Abstract
Background
Chloride channels are physiologically involved in cell division and motility. Chloride intracellular channel 1 (CLIC1) is overexpressed in a variety of human solid tumors compared with normal tissues, suggesting a potential involvement of CLIC1 in the regulation of tumorigenesis. This led us to investigate the role of CLIC1 in gliomagenesis.
Methods
We used the neurosphere system to isolate stem/progenitor cells from human glioblastomas (GBMs). CLIC1 targeting in GBM neurospheres was achieved by both lentiviral-mediated short-hairpin RNA transduction and CLIC1 antibody treatment, and its effect on stem-like properties was analyzed in vitro by proliferation and clonogenic assays and in vivo by orthotopic injection in immunocompromised mice. Channel activity was studied by perforated patch clamp technique. Differences in expression were analyzed by analysis of variance with Tamhane’s multiple comparison test. Kaplan–Meier analyses and log-rank test were used to assess survival. All statistical tests were two-sided.
Results
CLIC1 was statistically significantly overexpressed in GBMs compared with normal brain tissues (P < .001) with a better survival of patients with CLIC1 low-expressing tumors (CLIC1low vs CLIC1high survival: χ2 = 74.35; degrees of freedom = 1; log-rank P < .001). CLIC1 was variably expressed in patient-derived GBM neurospheres and was found enriched in the stem/progenitor compartment. CLIC1 silencing reduced proliferative (P < .01), clonogenic (P < .01), and tumorigenic capacity (P < .05) of stem/progenitor cells. The reduction of CLIC1 chloride currents with a specific CLIC1 antibody mirrored the biological effects of CLIC1 silencing in GBM patient–derived neurospheres.
Conclusions
Reduced gliomagenesis after CLIC1 targeting in tumoral stem/progenitor cells and the finding that CLIC1 expression is inversely associated with patient survival suggest CLIC1 as a potential target and prognostic biomarker.
Glioblastoma (GBM) is the most aggressive and lethal among brain tumors in adults, and it still represents a tremendous clinical challenge. Little is known about the molecular mechanisms underlying the genesis and the progression of GBM, which is characterized by the high propensity to infiltrate throughout the brain. The invasive nature of this type of tumor makes the neoplastic foci difficult to target and treat, with the result that tumor recurrence is inevitable despite aggressive surgery and adjuvant radiotherapy and/or chemotherapy (1).
As well as for other solid tumors, it has been demonstrated that the bulk of malignant cells in GBM is generated by a rare fraction of self-renewing, multipotent cancer stem cells (CSCs) responsible for tumor origin, progression, and recurrence (2,3). These subpopulations of cells have shown intrinsic resistance to therapy, being then capable to repopulate the tumor after treatment (4). Therefore, a new approach to cancer therapy should focus on specifically targeting the resistant CSC populations.
Several studies reported that glioma cells actively accumulate chloride ions and undergo a substantial volume decrease after chloride efflux due to an osmotic-driven, outward-directed water flow (5–7); glioma cells acquire an elongated, wedge-like shape and appear to shrink their cell volume, and this has been shown to be crucial for cell division and cell migration (8).
Chloride intracellular channel 1 (CLIC1) belongs to a class of chloride channels that does not fit the paradigm set by classical ion channel proteins (9–11). CLIC1 properties are enigmatic because CLIC1 can exist as both a soluble globular protein and as an integral membrane protein with ion channel function. Upon oxidative stress, CLIC1 translocates from the cytoplasm to the plasma membrane where it exerts its function as a chloride (Cl−) channel (12–14). The high level of conservation of CLIC1 protein among several species and its wide expression in mammalian cells argue for an important and conserved biological function; however, the understanding of its function is still incomplete. Recently, CLIC1 has been shown to be overexpressed in a variety of human solid tumors compared with normal tissues (15,16), including gliomas (17), suggesting a potential involvement of CLIC1 in the regulation of tumorigenesis. Its involvement in the cell cycle (10) and its functional expression during oxidative stress conditions (18–20) suggest CLIC1 as a candidate involved in the mechanism of glioma development. A possible role for CLIC1 in stem-like cellular subpopulations isolated from a GBM cell line has been recently reported (21).
Methods
Tumor Sample Preparation
Tumor specimens classified as GBM (World Health Organization [WHO] grade IV) were collected from consenting patients in the Department of Neurosurgery at Istituto Neurologico Carlo Besta, Milan, Italy. Tissues were enzymatically processed with papain (2mg/mL; Worthington Biochemical, Lakewood, NJ) at 37 °C and mechanically dissociated until achievement of single cell suspension.
Normal brain, WHO grade II, III and IV, were collected from consenting patients at the University of Freiburg, Germany, frozen, and processed for RNA isolation.
Neurosphere Culture and Clonogenic Assay
Human normal progenitor cells (NPCs) (Lonza, Amboise, France), human GBM neurospheres, normal murine stem cells, and tumoral murine neurospheres (GL261) (22) were grown as spheroid aggregates as previously described (23). To measure the clonogenicity, cells were resuspended in Dulbecco’s modified Eagle medium/F12 medium containing methylcellulose (StemCell Technologies, Vancouver, BC, Canada) and seeded on 35-mm culture plates (3000 cells per dish). A minimum of three plates per condition was used. Two weeks after plating, the number of clones was counted.
Western Immunoblotting
Primary antibodies used were CLIC1 (mouse monoclonal, 1:1000, clone CPTC-CLIC1-1; Millipore, Billerica, MA) and Vinculin (mouse monoclonal, 1:10000, clone HVIN-1; Sigma Aldrich, St. Louis, MO). Details are explained in the Supplementary Data (available online).
Immunofluorescence Analysis
Primary antibodies used were CLIC1 (mouse monoclonal, 1:1000, clone 356.1; Santa Cruz Biotechnology, Santa Cruz, CA), Sox2 (rabbit polyclonal, 1:500, ab15830; Abcam, Cambridge, UK), Nestin (rabbit polyclonal, 1:200, ABD69; Millipore), GFAP (rabbit polyclonal, 1:500, Z0334; DakoCytomation, Glostrup, Denmark), BrdU (mouse monoclonal, 5 µg/mL; BD Biosciences, Franklin Lakes, NJ), Cleaved Caspase-3 (rabbit polyclonal, 1:500; Cell Signaling, Danvers, MA). Confocal images and live-microscopy images were generated with a Leica SPII spectral confocal microscope (Leica Microsystems, Wetzlar, Germany). Details are explained in the Supplementary Data (available online).
Immunohystochemistry
Xenografted mouse brains were formalin-fixed and paraffin-embedded according to established procedures (24). Each brain was sliced into several coronal sections; the section with the largest tumor diameter was measured by ImageJ free software (http://rsbweb.nih.gov/ij/), and the intracranial tumor volume was estimated by the largest width squared × length × 0.5. Details are explained in the Supplementary Data (available online).
Electrophysiology
Cells were voltage-clamped as described in the Supplementary Data (available online).
Lentiviral-Mediated CLIC1 Silencing
Viral infection was performed according to established procedures (25). Cloning details are described in the Supplementary Data (available online).
In Vivo Assays
GBM-derived neurospheres were mechanically dissociated, and cells were resuspended in 2 µL of phosphate-buffered saline and stereotaxically injected into the nucleus caudatus (1mm posterior, 3mm left lateral, 3.5mm in depth from bregma) of 5-week-old female nu/nu CD1 mice (Charles River, Wilmington, MA). For CLIC1 antibody studies, cells were incubated with CLIC1 antibody (mouse monoclonal, 5 µg/mL, clone 356.1; Santa Cruz Biotechnology) or isotype control antibody for 72 hours before implantation (105 cells) into the brain of immunodeficient mice.
In vivo experiments in CD-1 nude mice were performed in accordance with Italian Law (D.L.vo 116/92 and following additions), which enforces EU 86/609 Directive (Council Directive 86/609/EEC of November 24, 1986, on the approximation of laws, regulations and administrative provisions of the Member States regarding the protection of animals used for experimental and other scientific purposes).
Statistical Analysis
To determine differences within group pairings, we used either the Bonferroni correction, when samples showed homogeneous variances, or the Tamhane test, when samples showed nonhomogeneous variances. Colocalization analysis was studied using the Cohen test to determine inter-rater agreement between categorical items (26). Statistical significance of Cohen’s kappa index was obtained by applying a χ2 test to positivity and negativity frequencies in compared conditions. In Kaplan–Meier curves, survival differences were compared by log-rank analysis. For the in vivo limiting dilution assay, tumor formation frequency and statistical significance were evaluated with the extreme limiting dilution analysis function (http://bioinf.wehi.edu.au/software/elda/). Statistical analysis was performed using Statistical Package for Social Science (SPSS) software. Statistical significance of differences for all parametric variables has been tested by means of analysis of variance (ANOVA) or generalized linear models (GLMs). Data are graphed as mean ± 95% confidence intervals (CIs). Differences were considered statistically significant when P was less than .05, .001, or .0001.
Results
Bioinformatic Analysis of CLIC1 Expression in Glioblastomas
To investigate a possible role for CLIC1 in human GBMs, we evaluated CLIC1 expression in the National Cancer Institute’s Repository for Molecular Brain Neoplasia Data (REMBRANDT) (27), and we found that CLIC1 expression was different between gliomas (grade II, III, IV) and control (nontumor) brain tissues (F test for dishomogeneous variances: F = 106.56; degrees of freedeom [df] = 2 and 385; P < .001) and, among gliomas, CLIC1 expression was higher in GBMs (WHO grade IV) (mean CLIC1 level in nontumor samples: 0.877, 95% CI = 0.783 to 0.970; mean CLIC1 level in grade II–III tumor samples: 1.814, 95% CI = 1.561 to 2.066; mean CLIC1 level in grade IV tumor samples: 4.891, 95% CI = 4.429 to 5.353; ANOVA with Tamhane multiple comparison test: non-tumor vs grade II–III: P < .0001, grade II–III vs grade IV: P < .0001) (Figure 1A).
Figure 1.
Chloride intracellular channel 1 (CLIC1) expression level in human gliomas. A) Box-plot showing CLIC1 mRNA levels in control (nontumor) brain tissues, astrocytomas (World Health Organization [WHO] grades II–III), and glioblastomas (GBM; WHO grade IV) derived from the National Cancer Institute’s Repository for Molecular Brain Neoplasia Data (REMBRANDT) database. B and C) Association of CLIC1 mRNA expression with patient prognosis. B) Kaplan–Meier survival plot based on patient data from REMBRANDT database. C) Kaplan–Meier survival plot based on subgroup from REMBRANDT database comprising only GBM patients. In each graph, patient samples have been divided into CLIC1 low-expressing tumors (CLIC1low, blue) and CLIC1 high-expressing tumors (CLIC1high, red) based on whether the tumors had CLIC1 mRNA levels that were less than or greater than median levels. D and E) Association of CLIC1 mRNA levels with GBM subtypes (proneural [PN], proliferative [Prolif], neural [N], classical [CL], and mesenchymal [MES]): microarray data set from Phillips et al. (26) (D) and from Verhaak et al. (27) (E) works were examined. In panels A, D, and E, the solid lines within the boxes represent the median value; the boxes show the 25th and 75th percentile range of CLIC1 mRNA levels; maximum and minimum values are depicted as horizontal bars; circles represent outliers. **P < .001 and ***P < .0001 of differences between means of indicated pairs calculated by analysis of variance with Tamhane multiple comparison test; NS = not significant.
Analysis of CLIC1 transcripts in relation to patient survival derived from REMBRANDT revealed that CLIC1 expression inversely associated with patient survival, suggesting a potential exploitation of CLIC1 as an outcome predictor (CLIC1low vs CLIC1high survival: χ2 = 74.35; df = 1; log-rank P < .001) (Figure 1B). Similar results were obtained when the analysis was restricted to the subgroup consisting of only GBM patients (χ2 = 10.99; df= 1; log-rank P < .01) (Figure 1C).
GBMs have been classified in molecular subtypes according to gene expression signatures (28,29). Phillips et al. (26) defined three subtypes (proneural [PN], proliferative [PROL], mesenchymal [MES]) similiar to those described by Verhaak et al. (27), who used data from the Cancer Genome Atlas (30) to describe four distinct subtypes (PN, neural [N], classic [CL], and MES). Thus, we analyzed CLIC1 expression using the Phillips’ microarray dataset and the three independent microarray datasets originated by Verhaak et al. (27). We found that CLIC1 expression was always statistically significantly higher in the MES subtype compared with the others (Figure 1, D and E; Supplementary Figure 1, available online) For the Phillips et al. (26) dataset, the mean CLIC1 level in PN subtype was 774 (95% CI = 596 to 953), the mean CLIC1 level in PROL subtype was 2207 (95% CI = 796 to 2617), and the mean CLIC1 level in MES subtype was 3196 (95% CI = 2804 to 3589) (ANOVA with Tamhane multiple comparison test: PN vs each of the others: always P < .001; PROL vs MES: P = .002). For the Verhaak dataset, the mean CLIC1 level in PN subtype was 1145 (95% CI = 980 to 1310), the mean CLIC1 level in N subtype aws 2129 (95% CI = 1829 to 2430), the mean CLIC1 level in CL subtype was 2237 (95% CI = 2049 to 2426), and the mean CLIC1 level in MES subtype was 3402, 95% CI = 3112 to 3692) (ANOVA with Tamhane multiple comparison test: N vs each of the others: always P < .001; MES vs each of the others: always P < .001; N vs CL: P = .990).
CLIC1 Expression in Patient-Derived GBM Neurospheres
To further evaluate the extent of CLIC1 overexpression in human gliomas, we examined CLIC1 expression level in a distinct set of normal brain tissues (n = 20) and astrocytic tumors of different grades (n = 13 WHO grade II; n = 28 WHO grade III; n = 20 WHO grade IV). CLIC1 was weakly expressed in normal brain specimens, and its expression increased with tumor grade, reaching the highest levels in GBMs (Figure 2A). Of note, GBM subgroups displayed a marked heterogeneity in CLIC1 expression (Figure 2A).
Figure 2.
Chloride intracellular channel 1 (CLIC1) expression in astrocytic tumors and in patient-derived glioblastoma (GBM) neurospheres. A) CLIC1 expression levels by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) in normal brain specimens (n = 20) and in astrocytic tumors of different grades (n = 13 for World Health Organization [WHO] grade II, n = 28 for WHO grade III, and n = 20 for WHO grade IV). The solid lines represent the mean value; Error bars represent 95% confidence intervals. ***P < .0001 of differences between means of indicated pairs calculated by analysis of variance with Tamhane multiple comparison test; NS = not significant. B) CLIC1 expression levels by qRT-PCR in different patient-derived GBM neurospheres. Experiments were performed in triplicate; error bars represent 95% confidence intervals. C) Representative images of CLIC1 immunostaining in GBM-derived neurospheres. Dissociated neurospheres were fixed and processed for immunofluorescence (CLIC1: red; Sox2, Nestin, and glial fibrillary acidic protein [GFAP]: green; 4’,6-diamidino-2-phenylindole [DAPI]: blue; merge: yellow). Scale bar = 50 μm. D) Statistical measures of inter-rater agreement of immunoreactive cells in (C) evaluated by Cohen’s kappa index. Cohen’s kappa is close to 1 for highly associated markers and close to 0 for unrelated markers. The statistical significance of observed kappa values has been evaluated by means of the χ2 test. Immunostained cells were counted at 20× magnification, five fields for each sample (mean cell number per field was 150). Three independent experiments were performed.
We isolated GBM stem/progenitor cells from surgically resected human GBM specimens and cultured them as neurospheres. We next assessed CLIC1 expression level in GBM-derived neurospheres. Similar to the results obtained for brain tissues (Figure 2A), GBM stem/progenitor cells expressed statistically significantly higher levels of CLIC1 mRNA compared with NPCs, with variable degrees among the tumor samples analyzed (Figure 2B). Considering that GBM-derived neurospheres are a mixed population of stem, progenitor, and differentiated cells, we investigated CLIC1 localization within the neurosphere. A specific CLIC1 antibody (Supplementary Figure 2, available online) revealed a colocalization between CLIC1 and putative stem/progenitor cell markers (Sox2, Nestin) (31), showing that CLIC1 is enriched in the stem/progenitor cell compartment of the neurosphere (Figure 2C). The frequency of CLIC1 colocalization with either Sox2 or Nestin has been quantified and evaluated by Cohen’s kappa index (32), which demonstrated that the frequency of colocalization observed was statistically significantly higher (Cohen’s kappa index close to 1.0) than what would be expected from stochastically behaving markers (Cohen’s kappa index close to 0.0) (Figure 2D; Supplementary Table 1, available online).
CLIC1 Subcellular Localization in Normal and Tumoral Neurospheres
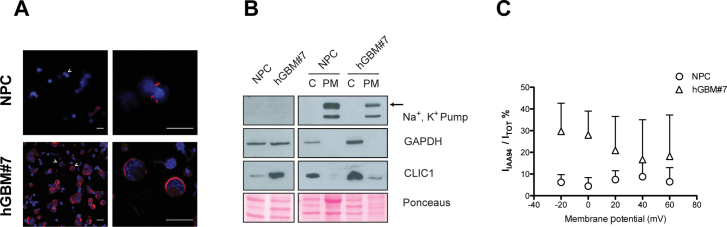
CLIC1 can exist as both soluble globular protein and integral membrane protein with ion channel function depending on the tissue and on the oxidative status. After oxidative stress, CLIC1 is able to translocate into plasma membrane, where it acts as a Cl− channel (12–14). Thus, we studied CLIC1 localization in normal human progenitor cells and GBM-derived neurospheres isolated from different patients. Immunofluorescent staining of nonpermeabilized cells revealed that CLIC1 is constitutively localized on the plasma membrane of GBM-derived neurospheres (Figure 3A; Supplementary Figure 3A, available online). In contrast, NPCs did not show plasma membrane staining (Figure 3A). Western blotting analysis of cell lysate fractions obtained from NPCs and neurospheres from GBM subject 7 (hGBM#7) cells showed CLIC1 enrichment in the plasma membrane of human tumoral neurospheres (Figure 3B), which is consistent with the findings of the immunofluorescence assay (Figure 3A). To unravel whether CLIC1 constitutively localized on cell plasma membrane functions as an ion channel, we measured CLIC1 ion channel activity by perforated patch clamp technique in NPCs and hGBM#7 cells. Cl− currents mediated by CLIC1 were isolated using the specific inhibitor indanyloxyacetic acid 94 (IAA94) and normalized to the total current (ITot) in the corresponding cell (IIAA94/ITot%). Interestingly, we found that CLIC1-mediated currents (IAA94-sensitive currents, IIAA94) were more represented in tumoral cells (GLM: P < .001 related to cell type; P = .43 related to membrane potential) (Figure 3C). Notably, the same results were obtained by analyzing tumoral murine neurospheres (GL261) and the normal counterpart (murine normal stem cells) (Supplementary Figure 3, B–D, available online).
Figure 3.
Chloride intracellular channel 1 (CLIC1) subcellular localization in normal and tumoral human neurospheres. A) Representative images of CLIC1 immunostaining in nonpermeabilized normal and tumoral human neurospheres. Dissociated neurospheres were fixed and processed for immunofluorescence (CLIC1: red; 4’,6-diamidino-2-phenylindole [DAPI]: blue). Cells were analyzed using confocal laser scanning microscopy, and a single optical x-y plane section is shown. Scale bar = 10 µm. hGBM#7 = neurospheres isolated from GBM patient 7; NPC = normal human progenitor cells. B) Western blotting analysis of CLIC1 expression levels in whole cell lysates (left panel) and plasma membrane and cytoplasm-containing fractions (right panel) derived from normal (NPC) and tumoral neurospheres (hGBM#7). Sodium–potassium pump and GAPDH expression were examined to assess the purity of plasma membrane and cytoplasmic fractions, respectively. Reversible Ponceau staining was used as a control for equal protein loading. C) CLIC1-mediated chloride (Cl−) currents measured by perforated patch clamp technique in normal (NPC) and tumoral (hGBM#7) neurospheres. Cl− currents mediated by CLIC1 (IIAA94) were isolated using the specific CLIC1 inhibitor indanyloxyacetic acid–94 (IAA94) and normalized to the total cell current (ITot) (IIAA94/ITot%). Mean values derived from five independent experiments were represented. Error bars represent 95% confidence intervals. Generalized linear model test of between-subjects effects: F for “potential” = 1.44, degrees of freedom (df) = 4, P = .24 (not significant); F for “cell type” = 36.17, df = 1, P = .001; F for variables interaction = 1.30, df = 8, P = .27 (not significant). No statistical significance for interaction means a similar pattern of IIAA94 / ITot change for different cell types at different membrane potential values, even if mean IIAA94 / ITot values are different between different cell types.
Taken together, these results demonstrate increased CLIC1 expression and activity in tumoral stem/progenitor cells compared with normal counterparts.
Effect of CLIC1 Silencing on Plasma Membrane Cl− Currents in Patient-Derived GBM Neurospheres
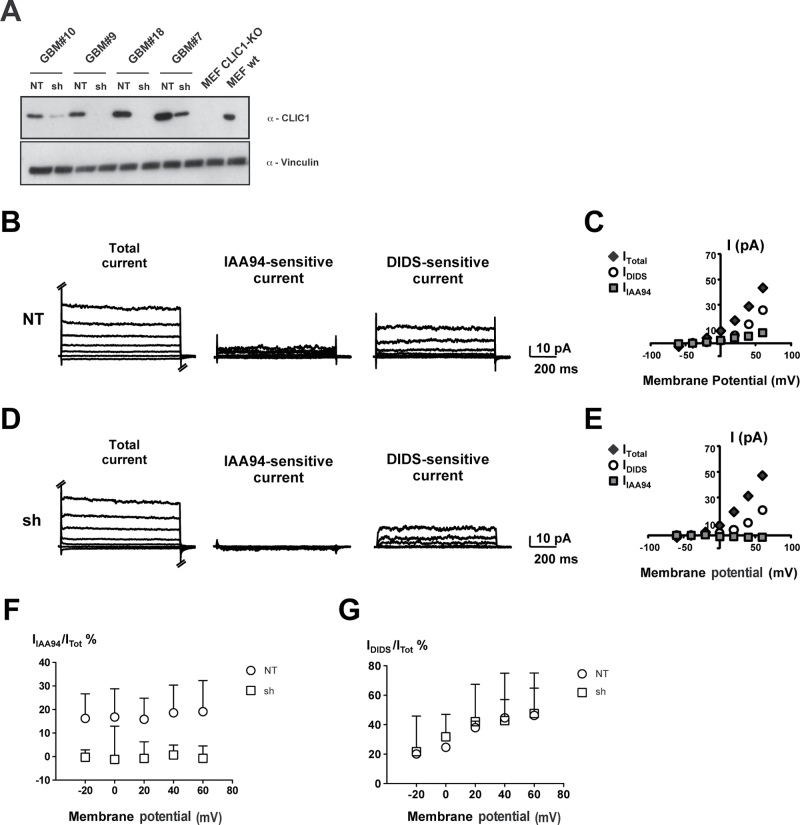
To disclose the role of CLIC1 in GBM stem/progenitor cells, we silenced CLIC1 expression in patient-derived GBM neurospheres by cloning short-hairpin RNA (shRNA) oligonucleotides specific against human CLIC1 mRNA (sh) in a lentiviral vector containing green fluorescent protein and the puromycin resistance gene as reporters. The same vector containing an shRNA targeting the luciferase mRNA sequence was used as control (nontargeting [NT]). Interference efficiency was confirmed by western blot: CLIC1 was silenced by nearly 90% in different samples (Figure 4A; Supplementary Figure 4A, available online).
Figure 4.
Effect of chloride intracellular channel 1 (CLIC1) silencing in glioblastoma (GBM) neurospheres. A) Western blotting analysis showing the efficiency of CLIC1 silencing in GBM neurospheres isolated from four patient samples. Dissociated neurospheres were transduced with lentivirus carrying either nontargeting short-hairpin RNA (shRNA) (NT) or CLIC1 shRNA (sh). Mouse embryonic fibroblasts derived from CLIC1 knockout mice (MEF CLIC1-KO) were used as negative controls. Vinculin was used as loading control. B and D) Representative current traces (total, indanyloxyacetic acid 94 [IAA94]–sensitive, and 4,4’-diisothiocyano-2,2’-stilbenedisulfonic acid [DIDS]–sensitive currents) from NT (B) and CLIC1-silenced (sh) (D) cells derived from GBM patient 10 (hGBM#10) NS and elicited by different potential steps (from −60 mV to 60 mV). C and E) The current–voltage relationships for the corresponding experiments in B and D. F) CLIC1-sensitive currents (IIAA94) were isolated using the specific CLIC1 inhibitor IAA94, and normalized to the total cell current (ITot) (IIAA94/ITot%). G) The other chloride (Cl−) currents in the cells were evaluated by the inhibitor DIDS (IDIDS) and normalized to the total cell current (ITot) (IDIDS/ITot%). Mean values derived from four independent experiments are represented. Error bars represent 95% confidence intervals. GLM test of between-subjects effects on IIAA94/ITot values: F for “potential” = 0.108, d. f. = 4, p = 0.979 (n. s.); F for “cell type” = 50.038, d. f. = 1, p < 0.001; F for variables interaction = 0.058, d. f. = 4, p = 0.993 n. s. No significance for interaction means a similar pattern of IIAA94/ITot change for different cell types at different membrane potential values, even if mean IIAA94/ITot values are different between different cell types. Generalized linear model test of between-subjects effects on IDIDS / ITot values: F for “potential” = 4.031, degrees of freedom (df) = 4, P = .01; F for “cell type” = 3.590, df = 1, P = .07; F for variables interaction = 0.085, df = 4, P = .99. No statistical significance for interaction means a similar pattern of IDIDS/ITot change for different cell types at different membrane potential values.
To sort out whether the reduction in total CLIC1 protein level was associated with a modification in the amount of the Cl− current mediated by this protein, we performed perforated patch clamp experiments in NT and CLIC1-silenced cells derived from two different patients (hGBM#7 and hGBM#10). Representative experiments from NT (Figure 4B; Supplementary Figure 4B, available online) and sh (Figure 4D; Supplementary Figure 4D, available online) cells are reported. Cell currents were measured before (total) and after IAA94 or 4,4’-diisothiocyano-2,2’-stilbenedisulfonic acid (DIDS) addition to the bath solution. The corresponding current/voltage relationships clearly show a IAA94-sensitive current in NT cells but not in sh cells (Figure 4, C and E; Supplementary Figure 4, C and E, available online). In all of the cells analyzed from both hGBM#7 and hGBM#10 neurospheres, we found that control cells always displayed an IAA94-sensitive current, whereas CLIC1-silenced cells consistently showed the absence of a detectable CLIC1-mediated Cl− current (GLM: P < .0001 related to cell type; P = .98 related to membrane potential) (Figure 4F; Supplementary Figure 4F, available online). The other Cl− currents in the cells, which were isolated using the inhibitor DIDS (DIDS-sensitive currents, IDIDS), were comparable (GLM: P = .07 related to cell type; P = .01 related to membrane potential) (Figure 4G; Supplementary Figure 4G, available online). These results corroborate the effects of CLIC1 silencing onto CLIC1 expression and ion channel activity, revealed as the lack of IAA94-sensitive current in CLIC1-silenced cells.
Effect of CLIC1 Silencing on Self-Renewal and Proliferation of Patient-Derived GBM Neurospheres
We next investigated the role of CLIC1 in regulating the maintenance and the growth of GBM neurospheres. In vitro self-renewal capacity of CLIC1-silenced and control cells was evaluated by methylcellulose assay. Single cells were plated in semisolid medium, single clones were counted after 15 days, and the clonogenic cells were calculated as the percentage of the total number of seeded cells. CLIC1-silenced cells formed statistically significantly fewer (hGBM#7 NT: 11.63±5.23%, sh: 3.83±1.75%; hGBM#9 NT: 14.86±3.37%, sh: 3.20±0.71%; hGBM#10 NT: 14.00±3.23%, sh: 3.35±0.98%; P < .01 in all experiments) (Figure 5, A and B; Supplementary Figure 5A, available online) and smaller colonies (NT: 502.5±56.85 µm; sh: 264.0±13.50 µm; P < .01; n = 5) (Figure 5C), with a lower cellular content compared with control cells (NT: 830.00±119.22 cells; sh: 483.33±85.68 cells; P < .01) (Figure 5D). When spheres generated at the first plating were dissociated and single cells were seeded on methyl-cellulose, control cells formed spheres with statistically significantly high efficiency, whereas CLIC1-silenced cells generated only a few small spheres, suggesting reduced self-renewal capacity (Supplementary Figure 5B, available online). Interestingly, there was no difference in clonogenic capacity between CLIC1-silenced and control cells at the third replating when CLIC1-silenced cells re-expressed the protein (Supplementary Figure 5, B and C, available online). Furthermore, CLIC1 silencing strongly reduced cellular growth kinetics in all patient-derived GBM neurospheres analyzed, as shown by 3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (hGBM#7: F = 233.5, df = 1, P < .001; hGBM#9: F=208.5, df = 1, P < .001; hGBM#10: F = 62.8, df = 1, P < .001) (Figure 5E; Supplementary Figure 5D, available online). Consistent with cell proliferation data, CLIC1 silencing strongly reduced the percentage of BrdU-positive cells in GBM-derived neurospheres (hGBM#7 NT: 64.32±3.93%, sh: 12.09±2.25%; hGBM#9 NT: 28.58±2.58%, sh: 14.48±0.18%; hGBM#10 NT: 41.39±0.80%, sh: 18.61±1.32%; P < .05 in all hGBM analyzed) (Figure 5F; Supplementary Figure 5E, available online). However, cell cycle analysis showed no alteration in cell cycle progression (Supplementary Figure 6A, available online). Moreover, no difference in the percentage of apoptotic cells between CLIC1-silenced cells and the control cells was detected (Supplementary Figure 6, B and C, available online). Together, these data indicate that CLIC1 downregulation affects the ability to steadily propagate GBM neurospheres.
Figure 5.
Effects of chloride intracellular channel 1 (CLIC1) silencing on clonogenicity and proliferation of glioblastoma (GBM) stem/progenitor cells. A) Representative microphotographs of control (NT) and CLIC1-silenced (sh) neurospheres formed in methilcellulose-containing medium after 15 days in culture. Scale bar = 300 µm. B) Neurosphere formation assay. The clonogenic capacity of control (NT) and CLIC1-silenced (sh) cells was evaluated by plating cells in methylcellulose-containing medium. After 15 days, each plate was examined under a light microscope, and the total number of neurospheres was determined. C) Quantification of the maximal diameters of control (NT) and CLIC1-silenced (sh) neurospheres from GBM patient 7 (hGBM#7). Ten neurospheres for each sample were analyzed. D) Quantification of hGBM#7 neurosphere cell number. Ten neurospheres for each sample were picked and dissociated, and the cell number was determined. E) The growth of control (NT) and CLIC1-silenced (sh) cells isolated from three patient samples was measured by 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay. Three independent experiments were performed; error bars represent 95% confidence intervals; ** P < .001. Generalized linear model tests of between-subjects effects showed statistically significant difference in all patients for the relative cell growth according to the time, the interference, and the interaction between those two variables. F) Control (NT) and CLIC1-silenced (sh) neurospheres isolated from three patient samples were subjected to BrdU incorporation assay: BrdU-positive cells were quantified by immunofluorescence. Immunostained cells were counted at 20× magnification, five fields for each sample (mean cell number per field was 150). For results in panels B, C, D, and F) an unpaired two-sided Student t test was used. Three independent experiments were performed; error bars represent 95% confidence intervals; *P < .05, **P < .001, ***P < .0001.
Effect of CLIC1 Antibody Treatment on Proliferation of Patient-Derived GBM Neurospheres
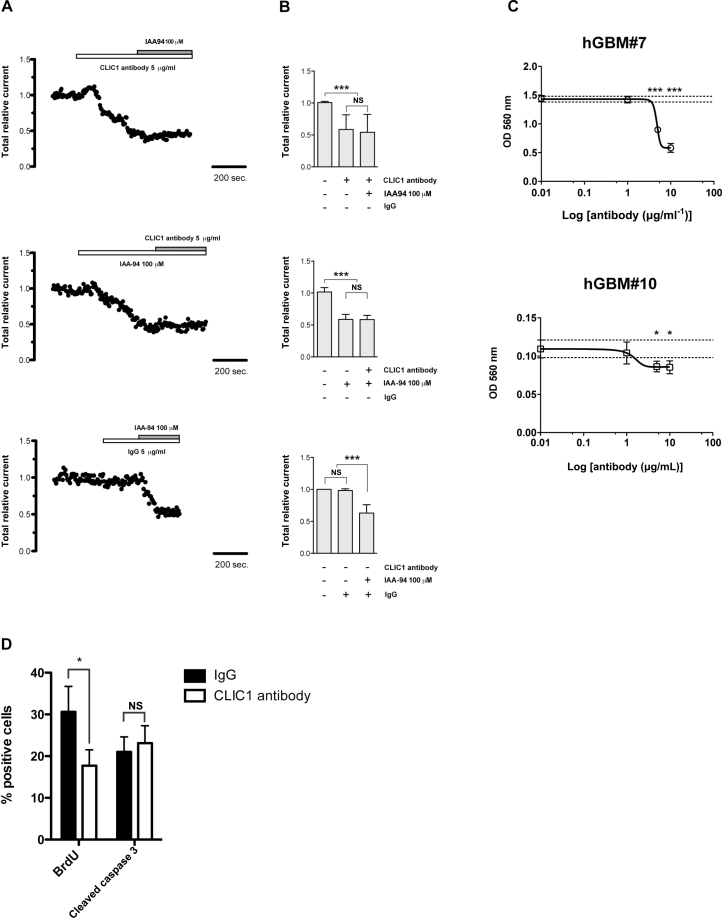
To determine whether the effect of CLIC1 silencing on GBM stem/progenitor growth is dependent on its function as an ion channel, we treated GBM neurospheres with a specific CLIC1 antibody. We performed electrophysiological recordings in perforated patch clamp configuration on cells derived by mechanically dissociated neurospheres to test the antibody efficacy in blocking CLIC1-mediated Cl− currents. Upon CLIC1 antibody addition, a reduction of total current was observed, but no further reduction was detected after IAA94 addition (Figure 6, A and B, upper panels); similar results were obtained by treating cells first with IAA94 and then with the specific CLIC1 antibody (Figure 6, A and B, middle panels) (ANOVA with Tamhane multiple comparison test for IAA94/CLIC1 antibody treatment: P < .0001). No alteration in total current was measured when the cells were treated with a mouse isotype antibody (immunoglobulin G [IgG]) as control (Figure 6, A and B, lower panels) (ANOVA with Tamhane multiple comparison test for IgG antibody treatment: P = .66). Overall, these data prove the efficacy and the specificity of CLIC1 antibody in blocking CLIC1-mediated Cl− currents.
Figure 6.
Effects of chloride intracellular channel 1 (CLIC1) antibody treatment on glioblastoma (GBM) neurospheres. A and B) The effect of CLIC1 antibody on CLIC1 currents was assessed by perforated patch clamp technique. A) Representative whole cell current traces recorded in the perforated patch configuration at 50 mV from control (NT) and CLIC1-silenced (sh) cells derived from cells from GBM patient 10 (hGBM#10) are shown. IAA94 = indanyloxyacetic acid 94; IgG = immunoglobulin G. B) Quantification of the different treatments as in (A) on whole cell current traces. Mean values derived from four independent experiments were represented. The statistical significance of the differences in relative total current after Tahmane test for dishomogeneous variances is shown: ***P < .0001; NS = not significant. C) Effect of CLIC1 antibody on GBM neurospheres derived from hGBM#7 and hGBM#10 patients. GBM neurospheres were treated with increasing concentrations of CLIC1 antibody (1, 5, and 10 µg/mL) for 72 hours, and cell viability was monitored by 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay; error bars represent 95% confidence intervals; three independent experiments were performed. The difference between cell viability at different antibody concentrations and reference mean viability in control conditions was evaluated by Bonferroni test. The statistical significance of the differences is shown: *P < .05, ***P < .0001. OD = optical density. D) Effect of CLIC1 antibody treatment on BrdU incorporation (left panel) and caspase 3 activation (right panel) in hGBM#7 neurospheres. BrdU- or cleaved caspase 3–positive cells were counted at 20× magnification, five fields for each sample (mean cell number per field was 150). Three independent experiments were performed. An unpaired two-sided Student t test was used: *P < .05; NS = not significant.
We next treated GBM neurospheres with different doses of CLIC1 antibody (1, 5, and 10 µg/mL) and measured the percentage of viable cells after 72 hours. The blockage of CLIC1 activity decreased cell growth in a dose-dependent manner (Figure 6C). The maximal biological effect was observed at the highest doses tested (10 µg/mL) in cells that express higher levels of CLIC1 (hGBM#7), whereas cells expressing lower levels of CLIC1 (hGBM#10) reached the maximal biological effect already at lower doses of CLIC1 antibody (5 µg/mL) (P < .05 at 5 and 10 μg/mL of CLIC1 antibody). BrdU uptake was reduced in both cell lines after CLIC1 antibody treatment (NT: 30.6±6.90%; sh: 17.7±4.30%; P < .05), whereas there were no differences in the percentages of apoptotic cells between treated and untreated cells (CLIC1 antibody: 23.1±4.2%; IgG: 21.0±3.6%; P = .42) (Figure 6D). These results recapitulate those obtained after CLIC1 silencing, demonstrating that CLIC1 ion channel activity is essential for the growth of GBM stem/progenitor cells.
Evaluation of CLIC1 Role on Glioblastoma Development
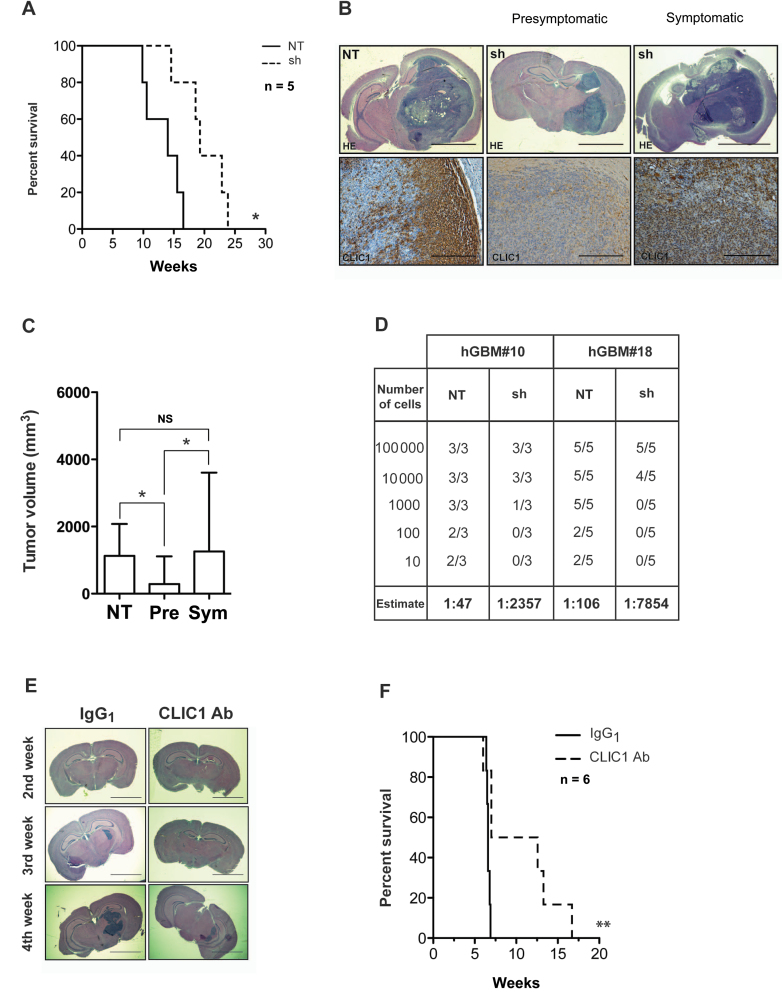
To determine the in vivo relevance of CLIC1 silencing, we performed an orthotopic transplantation assay. We stereotaxically implanted dissociated neurospheres infected with a lentivirus expressing either NT or shRNA specific for CLIC1 (sh) into the nucleus caudatus of immunodeficient mice. We monitored tumor formation and growth until the appearance of neurological signs. Survival of mice injected with CLIC1-silenced cells was prolonged in comparison with NT control mice (χ2 = 6.21; df = 1; P < .05) (Figure 7A; Supplementary Figure 7, available online). Both control and CLIC1-silenced mice eventually developed GBMs according to WHO classification. When we killed the transplanted mice at the same time (ie, at the appearance of the neurological signs in control mice), CLIC1-silenced mice (sh) were still presymptomatic (pre), and their tumors were statistically significantly smaller than those of the control mice (NT); however, when we analyzed CLIC1-silenced symptomatic (sym) mice, their tumors reached the size of control tumors (Figure 7, B and C) (ANOVA with Tamhane multiple comparison tests: NT vs pre: P < .05; pre vs sym: P < .05; NT vs sym: P = not significant). CLIC1-silenced xenografts lacked CLIC1 expression as detected by immunohistochemistry at the early time point, whereas CLIC1 expression level became comparable between CLIC1-silenced (sh) tumors and control tumors (NT) at the late time point (Figure 7B). Interestingly, when lower numbers of cells (102 and 10 for GBM#10 and 103, 102, and 10 for GBM#18) were injected into mice, none of the mice that received CLIC1-silenced cells developed tumors (Figure 7D). The calculated stem cell frequency by the extreme limiting dilution assay (ELDA)algorithm was statistically significantly lower in CLIC1-silenced cells (hGBM#10: χ2 = 17.5, df = 1, P < .0001; hGBM#18: χ2 = 34.2, df = 1, P < .0001) and was underestimated because of the observed CLIC1 re-expression in all formed tumors. Thus CLIC1 appears to be relevant for the formation of tumors in GBM neurospheres.
Figure 7.
Evaluation of chloride intracellular channel 1(CLIC1) role on glioblastoma development. A) Kaplan–Meier survival curve of mice intracranially transplanted with 105 control (NT) and CLIC1-silenced (sh) cells. Number of mice at risk expressed as weeks (number of mice at risk): 0 (5), 9.8 (5), 10.57 (4), 14 (3), 14.5 (2), 16.5 (1) for NT; 0 (5), 14.5 (5), 18.57 (4), 19.3 (3), 22.8 (2), and 23.8 (1) for sh group. Data are from one experiment with five mice per group. P value was calculated with log rank test: * P < .05; χ2 = 6.27; degrees of freedom (df) = 1. B) Representative brain images from mice intracranially injected with control (NT) and CLIC1-silenced (sh) cells stained with hematoxylin and eosin (HE) (top row; scale bar = 3mm) or CLIC1 (bottom row; scale bar = 300 µm). C) Tumor volume quantification, as indicated. Experiment was carried out using three mice per group. Error bars represent 95% confidence intervals; *P < .05. One-way analysis of variance with Bonferroni correction was used. Pre = presymptomatic; Sym = symptomatic. D) Table representing the incidence of tumor formation of tumor bearing mice and the cancer stem cell frequency calculated in the glioblastoma (GBM) neurospheres (estimate). hGBM#10: χ2 = 17.5; P < .0001; hGBM#18: χ2 = 34.2; P < .0001. E) Representative hematoxylin and eosin–stained histological images from mice intracranially injected with hGBM#7 cells treated with CLIC1 antibody (Ab) or isotype control antibody (scale bar = 3mm). Mice were killed at the second, third, and fourth week, as shown. IgG = immunoglobulin G. F) Kaplan–Meier survival analysis of mice intracranially implanted with 105 hGBM#7 cells treated with CLIC1 antibody or isotype control antibody. Number of mice at risk expressed as weeks (number of mice at risk): 0 (6), 6.4 (6), 96.5 (5), 6.6 (4), 6.8 (2), 6.9 (1) for IgG1 group; 0 (6), 6.6 (6), 7.0 (5), 12.6 (3), 13.3 (2), 16.7 (1) for CLIC1 group. Data are from one experiment with six mice per group. P value was calculated with log rank test: *P = .01; χ2= 6.36; df=1.
Given the ability of CLIC1 antibody to reduce the proliferation of patient-derived GBM neurospheres in vitro, we next explored a potentially translatable targeting of CLIC1 in vivo. To test this, we transplanted GBM-derived neurospheres treated with CLIC1 antibody into the brains of immunodeficient mice. We killed three mice every week, following tumor progression for a month. Cell treatment with CLIC1 antibody resulted in smaller tumors (Figure 7E) and statistically significantly improved overall mouse survival (Figure 7F) (n = 6; log-rank P < .01). Thus, transient exposure to CLIC1 antibody produces a statistically significant decrease in the in vivo tumorigenicity of GBM cells.
Discussion
In this study, we showed that CLIC1 plays a pivotal role in the tumorigenic potential of CSCs isolated from human GBM. We demonstrate that CLIC1 is essential for self-renewal and proliferation of GBM CSCs. Moreover, the demonstration of reduced gliomagenesis after CLIC1 targeting in tumoral stem/progenitor cells and the finding that CLIC1 expression inversely associates with patient survival suggest a potential exploitation of CLIC1 as a new molecular therapeutic target and a possible outcome predictor.
By investigating different publicly available expression microarray datasets, we identified CLIC1 to be overexpressed in brain tumors compared with normal brains, with expression increasing along with WHO tumor grades and reaching the highest expression level in GBMs. Moreover, among GBMs, we identified CLIC1 to cluster within the GBM MES subtype, which is considered to have poorer prognostic status because of high infiltration rate and marked vascularization (28,33), higher necrosis and associated inflammatory infiltrates (29), and increased treatment resistance. Importantly, our study also pointed out that CLIC1 expression is inversely associated with patient survival, and therefore it could be of potential prognostic value in monitoring glioma progression. CLIC1 overexpression has been demonstrated in a wide variety of tumor types (15,16), including glioma (17). Taken together, these studies demonstrate that CLIC1 overexpression confers proliferative advantage, is required for cancer cell migration and invasion, and sustains cancer cell tumorigenicity. Recently, chloride channels have been involved in the chemotherapeutic resistance of glioma stem-like cells (21). Here, we demonstrate that CLIC1 silencing negatively influences both proliferative capacity and self-renewal properties in vitro and impairs in vivo tumorigenic potential of stem/progenitor cells derived from GBM patients. GBMs are the most frequent brain tumors, and despite different treatment modalities, overall results have remained unchanged over the last 25 years. GBM patients have less than 30% probability of surviving more than 2 years, even with optimal therapy. Thus, the finding of a good target for patient-specific therapy would be of paramount importance from a clinical standpoint. GBMs contain a subpopulation of cancer stem cells with intrinsic resistance to therapy that can repopulate the tumor after treatment. Therefore, a new approach to cancer therapy might focus on specific targeting of the resistant CSC populations. In our study, we showed that CLIC1 is enriched in cancer stem/progenitor cells compared with the cells that make up the bulk of the tumor. In physiological conditions, CLIC1 exists usually in a soluble form in the cytoplasm, but after oxidative stimuli it translocates to the plasma membrane, where it acts as a chloride-selective ion channel (12–14). CLIC1 localization on the plasma membrane has been associated with cells in the G2/M stage of the cell cycle (10), and alteration of CLIC1 levels by RNA interference has been demonstrated to impair cell cycle progression in vitro (34). We found that CLIC1 is constitutively localized on the plasma membrane of GBM stem/progenitor cells compared with the normal counterpart. This different localization of CLIC1 in tumoral vs normal stem/progenitor cells could allow the specific targeting of cancer cells. Moreover, we demonstrate that CLIC1 silencing affects proliferation, clonogenicity, and tumorigenic potential of GBM stem/progenitor cells. Given that CLIC1 is constitutively expressed on the plasma membrane of GBM stem/progenitor cells, conferring them with a growth advantage, our results suggest CLIC1 as a molecular target for therapeutic purposes during the initiation and progression of the tumorigenic process. Notably, the treatment of human GBM stem/progenitor cells with the specific CLIC1 antibody mimics the biological effects of CLIC1 silencing, reducing tumor cell growth both in vitro and in vivo and demonstrating that CLIC1 biological effect is dependent on its function as an ion channel on the plasma membrane.
This study also had some limitations. Small molecules that specifically inhibit CLIC1 expression or functions are yet to be identified. The investigation of the molecular players mediating the functional effects of CLIC1 in GBM stem cells would permit new therapeutic strategies to block GBM. The investigation of these molecules for inhibiting GBM progression in human patients is therefore highly warranted.
In conclusion, these results open new perspectives for GBM molecular therapies: CLIC1 becomes an attractive target to test novel therapeutic approaches specifically directed to glioma stem/progenitor cells. Moreover, considering CLIC1 enrichment in GBM MES subtype, patients with a poor prognosis molecular profile (ie, mesenchymal glioblastoma patients) may be the subset that would gain particular benefit from CLIC1-dependent therapy.
Funding
This work was supported by the Foundation Veronesi (to PG and GP) and the Ministry of Health (to GP).
Supplementary Material
M. Mazzanti and G. Pelicci should be considered senior authors for their respective research fields.
We thank E. Cetti, the imaging, tissue culture, and mouse facilities at the IFOM-IEO Campus, for technical assistance; Dr. Samuel Breit (St Vincent’s Centre for Applied Medical Research, St Vincent’s Hospital) for the kind gift of MEF CLIC1 knock-out cells; and Federica Pisati for helpful assistance in immunohistochemistry analysis.
References
- 1. Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21(21):2683–2710 [DOI] [PubMed] [Google Scholar]
- 2. Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401 [DOI] [PubMed] [Google Scholar]
- 3. Galli R, Binda E, Orfanelli U, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64(19):7011–7021 [DOI] [PubMed] [Google Scholar]
- 4. Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760 [DOI] [PubMed] [Google Scholar]
- 5. Sontheimer H. Malignant gliomas: perverting glutamate and ion homeostasis for selective advantage. Trends Neurosci. 2003;26(10):543–549 [DOI] [PubMed] [Google Scholar]
- 6. Olsen ML, Schade S, Lyons SA, et al. Expression of voltage-gated chloride channels in human glioma cells. J Neurosci. 2003;23(13):5572–5582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Watkins S, Sontheimer H. Hydrodynamic cellular volume changes enable glioma cell invasion. J Neurosci. 2011;31(47):17250–17259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Habela CW, Ernest NJ, Swindall AF, et al. Chloride accumulation drives volume dynamics underlying cell proliferation and migration. J Neurophysiol 2009;101(2):750–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Valenzuela SM, Martin DK, Por SB, et al. Molecular cloning and expression of a chloride ion channel of cell nuclei. J Biol Chem. 1997;272(19):12575–12582 [DOI] [PubMed] [Google Scholar]
- 10. Valenzuela SM, Mazzanti M, Tonini R, et al. The nuclear chloride ion channel NCC27 is involved in regulation of the cell cycle. J Physiol. 2000;529(Pt 3):541–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Littler DR, Harrop SJ, Goodchild SC, et al. The enigma of the CLIC proteins: ion channels, redox proteins, enzymes, scaffolding proteins? FEBS Lett. 2010;584(10):2093–2101 [DOI] [PubMed] [Google Scholar]
- 12. Littler DR, Harrop SJ, Fairlie WD, et al. The intracellular chloride ion channel protein CLIC1 undergoes a redox-controlled structural transition. J Biol Chem. 2004;279(10):9298–9305 [DOI] [PubMed] [Google Scholar]
- 13. Goodchild SC, Howell MW, Cordina NM, et al. Oxidation promotes insertion of the CLIC1 chloride intracellular channel into the membrane. Eur Biophys J. 2009;39(1):129–138 [DOI] [PubMed] [Google Scholar]
- 14. Singh H, Ashley RH. Redox regulation of CLIC1 by cysteine residues associated with the putative channel pore. Biophys J. 2006;90(5):1628–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang JW, Peng SY, Li JT, et al. Identification of metastasis-associated proteins involved in gallbladder carcinoma metastasis by proteomic analysis and functional exploration of chloride intracellular channel 1. Cancer Lett. 2009;281(1):71–81 [DOI] [PubMed] [Google Scholar]
- 16. Petrova DT, Asif AR, Armstrong VW, et al. Expression of chloride intracellular channel protein 1 (CLIC1) and tumor protein D52 (TPD52) as potential biomarkers for colorectal cancer. Clin Biochem. 2008;41(14–15):1224–1236 [DOI] [PubMed] [Google Scholar]
- 17. Wang LHS, Tu Y, Ji P, et al. Elevated expression of chloride intracellular channel 1 is correlated with poor prognosis in human gliomas. J Exp Clin Cancer Res. 2012; 31(1):44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Milton RH, Abeti R, Averaimo S, DeBiasi S, et al. CLIC1 function is required for beta-amyloid-induced generation of reactive oxygen species by microglia. J Neurosci. 2008;28(45):11488–11499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Novarino G, Fabrizi C, Tonini R, et al. Involvement of the intracellular ion channel CLIC1 in microglia-mediated beta-amyloid-induced neurotoxicity. J Neurosci. 2004;24(23):5322–5330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Averaimo S, Milton RH, Duchen MR, et al. Chloride intracellular channel 1 (CLIC1): sensor and effector during oxidative stress. FEBS Lett. 2010;584(10):2076–2084 [DOI] [PubMed] [Google Scholar]
- 21. Kang MK, Kang SK. Pharmacologic blockade of chloride channel synergistically enhances apoptosis of chemotherapeutic drug-resistant cancer stem cells. Biochem Biophys Res Commun. 2008;373(4):539–544 [DOI] [PubMed] [Google Scholar]
- 22. Pellegatta S, Poliani PL, Corno D, et al. Neurospheres enriched in cancer stem-like cells are highly effective in eliciting a dendritic cell-mediated immune response against malignant gliomas. Cancer Res. 2006;66(21):10247–10252 [DOI] [PubMed] [Google Scholar]
- 23. Ortensi B, Osti D, Pellegatta S, et al. Rai is a new regulator of neural progenitor migration and glioblastoma invasion. Stem Cells. 2012;30(5):817–832 [DOI] [PubMed] [Google Scholar]
- 24. Goldstein M, Watkins S. Immunohistochemistry. Curr Protoc Mol Biol. Jan. 2008;chapter 14unit 14.6. 10.1002/0471142727.mb1406s81. [DOI] [PubMed] [Google Scholar]
- 25. Dull T, Zufferey R, Kelly M, et al. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72(11):8463–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cao H, Hripcsak G, Markatou M. A statistical methodology for analyzing co-occurrence data from a large sample. J Biomed Inform. 2007;40(3):343–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Madhavan S, Zenklusen JC, Kotliarov Y, Sahni H, Fine HA, Buetow K. REMBRANDT: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res. 2009;7(2):157–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173 [DOI] [PubMed] [Google Scholar]
- 29. Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. The Cancer Genome Atlas Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216): 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403 [DOI] [PubMed] [Google Scholar]
- 32. Carletta J. Assessing agreement on classification tasks: the kappa statistic. Computational Linguistics. 1996;22(2):249–254 [Google Scholar]
- 33. Carro MS, Lim WK, Alvarez MJ, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463(7279):318–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tung JJ, Kitajewski J. Chloride intracellular channel 1 functions in endothelial cell growth and migration. J Angiogenes Res. 2010;2:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.