Abstract
The Hippo pathway is an evolutionary conserved pathway that involves cell proliferation, differentiation, apoptosis and organ size regulation. Mst1 and Mst2 are central components of this pathway that are essential for embryonic development, though their role in controlling embryonic stem cells (ES cells) has yet to be exploited. To further understand the Mst1/Mst2 function in ES cell pluripotency and differentiation, we derived Mst1/Mst2 double knockout (Mst-/-) ES cells to completely perturb Hippo signaling. We found that Mst-/- ES cells express higher level of Nanog than wild type ES cells and show differentiation resistance after LIF withdrawal. They also proliferate faster than wild type ES cells. Although Mst-/- ES cells can form embryoid bodies (EBs), their differentiation into tissues of three germ layers is distorted. Intriguingly, Mst-/- ES cells are unable to form teratoma. Mst-/- ES cells can differentiate into mesoderm lineage, but further differentiation to cardiac lineage cells is significantly affected. Microarray analysis revealed that ligands of non-canonical Wnt signaling, which is critical for cardiac progenitor specification, are significantly repressed in Mst-/- EBs. Taken together our results showed that Mst1/Mst2 are required for proper cardiac lineage cell development and teratoma formation.
Introduction
The Hippo pathway was first discovered in Drosophila. Through genetic mosaic screens, core components of the Hippo pathway, such as Warts (Wts), Hippo (Hpo) and Salvador (Sav) were identified as tumor-suppressor genes [1-4]. These components restrict cell proliferation and promote apoptosis by repressing the downstream effector Yokie (Yki) in Drosophila. Depletion of core components of the Hippo pathway or overexpression of Yki results in enhanced cell proliferation and reduced apoptosis respectively [5]. This pathway is highly conserved in mammals. Serine/threonine kinases Mst1/Mst2 and Lats1/Lats2 in mammals are homologs of Hippo and Wts in Drosophila respectively. Together with an adaptor protein hMob1, they transmit signals to downstream effectors [6]. Through inhibiting the transcriptional co-activators and oncoproteins Yap (Yes kinase-associated protein) and Taz (transcriptional coactivator with PDZ-binding motif), the Hippo pathway promotes apoptosis and inhibits tumorigenesis in mammals [7-10].
Mst1 and Mst2 (Mammalian sterile 20-like kinases 1 and 2) are the core components of the Hippo pathway. They play important roles in early embryonic development, cell proliferation, apoptosis and organ size control. Mst1 null mice are viable and fertile but have a reduced number of mature naive T cells, while Mst2 null mice are also fertile but exhibit no developmental or immunological defects [11]. However, depletion of both Mst1 and Mst2 resulted in embryonic lethality at embryonic day 8.5, suggesting redundant roles of Mst1 and Mst2 [12]. One functional copy of either Mst1 or Mst2 is necessary and sufficient for early embryonic development [11,13].
Like other components of the Hippo pathway that promote apoptosis, Mst1/Mst2 are pro-apoptotic kinases [14,15]. Under oxidative stress, Mst1/Mst2 activate transcription factor Foxo and promote neuronal cell death [16,17]. Heart specific expression of Mst1 leads to dilated cardiomyopathy with reduction in cell density in heart [18]. Liver specific removal of Mst1/Mst2 in newborn mice results in liver enlargement and formation of hepatocellular carcinoma and cholangiocarcinoma [12,19-21]. Similarly, in mouse intestines and pancreas, inactivation of Mst1/Mst2 leads to intestinal stem cell overproliferation, colonic tumorigenesis and pancreas overgrowth [22-24], suggesting important roles of Mst1/Mst2 in organ size control and tumorigenesis.
Mst1/Mst2 activate Lats1 and Lats2 by phosphorylation, and in turn phosphorylate Yap and inhibit it from translocating into the nucleus [25]. Unphosphorylated Yap can be translocated into the nucleus to activate TEA-domain (TEAD) family members. The Yap/Taz-Tead complex further activates proliferation by a genome wide transcriptional program [26-28]. Ectopic expression of Yap in mammalian cells leads to a phenotype resembling that from ablation of core components of the Hippo pathway. Similar to the simultaneous removal of Mst1/Mst2, overexpression of Yap in mice results in a dramatic increase of liver mass with subsequent tumor formation. In addition previous research reveals that Yap is an important pluripotent factor. Expression of Yap enhances reprogramming of differentiated cells to induced pluripotent stem (iPS) cells [26,29,30]. In adults Yap is enriched in organs such as the small intestine and the developing brain and its expression is highly restricted to the progenitor or stem compartments, whereas in other tissues, such as skin and skeletal muscle, the expression of Yap is gradually decreased with regard to differentiation status [10,26,31,32]. Yap is therefore, a stemness gene in mammalian cells, while key components of Hippo pathway such as Mst1/Mst2, function to constrain this stemness gene in restricted compartments.
Mst1/Mst2 double knockout mice die at E8.5 with abnormalities in the placenta, vascular patterning and primitive hematopoiesis, suggesting that Mst1/Mst2 are not required for pluripotent inner cell mass (ICM) formation but are required for subsequent organ and tissue development. As an in vitro derivative of the pluripotent inner cell mass (ICM), ES cells retain the developmental characteristics of ICM and can self-renew and differentiate to all three germ layers. When ES cells are injected to the blastocysts, they can contribute to all of the animal cell types [33,34]. These unique properties make ES cells suitable for genetic modification and they have the potential to serve as a source of regenerative medicine for cell therapy. In addition, successful reprogramming of somatic cells into induced pluripotent stem cells opens a new gate for stem cell therapy that avoids various ethical issues [35-37]. Direct transplanting of ES cells into hosts can however lead to teratoma formation and this remains a clinical challenge for ES cell application. Harnessing this valuable tool for therapeutic use will require overcoming this problem through innovative exploitation of the mechanisms involved in ES cell pluripotency, differentiation and tumorigenesis. Many signaling pathways, including the Hippo pathway in ES cell pluripotency and lineage commitment, have yet to be well characterized. Mst1/Mst2 are however required for proper development of some organs and tissue in embryos, but it is not clear whether Mst1/Mst2 play any corresponding role during ES cell differentiation at cellular level that consequently leads to developmental defects. To further address this question, we generated Mst-/- ES cells from Mst1 and Mst2 mutant mice [12]. We found that the phosphorylation level of Yap was decreased in Mst-/- ES cells, whilst the pluripotency marker Nanog was increased significantly compared to wild type ES cells. Mst-/- ES cells also showed differentiation resistance for a relatively longer time compared to wild type ES cells under differentiation conditions. Consistent with the developmental defects of Mst1/Mst2 double knockout mice, Mst-/- ES cells showed lineage development distortion during embryoid body (EB) formation and obvious defects to differentiation to cardiac progenitor cells. Microarray analysis revealed that the non-canonical Wnt pathway ligands Wnt2b and Wnt5a, which are critical for cardiac progenitor cell differentiation, were significantly downregulated in Mst-/- EBs. Unlike wild type ES cells, Mst-/- ES cells could not form teratoma after subcutaneously injected into nude mice. Taken together, our data suggest that Mst1/Mst2 are required for teratoma formation. Their functions are also critical for proper cardiac lineage cell formation.
Results
Derivation of mouse Mst-/- embryonic stem cells
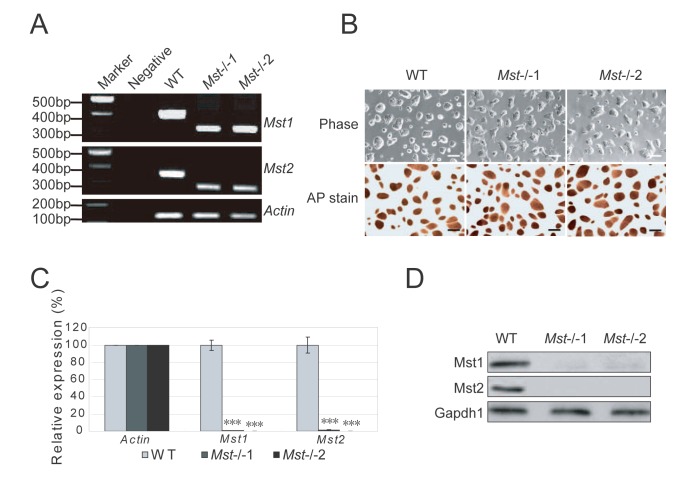
Based on the schematics of null alleles of Mst1 and Mst2 generated in a previous study [12], we crossed the Mst1 +/-Mst2-/- male and female mice, harvested the E3.5 embryos and derived ES cells on MEF feeder (Figure S1A and S1B). These cells were further adapted to form feeder-free ES cell lines under the 2i+LIF condition for genotyping. Exon 4 and 5 which encode kinase domain are deleted in Mst1 knockout, while exon 5 and 6 which encode kinase domain are deleted in Mst2 knockout (Figure S1C). With primers targeted to the adjacent sequence of the deleted regions of Mst1 (exon 4 and 5) and Mst2 (exon 5 and 6), PCR confirmed that respective regions of Mst1 and Mst2 genomic DNA were deleted in Mst1/Mst2 double knockout (Mst-/-) ES cell lines respectively (Figure 1A). Two Mst1/Mst2 double knockout lines, Mst-/-1 and Mst-/-2 ES cell lines were selected for further studies (Figure 1A). The Mst-/- ES cells still maintained dome-shaped colony morphology similar to that of the wild type ES cells (Figure 1B). They also expressed high levels of ES protein alkaline phosphatase (Figure 1B). To further confirm the absence of both Mst1 and Mst2 in the Mst-/- ES cells, primers targeting the corresponding deleted transcript region were used to do RT-PCR, the result confirmed that neither Mst1 nor Mst2 transcript was found in Mst-/- ES cells (Figure 1C). Further examination of the protein extracts with antibodies specifically against kinase domains of Mst1 and Mst2 protein showed that neither Mst1 nor Mst2 proteins were detected in the Mst-/- ES cells (Figure 1D). Taken together, these results showed that both Mst1 and Mst2 were functionally inactive in both Mst-/- ES cell lines.
Figure 1. Isolation of Mst-/- ES cells.
(A) Genotyping of wild type (WT) ES cells and Mst-/- ES cells derived from blastocysts by PCR amplification of genomic DNA. Wild type ES cells showed a larger band while Mst-/- ES cells displayed a smaller band. Actin was used as an internal control. (B) Phase contrast microscopy of wild type (WT) and two independent Mst-/- knockout ES cell lines (Mst-/-1 and Mst-/-2) grown on 0.2% gelatin in 2i+LIF medium (Upper). These cells were stained for alkaline phosphatase (Lower). Scale bar, 200 μm. (C) mRNA level of Mst1 and Mst2 in wild type ES cells and Mst-/- ES cells examined by quantitative real-time PCR using primers flanking the deleted region of Mst1 and Mst2. The data are shown as the mean ± S.D (n=3). Actin was normalized as an internal control. Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001). (D) Immunoblotting analysis of the expression of Mst1 and Mst2 in wild type ES cells and Mst-/- ES cells. Gapdh1 was used as a loading control.
Characterization of mouse Mst-/- embryonic stem cells
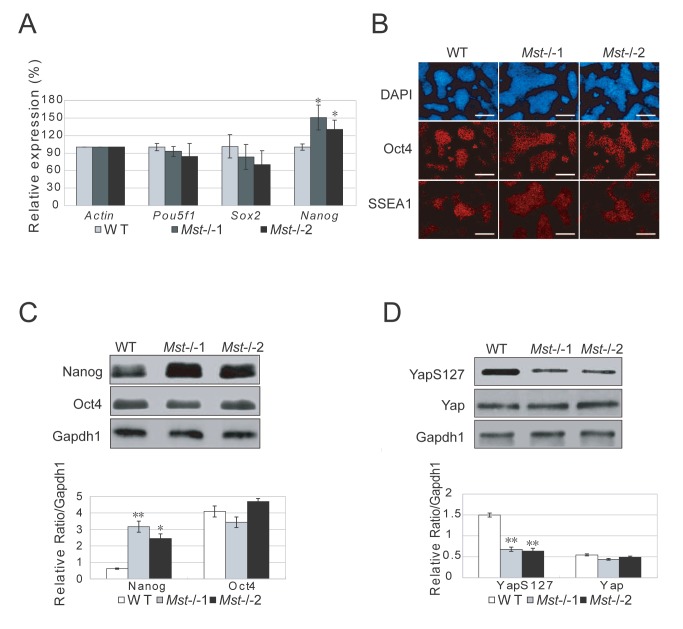
To examine whether deletion of Mst1 and Mst2 affects the integrity of ES cells, we examined the expression profiles of pluripotency markers Pou5f1, Nanog, and Sox2 by RT-PCR. There was an increase of Nanog transcripts in both Mst-/- ES cell lines, but the expression levels of Pou5f1 and Sox2 were similar in Mst-/- and wild type ES cells (Figure 2A). Immunofluorescence assay with antibodies against pluripotent markers Oct4 and SSEA1 revealed no significant difference between wild type ES cells and Mst-/- ES cells (Figure 2B). Further checking of the protein level of ES cell transcription factors, Nanog and Oct4, confirmed that Oct4 protein level was not affected by Mst1/Mst2 deletion, but Nanog protein was higher in Mst-/- ES cells than wild type ES cells (Figure 2C). This result suggests that the expression of Nanog may be regulated by Mst1/Mst2 kinases.
Figure 2. Characterization of Mst-/- ES cells.
(A) Quantitative real-time PCR to examine the mRNA level of pluripotent markers Pou5f1, Sox2 and Nanog in wild type ES cells and Mst-/- knockout ES cells. Actin was analyzed as an internal control. The data are shown as the mean ± S.D (n=3). Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001). (B) Immunofluorescence staining of the pluripotent protein Oct4 and SSEA1 expression in wild type ES cells and Mst-/- knockout ES cells. Neuclei were stained with DAPI. Scale bar, 200μm. (C) Immunoblotting and densitometric analysis of Nanog and Oct4 in wild type ES cells and Mst-/- ES cells. Gapdh1 was analyzed as an internal control. The data are shown as the mean ± S.D (n=2). Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001). (D) Immunoblotting and densitometric analysis of the expression of Yap and phosphorylated Yap (YapS127) in wild type ES cells and Mst-/- ES cells. Gapdh1 was analyzed as an internal control. The data are shown as the mean ± S.D (n=2). Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001).
As a downstream effector of the Hippo pathway, Yap is also a pluripotent factor in ES cells and expression of Yap promotes reprogramming efficiency of mouse iPS cells. We therefore examined the expression of Yap by RT-PCR. The expression of Yap is similar in wild type ES cells and Mst-/- ES cells (Figure S2A). Further examination of Yap and phosphorylated Yap by western blot and immunofluorescence stain revealed that total Yap is not changed in Mst-/- ES cells, but phosphorylated Yap is significantly reduced in Mst-/- ES cells (Figure 2D and S2B). This observation is consistent with previous reports that Mst1/Mst2 phosphorylate Lats1/Lats2, which in turn phosphorylate Yap. As unphosphorylated Yap actively promotes cell proliferation, these data confirmed that the Hippo pathway, which is active in ES cells, is significantly downregulated in Mst-/- ES cells. As the effect of upregulation of active Yap, Ctgf and Cyr61, the downstream targets of Yap [38], were significantly increased in Mst-/- ES cells (Figure S2C).
Differentiation resistance of mouse Mst-/- embryonic stem cells
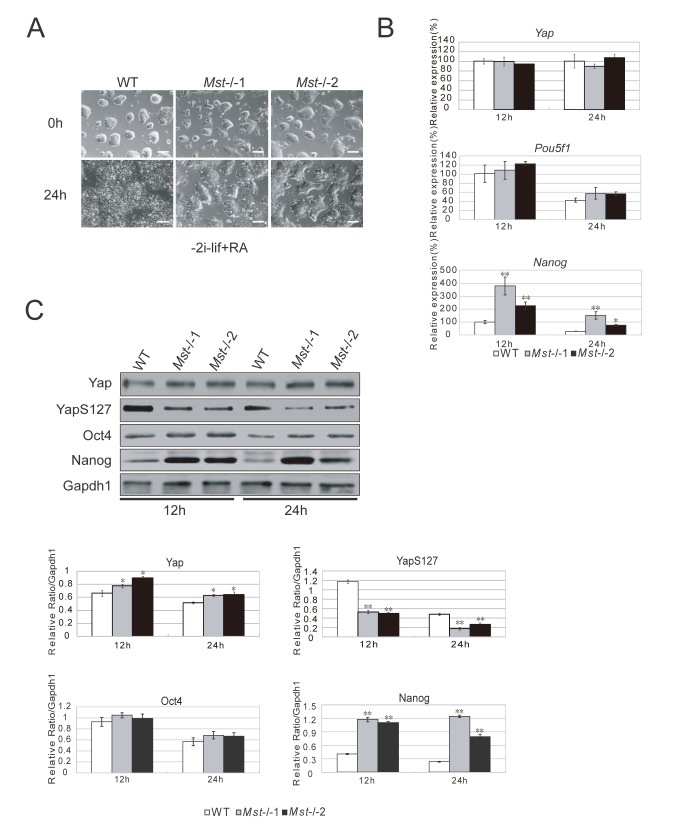
Based on above observations, we examined whether the maintenance of pluripotency was altered in ES cells by Mst1/Mst2 deletion. We withdrew the molecular chemicals 2i (CHIR99021 and PD0325901) and LIF from the ES cell culture medium, and added retinoic acid (RA) to promote ES cell differentiation. We found that wild type ES cells completely lost colony morphology and were differentiated after 24 hours, while Mst-/- ES cells still maintained a certain level of colony morphology (Figure 3A). RT-PCR examination of the pluripotent markers showed that the mRNA level of Nanog was significant higher in Mst-/- ES cells than wild type ES cells, while the mRNA level of Yap and Pou5f1 in Mst-/- ES cells and wild type ES cells was similar (Figure 3B). We further measured the protein level by western blot. Mst-/- ES cells expressed slightly more Yap protein than wild type ES cells from 12 hours after LIF withdrawal, but phosphorylated Yap (S127) was significantly decreased in Mst-/- ES cells compared to wild type ES cells, suggesting an increase of active unphosphorylated Yap in Mst-/- ES cells. Consistent with the upregulation of Nanog transcript, Nanog protein was also significantly increased in Mst-/- ES cells. And there was no obvious change of Oct4 between Mst-/- ES cells and wild type ES cells (Figure 3C). Taken together, these data showed that deletion of Mst1 and Mst2 increased the barrier of ES cell differentiation through upregulating the pluripotent marker unphosphorylated Yap and Nanog.
Figure 3. Differentiation resistance of Mst-/- ES cells.
(A) Morphology of wild type ES cells and Mst-/- ES cells initially and 24 hour after growing in ES cell differentiation medium supplemented with RA, but not 2i and LIF. Scale bar, 200 μm. (B) Quantitative real-time PCR to examine the mRNA level of Yap, Pou5f1 and Nanog in wild type ES cells and Mst-/- ES cells during ES cell differentiation medium for 12 hours and 24 hours. Actin was analyzed as an internal control. The data are shown as the mean ± S.D (n=3). Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001). (C) Immunoblotting and densitometric analysis of Yap, YapS127, Oct4 and Nanog in wild type ES cells and Mst-/- ES cells in ES cell differentiation medium for 12 hours and 24 hours. Gapdh1 was analyzed as an internal loading control. The data are shown as the mean ± S.D (n=2). Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001).
Enhanced cell proliferation of mouse Mst-/- embryonic stem cells
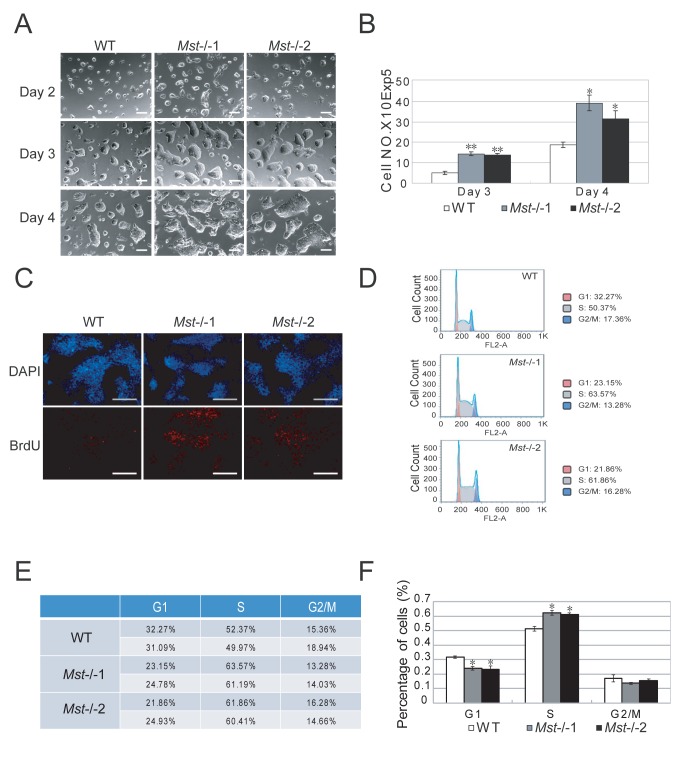
To assess the effects of Mst1 and Mst2 on ES cell proliferation, the same number of wild type and Mst-/- ES cells were trypsinized to single cells and plated. After 2 days, dome-shaped colonies were formed from single cells of both cell types. Interestingly, Mst-/- ES cell colony size was generally bigger than wild type ES cells (Figure 4A). Meanwhile, the cell numbers of Mst-/- ES cells in day 3 and day 4 cultures were significantly greater than wild type ES cells, suggesting Mst-/- ES cells have a higher proliferation capacity than wild type ES cells (Figure 4B). We then measured the expression of cell cycle related genes Ccnd2 and Ccnd3 by RT-PCR and found that Mst-/- ES cells expressed significantly higher Ccnd2 and Ccnd3 than wild type ES cells (Figure S6A).To further substantiate our observation, wild type and Mst-/- ES cells were pulse incorporated with BrdU for 1 hour after serum starvation and quantified by immunofluorescence staining and flow cytometry respectively. We observed significantly fewer BrdU-positive cells in the wild type ES cells, compared to the Mst-/- ES cells (Figure 4C, S6B and S6C). Cell cycle analysis by Propidium Iodide staining revealed that more than 60% of Mst-/- ES cells were at S phase, while about 50% wild type ES cell were at S phase (Figure 4D, 4E and 4F), suggesting that DNA is more actively synthesized in Mst-/- ES cells than wild type ES cells.
Figure 4. Mst-/- ES cells proliferate faster than wild type ES cells.
(A) Morphology of 1x105 wild type ES cells or Mst-/- ES cells grown in 2i+LIF ES medium for 2 days, 3 days and 4 days respectively. Scale bar, 200 μm. (B) Statistical analysis of the growth rate of wild type ES cells and Mst-/- ES cells on day 3 and day 4 culture. The data were shown as the mean ± S.D (n=3). Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001). (C) Immunofluorescence staining with BrdU antibodies to examine BrdU integration in wild type ES cells and Mst-/- ES cells after serum starvation for 12 hours. Cells are pulsed labeled with BrdU for 45 minutes. The nuclei were stained with DAPI. Scale bar, 200 μm. (D) Representative histograms of cell cycle distribution in Mst-/- ES cells and wild type ES cells. (E) Table of the cell cycle distribution in Mst-/- ES cells and wild type ES cells from two independent experiments. (F) Statistical analysis of cell cycle distribution in Mst-/- ES cells and wild type ES cells from two independent experiments. (*, P<0.05).
Defects of mouse Mst-/- embryonic stem cells in the formation of teratomas
Mice with Mst1 or Mst2 single gene knockout were viable and fertile, whilst Mst1/Mst2 double knockout mice died early in gestation, suggesting that Mst kinases are essential for the early developmental program.
To examine whether Mst-/- ES cells maintain the properties of ICM in vivo, we labeled the Mst-/- ES cells with GFP by lentivirus and injected them into 8-cell embryos. All Mst-/- ES cells successfully integrated into the inner cell mass (ICM), indicating that Mst deletion doesn’t affect cell surface identity at the ES cell state (Figure S3A).
Then we injected wild type ES cells and Mst-/- ES cells subcutaneously into nude mice to check for teratoma formation. Wild type ES cells could form teratoma with tissue from all three germ layers within 6 weeks, but no tumor tissue was detected 6 weeks after Mst-/- ES cells were subcutaneously injected into nude mice. Even 8 weeks later, still no tumor was formed in Mst-/- ES cell injected mice (Figure S3B). RT-PCR examination of ES cells and teratoma revealed that the expression of Yap, Mst1 and Mst2 were higher in teratoma than ES cells (Figure S3C), suggesting that Hippo pathway may play an important role in teratoma formation.
To explore the reason why Mst-/- ES cells cannot form teratoma, we compared the expression of genes involved in apoptosis and tumorigenesis between wild type EBs and Mst-/- EBs by microarray experiment. FoxO family genes FoxO1, FoxO3 and FoxO4, were slightly increased in Mst-/- EBs (Figure S3D). However, not all apoptosis related genes were upregulated. Some pro-apoptotic genes, such as Bid and Bax were increased in Mst-/- EBs; while other pro-apoptosis genes such as Bmf and Bcl2112 were downregulated in Mst-/- EBs. Similarly, some anti-apoptosis genes such as Bcl2l1 and Birc5 were increased in Mst-/- EBs, while another anti-apoptosis gene Bcl2 was decreased. Although it was reported that greater number of apoptotic cells were detected in Mst1/Mst2 double knockout embryos than Mst1/Mst2 single copy gene knockout embryos during E8.5 to E9.5[13], it was not for sure whether apoptosis were enhanced after ES cells were injected for teratoma. Interestingly, some tumorigenesis related genes Afp, CD34, Eno2 and Nes were obviously reduced in Mst-/- EB, suggesting a reduced tumorigenesis capacity during Mst-/- ES cell differentiation. This may be one of the reasons that Mst-/- ES cell cannot form teratoma.
Deletion of Mst1/Mst2 distorts embryonic stem cell differentiation in vitro
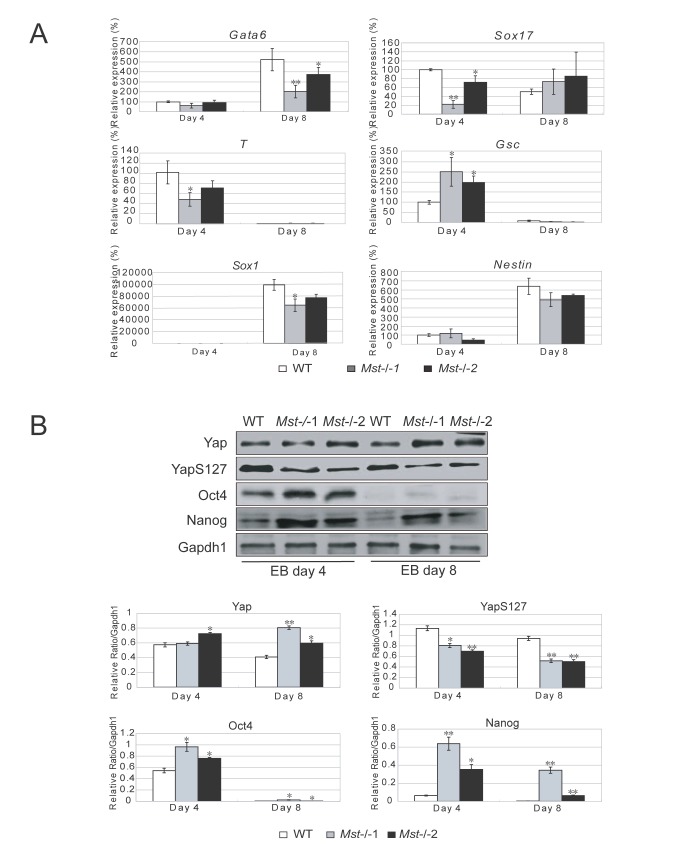
To further examine the effect of depletion of Mst1/Mst2 in ES cell differentiation, we examined the expression of multiple lineage markers in wild type ES cells and Mst-/- ES cells at both day 4 and day 8 during EB formation by RT-PCR. As shown in Figure 5A, endodermal markers, Gata6 and Sox17, were affected in Mst-/- EBs. Compared to the wild type EBs the Sox17 level was significantly lower in day 4 Mst-/- EBs, whilst the Gata6 level was lower in day 8 Mst-/- EBs. The expression of ectodermal markers Sox1 and Nestin and mesoderm markers T and Gsc were also slightly disturbed in Mst-/- EBs (Figure 5A). We also selectively examined protein level of three germ layer markers by western blot. Ectoderm marker Pax6 was increased in day 4 Mst-/- EBs and mesoderm marker T was upregulated in day 8 Mst-/- EBs, while endoderm marker Gata6 was decreased in day 8 Mst-/- EB (Figure S4A). To find out what other genes are affected by Mst deletion during EB formation, expression profile of day 4 and day 8 Mst-/- EBs and wild type EBs were compared by microarray experiment. In keeping with the quantitative RT-PCR results, microarray data showed that the expression of multiple pluripotency-related genes and lineage associated genes was distorted. Multiple pluripotent markers, such as Dppa2, Dppa3, Dppa4, Oct4, Sox2, Nanog, Esrrb, Tcl1, Lefty1 and Fgf4 were higher in Mst-/- EBs than wild type EBs (Figure S4B). Many Ectoderm markers Tubb3, Fgf8, Notch1 and Fgf5 were also significantly higher in Mst-/- EBs than wild type EBs; whilst endoderm markers Gata6, Gata4, Cxcr4 and Sox17, were significantly lower in day4 Mst-/- EBs than wild type EBs. Mesoderm markers T and Mixl1 were similarly expressed in day 4 Mst-/- EBs and wild type EBs, but higher in day 8 Mst-/- EBs.
Figure 5. Depletion of Mst1/Mst2 affects proper EB differentiation.
(A) Quantitative real time PCR to reveal the mRNA level of endoderm markers Gata6 and Sox17, mesoderm markers T and Gcs, and ectoderm markers Sox1 and Nestin in wild type EBs and Mst-/- EBs at day 4 and day 8 during EB formation. Actin was analyzed as an internal control. The data were shown as the mean ± S.D (n=3). Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001). (B) Immunoblotting and densitometric analysis to check the protein level of Yap, YapS127, Oct4 and Nanog in day 4 and day 8 wild type EBs and Mst-/- EBs. Gapdh1 was analyzed as an internal control. The data are shown as the mean ± S.D (n=2). Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001).
To further investigate the differences between Mst-/- EBs and wild type EBs, we also measured the protein levels of Yap, Oct4 and Nanog during EB formation at day 4 and day 8. An evidently higher protein level of Oct4 and Nanog was detected in day 4 Mst-/- EBs compared to wild type EBs. At day 8, both Oct4 and Nanog were undetectable in wild type EBs, but Nanog was still expressed in Mst-/- EBs (Figure 5B), whilst inactivated phosphorylated Yap was consistently expressed at a lower level in Mst-/- EBs than wild type EBs at both day 4 and day 8. There was no obvious difference in the total Yap protein level between Mst-/- EBs and wild type EBs at day 4, but Yap protein level was higher in Mst-/- EBs than wild type EBs at day 8. Therefore, unphosphorylated active Yap was higher in Mst-/- EBs than in wild type EBs. Taken together, pluripotent markers Oct4, Nanog and active Yap were expressed at a higher level in Mst-/- EBs than wild type EBs, indicating that Mst-/- ES cells display differentiation resistance during EB formation (Figure 5B). This finding demonstrates that Mst-/- ES cells show distorted early germ layer marker expression, which may be due to sustained higher expression of pluripotent markers during Mst-/- EB formation.
Mst1/Mst2 are required for embryonic stem cells to differentiate into cardiac progenitor cells
Besides studying the role of Mst1/Mst2 in early germ layer formation, we also examined the potential of Mst-/- ES cells at later lineage cell differentiation. To generate neural progenitor cells, day 4 wild type and Mst-/- EBs were attached to tissue culture plates in N2 medium. At day 2, epithelium-like cells were formed in both cell lines. And at day 8, neurosphere-like colonies were observed both in wild type and Mst-/- cells. These cells could be passaged on 0.01% Poly-L-ornithine coated culture dishes and showed proliferation in VEGF and BFGF medium (Figure S5A). The identity of NSCs was further confirmed through immunostaining with antibody against the NSC marker Pax6 (Figure S5B). Microarray analysis also revealed that many neurongenesis related genes such as Mapt, Nrsn1, Neurod1 and Nefm, were expressed at a higher level in Mst-/- EBs than wild type EBs at day 4, indicating that Mst1/Mst2 may involve in repressing ES cell to neural lineage differentiation. Meanwhile, the expression of skin development related genes was similar in wild type EBs and Mst-/- EBs, suggesting the repressive role of Mst1/Mst2 in later ectoderm tissue differentiation is specific (Figure S5C).
Next, we compared the expression of some later endoderm tissue markers in Mst-/- EBs and wild type EBs. Although the expression of some genes is disturbed due to Mst1/Mst2 depletion, there is no tissue specific pattern change (Figure S5C). Liver specific markers Alb and AAT protein are at the similar level in wild type EBs and Mst-/- EBs (Figure S5D).
Meanwhile, we also examined Mst-/- ES cells for later mesoderm tissue differentiation potential. Skeletal muscle associated gene showed no changes between Mst-/- EBs and wild type EBs (Figure S7A). Although the expression level of some genes related to smooth muscle, endothelial cells and mesenchymal stem cells (MSCs) was different between Mst-/- EBs and wild type EBs, no lineage specific expression change pattern was observed. Interestingly, multiple hematopoietic stem cell (HSC) associated genes such as CD34, CD38, Esam and Ngfr were significantly lower in day 8 Mst-/- EBs than wild type EBs, suggesting a developmental delay of this lineage. Besides, cardiac stem cell genes Tbx5, Smarcd3, Isl1, Mesp1, Kdr, Etv2 and Nkx2.5 were dramatically decreased in day 8 Mst-/- EBs. Cardiomyocyte related genes, such as Igf1, Ankrd1, Fli1 and Myh6 were also downregulated in day 8 Mst-/- EBs. Representative genes of later mesoderm tissues were also selected to validate by RT-PCR, the results matched the microarray profile and also confirmed the downregulation of cardiac lineage genes in Mst-/- EBs (Figure S7B).
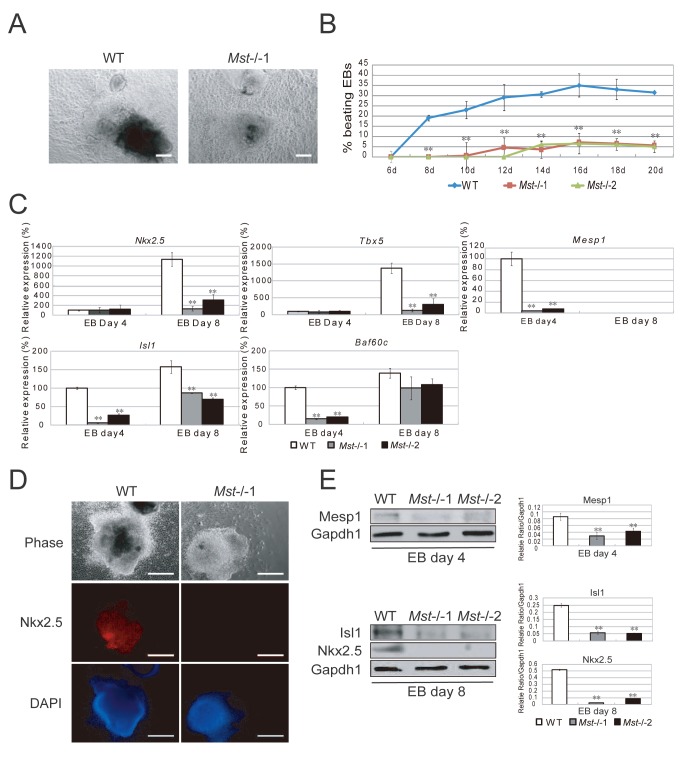
To validate whether Mst-/- ES cells have defect in cardiac lineage differentiation, day 6 wild type and Mst-/- EBs were plated on human fibronectin treated dishes to induce cardiac lineage cells. On day 3, a lot of beating cell clumps were observed in wild type sample but no beating cell clumps were observed in Mst-/- samples (Figure 6A, Movie S1 and Movie S2). Longer culture time revealed that a few beating cell clumps emerged from day 12 in Mst-/- samples. However, the beating cell clumps in Mst-/- samples were about 6 times less than wild type EBs even after culturing for 20 days (Figure 6B). RT-PCR also confirmed that cardiac progenitor markers Nkx2.5, Tbx5, Mesp1, Isl1 and Baf60c were significantly repressed in Mst-/- samples as compared to wild type samples (Figure 6C). Immunofluorescence revealed that Nkx2.5 was expressed in the wild type samples but not in the Mst-/- samples (Figure 6D). Western blot analysis of the protein lysates also confirmed expression of Mesp1, Isl1 and Nkx2.5 in the wild type samples, but not in the Mst-/- samples (Figure 6E). All these data demonstrated that Mst1 and Mst2 were required for ES cells to differentiate into cardiac lineage cells.
Figure 6. ES cell to cardiac progenitor cell differentiation is disturbed by Mst1/Mst2 depletion.
(A) Phase contrast pictures of differentiated wild type EBs and Mst-/- EBs in cardiac differentiation medium. Scale bar, 200 μm. (B) Percentage of spontaneously beating EBs determined from day 6 to day 20 during differentiation (n>100 per time point). Mst-/-EBs showed a significant less beating EBs than wild type EBs. Experiments were performed in triplicate, and error bars represent SD. Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001). (C) Relative mRNA levels of cardiac progenitor cell markers Nkx2.5, Tbx5, Mesp1, Isl1 and Baf60c in wild type and Mst-/- EBs at day 4 and day 8 during EB formation. Actin was used as an internal control. The data are shown as the mean ± S.D (n=3). Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001). (D) Immunofluorescence stain with antibody against Nkx2.5 to examine cardiac progenitor marker Nkx2.5 expression in the wild type EBs and Mst-/- EBs in cardiac differentiation medium for 8 days. Scale bar, 200 μm. (E) Immunoblotting and denstitometric analysis with antibody against Mesp1, Isl1 and Nkx2.5 to check their expression in wild type EBs and Mst-/- EBs in cardiac differentiation medium for 4 days or 8 days. Gapdh1 was analyzed as an internal control. The data are shown as the mean ± S.D (n=2). Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001).
Mst1/Mst2 are involved in cardiogenesis through regulating non-canonical wnt ligands
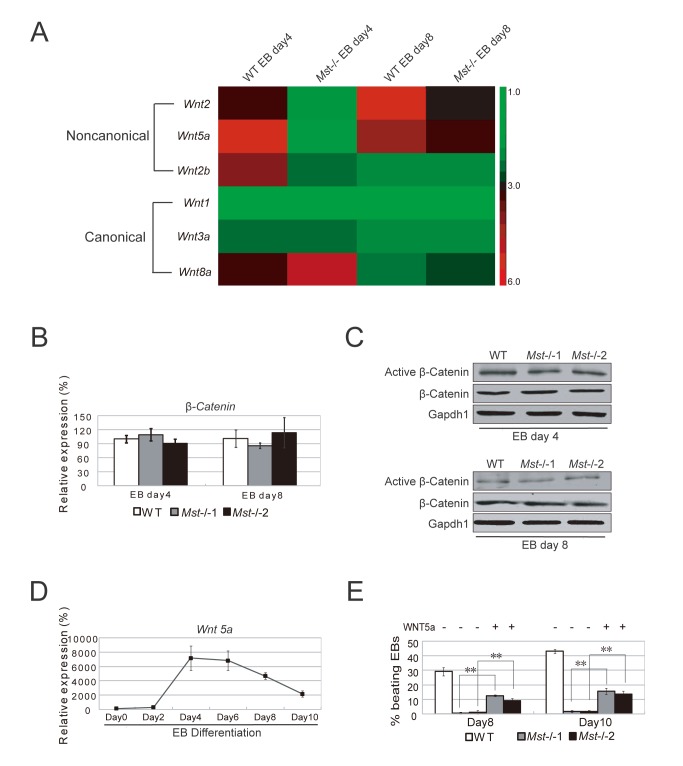
Wnt signaling plays an important role in regulating cardiac progenitor cell specification. Canonical Wnt3a/β-catenin signaling is important for cardiomyogenesis during development. While non-canonical Wnt ligand Wnt2 accelerates cardiac myocyte differentiation from ES-cell derived mesodermal cells and Wnt5a is essential for second heart field progenitor development.
To check whether there is any link between the defect of Mst-/- ES cell cardiac lineage differentiation and the Wnt signaling abnormal regulation, we examined the expression of Wnt ligand genes in day 4 and day 8 wild type and Mst-/- EBs. The expression of canonical Wnt ligand gene Wnt1 and Wnt3a showed no change between in Mst-/- EBs and wild type EBs, while another canonical Wnt ligand gene Wnt8a was slightly increased (Figure 7A). RT-PCR and western blot to examine their downstream mediator β-catenin revealed that total and active β-catenin were at the similar level in wild type EBs and Mst-/- EBs, indicating the defect of Mst-/- ES cell to cardiac lineage differentiation may not be related to canonical Wnt signaling (Figure 7B and 7C). On the other hand, several non-canonical Wnt ligand genes, such as Wnt2, Wnt2b, and Wnt5a, were significantly downregulated in Mst-/- EBs (Figure 7A). Exogenous Wnt2 can enhance ES cell to cardiomyocyte differentiation. To validate whether Wnt5a can also enhance the differentiation, we first examined Wnt5a expression during wild type EB differentiation. We found that Wnt5a was dramatically increased in day 4 EBs (Figure 7D). Therefore we added Wnt5a recombinant protein to day 2 Mst-/- EB culture. Notably, Mst-/- EBs grown in medium supplemented with Wnt5a showed increase number of beating EBs compared to Mst-/- EBs grown in non-Wnt5a medium on day 8 and day 10, although less than wild type EBs, indicating that Wnt5a can partially rescue Mst-/- EB defect for cardiomyocyte differentiation (Figure 6B and 7E). Taken together, these data suggest that Mst1/Mst2 may involve in cardiomyocyte differentiation through crosstalking with non-canonical Wnt signaling.
Figure 7. ES cell to cardiac progenitor cell differentiation is disturbed by Mst1/Mst2 depletion.
(A) Heatmap of the expression of non-canonical Wnt signaling ligands (Wnt2, Wnt2b and Wnt5a) and canonical Wnt ligands (Wnt1, Wnt3a, Wnt8a and Wnt11) in day 4 and day 8 wild type EBs and Mst-/- EBs. (B) Relative mRNA levels of β-catenin in wild type and Mst-/- EBs at day 4 and day 8 during EB formation. Actin was used as an internal control. The data are shown as the mean ± S.D (n=3). Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001). (C) Immunoblotting analysis with antibodies against Active β-catenin and total β-catenin to check its expression in day 4 and day 8 wild type EBs and Mst-/- EBs. Gapdh1 was analyzed as an internal control. (D) Relative mRNA levels of Wnt5a during EB formation from day0 to day10. Actin was used as an internal control. The data are shown as the mean ± S.D (n=3). Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001). (E) Recombinant Wnt5 were supplemented to the Mst-/-EB culture from day 2 and day 10. Wild type EBs and Mst-/- EBs were grown in non-Wnt5a supplemented medium as controls. The percentage of beating EBs was profiled on day 8 and day 10 after initiating EBs culture. The data are shown as the mean ± S.D (n=3). Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001).
Discussion
Mst1 and Mst2, the key components of the Hippo pathway in mammals, have been widely studied in different cell types, such as liver, neural, heart and intestine cells, in humans and mice. In these cell types, Mst1 and Mst2 are mainly involved in restricting cell proliferation, promoting apoptosis and regulating tumorigenesis [12,20,22-24,39]. Mst1 and Mst2 also play critical roles in mouse embryo development. Single Mst1 or Mst2 deletion gives rise to only minor defects in mice after birth, but deletion of both Mst1 and Mst2 simultaneously leads to embryonic lethality at E8.5 as a result of obvious defects in placental development and vascular patterning, etc [11-13].
To dissect the role of Mst1/Mst2 in mouse embryonic stem cells, we established Mst-/- ES cells. We found that phosphorylated Yap was decreased whilst unphosphorylated Yap was increased in Mst-/- ES cells. Consistently, Yap downstream targets, Ctrf and Cyr61 were also significantly upregulated in Mst-/- ES cells. This confirms that the Hippo pathway is active in mouse ES cells and that Mst1/Mst2 contributes to deactivate Yap. But it is interesting that residual phosphorylation of Yap can be detected in Mst-/- ES cells, indicating that Mst kinases may not be the exclusive components that lead to Yap inactivation and alternative pathways may exist to inactivate Yap. Actually, it was reported that Akt kinase can phosphorylate Yap at Serine 127 to regulate Yap translocation [40,41]. Besides, angiomotin (Amot) and angiomoti-like1 (AmotL1) physically interact with Yap and restrict Yap activity in a Hippo pathway-independent manner [42,43]. Therefore, Yap regulation in vivo is a multi-level complicated procedure. Unphosphorylated Yap is an important pluripotent factor which plays a critical role in maintaining embryonic stem cell pluripotency and improves the efficiency of iPSC formation. Yap directly binds to the regulatory regions of Nanog in mouse ES cells based on the ChIP-seq data published [26]. We consistently detected higher level of Nanog in Mst-/- ES cells and early Mst-/- EB than wild type control, suggesting that unphosphorylated Yap may directly binds to Nanog and activate its expression. Since ES cells with high level of Nanog show differentiation resistance, Mst-/- ES cells also show differentiation resistance due to upregulation of Nanog.
It is very intriguing that Mst-/- Es cells show enhanced cell proliferation (Figure 4, S6A and S6B), but cannot form teratomas in nude mice. Mst-/- ES cells obviously maintained ES cell surface identity, as Mst-/- ES cells can integrate into ICMs (Figure S4). But Mst-/- EB differentiation capacity is somehow distorted and weakened. For example, Mst-/- ES cells show great defect in cardiac lineage differentiation. It is unclear whether the differentiation distortion of ES cells links to their teratoma formation capacity. As embryonic carcinoma cells (EC cells) are also pluripotent, but with lower differentiation capacity as compared to ES cells, they can form teratocarcinomas, a malignant teratoma. Although Mst-/- embryos show greater number of cell apoptosis than the control [13]. Our Mst-/- EB microarray data show no strong evidence of enhance apoptosis during differentiation. Whereas relative lower expression level of tumorigenesis genes Eno2, Nes, CD34 and Afp in Mst-/- EBs than wild type EBs may be one of the reasons that Mst-/- ES cells cannot grow teratoma. Besides, it is reported that natural killer (NK) cells which are still active in nude mice preferentially involved in rejecting undifferentiated ES cells [44]. Mst-/- ES cells show differentiation resistance. Hence, they may be more susceptible for elimination by NK cells than wild type ES cells in nude mice. This may account for another reason for the failure of teratoma formation of Mst-/- ES cells. Above all, the inability of ES cells to form teratomas after Mst1/Mst2 deletion is quite interesting. It indicates that we can prevent teratoma formation from ES cells through inhibition of Mst1/Mst2 kinase activities, though further investigations are required to elucidate the mechanism of this. Although it is premature to translate this idea into an application, the transplantation of ES cells, combined with Mst kinase inhibition, may be worthy of trials in the repair of certain lineage cells, such as neural progenitor cells, in which Mst1/Mst2 do not play essential roles. Such treatments will however be limited by the fact that Mst1/Mst2 are required for differentiation into cardiac progenitor cells and possibly other cell lineages.
To dissect the role of Mst1/Mst2 in ES cell differentiation, in this study, we systematically compared the transcriptome between Mst-/- ES cells and wild type ES cells during their EB formation. We found that Mst-/- ES cells display some lineage differentiation distortion. For example, the expression of early endoderm gene is obviously decreased in day 4 Mst-/- EBs, while the expression of most early ectoderm genes is upregulated, suggesting Hippo signaling may play an important role in early germ layer determination. There is no obvious disturbance in early mesoderm formation in Mst-/- EBs. But Microarray comparison of the mesoderm derived tissue expression profile revealed that markers of cardiac lineage and hematopoietic stem cells (HSC) are synergistically affected, suggesting an important role of Mst1/Mst2 in the differentiation of mesoderm to these lineages.
Mst1/Mst2 double knockout mice can form heart in vivo, however, the organization and morphology were abnormal, suggesting Mst1/Mst2 play an essential role in heart formation. Recent research reported crosstalk between the Hippo pathway and canonical Wnt signaling in regulating cardiomyocyte proliferation [45-47]. In order to gain insight into the detailed mechanism, we analyzed the microarray data of day 4 and 8 EBs and found that Wnt ligands show different expression between Mst-/- EBs and wild type EBs. As canonical Wnt signaling downstream mediator β-catenin show no obvious change in Mst-/- EBs, canonical Wnt signaling therefore is not responsible for the cardiac lineage differentiation defect in the Mst-/- ES cells. Interestingly, non-canonical Wnt signaling ligands Wnt2, Wnt2b and Wnt5a were significantly decreased in Mst-/- EBs compared to wild type cells. Wnt2b null ES cells form no beating EBs, with much lower levels of cardiac genes than wild type EBs [48]. Exogenous Wnt2 can promote ES cell to cardiac lineage cell differentiation [49]. The Wnt5a signal is instructive for the differentiation of cardiac neural crest (CNC) cells during formation of the aortopulmonary septum through a non-canonical Wnt/Ca2+ pathway and hence essential for second heart field progenitor development [50-53]. We found that addition of Wnt5a recombinant proteins can partially rescue cardiac differentiation defect of Mst-/- EBs, which further substantiate that Hippo pathway regulates cardiac lineage differentiation through crosstalk with non-canonical Wnt signaling. Our studies indicate however that there might be connections between these two pathways. This area requires further exploration.
Materials and Methods
ES cell derivation and cell culture
All mouse experiments are performed under the approval from Animal Experimentation Ethics Committee (AEEC) in the Chinese University of Hong Kong. Mst-/- knockout mouse embryos were obtained by crossing Mst1+/-, Mst2-/- (C57BL/6) female and male mice. E3.5 blastocysts were flushed out from the uterus and cultured on mitomycin treated MEF feeder in a 96-well plate with N2B27 medium with 2i (PD0325901, 0.4mM: Stemgent, San Diego, CA; CHIR99021, 3mM: Stemgent) and LIF (1000 U/ml). The ICM outgrowths were treated with 0.05% Trypsin (Invitrogen) and passaged on a 24-well plate until stable ES cell lines were obtained. The ES cells were maintained on feeders under the normal ES medium (DMEM supplemented with 15% FBS, 0.1 mM non-essential amino acids, 0.1mM 2-mercaptoethanol, 2mM Glutamine, 100U/ml penicillin/streptomycin and 1000U/ml LIF). To obtain feeder free ES cells, the ES cells were grown on a 0.2% Gelatin coated dish in 2i+LIF medium. For ES cell differentiation, 2i and LIF were removed from the ES medium and 2μM RA was added to enhance differentiation. For EB formation, wild type ES cells and Mst-/- ES cells were trypsinized into single cells and then replaced at 1x106 cells in a 10 cm non-adherent dish in ES cell medium devoid of 2i and LIF. Floating EBs were harvested at 4 days or 8 days for analysis. For cell proliferation assay, 1x105 cells were plated in 6-well dishes. The cells were trypsinized to single cells and counted on day 3 and day 4. For neural stem cell differentiation and cardiac progenitor cell differentiation, previously described methods were followed [54,55]. Briefly, for neural stem cell differentiation, day 4 EBs were cultured in DMEM+10%FBS medium for 24 hours on tissue culture plate to allow EBs to attach to the plate. The medium was subsequently switched to DMEM/F12 (1:1) supplemented with N2 and changed every 2 days. For passage, cells were dissociated by 0.25% trypsin + 0.04% EDTA and then replated in DMEM/F12 (1:1) supplemented with N2 and bFGF (5ng/ml) medium. For cardiac progenitor cell differentiation, day 6 EBs were plated onto a tissue culture plate treated with human fibronectin for further analysis. For the rescue experiment, WNT5a recombinant protein were purchased form R&D company, 200ug/ml WNT5a were added to Mst-/- EB samples on day 2.
Quantitative real-time PCR
Total RNAs were extracted from ESC or EB samples using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. Aliquots of 1μg of RNA were used as templates for reverse transcription with the PrimeScript RT reagent Kit (TaKaRa) according to the instructions. Real-time PCR analysis was performed using the ABI Prism 7900HT machine (Applied Biosystems) with the SYBR Premix Ex Taq (TaKaRa). The generated threshold cycle (CT) value for each transcript was normalized against the CT value of an internal control, Actin, and subsequently normalized against the CT value of corresponding transcripts of the control sample. The sequences of RT primers are listed in Table S1. For each primer used, only one correct size band was observed. All experiments were repeated at least three times with samples from independent experiments.
Western blot analysis
ESC or EB samples were collected and washed twice with PBS. RIPA buffer (150 mM NaCl, 10 mM Tris, pH 7.2, 0.1% SDS, 1.0% Triton X-100, 1.0% deoxycholate, 5 mM EDTA, and protease inhibitors) was used to lyse the cells. Protein samples were separated by SDS-polyacrylamide gel electrophoresis and transferred to PVDF membranes (Pall corporation). The membranes were blocked with 5% nonfat dry milk (BD company) in TBST+0.1% Tween and incubated with primary antibody in TBST+0.1% Tween overnight at 4°C. The primary antibodies and dilutions used were mouse anti-Gapdh (A-3) (sc-137179; Santa Cruz Biotechnology) at 1/2,000, goat anti-Oct3/4 (N-19) (sc-8628; Santa Cruz Biotechnology) at 1/2,000, rabbit anti-Nanog (RCAB0002P-F; Cosmo bio co) at 1/2000, rabbit anti-Mst1 (catalog no. 3682; Cell Signaling Technologies) at 1/1,000, rabbit anti-Mst2 (catalog no. 3952; Cell Signaling Technologies) at 1/1,000, rabbit anti-Yap (catalog no. 4912; Cell Signaling Technologies) at 1/1,000, rabbit anti-Phospho-Yap (Ser127) (catalog no. 4911; Cell Signaling Technologies) at 1/1,000, rabbit anti-Pax6 (Ab2237; Milipore) at 1/1,000, goat anti-Brachyury (N-19) (sc-17743; Santa Cruz Biotechnology) at 1/1,000, rabbit anti-Gata6 (AF1700; R&D system) at 1/1,000, goat anti-Mesp1 (T-15) (sc-163078; Santa Cruz Biotechnology) at 1/1,000, goat anti-Nkx2.5 (N-19) (sc-8697; Santa Cruz Biotechnology) at 1/1,000, rabbit anti-Islet1 (T-15) (ab-20670; Abcam) at 1/1,000, mouse anti active-β-Catenin (05-665; Milipore) at 1/1,000, rabbit anti β-Catenin (95875; Cell signaling) at 1/1,000, goat anti-AAT (G-17) (sc-14586; Santa Cruz Biotechnology) at 1/1,000, goat anti-Albumin (A90-134A; Bethyl) at 1/1,000Signals were detected with ECL detection reagents (Abcam). Densitometric analysis was performed with software ImageJ. The relative ratio of protein dentisty was calculated by normalizing with Gapdh1.
Alkaline Phosphatase staining
ES cells were fixed with a fixative solution (90% Methanol and 10% Formaldehyde) for 20 minutes at room temperature, and then rinsed with 1x Rinse buffer (20mM Tris-HCl, pH 7.4, 0.15M NaCl, 0.05% Tween-20), followed by incubating the cells in Alkaline Phosphatase staining solution (Fast Red violet/Naphthol AS-BI phosphate solution/Water=2:1:3) in the dark at room temperature for 15 minutes. The cells were then washed with PBS buffer and photographed with an Olympus microscope (FV1000, Olympus, Tokyo, Japan).
Immunofluorescence stain
Immunofluorescence staining of ES and differentiated cells was performed with the following standard protocols. Cells were fixed in 4% paraformaldehyde at room temperature for 10 minutes and permeabilized with 0.5% Triton X-100 (for nuclear stain), followed by blocking with 1% BSA in PBS for 1 hour and then primary antibody overnight at 4°C. The primary antibodies and dilutions used were anti-SSEA1 antibody (SC-21702, Santa Cruz Biotechnology) at 1:200, goat anti-Oct3/4 (N-19) (sc-8628; Santa Cruz Biotechnology) at 1/200, rabbit anti-Yap (catalog no. 4912; Cell Signaling Technologies) at 1/100, rabbit anti-Phospho-Yap (Ser127) (catalog no. 4911; Cell Signaling Technologies) at 1/100, rabbit anti-Pax6 (Ab2237; Milipore) at 1/200, goat anti-Nkx2.5 (N-19) (sc-8697; Santa Cruz Biotechnology) at 1/200, After washing with PBS, the samples were incubated with the appropriate secondary antibodies, conjugated with Alexa Fluor 594 (Molecular Probes) in PBS, for 1 hour at room temperature. Nuclei were stained with 4, 6-diamidino-2-phenylindole dilactate (DAPI; Molecular Probes, D3571) for 10min. Images were captured with an Olympus fluorescence microscope (FV1000, Olympus, Tokyo, Japan).
Teratoma Assays
2x106 wild type ES cells and Mst-/- ES cells respectively were harvested with 0.25% Trypsin, suspended in 0.9% NaCl and injected subcutaneously into nude mice. Six weeks after injection, the nude mice were euthanized and tumors removed were fixed in 4% paraform, and then subjected to hematoxylin and eosin stain for histological analysis.
Microarray
Total RNA derived from wild type and Mst-/- ES cell and EB samples (day 4 and 8) was extracted with Trizol (Invitrogen) and purified with an RNeasy mini-kit (Qiagen). The purified RNAs were reversed-transcribed, labeled and hybridized to Affymetrix mouse exon 1.0 ST Array. The arrays were processed following the manufacturer’s instruction. The data were analyzed with software Partek and Genespring. The threshold for gene expression was set at >1.5-fold. Miscroarray data are accessible under GEO accession number GSE50219.
Cell cycle analysis
Wild type ES cells and Mst-/- ES cells were trypsinized into single cells and then fixed in cold 70% ethanol overnight. RNA was removed by 100 µg/ml Rnase (Sigma) treatment at room temperature for 20 minutes. The cells were then stained with 5 mg/ml propidium iodide (PI, Sigma) at 37 degree for 1 hour. Flow cytometric analysis was carried out on 10,000 gated events using FACSCalubur (BD Biosciences). Then cell cycle phase distribution was analyzed using software Flowjo (version 7.6).
Brdu cell proliferation assay
Wild type and Mst-/- ES cells were pulse-labelled with BrdU (1:1000; Cell signaling) for 45 minutes. ES cells were dispersed into single cells; the cells were fixed overnight in 70% ethanol at 4 degree. DNA denaturation was subsequently performed by incubation in 1.5N HCl for 20 minutes at room temperature. The cells then washed and incubated with 0.1 M sodium tetraborate for 10 minutes at room temperature. The cells were incubated with Alexa Fluor 488- conjugated mouse anti-BrdU antibody (1:100; Molecular Probes) in 2% BSA-PBS for 2 hours at 4 degree. The cells were then incubated with 100 mg/ml RNase (Sigma) for 15 minutes and then subject to FACSCalibur (BD Biosciences) to record the data. The data was analysed by CellQuest program.
Supporting Information
Mst-/- ES cell derivation. (A) 3.5 day blastocysts obtained by crossing Mst1+/-, Mst2-/- male and female mice. Scale bar, 200μm. (B) ICM outgrowth formed 5 days after a single blastocyst was seeded on MEF feeder in 2i+LIF ES medium. Scale bar, 200μm. (C) Schematics of targeted deletion loci (kinase domain) of Mst1 and Mst2. Boxes denote exons and lines denote intron.
(TIF)
The expression of Yap and Yap targets in Mst-/- ES cells. (A) Quantitative RT-PCR to check mRNA level of Yap in wild type and Mst-/- ES cells. Actin was analyzed as an internal control. The data are shown as the mean ± S.D (n=3). Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001). (B) Immunofluorescence staining of Yap and phosphorylated YapS127 in wild type ES cells and Mst-/- ES cells. Scale bar, 200 μm. (C) Quantitative RT-PCR to check mRNA level of Ctgf and Cyr61 in wild type and Mst-/- ES cells. Actin was analyzed as an internal control. The data are shown as the mean ± S.D (n=3). Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001).
(TIF)
Examination of Mst-/- ES cell pluripotency by embryo injection and teratoma formation. (A) GFP labeled Mst-/- ES cells were integrated into ICM (arrow indicated in right panel) of blastocyst after aggregation with 8-cell stage embryos. Scale bar, 200 μm. (B) Teratoma formed by wild type ES cells 6 weeks after subcutaneous injection of wild type ES cells into nude mice (indicated by a white box). H&E staining showed tissue of three germ layers (ectoderm, mesoderm and endoderm). No teratomas were formed by subcutaneous injection of Mst-/- ES cells. Scale bar, 200 μm. (C) Quantitative RT-PCR to check mRNA level of Mst1, Mst2 and Yap in ES cells and teratomas. Actin was analyzed as an internal control. The data are shown as the mean ± S.D (n=3). Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001). (D) Heatmap to show the expression of Foxo genes (Foxo1, 3a and 4), proapoptotic, antiapoptotic genes and tumor marker genes in day 8 wild type EBs and Mst-/- EBs.
(TIF)
Pluripotency and lineage marker expression during Mst-/- EB formation. (A) Immunoblotting and densitometric analysis to check the protein level of Pax6, Gata6 and T in day 4 and day 8 wild type EBs and Mst-/- EBs. Gapdh1 was analyzed as an internal control. The data are shown as the mean ± S.D (n=2). Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001). (B) Heatmap to show the expression of pluripotent genes and lineage genes (Ectoderm, Mesoderm and Endoderm) in day 4 and day 8 wild type EBs and Mst-/- EBs.
(TIF)
The expression of ectoderm and endoderm lineage markers during Mst-/- EB formation. (A) Phase contrast pictures of differentiated neural progenitor cells grown from wild type EBs and Mst-/- EBs at day 2 (top) and day 8 (middle) after attaching day 4 EBs to the plate (top) and the 2nd passage culture of neural progenitor cells (bottom). Scale bar, 200 μm. (B) Immunofluorescence staining with antibody against Pax6 to examine the expression of Pax6 in wild type ES cells and Mst-/- ES cells cultured in neural differentiation medium for 8 days. Scale bar, 200 μm. (C) Heatmap to show the expression of cell markers of ectoderm and endoderm differentiated tissue cells in day 4 and day 8 wild type EBs and Mst-/- EBs. (D) Immunoblotting analysis to check hepatocyte markers (Albumin and AAT) in day 4 and day 8 wild type EBs and Mst-/- EBs. Gapdh1 was analyzed as an internal control.
(TIF)
Comparison of proliferation difference between wild type ES cells and Mst-/- ES cells. (A) Quantitative RT-PCR to check mRNA level of Ccnd2 and Ccnd3, in wild type ES cells and Mst-/- ES cells. Actin was analyzed as an internal control. The data are shown as the mean ± S.D (n=3). Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001). (B) Flow diagram of BrdU labeled wild type ES cells and Mst-/- ES cells. The percentage of BrdU positive cell was marked in the diagram. (C) Percentage of BrdU positive cells in wild type ES cells and Mst-/- ES cells. Results represent the mean± S.D (n=2). Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001).
(TIF)
The expression of mesoderm lineage markers during Mst-/- EB formation. (A) Heatmap to show the expression of mesoderm differentiated tissue genes in day 4 and day 8 wild type EBs and Mst-/- EBs. (B) Quantitative RT-PCR to validate microarray results and check mRNA level of cardiomyocyte marker genes (Tnnt2 and Mef2c), endothelial cell marker genes (Pecam1 and Cdh5), Smooth muscle cell marker gene (Acta2), skeletal muscle cell marker gene (MyoD1) and hematopoietic stem cell marker genes (Tal1 and CD34) in wild type and Mst-/- EB cells. Actin was analyzed as an internal control. The data are shown as the mean ± S.D (n=3). Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001).
(TIF)
Wild type EBs grown in cardiac differentiation medium.
(AVI)
Mst-/- EBs grown in cardiac differentiation medium.
(AVI)
Genotyping and quantitative RT-PCR primer list.
(XLS)
Acknowledgments
We are grateful to Dennis Lo for advice. We thank Carol Yi-Ki Szento for providing microarray assay service. We thank Xiang Sun and Jiangxue Li for critical comments on the manuscript.
Funding Statement
The authors have no support or funding to report.
References
- 1. Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ (1995) The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev 9: 534-546. doi: 10.1101/gad.9.5.534. PubMed: 7698644. [DOI] [PubMed] [Google Scholar]
- 2. Pantalacci S, Tapon N, Léopold P (2003) The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat Cell Biol 5: 921-927. doi: 10.1038/ncb1051. PubMed: 14502295. [DOI] [PubMed] [Google Scholar]
- 3. Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA et al. (2002) salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 110: 467-478. doi: 10.1016/S0092-8674(02)00824-3. PubMed: 12202036. [DOI] [PubMed] [Google Scholar]
- 4. Wu S, Huang J, Dong J, Pan D (2003) hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 114: 445-456. doi: 10.1016/S0092-8674(03)00549-X. PubMed: 12941273. [DOI] [PubMed] [Google Scholar]
- 5. Huang J, Wu S, Barrera J, Matthews K, Pan D (2005) The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122: 421-434. doi: 10.1016/j.cell.2005.06.007. PubMed: 16096061. [DOI] [PubMed] [Google Scholar]
- 6. Saucedo LJ, Edgar BA (2007) Filling out the Hippo pathway. Nat Rev Mol Cell Biol 8: 613-621. doi: 10.1038/nrm2221. PubMed: 17622252. [DOI] [PubMed] [Google Scholar]
- 7. Dong J, Feldmann G, Huang J, Wu S, Zhang N et al. (2007) Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130: 1120-1133. doi: 10.1016/j.cell.2007.07.019. PubMed: 17889654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hao Y, Chun A, Cheung K, Rashidi B, Yang X (2008) Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem 283: 5496-5509. PubMed: 18158288. [DOI] [PubMed] [Google Scholar]
- 9. Lei QY, Zhang H, Zhao B, Zha ZY, Bai F et al. (2008) TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol Cell Biol 28: 2426-2436. doi: 10.1128/MCB.01874-07. PubMed: 18227151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang H, Pasolli HA, Fuchs E (2010) Yes-associated protein (YAP) transcriptional coactivator functions in balancing growth and differentiation in skin. Proc Natl Acad Sci U S A 108: 2270-2275. PubMed: 21262812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou D, Medoff BD, Chen L, Li L, Zhang XF et al. (2008) The Nore1B/Mst1 complex restrains antigen receptor-induced proliferation of naive T cells. Proc Natl Acad Sci U S A 105: 20321-20326. doi: 10.1073/pnas.0810773105. PubMed: 19073936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Song H, Mak KK, Topol L, Yun K, Hu J et al. (2010) Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc Natl Acad Sci U S A 107: 1431-1436. doi: 10.1073/pnas.0911409107. PubMed: 20080598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oh S, Lee D, Kim T, Kim TS, Oh HJ et al. (2009) Crucial role for Mst1 and Mst2 kinases in early embryonic development of the mouse. Mol Cell Biol, 29: 2009/09/30 ed. pp. 6309-6320 PubMed: 19786569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. O'Neill E, Rushworth L, Baccarini M, Kolch W (2004) Role of the kinase MST2 in suppression of apoptosis by the proto-oncogene product Raf-1. Science 306: 2267-2270. doi: 10.1126/science.1103233. PubMed: 15618521. [DOI] [PubMed] [Google Scholar]
- 15. Matallanas D, Romano D, Al-Mulla F, O'Neill E, Al-Ali W et al. (2011) Mutant K-Ras activation of the proapoptotic MST2 pathway is antagonized by wild-type K-Ras. Mol Cell 44: 893-906. doi: 10.1016/j.molcel.2011.10.016. PubMed: 22195963. [DOI] [PubMed] [Google Scholar]
- 16. Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villén J et al. (2006) A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell 125: 987-1001. doi: 10.1016/j.cell.2006.03.046. PubMed: 16751106. [DOI] [PubMed] [Google Scholar]
- 17. Yuan Z, Lehtinen MK, Merlo P, Villén J, Gygi S et al. (2009) Regulation of neuronal cell death by MST1-FOXO1 signaling. J Biol Chem 284: 11285-11292. PubMed: 19221179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao Q, Beck AJ, Vitale JM, Schneider JS, Chang C et al. (2011) Injection of wild type embryonic stem cells into Mst1 transgenic blastocysts prevents adult-onset cardiomyopathy. Stem. Cell Res 7: 326-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu AM, Xu MZ, Chen J, Poon RT, Luk JM (2010) Targeting YAP and Hippo signaling pathway in liver cancer. Expert Opin Ther Targets 14: 855-868. doi: 10.1517/14728222.2010.499361. PubMed: 20545481. [DOI] [PubMed] [Google Scholar]
- 20. Lee KP, Lee JH, Kim TS, Kim TH, Park HD et al. (2010) The Hippo-Salvador pathway restrains hepatic oval cell proliferation, liver size, and liver tumorigenesis. Proc Natl Acad Sci U S A 107: 8248-8253. doi: 10.1073/pnas.0912203107. PubMed: 20404163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Song JY, Lee JH, Joe CO, Lim DS, Chung JH (2010) Retrotransposon-specific DNA hypomethylation and two-step loss-of-imprinting during WW45 haploinsufficiency-induced hepatocarcinogenesis. Biochem Biophys Res Commun 404: 728-734. PubMed: 21163252. [DOI] [PubMed] [Google Scholar]
- 22. Zhou D, Zhang Y, Wu H, Barry E, Yin Y et al. (2011) Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proc Natl Acad Sci U S A, 108: 1312-1320. PubMed: 22042863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cai J, Zhang N, Zheng Y, de Wilde RF, Maitra A et al. (2010) The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev 24: 2383-2388. doi: 10.1101/gad.1978810. PubMed: 21041407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. George NM, Day CE, Boerner BP, Johnson RL, Sarvetnick NE (2012) Hippo signaling regulates pancreas development through inactivation of Yap. Mol Cell Biol 32: 5116-5128. doi: 10.1128/MCB.01034-12. PubMed: 23071096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chan SW, Lim CJ, Chen L, Chong YF, Huang C et al. (2011) The Hippo pathway in biological control and cancer development. J Cell Physiol 226: 928-939. doi: 10.1002/jcp.22435. PubMed: 20945341. [DOI] [PubMed] [Google Scholar]
- 26. Lian I, Kim J, Okazawa H, Zhao J, Zhao B et al. (2010) The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev 24: 1106-1118. doi: 10.1101/gad.1903310. PubMed: 20516196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhao B, Wei X, Li W, Udan RS, Yang Q et al. (2007) Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev 21: 2747-2761. doi: 10.1101/gad.1602907. PubMed: 17974916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ramos A, Camargo FD (2012) The Hippo signaling pathway and stem cell biology. Trends Cell Biol 22: 339-346. doi: 10.1016/j.tcb.2012.04.006. PubMed: 22658639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M et al. (2009) The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell 16: 398-410. doi: 10.1016/j.devcel.2009.02.003. PubMed: 19289085. [DOI] [PubMed] [Google Scholar]
- 30. Tamm C, Böwer N, Annerén C (2011) Regulation of mouse embryonic stem cell self-renewal by a Yes-YAP-TEAD2 signaling pathway downstream of LIF. J Cell Sci 124: 1136-1144. doi: 10.1242/jcs.075796. PubMed: 21385842. [DOI] [PubMed] [Google Scholar]
- 31. Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW et al. (2007) YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol 17: 2054-2060. doi: 10.1016/j.cub.2007.10.039. PubMed: 17980593. [DOI] [PubMed] [Google Scholar]
- 32. Watt KI, Judson R, Medlow P, Reid K, Kurth TB et al. (2010) Yap is a novel regulator of C2C12 myogenesis. Biochem Biophys Res Commun 393: 619-624. doi: 10.1016/j.bbrc.2010.02.034. PubMed: 20153295. [DOI] [PubMed] [Google Scholar]
- 33. Rossant J (2001) Stem cells from the Mammalian blastocyst. Stem Cells 19: 477-482. doi: 10.1634/stemcells.19-6-477. PubMed: 11713338. [DOI] [PubMed] [Google Scholar]
- 34. Smith AG (2001) Embryo-derived stem cells: of mice and men. Annu Rev Cell Dev Biol 17: 435-462. doi: 10.1146/annurev.cellbio.17.1.435. PubMed: 11687496. [DOI] [PubMed] [Google Scholar]
- 35. Keller G (2005) Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev 19: 1129-1155. doi: 10.1101/gad.1303605. PubMed: 15905405. [DOI] [PubMed] [Google Scholar]
- 36. Tokuzawa Y, Maruyama M, Yamanaka S (2006) Utilization of digital differential display to identify novel targets of Oct3/4. Methods Mol Biol 329: 223-231. PubMed: 16845994. [DOI] [PubMed] [Google Scholar]
- 37. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T et al. (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861-872. doi: 10.1016/j.cell.2007.11.019. PubMed: 18035408. [DOI] [PubMed] [Google Scholar]
- 38. Pobbati AV, Hong W (2013) Emerging roles of TEAD transcription factors and its coactivators in cancers. Cancer Biol Ther 14: 390-398. doi: 10.4161/cbt.23788. PubMed: 23380592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lu L, Li Y, Kim SM, Bossuyt W, Liu P et al. (2010) Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci U S A 107: 1437-1442. doi: 10.1073/pnas.0911427107. PubMed: 20080689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Basu S, Totty NF, Irwin MS, Sudol M, Downward J (2003) Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell 11: 11-23. doi: 10.1016/S1097-2765(02)00776-1. PubMed: 12535517. [DOI] [PubMed] [Google Scholar]
- 41. Zhang H, Wu S, Xing D (2012) Inhibition of Abeta(25-35)-induced cell apoptosis by low-power-laser-irradiation (LPLI) through promoting Akt-dependent YAP cytoplasmic translocation. Cell Signal 24: 224-232. doi: 10.1016/j.cellsig.2011.09.004. PubMed: 21945154. [DOI] [PubMed] [Google Scholar]
- 42. Zhao B, Li L, Lu Q, Wang LH, Liu CY et al. (2011) Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev 25: 51-63. doi: 10.1101/gad.2000111. PubMed: 21205866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chan SW, Lim CJ, Chong YF, Pobbati AV, Huang C et al. (2011) Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J Biol Chem 286: 7018-7026. doi: 10.1074/jbc.C110.212621. PubMed: 21224387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dressel R, Nolte J, Elsner L, Novota P, Guan K et al. (2010) Pluripotent stem cells are highly susceptible targets for syngeneic, allogeneic, and xenogeneic natural killer cells. FASEB J 24: 2164-2177. doi: 10.1096/fj.09-134957. PubMed: 20145206. [DOI] [PubMed] [Google Scholar]
- 45. Vincent SD, Buckingham ME (2010) How to make a heart: the origin and regulation of cardiac progenitor cells. Curr Top Dev Biol 90: 1-41. doi: 10.1016/S0070-2153(10)90001-X. PubMed: 20691846. [DOI] [PubMed] [Google Scholar]
- 46. Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E et al. (2011) Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science 332: 458-461. doi: 10.1126/science.1199010. PubMed: 21512031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gessert S, Kühl M (2010) The multiple phases and faces of wnt signaling during cardiac differentiation and development. Circ Res 107: 186-199. doi: 10.1161/CIRCRESAHA.110.221531. PubMed: 20651295. [DOI] [PubMed] [Google Scholar]
- 48. Wang H, Gilner JB, Bautch VL, Wang DZ, Wainwright BJ et al. (2007) Wnt2 coordinates the commitment of mesoderm to hematopoietic, endothelial, and cardiac lineages in embryoid bodies. J Biol Chem 282: 782-791. PubMed: 17098737. [DOI] [PubMed] [Google Scholar]
- 49. Onizuka T, Yuasa S, Kusumoto D, Shimoji K, Egashira T et al. (2012) Wnt2 accelerates cardiac myocyte differentiation from ES-cell derived mesodermal cells via non-canonical pathway. J Mol Cell Cardiol 52: 650-659. doi: 10.1016/j.yjmcc.2011.11.010. PubMed: 22146296. [DOI] [PubMed] [Google Scholar]
- 50. Chen L, Fulcoli FG, Ferrentino R, Martucciello S, Illingworth EA et al. (2012) Transcriptional control in cardiac progenitors: Tbx1 interacts with the BAF chromatin remodeling complex and regulates Wnt5a. PLOS Genet 8: e1002571 PubMed: 22438823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cohen ED, Miller MF, Wang Z, Moon RT, Morrisey EE (2012) Wnt5a and Wnt11 are essential for second heart field progenitor development. Development 139: 1931-1940. doi: 10.1242/dev.069377. PubMed: 22569553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Onizuka T, Yuasa S, Kusumoto D, Shimoji K, Egashira T et al. (2011) Wnt2 accelerates cardiac myocyte differentiation from ES-cell derived mesodermal cells via non-canonical pathway. J Mol Cell Cardiol 52: 650-659. PubMed: 22146296. [DOI] [PubMed] [Google Scholar]
- 53. Schleiffarth JR, Person AD, Martinsen BJ, Sukovich DJ, Neumann A et al. (2007) Wnt5a is required for cardiac outflow tract septation in mice. Pediatr Res 61: 386-391. doi: 10.1203/pdr.0b013e3180323810. PubMed: 17515859. [DOI] [PubMed] [Google Scholar]
- 54. Christoforou N, Miller RA, Hill CM, Jie CC, McCallion AS et al. (2008) Mouse ES cell-derived cardiac precursor cells are multipotent and facilitate identification of novel cardiac genes. J Clin Invest 118: 894-903. PubMed: 18246200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Okabe S, Forsberg-Nilsson K, Spiro AC, Segal M, McKay RD (1996) Development of neuronal precursor cells and functional postmitotic neurons from embryonic stem cells in vitro. Mech Dev 59: 89-102. doi: 10.1016/0925-4773(96)00572-2. PubMed: 8892235. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mst-/- ES cell derivation. (A) 3.5 day blastocysts obtained by crossing Mst1+/-, Mst2-/- male and female mice. Scale bar, 200μm. (B) ICM outgrowth formed 5 days after a single blastocyst was seeded on MEF feeder in 2i+LIF ES medium. Scale bar, 200μm. (C) Schematics of targeted deletion loci (kinase domain) of Mst1 and Mst2. Boxes denote exons and lines denote intron.
(TIF)
The expression of Yap and Yap targets in Mst-/- ES cells. (A) Quantitative RT-PCR to check mRNA level of Yap in wild type and Mst-/- ES cells. Actin was analyzed as an internal control. The data are shown as the mean ± S.D (n=3). Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001). (B) Immunofluorescence staining of Yap and phosphorylated YapS127 in wild type ES cells and Mst-/- ES cells. Scale bar, 200 μm. (C) Quantitative RT-PCR to check mRNA level of Ctgf and Cyr61 in wild type and Mst-/- ES cells. Actin was analyzed as an internal control. The data are shown as the mean ± S.D (n=3). Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001).
(TIF)
Examination of Mst-/- ES cell pluripotency by embryo injection and teratoma formation. (A) GFP labeled Mst-/- ES cells were integrated into ICM (arrow indicated in right panel) of blastocyst after aggregation with 8-cell stage embryos. Scale bar, 200 μm. (B) Teratoma formed by wild type ES cells 6 weeks after subcutaneous injection of wild type ES cells into nude mice (indicated by a white box). H&E staining showed tissue of three germ layers (ectoderm, mesoderm and endoderm). No teratomas were formed by subcutaneous injection of Mst-/- ES cells. Scale bar, 200 μm. (C) Quantitative RT-PCR to check mRNA level of Mst1, Mst2 and Yap in ES cells and teratomas. Actin was analyzed as an internal control. The data are shown as the mean ± S.D (n=3). Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001). (D) Heatmap to show the expression of Foxo genes (Foxo1, 3a and 4), proapoptotic, antiapoptotic genes and tumor marker genes in day 8 wild type EBs and Mst-/- EBs.
(TIF)
Pluripotency and lineage marker expression during Mst-/- EB formation. (A) Immunoblotting and densitometric analysis to check the protein level of Pax6, Gata6 and T in day 4 and day 8 wild type EBs and Mst-/- EBs. Gapdh1 was analyzed as an internal control. The data are shown as the mean ± S.D (n=2). Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001). (B) Heatmap to show the expression of pluripotent genes and lineage genes (Ectoderm, Mesoderm and Endoderm) in day 4 and day 8 wild type EBs and Mst-/- EBs.
(TIF)
The expression of ectoderm and endoderm lineage markers during Mst-/- EB formation. (A) Phase contrast pictures of differentiated neural progenitor cells grown from wild type EBs and Mst-/- EBs at day 2 (top) and day 8 (middle) after attaching day 4 EBs to the plate (top) and the 2nd passage culture of neural progenitor cells (bottom). Scale bar, 200 μm. (B) Immunofluorescence staining with antibody against Pax6 to examine the expression of Pax6 in wild type ES cells and Mst-/- ES cells cultured in neural differentiation medium for 8 days. Scale bar, 200 μm. (C) Heatmap to show the expression of cell markers of ectoderm and endoderm differentiated tissue cells in day 4 and day 8 wild type EBs and Mst-/- EBs. (D) Immunoblotting analysis to check hepatocyte markers (Albumin and AAT) in day 4 and day 8 wild type EBs and Mst-/- EBs. Gapdh1 was analyzed as an internal control.
(TIF)
Comparison of proliferation difference between wild type ES cells and Mst-/- ES cells. (A) Quantitative RT-PCR to check mRNA level of Ccnd2 and Ccnd3, in wild type ES cells and Mst-/- ES cells. Actin was analyzed as an internal control. The data are shown as the mean ± S.D (n=3). Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001). (B) Flow diagram of BrdU labeled wild type ES cells and Mst-/- ES cells. The percentage of BrdU positive cell was marked in the diagram. (C) Percentage of BrdU positive cells in wild type ES cells and Mst-/- ES cells. Results represent the mean± S.D (n=2). Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001).
(TIF)
The expression of mesoderm lineage markers during Mst-/- EB formation. (A) Heatmap to show the expression of mesoderm differentiated tissue genes in day 4 and day 8 wild type EBs and Mst-/- EBs. (B) Quantitative RT-PCR to validate microarray results and check mRNA level of cardiomyocyte marker genes (Tnnt2 and Mef2c), endothelial cell marker genes (Pecam1 and Cdh5), Smooth muscle cell marker gene (Acta2), skeletal muscle cell marker gene (MyoD1) and hematopoietic stem cell marker genes (Tal1 and CD34) in wild type and Mst-/- EB cells. Actin was analyzed as an internal control. The data are shown as the mean ± S.D (n=3). Statistically significant differences are indicated (*, P<0.05; **, P<0.01; ***, P<0.001).
(TIF)
Wild type EBs grown in cardiac differentiation medium.
(AVI)
Mst-/- EBs grown in cardiac differentiation medium.
(AVI)
Genotyping and quantitative RT-PCR primer list.
(XLS)