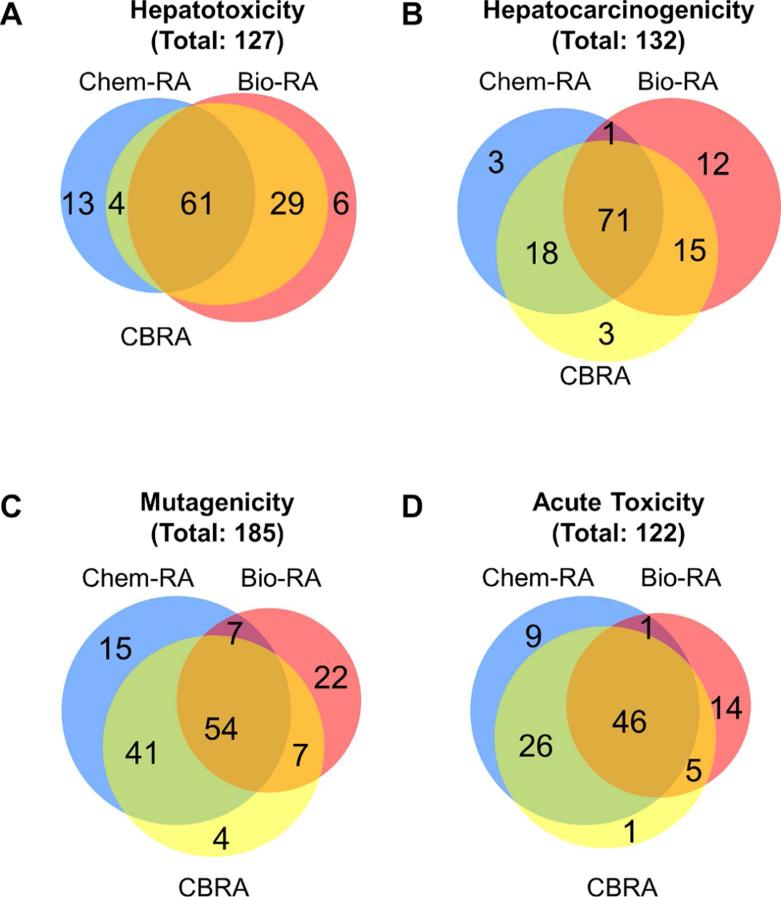
While chemical read-across (white bars) exhibited highest balanced accuracy of classification for some endpoints (i.e., mutagenicity and rat acute toxicity), biological read-across (black bars) had greater balanced accuracy for data sets 1 and 2 of rat hepatotoxicity and carcinogenicity, respectively, where biological descriptors represented gene expression data. However, biological read-across based on in vitro cytotoxicity screening data alone in data sets 3 and 4 exhibited the poorest classification accuracy (close to 50%), a result similar to that reported previously16. Importantly, the balanced accuracy of CBRA (medium gray bars) was consistently among the highest across all types of read-across models. Still, in three data sets (rat hepatotoxicity, mutagenicity, and rat acute toxicity), CBRA's performance, though among the best, did not surpass that of the simpler chemical read-across (white bars) or biological read-across (black bars). Similar outcomes were obtained when a comparison was made of the number of compounds correctly predicted by chemical read-across, biological read-across, and CBRA (Figure 7). Thus, we posit that given CBRA's consistently good performance, it should be employed where possible because it often offers the best chance of improving classification accuracy and model interpretation.
Figure 7.
Venn diagrams depicting the number of compounds correctly predicted by chemical RA (blue circles), biological RA (red circles) and CBRA (yellow circles). Analysis was conducted on four data sets as detailed in Methods. Four datasets were analyzed as detailed in Materials and Methods: rat hepatotoxicity (A), rat hepatocarcinogenicity (B), mutagenicity (C), and rat acute oral toxicity (D).
Y-randomization test
The prediction performance of all models presented in this study is given in Supporting Information Table 1. Most models built with real data significantly outperformed those generated by y-randomization (p-value < 0.05) and hence, were unlikely to be fitted by chance. There were two exceptions, however, i.e., models whose balanced accuracies were very poor (50%, 52%), i.e., indistinguishable from the random baseline of 50% (Supporting Information Table 1).
DISCUSSION
Improvements due to ensemble modeling and enhanced aggregation
Ensemble models have been shown to be more accurate than their constituent models.28 The CBRA approach, effectively an ensemble model, utilizes two distinct descriptor types, i.e., chemical and biological, to increase classification accuracy and uncover associations between different types of descriptors that may characterize each compound. Enhanced aggregation employed by CBRA ensures that the more similar neighbors have higher weights, regardless of whether they are biological or chemical neighbors.
Our results show that simple ensemble modeling, which gives equal weights to both chemical and biological models, is insufficient to achieve high classification accuracy, as illustrated by the modest results of the simple ensemble model (dark gray, Figure 6). Instead, enhanced aggregation employed by CBRA ensures that the more similar neighbors have higher weights, regardless of whether they are biological or chemical neighbors. This feature of CBRA is perhaps best exemplified by the following three case studies and their radial plots (Figures 1-3) illustrating how highly similar neighbors drive the prediction outcome. These case studies were selected to represent: 1) prediction driven by biological neighbors, 2) prediction driven by chemical neighbors, and 3) concordant predictions by biological and chemical neighbors.
Case study: Chloramphenicol (biological space-based prediction)
Chloramphenicol (Figure 1 and Supporting Information Table 3) is an anti-bacterial drug whose hepatotoxicity was linked to oxidative stress initiated by reactive metabolites29. Chloramphenicol increased the level of serum enzymes, as well as caused liver hypertrophy and necrosis in treated rats in the TG-GATES studies.18 There is greater similarity in toxicogenomics profiles between chloramphenicol and its several “toxic” biological neighbors (0.83-0.86) than that with its nontoxic chemical neighbors identified using inherent chemical properties (0.65-0.67).
The gene expression profiles of chloramphenicol and its highly similar biological neighbors across 30 genes showed a consistent gene signature (Supporting Information Figure 1A). In contrast, chloramphenicol and its chemical neighbors were characterized by relatively dissimilar descriptor profiles (Supporting Information Figure 1B). Several genes critical to the prediction of chloramphenicol's activity (Abce1, Tomm22 and Bmf) are known to be implicated in mitochondrial and cell cycle regulation processes (Supporting Information Table 4). Such deregulation is consistent with the known oxidative stress mediated hepatotoxicity of chloramphenicol. More importantly, this analysis indicates that statistically significant features elucidated by the CBRA model agree with the existing mechanistic knowledge of the compound toxicity. Thus, such model interpretation by CBRA may generate hypotheses about a compound's possible mechanism when only short-term assays are available.
Case study: Carbamazepine (chemical space-based prediction)
Carbamazepine is an anti-convulsant drug that acts on neuronal voltage-gated sodium channels. In the TG-GATES data set, it was classified as non-hepatotoxic in the rat. The case of carbamazepine (Figure 2) contrasts with that of chloramphenicol. Whereas biological RA afforded more accurate prediction than chemical RA for chloramphenicol, the opposite was found to be true for carbamazepine. Nonetheless, CBRA, in taking a similarity-weighted aggregate of the activities of kbio=5 biological neighbors and kchem=5 chemical neighbors, correctly predicted carbamazepine as nontoxic.
Carbamazepine and its highly similar chemical neighbors, in addition to sharing several chemical features, also exert similar pharmacological effects. Carbamazepine's nearest chemical neighbors (phenytoin, pemoline, phenylbutazone and phenobarbital) are also anti-convulsant drugs that share a tricyclic scaffold with a polar amide group in the middle (Figure 2). This common chemical motif is also responsible for their anti-convulsant effects. The associated pharmacophore involves an amide moiety in the middle and a side lipophilic aryl ring for interaction with the sodium channel in order to exert the drug's anti-convulsant effects.30, 31
Carbamazepine's nearest biological neighbors (bendazac, flutamide, chloramphenicol, disulfiram and phenylanthranilic acid) have few obvious commonalities. Their gene expression profiles across the 30 predictor genes showed considerable heterogeneity (Supporting Information Figure 1C). They also induce liver injury via different mechanisms and exhibit different histopathology and blood chemistry in the TG-GATES database. In this instance, biological similarity determined by 24-hour gene expression may not suffice to signal 28-day liver injury.
Case study: Benzbromarone (concordant chemical and biological predictions)
Benzbromarone is an anti-gout agent withdrawn from the market in 2003 due to hepatotoxicity concerns.32 In the TG-GATES data set, both its biological and chemical neighbors were predictive of its hepatotoxicity. Here, we show how CBRA can provide a prediction outcome bolstered by concordant predictions as well as postulate associations between the biological and chemical neighbors for subsequent analysis (Figure 3).
Benzbromarone-induced hepatotoxicity is attributed to disruptions in the mitochondrial β-oxidation of fatty acids, possibly mediated by peroxisome proliferator-activated receptor-alpha (PPARα) activation.33 It exhibits a gene expression profile similar to those of its biological neighbors (fenofibrate, benziodarone, clofibrate and WY-14643), all known PPARα activators. Furthermore, the genes important for predicting benzbromarone's activity (Bcs1l, Tomm20, Abce1 and LOC100360017, Supporting Information Table 4), relate to mitochondrial functions, indicative of the mitochondrial-mediated hepatotoxicity observed in benzbromarone.
In this case, benzbromarone's biological and chemical neighborhoods overlap and provide concordant predictions, possibly indicative of common biological-chemical associations between the two neighborhoods. The overlapping neighbor, benziodarone, exhibits PPAR activity similar to its biological neighbors and a lipophilic, planar structure similar to its chemical neighbors. In addition, the cross-talk between estrogen and other sex hormones and PPAR-mediated signaling is well recognized,34 which makes the association of its chemical analog, ethinyl estradiol, plausible. Hence, such cross-inference from one neighborhood to another can still provide useful clues for formulating hypotheses about biological-chemical associations, the strength of which are dependent on the extent of the overlapping neighborhoods. Furthermore, such analysis provides a novel way for the concurrent study of chemical and biological features and their underlying interactions.
Advantages and Limitations of CBRA
Read-across method is based on the expectation that chemically similar molecules should elicit similar biological responses. It is worth noting, however, that whichever way the chemical similarity is defined, it always has relative meaning; that is, the similarity search exercise identifies the most similar compounds in a given set of compounds, and not necessarily the most similar chemically feasible structures. Further, structure-activity relationship landscapes are known to be “rough,” with many molecules appearing chemically similar but nevertheless having rather different biological activities. The latter observation is best illustrated by the frequent presence of so called “activity cliffs”35 in many chemical data sets. It is for this reason that we observed different chemical and biological neighbors for many compounds across four data sets that were evaluated. Thus, we argue that it is critical to weigh in both chemical and biological neighbor's contribution in predicting every compound's activity. Such “enhanced” aggregation underlying CBRA exploits the complementary information inherent in both the chemical and biological neighbors to arrive at the most optimal prediction.
It is due to the relative power of similarity that CBRA may yield incorrect predictions by either or both set(s) of neighbors. In the latter case, neither biological nor chemical neighbors are instructive for model prediction or interpretation. In the former case, using either biological or chemical neighbors (instead of both as in CBRA) would yield better predictions for certain compounds. However, such accuracy provided by one set of neighbors may be limited to certain compounds and not the entire data set, as evident by the slightly smaller fraction of compounds correctly predicted by biological or chemical read-across models (50-77% overall accuracy, mean=65%, SD=9%) vs. that by CBRA (57-80% overall accuracy, mean=69%, SD=10%, Supporting Information Table 1). In other words, CBRA's predictions may be less accurate than either biological or chemical read-across for a minority of compounds but the CBRA approach succeeds overall showing higher accuracy on average as compared to other read-across methods.
As with most ensemble models, the decreased interpretability and increased computational cost may outweigh the gains in accuracy.36, 37 CBRA, like other instance-based learners, is better suited to data sets where variable selection has already been performed to reduce noise due to irrelevant variables.38 This variable selection step is necessary because, unlike models employing variable-specific weights, all variables in CBRA, including irrelevant ones, are given equal consideration when calculating similarity.
Despite certain limitations we argue that CBRA remains transparent and interpretable since neighbors of each compound can be easily identified and important variables (chemical features or specific genes) can be elicited as illustrated in our case studies. The important variables not only suggest mechanisms of action for closer toxicological examination but may also act as markers for a particular mechanism. Such marker profiles may help to uncover the toxicity mechanisms of new compounds whose toxicities were previously unknown. Additional studies may be undertaken to investigate if there are relationships between classes of chemicals and the selection of significant genes as defined by the CBRA method common to compounds in each chemical class.
In addition, the consideration of toxicokinetics may be essential for constructing a read-across argument, but CBRA does not yet take this into account in the present format. The similarity of toxicokinetic profiles, especially metabolism, is often considered before weighing the similarity in the mechanism of action. It is therefore reasonable to conclude that the inclusion of toxicokinetic descriptors may ultimately help in improving prediction accuracy and acceptance of read-across. This is especially relevant when in vitro data is used to predict in vivo endpoints.39 Indeed, CBRA may be extended to more than two spaces to accommodate toxicokinetic considerations although additional visualization techniques40 may be required.
Recommendations for chemical-biological modeling and its application in hazard assessment
Our experience and observations with using both chemical and biological descriptors suggest the following methodological implications for predicting chemical hazards. First, biological assays such as gene expression are expected to be more predictive than more simple assays measuring binary biological responses in vitro (e.g., binding/nonbinding to a target protein). Second, it is advantageous to consider biological variables relevant to the prediction target. The bioassays may be selected rationally according to biological pathways.41 Third, variable selection and rigorous model validation prevents selection bias towards overly optimistic models.39 Thus, careful modeling and validation according to OECD (Q)SAR principles8 is necessary to ensure robust and accurate models.42 Lastly, irrelevant variables may affect some classification methods more than others. For example, as explained earlier, instance-based methods including CBRA are more susceptible to irrelevant variables while others such as random forest can better tolerate noisy variables.27
In addition, our work has potential practical applications for the use of read-across under REACH and other regulatory initiatives. Read-across approach guidance under REACH (http://www.reachonline.eu/REACH/EN/REACH_EN/articleXI.html) stipulates that “the group concept requires that physicochemical properties, human health effects and environmental effects or environmental fate may be predicted from data for reference substance(s) within the group by interpolation to other substances in the group.” In this sense, even though typical interpretation of “similarity” is focused on a common functional group, or common precursors/breakdown products, the biological data provides additional confidence for the consistent pattern in the “potency” of the compounds to elicit toxicity across the category. CBRA's transparency in displaying the compounds selected for read-across allows users to examine the suitability of the neighbors before relying on them for subsequent prediction. As such, CBRA satisfies the requirement for “adequate and reliable documentation of the applied method” by providing a defined process for analog selection and prediction, as well as enabling visual interpretation of the similarities across several data domains.
Conclusions
Given the complex biological processes mediating chemical toxicity, hazard prediction will benefit from the inclusion of biological data in addition to chemical information. Previously, we have demonstrated that hybrid models of hepatotoxicity pooling biological (gene expression profiles) and chemical features could not achieve higher accuracy than biological models.17 Herein, we have developed CBRA as an alternative method combining the same biological and chemical descriptors and demonstrated that its balanced accuracy was among the best when compared with other models using biological and/or chemical descriptors. This result was also replicated in three other data sets.
One reason for the success of CBRA is that, as a local modeling technique incorporating relative similarity weighting scheme, it relies on objective metrics to predict toxicity class of a compound when predictions made with chemical vs. biological neighbors disagree. Additionally, since prediction is based on a small number of similar compounds, both the modeling process and its outcome are transparent. All neighbors are displayed using star plots that also show relative similarities between such neighbors and the target molecule such that users can examine the arguments made by the model when assigning a specific call (toxic or non-toxic) to the target compound. CBRA also highlights key biological and chemical features for further mechanistic interpretation. In summary, CBRA represents a novel hybrid read-across method that is both predictive and interpretable. It combines the simplicity and transparency of read-across methods with the benefits afforded by more sophisticated techniques such as ensemble modeling and instance-based learning while incorporating modern diverse data streams, making CBRA a potentially appealing tool for chemical hazard assessment.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Takeki Uehara for facilitating access the TG-GATES data set.
FUNDING SOURCES
The work was supported in part by grants from NIH (GM076059, GM066940) and EPA (RD83272001, RD83382501).
ABBREVIATIONS
- AD
applicability domain
- ADME
absorption, distribution, metabolism, excretion
- AUC
area under curve
- CBRA
chemical-biological read-across
- kNN
k nearest neighbors
- PPARα
peroxisome proliferator activated receptor alpha
- QSAR
quantitative structure activity relationship
Footnotes
SUPPORTING INFORMATION AVAILABLE:
Data sets, code to generate radial plots, prediction performance of models, prediction outcomes and importance rank of chemical descriptors and biological assays. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare that there are no conflicts of interest.
REFERENCES
- 1.Johnson MA, Maggiora GM. Concepts and Applications of Molecular Similarity. Wiley-Interscience; New York, NY: 1990. [Google Scholar]
- 2.Willett P, Barnard JM, Downs GM. Chemical similarity searching. J. Chem. Inf. Comput. Sci. 1998;38:983–996. [Google Scholar]
- 3.Enoch SJ, Cronin MT, Schultz TW, Madden JC. Quantitative and mechanistic read across for predicting the skin sensitization potential of alkenes acting via Michael addition. Chem. Res. Toxicol. 2008;21:513–520. doi: 10.1021/tx700322g. [DOI] [PubMed] [Google Scholar]
- 4.Hewitt M, Ellison CM, Enoch SJ, Madden JC, Cronin MT. Integrating (Q)SAR models, expert systems and read-across approaches for the prediction of developmental toxicity. Reprod. Toxicol. 2010;30:147–160. doi: 10.1016/j.reprotox.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Schuurmann G, Ebert RU, Kuhne R. Quantitative read across for predicting the acute fish toxicity of organic compounds. Environ. Sci. Technol. 2011;45:4616–4622. doi: 10.1021/es200361r. [DOI] [PubMed] [Google Scholar]
- 6.Wang NC, Jay Zhao Q, Wesselkamper SC, Lambert JC, Petersen D, Hess-Wilson JK. Application of computational toxicological approaches in human health risk assessment. I. A tiered surrogate approach. Regul. Toxicol. Pharmacol. 2012;63:10–19. doi: 10.1016/j.yrtph.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Wu S, Blackburn K, Amburgey J, Jaworska J, Federle T. A framework for using structural, reactivity, metabolic and physicochemical similarity to evaluate the suitability of analogs for SAR-based toxicological assessments. Regul.Toxicol. Pharmacol. 2010;56:67–81. doi: 10.1016/j.yrtph.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Gleeson MP, Modi S, Bender A, Robinson RL, Kirchmair J, Promkatkaew M, Hannongbua S, Glen RC. The challenges involved in modeling toxicity data in silico: a review. Curr. Pharm. Des. 2012;18:1266–1291. doi: 10.2174/138161212799436359. [DOI] [PubMed] [Google Scholar]
- 9.Voutchkova AM, Osimitz TG, Anastas PT. Toward a comprehensive molecular design framework for reduced hazard. Chem. Rev. 2010;110:5845–5882. doi: 10.1021/cr9003105. [DOI] [PubMed] [Google Scholar]
- 10.Zvinavashe E, Murk AJ, Rietjens IM. Promises and pitfalls of quantitative structure activity relationship approaches for predicting metabolism and toxicity. Chem. Res. Toxicol. 2008;21:2229–2236. doi: 10.1021/tx800252e. [DOI] [PubMed] [Google Scholar]
- 11.Nikolova N, Jaworska J. Approaches to measure chemical similarity – a review. QSAR Comb. Sci. 2003;22:1006–1026. [Google Scholar]
- 12.Judson RS, Martin MT, Egeghy P, Gangwal S, Reif DM, Kothiya P, Wolf M, Cathey T, Transue T, Smith D, Vail J, Frame A, Mosher S, Cohen Hubal EA, Richard AM. Aggregating Data for Computational Toxicology Applications: The U.S. Environmental Protection Agency (EPA) Aggregated Computational Toxicology Resource (ACToR) System. Int. J. Mol. Sci. 2012;13:1805–1831. doi: 10.3390/ijms13021805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rusyn I, Sedykh A, Low Y, Guyton KZ, Tropsha A. Predictive modeling of chemical hazard by integrating numerical descriptors of chemical structures and short-term toxicity assay data. Toxicol. Sci. 2012;127:1–9. doi: 10.1093/toxsci/kfs095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valerio LG, Jr., Choudhuri S. Chemoinformatics and chemical genomics: potential utility of in silico methods. J. Appl. Toxicol. 2012;32:880–889. doi: 10.1002/jat.2804. [DOI] [PubMed] [Google Scholar]
- 15.Sedykh A, Zhu H, Tang H, Zhang L, Richard A, Rusyn I, Tropsha A. Use of in vitro HTS-derived concentration response data as biological descriptors improves the accuracy of QSAR models of in vivo toxicity. Environ. Health Perspect. 2011;119:364–370. doi: 10.1289/ehp.1002476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu H, Rusyn I, Richard A, Tropsha A. Use of cell viability assay data improves the prediction accuracy of conventional quantitative structure-activity relationship models of animal carcinogenicity. Environ. Health Perspect. 2008;116:506–513. doi: 10.1289/ehp.10573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Low Y, Uehara T, Minowa Y, Yamada H, Ohno Y, Urushidani T, Sedykh A, Muratov E, Kuz'min V, Fourches D, Zhu H, Rusyn I, Tropsha A. Predicting drug-induced hepatotoxicity using QSAR and toxicogenomics approaches. Chem. Res. Toxicol. 2011;24:1251–1262. doi: 10.1021/tx200148a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uehara T, Ono A, Maruyama T, Kato I, Yamada H, Ohno Y, Urushidani T. The Japanese toxicogenomics project: application of toxicogenomics. Mol. Nutr. Food Res. 2010;54:218–227. doi: 10.1002/mnfr.200900169. [DOI] [PubMed] [Google Scholar]
- 19.Fielden MR, Brennan R, Gollub J. A gene expression biomarker provides early prediction and mechanistic assessment of hepatic tumor induction by nongenotoxic chemicals. Toxicol. Sci. 2007;99:90–100. doi: 10.1093/toxsci/kfm156. [DOI] [PubMed] [Google Scholar]
- 20.Natsoulis G, Pearson CI, Gollub J, B PE, Ferng J, Nair R, Idury R, Lee MD, Fielden MR, Brennan RJ, Roter AH, Jarnagin K. The liver pharmacological and xenobiotic gene response repertoire. Mol. Syst. Biol. 2008;4:175. doi: 10.1038/msb.2008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lock EF, Abdo N, Huang R, Xia M, Kosyk O, O'Shea SH, Zhou YH, Sedykh A, Tropsha A, Austin CP, Tice RR, Wright FA, Rusyn I. Quantitative high-throughput screening for chemical toxicity in a population based in vitro model. Toxicol. Sci. 2012;126:578–588. doi: 10.1093/toxsci/kfs023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mortelmans K, Zeiger E. The Ames Salmonella/microsome mutagenicity assay. Mutat. Res. 2000;455:29–60. doi: 10.1016/s0027-5107(00)00064-6. [DOI] [PubMed] [Google Scholar]
- 23.United Nations Economic Commission for Europe . Globally Harmonized System of Classification and Labelling of Chemicals (GHS) United Nations Economic Commission for Europe; Brussels: 2009. Health hazards; pp. 109–111. [Google Scholar]
- 24.Fourches D, Muratov E, Tropsha A. Trust, but verify: on the importance of chemical structure curation in cheminformatics and QSAR modeling research. J. Chem. Inf. Model. 2010;50:1189–1204. doi: 10.1021/ci100176x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng W, Tropsha A. Novel variable selection quantitative structure--property relationship approach based on the k-nearest neighbor principle. J. Chem. Inf. Comput. Sci. 2000;40:185–194. doi: 10.1021/ci980033m. [DOI] [PubMed] [Google Scholar]
- 26.Efron B, Tibshirani R. Bootstrap methods for standard errors, confidence intervals, and other measures of statistical accuracy. Statist. Sci. 1986;1:54–75. [Google Scholar]
- 27.Breiman L. Random forests. Machine Learning J. 2001;45:5–32. [Google Scholar]
- 28.Zhu H, Tropsha A, Fourches D, Varnek A, Papa E, Gramatica P, Oberg T, Dao P, Cherkasov A, Tetko IV. Combinatorial QSAR modeling of chemical toxicants tested against Tetrahymena pyriformis. J. Chem. Inf. Model. 2008;48:766–784. doi: 10.1021/ci700443v. [DOI] [PubMed] [Google Scholar]
- 29.Farombi EO, Adaramoye OA, Emerole GO. Influence of chloramphenicol on rat hepatic microsomal components and biomarkers of oxidative stress: protective role of antioxidants. Pharmacol. Toxicol. 2002;91:129–134. doi: 10.1034/j.1600-0773.2002.910307.x. [DOI] [PubMed] [Google Scholar]
- 30.Lipkind GM, Fozzard HA. Molecular model of anticonvulsant drug binding to the voltage gated sodium channel inner pore. Mol. Pharmacol. 2010;78:631–638. doi: 10.1124/mol.110.064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sridhar SK, Pandeya SN, Stables JP, Ramesh A. Anticonvulsant activity of hydrazones, Schiff and Mannich bases of isatin derivatives. Eur. J. Pharm. Sci. 2002;16:129–132. doi: 10.1016/s0928-0987(02)00077-5. [DOI] [PubMed] [Google Scholar]
- 32.Lee MH, Graham GG, Williams KM, Day RO. A benefit-risk assessment of benzbromarone in the treatment of gout. Was its withdrawal from the market in the best interest of patients? Drug Saf. 2008;31:643–665. doi: 10.2165/00002018-200831080-00002. [DOI] [PubMed] [Google Scholar]
- 33.Kunishima C, Inoue I, Oikawa T, Nakajima H, Komoda T, Katayama S. Activating effect of benzbromarone, a uricosuric drug, on peroxisome proliferator-activated receptors. PPAR Res. 2007;2007:36092. doi: 10.1155/2007/36092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Komar CM. Peroxisome proliferator-activated receptors (PPARs) and ovarian function-implications for regulating steroidogenesis, differentiation, and tissue remodeling. Reprod. Biol. Endocrinol. 2005;3:41. doi: 10.1186/1477-7827-3-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maggiora GM. On outliers and activity cliffs--why QSAR often disappoints. J. Chem. Inf. Model. 2006;46:1535. doi: 10.1021/ci060117s. [DOI] [PubMed] [Google Scholar]
- 36.Elder JF. The generalization paradox of ensembles. J. Comp. Graph. Stat. 2003;12:853–864. [Google Scholar]
- 37.Hewitt M, Cronin MT, Madden JC, Rowe PH, Johnson C, Obi A, Enoch SJ. Consensus QSAR models: do the benefits outweigh the complexity? J. Chem. Inf. Model. 2007;47:1460–1468. doi: 10.1021/ci700016d. [DOI] [PubMed] [Google Scholar]
- 38.Aha DW, Kibler D, Albert MK. Instance-based learning algorithms. Mach. Learn. 1991;6:37–66. [Google Scholar]
- 39.Thomas RS, Black MB, Li L, Healy E, Chu TM, Bao W, Andersen ME, Wolfinger RD. A comprehensive statistical analysis of predicting in vivo hazard using high-throughput in vitro screening. Toxicol. Sci. 2012;128:398–417. doi: 10.1093/toxsci/kfs159. [DOI] [PubMed] [Google Scholar]
- 40.Reif DM, Sypa M, Lock EF, Wright FA, Wilson A, Cathey T, Judson RR, Rusyn I. ToxPi GUI: an interactive visualization tool for transparent integration of data from diverse sources of evidence. Bioinformatics. 2013;29:402–403. doi: 10.1093/bioinformatics/bts686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Judson RS, Kavlock RJ, Setzer RW, Cohen Hubal EA, Martin MT, Knudsen TB, Houck KA, Thomas RS, Wetmore BA, Dix DJ. Estimating Toxicity-Related Biological Pathway Altering Doses for High-Throughput Chemical Risk Assessment. Chem. Res. Toxicol. 2011;24:451–462. doi: 10.1021/tx100428e. [DOI] [PubMed] [Google Scholar]
- 42.Tropsha A. Best practices for QSAR model development, validation, and exploitation. Mol. Inf. 2010;29:476–488. doi: 10.1002/minf.201000061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.