Abstract
Background
RAS-coupled MAPK and PI3K pathways play a fundamental role in thyroid tumorigenesis, and classical genetic alterations upregulating these pathways are well characterized. We hypothesized that gene abnormality of negative modulators of these signaling pathways might be an important alternative genetic background for thyroid cancer.
Methods
By examining gene expression patterns of negative modulators of RAS signaling, we attempted to identify potential tumor suppressor genes. We then analyzed the methylation and mutation patterns of the identified gene in 101 thyroid tumors and tested its functions in vitro and in vivo to establish the tumor suppressor role in thyroid cancer.
Results
Among 13 negative modulators of the RAS pathway screened, RASAL1, encoding a RAS GTPase-activating protein, was frequently hypermethylated in thyroid cancers, which was coupled to its silencing in thyroid cancer cells. We also, for the first time, identified the presence of RASAL1 mutations, with a prevalence of 4.88% (n = 2 of 41) in follicular thyroid cancer (FTC) and 16.67% (n = 5 of 30) in anaplastic thyroid cancer (ATC). RASAL1 displayed MAPK- and PI3K-suppressing and thyroid tumor–suppressing activities, which were all impaired by the mutations. Hypermethylation and mutations of RASAL1 were mutually exclusive and collectively found in zero of 20 benign thyroid tumors, 3.22% (n = 1 of 31) of papillary thyroid cancers, 31.70% (n = 13 of 41) of FTCs, and 33.33% (n = 10 of 30) of ATCs. A rate of 20.83% (n = 5 of 24) of tumors carrying RASAL1 mutation or methylation at high levels (>50%) vs 44.16% (n = 34 of 77) of tumors carrying no RASAL1 mutation or methylation at low levels (< 50%) harbored any of the classical mutations (two-sided P = .02, Fisher exact test) in RAS, BRAF, PTEN, and PIK3CA genes in the MAPK and PI3K pathways, revealing a largely mutually exclusive relationship.
Conclusions
We identified RASAL1 as a major tumor suppressor gene that is frequently inactivated by hypermethylation and mutations, providing a new alternative genetic background for thyroid cancer, particularly FTC and ATC.
RAS proteins control many cellular processes, including cell migration, proliferation, differentiation, and survival (1). Extracellular signals are transduced intracellularly through the RAS pathway, and the amplitude and duration of this signaling are critical to appropriate biological responses of the cell. The normal signaling of this pathway is delicately regulated by various modulators that function as enhancers or inhibitors of RAS and its downstream pathways. The main pathways downstream of RAS in human tumorigenesis include RAF→ MEK → ERK (MAPK) and PI3K→AKT→mTOR (PI3K) pathways. The RAS GTPase-activating protein (RasGAP) RASAL1 that catalyzes RAS inactivation (2–5) and other negative modulators of the RAS signaling system (6–17) are presumably tumor suppressor genes (TSGs), and their aberrant silencing has been observed in various cancers. Germline genetic alterations in some of these negative modulators, such as NF1, SPRED1, PTEN, LKB1, TSC1, and TSC2, are associated with inherited cancer syndromes and development of a variety of cancers (18,19). Thus, like the genetic activation of the classical components of RAS signaling–coupled pathways, such as RAS and BRAF in the MAPK pathway and RAS and PIK3CA in the PI3K pathway, genetic or epigenetic inactivation of the negative modulators of these pathways is presumably an important alternative mechanism in human tumorigenesis.
Follicular cell–derived thyroid cancer is a common endocrine malignancy with a rapidly rising incidence globally (20,21). Thyroid cancer histologically consists mainly of papillary thyroid cancer (PTC), follicular thyroid cancer (FTC), and anaplastic thyroid cancer (ATC) (22). RAS-coupled MAPK and PI3K pathways play a fundamental role in thyroid tumorigenesis (23). Genetic alterations are the fundamental driving force for the aberrant activation of these two pathways in thyroid cancer and occur in a thyroid tumor type–predilected manner. BRAF mutation that activates the MAPK pathway is most common in PTC (24,25), whereas RAS, PTEN, and PIK3CA mutations that activate the PI3K pathway are most common in FTC and ATC (26). These classical genetic alterations activating the MAPK and PI3K pathways account for about 65% to 70% of thyroid cancers, leaving 30% to 35% of thyroid cancers with no known genetic background responsible for aberrant signaling of the RAS-coupled MAPK and PI3K pathways (23).
In this study, we investigated alternative RAS signaling–related genes that may play an important role in thyroid tumorigenesis through the RAS-coupled MAPK and PI3K pathways. We focused on negative modulators of RAS signaling that are potential TSGs and, for the first time, specifically identified RASAL1 as a major TSG with common epigenetic and genetic alterations in thyroid cancer.
Methods
Human Thyroid Tissues and Cell Lines
The study included 101 human thyroid tumors. Protocols using human thyroid tumor tissues were approved by the institutional review board of the Johns Hopkins Medical Institution. Written informed consent was provided from patients where required. All follicular thyroid cancer tumors used in this study were conventional type. Some genomic DNA samples were from our previous studies (27).
Thyroid cancer cell lines and their genetic backgrounds are summarized in Supplementary Table 1 (available online), which we gratefully received as acknowledged previously (28). They were all grown in Roswell Park Memorial Institute 1640 medium with 10% fetal bovine serum, except for FTC133, which was cultured in Dulbecco’s modified Eagle’s medium/Ham’s F-12 medium as described previously (28).
Methylation-Specific Polymerase Chain Reaction (MSP)
DNA bisulfite treatment was performed as previously described (29). See Supplementary Table 2 (available online) for MSP primer sequences. The relative level of methylation was normalized using the ratio obtained from the values of the gene of interest over the values of the internal reference gene (β-actin). The relative methylation level was calculated by the formula [M/(M+U)] × 100%, in which M and U represent the density of the methylation and unmethylation band, respectively. Results represent the percentage of allelic methylation of the tumor.
Mutational Analysis by Genomic Sequencing
Exons 2 to 22 that span the whole coding region of RASAL1 were polymerase chain reaction (PCR) amplified, and the sequences were analyzed by Sanger sequencing (see Supplementary Table 2, available online, for primer sequences). The sequencing and primers for mutation analysis of BRAF, PTEN, PIK3CA, N2-RAS, H2-RAS, and K1-RAS genes are as described previously (27).
Tumor Formation in Nude Mice
The animals’ care was in accord with the guidelines of Johns Hopkins Medical Institutions. K1 cells stably transfected with doxcycycline-inducible wild-type RASAL1 or RASAL1 mutants constructed as described above were injected (1×107 cells/mouse) subcutaneously into flanks of female nude mice at the age of 4 weeks (Harlan Sprague Dawley, Indianapolis, IN). Mice were fed vehicle or 0.2mg/mL doxycycline, which was stored in a 0.5% sucrose solution in light-proof bottles, beginning when the tumor grew to a size of 2mm at 1.5 weeks after the inoculation of cells. Tumor size was measured, and volume was calculated using the formula (length × width2)/2. After 5 weeks, mice were sacrificed, and tumors were surgically removed, photographed, and weighed.
Detailed methods for Western blot analysis, reverse-transcription (RT)–PCR, plasmids and cell tranfection, colony-formation assays, and RAS pull-down assays can be found in the Supplementary Methods (available online).
Statistical Analysis
For continuous data with a normal distribution or an abnormal distribution, Student t test and Wilcoxon Mann–Whitney test was used to analyze the statistical significance of differences between two groups, respectively. For categorical data, Fisher exact test was used. All reported P values were two-sided, and P less than .05 was considered statistically significant.
Results
Screening for Potential Candidate TSGs in Thyroid Cancer Cells
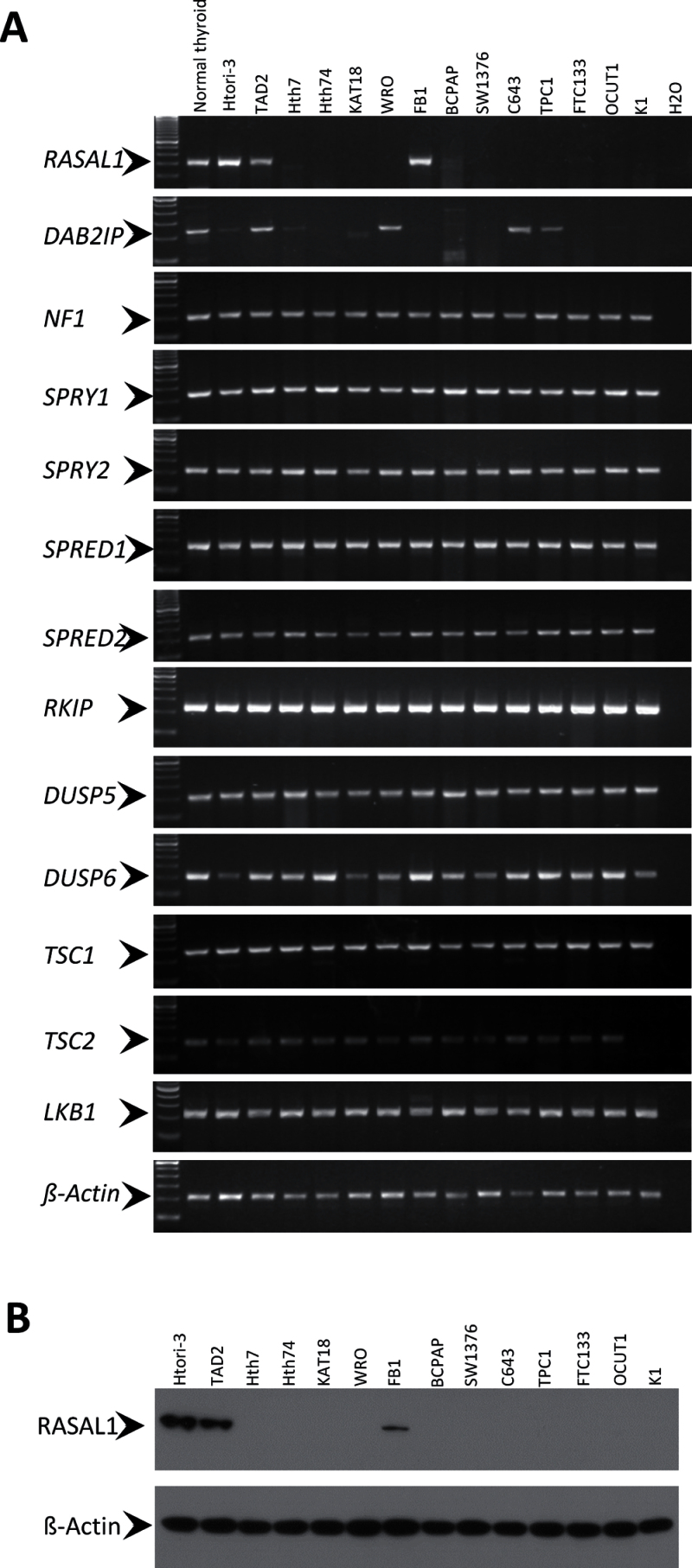
A common feature of TSGs in human cancer is aberrant silencing, so we initially screened for potential candidate TSGs involved in the modulation of RAS signaling by examining the expression pattern of the negative modulators of the pathway. RASSF1A and PTEN are two negative modulators that are known to be downregulated in thyroid cancer (26,30,31). Here we examined the expression of another 13 negative modulators of the RAS signaling pathway that are candidate human TSGs (Supplementary Figure 1, available online) by RT-PCR analysis of a cDNA panel derived from 12 human thyroid cancer cell lines (Supplementary Table 1, available online). Among these, the expression of two RasGAP genes, RASAL1 and DAB2IP, was silenced in 11 and seven of the 12 thyroid cancer cell lines, respectively, whereas they were abundantly expressed in a normal human thyroid tissue pool derived from five persons and two immortalized normal human thyroid epithelial cell lines (Htori-3 and TAD2) (Figure 1A). In contrast, the genes for the remaining 11 negative modulators of the RAS signaling pathway (NF1, SPRY1, SPRY2, SPRED1, SPRED2, RKIP, DUSP5, DUSP6, TSC1, TSC2, LKB1) were expressed in all of the thyroid cancer cell lines, except for K1 cells, in which TSC2 was not expressed. We were particularly interested in RASAL1 because its mRNA expression was completely lost in all of the thyroid cancer cell lines except for FB1 (Figure 1A), which was confirmed by the corresponding loss of the expression of RASAL1 protein (Figure 1B). Like mRNA, normal expression of RASAL1 protein was also seen in normal thyroid cell lines Htori-3 and TAD2 and thyroid cancer cell line FB1.
Figure 1.
Expression of candidate tumor-suppressor genes that negatively regulate RAS signaling in thyroid cancer cell lines. A) Reverse-transcription analysis of a cDNA panel derived from 12 human thyroid cancer cell lines as indicated. The cDNA from normal human thyroid tissue and two immortalized normal human thyroid epithelial cell lines (Htori-3 and TAD2) were used as positive controls. ß-Actin cDNA was used to show the integrity and quantity of cDNA samples. The primer sequences are presented in Supplementary Table 2 (available online). B) Detection of RASAL1 protein in thyroid cell lines by western blotting assay. ß-Actin protein level was used to show the quantity and integrity of protein samples.
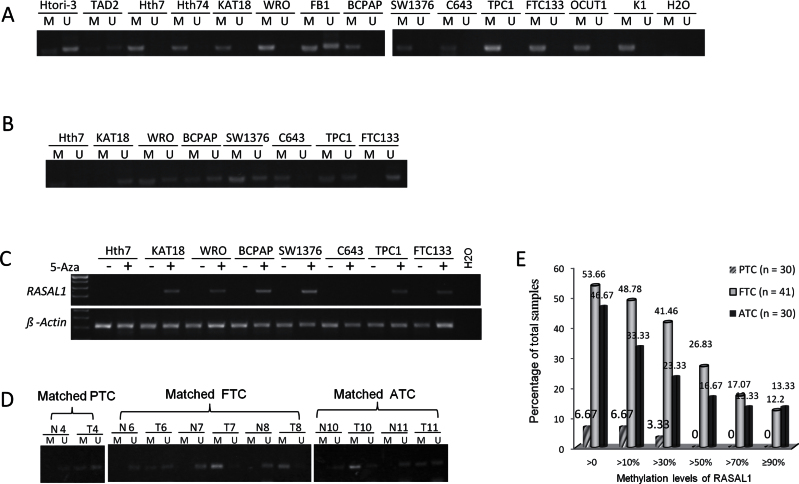
Another common feature of TSGs is their aberrant hypermethylation in the promoter and 5’ regions, as exemplified by PTEN and RASSF1A in thyroid cancer (30,31). We found that the RASAL1promoter region was completely methylated in 11 thyroid cancer cell lines that lost the expression of RASAL1, whereas it was only partially methylated in the two normal thyroid cell lines and cancer cell line FB1 that expressed RASAL1 (Figure 2A). Treatment of cells with demethylating agent 5-aza-2-deoxycytidine (5-Aza) induced demethylation of RASAL1 (Figure 2B) and restored its expression (Figure 2C) in six of the eight thyroid cancer cell lines tested, suggesting that promoter methylation was a mechanism for the loss of RASAL1 expression in thyroid cancer cells.
Figure 2.
Hypermethylation of RASAL1 and its re-expression induced by demethylation in thyroid cancer cells. A) Methylation-specific polymerase chain reaction (MSP) analysis revealed hypermethylation of the promoter area of RASAL1 in most of the 14 thyroid cell lines. B) Partial demethylation of RASAL1 revealed by MSP analysis in thyroid cancer cell lines after 5-Aza-dC treatment. After cells were treated with 5 µM 5-Aza-dC for 24 hours, RNA and genomic DNA were extracted and used for reverse-transcription polymerase chain reaction (RT-PCR) and MSP analysis, respectively. C) RT-PCR analysis of RASAL1 expression in thyroid cancer cell lines treated with or without demethylating agent 5-Aza-dC. D) MSP analysis of RASAL1 in matched normal and cancerous thyroid tissues. Six matched sample pairs are presented that had different methylation patterns between normal and matched tumor tissues. E) RASAL1 methylation levels represented by the indicated allelic methylation percentages (x-axis) and their frequencies (y-axis) in different types of thyroid cancers. ATC = anaplastic thyroid cancer; FTC = follicular thyroid cancer; PTC = papillary thyroid cancer.
We next investigated the methylation status of RASAL1 in thyroid tumors. We first examined 13 primary thyroid cancers with matched normal thyroid tissues. We found RASAL1 methylation in six thyroid cancers (n = 1 of 5 PTCs, n = 3 of 4 FTCs, and n = 2 of 4 ATCs) but no methylation in any of the matched normal thyroid tissues (Figure 2D). MSP analysis of an additional 88 thyroid tumor samples showed that RASAL1 was hypermethylated predominantly in FTC and ATC no matter what methylation levels were used as cutoff values (Figure 2E). If a level of greater than 50% allelic methylation was used, which means that at least in some cells of the tumor both alleles of RASAL1 are methylated, 26.83% (n = 11 of 41) of FTCs and 16.67% (n = 5 of 30) ATCs met this cutoff point but no PTCs did. No RASAL1 methylation at all was seen in 20 benign thyroid tumors, and the average methylation level of RASAL1 in PTC was statistically significantly lower than that in ATC (1.59±6.88 vs 19.32±33.79; P = .007) or FTC (1.59±6.88 vs 27.97±34.75; P < .001) (Figure 3B). Thus, epigenetic impairment of RASAL1 was a common event in thyroid cancers, particularly in FTC and ATC.
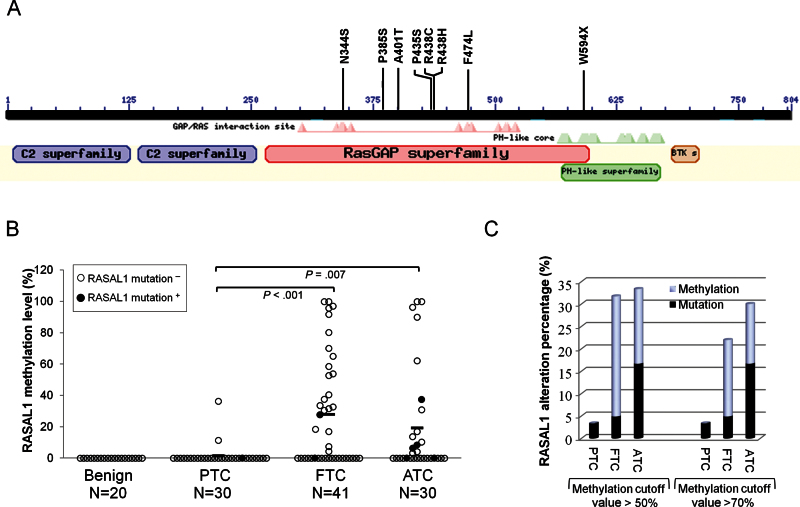
Figure 3.
Analysis of the relationship between mutations and hypermethylation of RASAL1 and the collective rates of the two events in thyroid cancer. A) Shown are the functional domains of RASAL1, including the RasGAP domain where all eight RASAL1 mutations are located. The amino acid numbers of the RASAL1 protein (shown as a black bar) are shown above the black bar. The annotation of RASAL1 domains, which was generated by the National Center for Biotechnology Information (NCBI) online tool Conserved Domain Search (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), is presented under the black bar at corresponding sites. B) Mutation and methylation of RASAL1 in individual cases of various types of thyroid tumor. The y-axis represents the allelic percentage of RASAL1 methylation of an individual tumor. Each individual case of thyroid tumor is represented by a circle, with solid circles representing the cases positive for RASAL1 mutation. Mutation-positive cases generally had low methylation levels, indicating an inverse relationship between the genetic and epigenetic alterations of RASAL1. P values were calculated by Wilcoxon Mann–Whitney test for comparison of the methylation levels between different types of thyroid cancer. C) Rates (percentage of total samples of each tumor type) of collective RASAL1 alterations (mutation and hypermethylation). Two cutoff values (50% and 70%) were used to define the level of RASAL1 methylation. ATC = anaplastic thyroid cancer; FTC = follicular thyroid cancer; PTC = papillary thyroid cancer.
Identification of RASAL1 Mutations in Thyroid Cancers
The above findings strongly suggest that RASAL1 is an important TSG in thyroid cancer. Because mutations are a classical mechanism in the impairment of TSGs and no RASAL1 mutations are currently known in human cancers, we next sought RASAL1 mutations as genetic support for the TSG candidacy of RASAL1 in thyroid cancer. We sequenced all 21 exons of RASAL1 that span its whole coding region on genomic DNA from 101 primary thyroid cancer samples. We found seven missense mutations and one nonsense mutation in eight thyroid cancers, including one of 30 PTCs (3.33%), two of 41 (4.88%) FTCs, and five of 30 (16.67%) ATCs, which were all located in the RAS GTPase-activating domain of RASAL1 (Figure 3A; Table 1; Supplementary Figure 2 and Supplementary Table 3, available online). All the mutations were confirmed by repeating the amplification PCR and bidirectional sequencing. No RASAL1 mutation was found in 20 benign thyroid tumors. Six of the eight mutations were C→T or G→A transitions, including three (37.5%) transitions that occurred at CpG dinucleotide sequences (Table 1). In addition, six of the eight mutations occurred at the site of short direct repeats.
Table 1.
Mutations and associated sequence motifs of RASAL1 in thyroid cancer
| Nucleotide change* | Codon change | Amino acid change | Exon of Location† | Sequence‡ | Sequence Motif | |
|---|---|---|---|---|---|---|
| CpG | Direct repeats | |||||
| A1031G | AAC → AGC | N344S | Exon 13 | CGTTCTAACTCCCTG | ||
| C1153T | CCC → TCC | P385S | Exon 14 | TGGATCCCTGCAAG | Yes | |
| G1201A | GCA → ACA | A401T | Exon 14 | TCAAAGGCGCACTCTC | Yes | Yes |
| C1303T | CCC → TCC | P435S | Exon 14 | CTGCCCGCCCGCCAT | Yes | |
| C1312T | CGC → TGC | R438C | Exon 14 | GCCATGCGCCTCGCC | Yes | Yes |
| G1313A | CGC → CAC | R438H | Exon 14 | GCCATGCGCCTCGCC | Yes | Yes |
| C1422A | TTC → TTA | F474L | Exon 15 | CGATTCTTCGCACC | Yes | Yes |
| G1782A | TGG → TGA | W594X | Exon 17 | ACGTCTGGCTCAGCG | ||
* Nucleotide number 1 is defined as A of the ATG translation initiation codon (GeneBank accession No. NM_004658).
† The exon structure is based on the RASAL1 cDNA sequence (GeneBank accession No. NM_004658).
‡ The mutated nucleotides are underlined and all the motif sequences containing the mutation are in bold. The direct repeat sequences are further italicized to distinguish them from the CpG motif. Only direct repeats with less than two nucleotides in-between are taken into account in this study. A single mutation could be classified into more than one specific sequence (eg, CpG motif or direct repeat), because a mutation might be caused independently by more than one mechanism (40).
Amino acid sequence alignment analysis of the RAS GTPase-activating domains derived from six human RasGAPs showed that six of the seven missense mutations of RASAL1 were located at conserved sites, which is similar to the missense mutations of NF1, another RasGAP, in human cancers (Figure 4A). On the similarity plot that was generated on the basis of the multiple alignment of RASAL1 homologues from 18 species, we observed that the RASAL1 missense mutations were situated on top of the similarity peaks in regions of high similarity (Figure 4B).
Figure 4.
Conservativeness of the amino acid sites of RASAL1 mutations. A) Multiple sequence alignment of the RAS GTPase-activating domain sequences derived from six human RasGAPs. The protein homologues were aligned by Constraint-based Multiple Alignment Tool of the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/tools/cobalt/). Sequence identity is shown in black boxes, dark gray boxes, and light gray boxes that represent different percentages of sequence identity, as indicated. The sequences that are substantially homologous among the RasGAP proteins are boxed and shown as blocks (39). The locations of RASAL1 mutations in the alignment sequence are indicated with arrows (for missense mutation) or stars (for nonsense mutation). Two missense mutations (R438C and R438H) occur at the same site and are thus indicated by one arrow. The locations of NF1 missense mutations in the alignment sequence are indicated with triangles. The NF1 mutation data was retrieved from databases HGMD (http://www.hgmd.cf.ac.uk) and COSMIC (http://www.sanger.ac.uk/genetics/CGP/cosmic/). B) Missense mutations in the similarity plot of RASAL1 homologues. The program Vector NTI (Invitrogen, Grand Island, NY) was used to generate the amino acid sequence similarity plot of RASAL1 homologues from 18 species, including Homo sapien (NP_004649.2), Pan troglodyte (XP_509394.2), Macaca mulatta (XP_002808039.1), Callithrix jacchus (XP_002753077.1), Bos taurus (XP_590469.5), Sus scrofa (XP_003132956.2), Equus caballus (XP_001915233.1), Ailuropoda melanoleuca (XP_002925340.1), Canis familiaris (XP_543403.2), Oryctolagus cuniculus (XP_002719824.1), Rattus norvegicus (NP_001101805.1), Mus musculus (NP_038860.2), Monodelphis domestica (XP_001378640.1), Meleagris gallopavo (XP_003211173), Tetraodon nigroviridis (CAF93132), Anolis carolinensis (XP_003226069.1), Saccoglossus kowalevskii (XP_002734139.1), and Xenopus tropicalis (NP_001008049.1). A sliding window of five amino acids was chosen. The x-axis represents coordinates on the amino acids and the y-axis shows similarity scores, with score 1 representing 100% sequence identity. The three dashed lines represent similarity scores of 1, 0.8, and 0.6, respectively. The RASAL1 missense mutations identified in this study are marked with red lines at the corresponding locations. All of the mutations are located in regions with similarity scores greater than 0.8.
There was a reciprocal relationship between mutations and the methylation of RASAL1. None of the thyroid cancers harboring RASAL1 mutations had RASAL1 methylation level higher than 50%; in fact, most of the RASAL1 mutation–positive cancers had RASAL1 methylation levels lower than 20% (Figure 3B). This was in contrast with the RASAL1 mutation–negative cancers, in which high levels of RASAL1 methylation were common and many had a methylation level greater than 50%. Thus, mutation and hypermethylation of RASAL1 were mutually exclusive, suggesting that either the genetic or epigenetic alteration of this gene was sufficient for its role in tumorigenesis. When both RASAL1 mutation and allelic methylation level of greater than 50% were counted, the genetic and epigenetic alterations were collectively found in 3.22% (n = 1 of 31) of PTCs, 31.70% (n = 13 of 41) of FTCs, and 33.33% (n = 10 of 30) of ATCs (Figure 3C). This FTC- and ATC-preferential distribution pattern of genetic and epigenetic alterations of RASAL1 remained even under a methylation cutoff value of greater than 70%
Relationship Between RASAL1 Alterations and Classical Mutations in RAS-Coupled Signaling Pathways
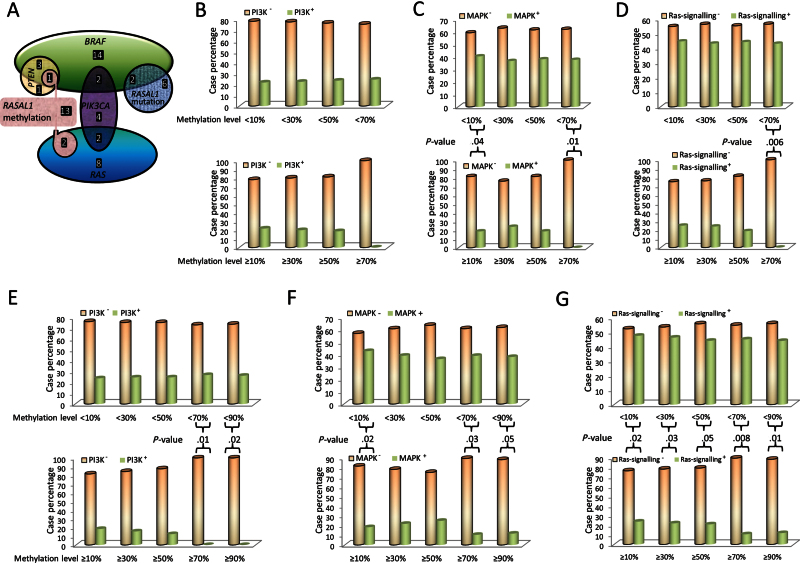
We examined the relationship between RASAL1 alterations and hotspot mutations of six classical genes in RAS-coupled MAPK and PI3K pathways, including BRAF, PTEN, PIK3CA, and the three RASs in 101 thyroid cancer samples. We found that five of 24 (20.83%) tumors carrying RASAL1 mutation or methylation at high levels (>50%) vs 34 of 77 (44.16%) tumors carrying no RASAL1 mutation or methylation at low levels (<50%) harbored any of the classical mutations in the six genes (P = .02, Fisher Exact test) (Supplementary Table 3, available online). This preferential distribution pattern revealed a largely mutually exclusive relationship between RASAL1 alterations and the classical mutations in RAS pathways. This relationship is also illustrated in Figure 5A. We further classified the six classical RAS-signaling pathway–related genes into three pathway groups, including PI3K, MAPK, and RAS pathways, as defined in the legend to Figure 5, and analyzed the relationship between RASAL1 alterations and the genetic alterations in the three pathway groups. As shown in Figure 5, B–D, with increasing the cutoff values of methylation level, tumors carrying RASAL1 methylation showed a decreasing rate of concurrence with mutations in all of the three pathways. A statistically significant mutual exclusivity was always achieved between the RASAL1 methylation and the alterations in MAPK and RAS pathways at a 90% cutoff value of methylation (P = .01 and .005, respectively). A similar mutually exclusive relationship was also observed between the collective RASAL1 alterations (either methylation at 90% cutoff value or mutation) and the mutations in the PI3K, MAPK, and RAS pathways (P = .02, .05, and .01, respectively at 90% cutoff value for methylation level) (Figure 5, E–G).
Figure 5.
Analysis of the relationship between RASAL1 alterations and mutations of classical genes in the RAS signaling pathway. A) Schematic illustration of the mutual exclusion of hypermethylation and mutations of RASAL1 with mutations of classical genes (RAS, BRAF, PIK3CA, and PTEN) in the RAS signaling pathway analyzed on 101 thyroid cancer samples (30 papillary thyroid cancer, 41 follicular thyroid cancer, and 30 anaplastic thyroid cancer). A methylation level greater than 50% was the cutoff value for RASAL1 methylation. Each shape with different color represents one gene alteration as indicated inside the shape. The overlap regions of the shapes represent the samples that contain two or more indicated gene alterations. All of the gene alterations are mutations, except for RASAL1, which also has hypermethylation as indicated. The number of tumor samples with the indicated gene alteration is represented by white printed numbers in each case. B) Association of RASAL1 methylation with mutations of genes in the PI3K pathway. Four cutoff allelic methylation values, as indicated on the x-axis, were used to define RASAL1 methylation levels. The y-axis represents the percentages of cancer samples that reached the indicated RASAL1 methylation level indicated on the x-axis. The empty bars (PI3K−) represent samples negative for mutations in RAS, PIK3CA, or PTEN in the PI3K pathway, and the solid bars (PI3K+) represent samples positive for mutations in RAS, PIK3CA, or PTEN in the PI3K pathway. C) Inverse association of RASAL1 methylation with mutations of genes (RAS or BRAF) in the MAPK pathway. The empty bars (MAPK−) represent samples negative for mutations in RAS or BRAF in the MAPK pathway, and the solid bars (MAPK+) represent samples positive for mutations in RAS or BRAF in the MAPK pathway. The remaining definitions are the same as those in (B). D) Inverse association of RASAL1 methylation with mutations of genes (RAS, PIK3CA, PTEN, or BRAF) in the RAS-signaling pathway (including both PI3K and MAPK pathways). The empty bars (RAS signaling−) represent samples negative for mutations in RAS, PIK3CA, PTEN, or BRAF in the RAS signaling pathway, and the solid bars (RAS signaling+) represent samples positive for mutations in RAS, PIK3CA, PTEN, or BRAF in the RAS signaling pathway. The remaining definitions are the same as those in (B). E–G) Inverse association of RASAL1 alterations (collectively including both RASAL1 methylation and mutations) with gene mutations in the PI3K pathway (E), MAPK pathway (F), or RAS signaling pathway (G) as defined for (B), (C), and (D), respectively. The y-axis in (E), (F), and (G) represents the percentage of tumor samples that were collectively positive for RASAL mutations or the methylation levels indicated on the x-axis. The remaining definitions are the same as those in (B). In each panel of Figure 5, the upper portion shows the cases with RASAL1 methylation less than the level indicated on the x-axis, and the lower portion shows the cases with RASAL1 methylation equal to or greater than the level indicated on the x-axis. Comparisons of the upper and lower groups in each of panels (C–G) were performed using two-tailed Fisher exact test, and those that were statistically significant are indicated with specific P values in the figure.
Functional Characterization of RASAL1 and Its Mutations in Thyroid Tumorigenesis
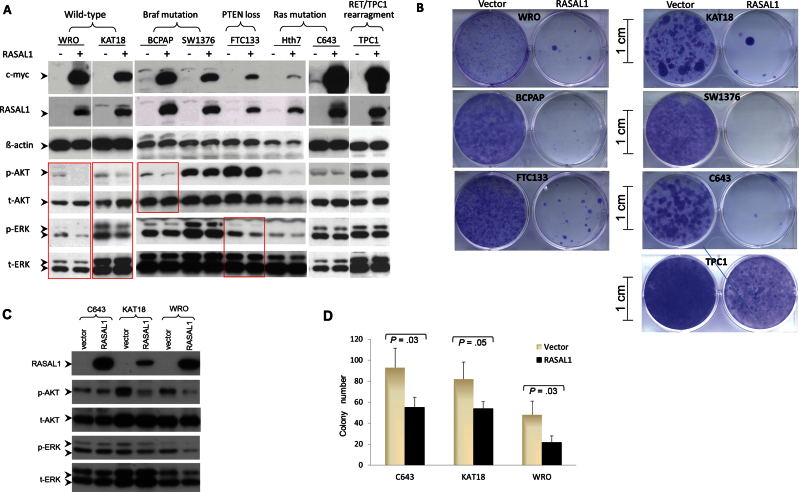
To further test the role of RASAL1 in thyroid tumorigenesis, we investigated its biological functions in various thyroid cancer cell lines. We cloned RASAL1 cDNA and constructed expression vectors to test the impact of induced expression of RASAL1 on the MAPK and PI3K signaling pathways and cellular behaviors. Figure 6A shows the effects of RASAL1 on the phosphorylation of the downstream signaling molecules ERK and AKT of the MAPK and PI3K pathways, respectively; these effects of RASAL1 were cell type and classical mutation type dependent. Specifically, acute expression of RASAL1 decreased both pERK and pAkt levels in WRO and KAT18 cells, which harbored wild-type RAS, BRAF, PIK3CA, and PTEN, whereas expression of RASAL1 only reduced pAKT but not pERK in BCPAP cells, which harbored BRAF mutation, resulting in activation of the MAPK pathway, and reduced pERK but not pAKT in FTC-133 cells, which harbored PTEN mutation, resulting in activation of the PI3K pathway. No effect was observed in C643 and Hth7 cells, which harbored RAS mutation that could activate both MAPK and PI3K pathways. There was also no effect of RASAL1 expression in TPC1 cells harboring RET-PTC1 rearrangement that could also activate both MAPK and PI3K pathways (32). No effect was observed in SW1736 cells, which harbored BRAF mutation. Whether SW1736 also harbors PI3K pathway–activating genetic alterations is unknown. Thus, overall, RASAL1 suppressed the MAPK pathway or PI3K pathway when it was not activated by classical genetic alterations in thyroid cancer cells.
Figure 6.
Examination of the effects of reintroduction of RASAL1 on RAS signaling and growth of thyroid cancer cells. A) A representative result of Western blotting analysis of RASAL1 expression and phosphorylation of ERK and Akt as well as other related proteins as indicated in various thyroid cancer cell lines that harbor different genetic alterations. B) A representative result of colony formation of thyroid cancer cells in monolayer culture after transfection of plasmids with or without insertion of RASAL1 cDNA followed by drug selection for 2 weeks. Cell colonies were stained with crystal violet. Scale bar = 1cm. C) Western blotting analysis of RASAL1 expression and phosphorylation of ERK and Akt and related proteins as indicated in three selected thyroid cancer cell lines stably transfected with RASAL1. D) Bar graph presentation of the colony numbers (mean ± standard deviation) from three experiments for cells stably transfected with empty vectors or RASAL1 as in (C). Only cell colonies containing more than 50 cells were included. Statistically significant P values (two-sided Student t test) are shown where indicated in the figure.
Compared with the control vector, transient reintroduction of RASAL1 dramatically inhibited cell colony growth in monolayer culture in six of sevent thyroid cancer cell lines tested (Figure 6B). We also generated three cell pools with stable expression of RASAL1 (Figure 6C). Among these, inhibition of pERK and pAKT was observed in KAT18 and WRO cells (Figure 6C), as seen with transient expression of RASAL1 (Figure 6A). Anchorage-independent cell growth was statistically significantly suppressed in all three stably transfected thyroid cancer cell lines. Specifically, the colony numbers for control vector vs RASAL1 transfection were 92.67±18.15 vs 55.33±9.71, (P = .03) for C643; 81.67±16.29 vs 54.00±6.56 (P = .05) for KAT18; and 48.00±13.45 vs 21.67±6.35 (P = .03) for WRO cells (n = 3 in all cases) (Figure 6D).
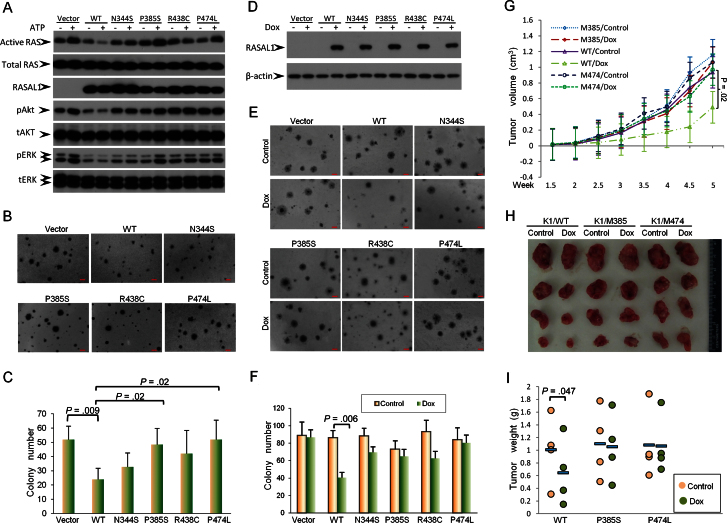
To test the biological relevance of the RASAL1 mutations in thyroid tumorigenesis, we selectively generated four RASAL1 mutants by site-directed in vitro mutagenesis and established WRO cell pools that stably expressed these mutants. Although wild-type RASAL1 suppressed the activation of RAS and the increase in pAKT and pERK stimulated by ATP treatment of cells, RASAL1 mutants P385S and F474L lost such inhibitory functions (Figure 7A). Correspondingly, these two mutants failed to inhibit the colony formation of WRO cells, which, in contrast, was suppressed by the wild-type RASAL1 (colony number: vector 52.00±9.54 vs wild-type 24.00±7.81; P = .009) (Figure 7, B and C). The other two mutants, N344S and R438C, showed a slight inhibitory effect on RAS activation (Figure 7A) and cell colony formation (Figure 7, B and C), which was weaker than the inhibitory effect of the wild-type RASAL1, suggesting that these two mutations partially impaired the function of RASAL1. To further test the impact of RASAL1 mutations on the function of RASAL1, we established K1 thyroid cancer cell pools with doxcycline-inducible expression of RASAL1 proteins. As shown in Figure 7D, wild-type RASAL1 and its mutants could be abundantly expressed upon doxcycline treatment of cells. Again, compared with wild-type RASAL1 (colony number: control 86.00±8.54 vs Dox 40.33±6.03; P = .006), the four mutants displayed varying impairment in their inhibitory effects on K1 cell colony formation, with P474L having almost completely lost the inhibitory function (Figure 7, E and F). We finally performed in vivo studies to test the impact of RASAL1 and its mutants on xenograf thyroid tumor growth. Upon doxycycline-induced expression of wild-type RASAL1, the growth of K1 cell-derived xenograft tumors in nude mice was statistically significantly suppressed both in tumor volume (0.94±0.52cm3 vs 0.49±0.4cm3; P = .02) (Figure 7G) and tumor weight (1.01±0.54g vs 0.66±0.53g; P = .047) (Figure 7, H and I). In contrast, RASAL1 mutants P385S and F474L showed no effect on tumor growth.
Figure 7.
Examination of tumor-suppressing functions of the wild-type and mutant RASAL1 in vitro and in vivo. A) Effects of wild-type RASAL1 and various indicated RASAL1 mutants on the activation of RAS in WRO cells. Cells stably transfected with various indicated RASAL1 constructs were treated with or without ATP, and the active RAS (RAS-GTP) was pulled down and analyzed by Western blotting using an RAS Activation Assay Kit as described in the Methods. Cell lysates from the same treatment were also used for Western blotting analysis of pAkt, pERK, and other related proteins as indicated. B) Effects of wild-type and mutant RASAL1 on the colony formation of WRO cells in soft agar. Scale bar = 100 μm. C) Bar graph presentation of colony numbers from three experiments (mean ± standard deviation) corresponding to (B). Two-tailed Student t test was used for the statistical analysis. D) Western blotting analysis of inducible expression of RASAL1 in K1 cells. The procedures for the construction of doxycycline (Dox)–inducible expression device for RASAL1 and transfection of K1 cells are as described in the Supplementary Methods (available online). E) Effects of Dox-induced expression of wild-type and mutant RASAL1 on the colony formation of K1 cells. Scale bar = 100 μm. F) Bar graph presentation of cell colony numbers from three experiments (mean ± standard deviation) corresponding to (E). Two-tailed Student t test was used for statistical analysis. G) Time course of growth of xenograft thyroid tumors that developed in mice after subcutaneous inoculation with K1 cell clones transfected with wild-type RASAL1 or the indicated RASAL1 mutants. Each time point represents the average ± standard deviation of the values obtained from four mice in each group. Two-tailed paired Student t test was used to compare tumor sizes at week 5. H) Photographs of the tumors that were surgically removed from mice in each group after they were killed at the end of 5 weeks from the cell inoculation. I) Weight of individual tumors surgically removed from the animals in each group corresponding to (H). The average weight of the tumors from each group is indicated with a short horizontal bar. A statistically significant inhibitory effect of wild-type RASAL1 on tumor growth was observed (P = .047 on two-tailed paired Student t test), but no such effect was observed for the two indicated RASAL1 mutants.
Discussion
As a negative modulator of the RAS signaling pathway by functioning as a RasGAP that catalyzes RAS inactivation, RASAL1 has been suggested to be a candidate TSG in recent years (3–5). However, direct evidence to demonstrate its tumor suppressor function is lacking. Moreover, there has been no genetic evidence to support RASAL1 as a typical TSG. The relative role of RASAL1 with respect to that of classical genes in MAPK- and PI3K-promoted tumorigenesis is also unknown. Thus, whether RASAL1 is truly a major tumor suppressor that plays an important role in human tumorigenesis has not been definitively established.
Our in vitro and in vivo data, at various levels, establish RASAL1 as a true TSG and, in particular, a major TSG in thyroid cancer. The evidence to support this conclusion includes the following: 1) RASAL1 inhibited both in vitro thyroid cancer cell growth and in vivo thyroid tumor growth; 2) RASAL1 was commonly hypermethylated in the 5’ region in thyroid cancers, causing its silencing in thyroid cancer cells; 3) nonrecurrent impairing mutations were identified in the RAS GTPase-activating domain of RASAL1; and 4) RASAL1 mutations and hypermethylation were mutually exclusive, suggesting equal importance of either genetic or epigenetic inactivation of this gene. These characteristics of RASAL1 fully meet the classical criteria of TSG. Moreover, the ability of RASAL1 to suppress both RAS-coupled MAPK and PI3K pathways when there was no classical genetic alteration in the two pathways and the mutual exclusivity of the inactivating genetic and epigenetic alterations of RASAL1 with the classical mutations in these pathways also strongly support RASAL1 being an important TSG.
All our results on RASAL1 can be explained by its function as a classical RasGAP except for its inhibitory effect on C643 cell growth (Figure 6). This cell harbors mutant H-RAS, which is constitutively activated and hence is independent of RASAL1 regulation in signaling through the MAPK and PI3K pathways (5); in fact, we found that introduction of RASAL1 had no effect on pERK and pAKT in C643 cells (Figure 5). This raises the possibility that RASAL1 may be able to negatively regulate cell growth through an unidentified RAS-independent mechanism. This possibility is suggested by the case of NF1, another RasGAP, which could suppress cells through a RAS-independent mechanism (33–35).
MAPK and PI3K pathways play a fundamental role in thyroid tumorigenesis, in which the known classical genetic alterations in the two pathways, such as mutations in RAS, BRAF, PIK3CA, and PTEN, account for 65% to 70% of thyroid cancers (23). The inactivating genetic and epigenetic alterations of RASAL1 in the MAPK and PI3K pathways provide an alternative genetic background for thyroid cancers that do not harbor classical genetic alterations in the two pathways. It is interesting to note that hypermethylation and mutations of RASAL1 were particularly common in FTC and ATC but uncommon in PTC. Because the BRAF/MAPK pathway plays a fundamental role in the tumorigenesis of PTC (23,36–38), whereas the PI3K/AKT pathway plays a fundamental role in FTC and ATC (26), it seems that impairment of RASAL1 may preferentially result in upregulation of the PI3K pathway over the MAPK pathway in thyroid tumorigenesis and promote the development of FTC and ATC over PTC. This possibility is consistent with the fact that in thyroid cancer RAS preferentially activates the PI3K pathway over the MAPK pathway (23).
Although this study definitely establishes RASAL1 as a TSG in thyroid cancer, it is limited by leaving several issues undefined. For example, what is the mechanism of the RAS-independent function of RASAL1 in some cells such as C643 cells? What is the clinical implication of genetic and epigenetic findings of this study? Could germline genetic alterations of RASAL1, like those in other TSGs in the RAS pathway, exist and cause inherited cancer syndromes? These important questions need future studies to answer.
In summary, we provide strong evidence demonstrating that RASAL1 is a prominent TSG that is frequently inactivated by hypermethylation and mutations in thyroid cancer, particularly FTC and ATC. Impairment of RASAL1 is an alternative genetic background in thyroid cancers that do not harbor classical genetic alterations in the RAS-coupled MAPK and PI3K pathways.
Funding
This study was supported by a grant from the National Institutes of Health (RO1 CA134225 to MX).
Supplementary Material
The authors have no conflict of interest to disclose.
The National Institutes of Health had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
References
- 1. Rebollo A, Martinez A. Ras proteins: recent advances and new functions. Blood. 1999;94(9):2971–2980. [PubMed] [Google Scholar]
- 2. Vigil D, Cherfils J, Rossman KL, et al. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10(12):842–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kolfschoten IG, van Leeuwen B, Berns K, et al. A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity. Cell. 2005;121(6):849–858. [DOI] [PubMed] [Google Scholar]
- 4. Jin H, Wang X, Ying J, et al. Epigenetic silencing of a Ca(2+)-regulated Ras GTPase-activating protein RASAL defines a new mechanism of Ras activation in human cancers. Proc Natl Acad Sci U S A. 2007;104(30):12353–12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ohta M, Seto M, Ijichi H, et al. Decreased expression of the RAS-GTPase activating protein RASAL1 is associated with colorectal tumor progression. Gastroenterology. 2009;136(1):206–216. [DOI] [PubMed] [Google Scholar]
- 6. Yohay KH. The genetic and molecular pathogenesis of NF1 and NF2. Semin Pediatr Neurol. 2006;13(1):21–26. [DOI] [PubMed] [Google Scholar]
- 7. Johannessen CM, Reczek EE, James MF, et al. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc Natl Acad Sci U S A. 2005;102(24):8573–8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Viskochil D. Genetics of neurofibromatosis 1 and the NF1 gene. J Child Neurol. 2002;17(8):562–570. [DOI] [PubMed] [Google Scholar]
- 9. Wakioka T, Sasaki A, Kato R, et al. Spred is a Sprouty-related suppressor of Ras signalling. Nature. 2001;412(6847):647–651. [DOI] [PubMed] [Google Scholar]
- 10. Sasaki A, Taketomi T, Wakioka T, et al. Identification of a dominant negative mutant of Sprouty that potentiates fibroblast growth factor- but not epidermal growth factor-induced ERK activation. J Biol Chem. 2001;276(39):36804–36808. [DOI] [PubMed] [Google Scholar]
- 11. Granovsky AE, Rosner MR. Raf kinase inhibitory protein: a signal transduction modulator and metastasis suppressor. Cell Res. 2008;18(4):452–457. [DOI] [PubMed] [Google Scholar]
- 12. Ueda K, Arakawa H, Nakamura Y. Dual-specificity phosphatase 5 (DUSP5) as a direct transcriptional target of tumor suppressor p53. Oncogene. 2003;22(36):5586–5591. [DOI] [PubMed] [Google Scholar]
- 13. Furukawa T, Sunamura M, Motoi F, et al. Potential tumor suppressive pathway involving DUSP6/MKP-3 in pancreatic cancer. Am J Pathol. 2003;162(6):1807–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wrighton KH. Tumour suppressors: Role of nuclear PTEN revealed. Nat Rev Cancer. 2011;11(3):154. [DOI] [PubMed] [Google Scholar]
- 15. Hezel AF, Bardeesy N. LKB1; linking cell structure and tumor suppression. Oncogene. 2008;27(55):6908–6919. [DOI] [PubMed] [Google Scholar]
- 16. van Veelen W, Korsse SE, van de LL, et al. The long and winding road to rational treatment of cancer associated with LKB1/AMPK/TSC/mTORC1 signaling. Oncogene. 2011;30(20):2289–2303 [DOI] [PubMed] [Google Scholar]
- 17. Richter AM, Pfeifer GP, Dammann RH. The RASSF proteins in cancer; from epigenetic silencing to functional characterization. Biochim Biophys Acta. 2009;1796(2):114–128. [DOI] [PubMed] [Google Scholar]
- 18. Bentires-Alj M, Kontaridis MI, Neel BG. Stops along the RAS pathway in human genetic disease. Nat Med. 2006;12(3):283–285. [DOI] [PubMed] [Google Scholar]
- 19. Brems H, Chmara M, Sahbatou M, et al. Germline loss-of-function mutations in SPRED1 cause a neurofibromatosis 1-like phenotype. Nat Genet. 2007;39(9):1120–1126. [DOI] [PubMed] [Google Scholar]
- 20. Howlader N, Noone AM, Krapcho M, et al. SEER Cancer Statistics Review, 1975–2009 (Vintage 2009 Populations). http://seer.cancer.gov/csr/1975_2009_pops09 Accessed August 13, 2013.
- 21. Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. [DOI] [PubMed] [Google Scholar]
- 22. Hundahl SA, Fleming ID, Fremgen AM, et al. A National Cancer Data Base report on 53,856 cases of thyroid carcinoma treated in the U.S., 1985–1995 [see comments]. Cancer. 1998;83(12):2638–2648. [DOI] [PubMed] [Google Scholar]
- 23. Xing M. Molecular pathogenesis and mechanisms of thyroid cancer. Nat Rev Cancer. 2013;13(3):184–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xing M. BRAF mutation in thyroid cancer. Endocr Relat Cancer. 2005;12(2):245–262. [DOI] [PubMed] [Google Scholar]
- 25. Xing M. BRAF mutation in papillary thyroid cancer: pathogenic role, molecular bases, and clinical implications. Endocr Rev. 2007;28(7):742–762. [DOI] [PubMed] [Google Scholar]
- 26. Xing M. Genetic alterations in the phosphatidylinositol-3 kinase/Akt pathway in thyroid cancer. Thyroid. 2010;20(7):697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hou P, Liu D, Shan Y, et al. Genetic alterations and their relationship in the phosphatidylinositol 3-kinase/Akt pathway in thyroid cancer. Clin Cancer Res. 2007;13(4):1161–1170. [DOI] [PubMed] [Google Scholar]
- 28. Liu D, Hou P, Liu Z, et al. Genetic alterations in the phosphoinositide 3-kinase/Akt signaling pathway confer sensitivity of thyroid cancer cells to therapeutic targeting of Akt and mammalian target of rapamycin. Cancer Res. 2009;69(18):7311–7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu D, Hu S, Hou P, et al. Suppression of BRAF/MEK/MAP kinase pathway restores expression of iodide-metabolizing genes in thyroid cells expressing the V600E BRAF mutant. Clin Cancer Res. 2007;13(4):1341–1349. [DOI] [PubMed] [Google Scholar]
- 30. Hou P, Ji M, Xing M. Association of PTEN gene methylation with genetic alterations in the phosphatidylinositol 3-kinase/AKT signaling pathway in thyroid tumors. Cancer. 2008;113(9):2440–2447. [DOI] [PubMed] [Google Scholar]
- 31. Xing M, Cohen Y, Mambo E, et al. Early occurrence of RASSF1A hypermethylation and its mutual exclusion with BRAF mutation in thyroid tumorigenesis. Cancer Res. 2004;64(5):1664–1668. [DOI] [PubMed] [Google Scholar]
- 32. Ishizaka Y, Ushijima T, Sugimura T, et al. cDNA cloning and characterization of ret activated in a human papillary thyroid carcinoma cell line. Biochem Biophys Res Commun. 1990;168(2):402–408. [DOI] [PubMed] [Google Scholar]
- 33. Shapira S, Barkan B, Friedman E, et al. The tumor suppressor neurofibromin confers sensitivity to apoptosis by Ras-dependent and Ras-independent pathways. Cell Death Differ. 2007;14(5):895–906. [DOI] [PubMed] [Google Scholar]
- 34. Nur-E-Kamal MS, Varga M, Maruta H. The GTPase-activating NF1 fragment of 91 amino acids reverses v-Ha-Ras-induced malignant phenotype. J Biol Chem. 1993;268(30):22331–22337. [PubMed] [Google Scholar]
- 35. Gutmann DH, Collins FS. The neurofibromatosis type 1 gene and its protein product, neurofibromin. Neuron. 1993;10(3):335–343. [DOI] [PubMed] [Google Scholar]
- 36. Nucera C, Porrello A, Antonello ZA, et al. B-Raf(V600E) and thrombospondin-1 promote thyroid cancer progression. Proc Natl Acad Sci U S A. 2010;107(23):10649–10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chakravarty D, Santos E, Ryder M, et al. Small-molecule MAPK inhibitors restore radioiodine incorporation in mouse thyroid cancers with conditional BRAF activation. J Clin Invest. 2011;121(12):4700–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hu S, Liu D, Tufano RP, et al. Association of aberrant methylation of tumor suppressor genes with tumor aggressiveness and BRAF mutation in papillary thyroid cancer. Int J Cancer. 2006;119(10):2322–2329. [DOI] [PubMed] [Google Scholar]
- 39. Scheffzek K, Lautwein A, Kabsch W, et al. Crystal structure of the GTPase-activating domain of human p120GAP and implications for the interaction with Ras. Nature. 1996;384(6609):591–596. [DOI] [PubMed] [Google Scholar]
- 40. Todorova A, Danieli GA. Large majority of single-nucleotide mutations along the dystrophin gene can be explained by more than one mechanism of mutagenesis. Hum Mutat. 1997;9(6):537–547. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.