Abstract
Singlet oxygen (1O2) is the main agent of photooxidative stress and is generated by photosensitizers as (bacterio)chlorophylls. It leads to the damage of cellular macromolecules and therefore photosynthetic organisms have to mount an adaptive response to 1O2 formation. A major player of the photooxidative stress response in Rhodobacter sphaeroides is the alternative sigma factor RpoE, which is inactivated under non-stress conditions by its cognate anti-sigma factor ChrR. By using random mutagenesis we identified RSP_1090 to be required for full activation of the RpoE response under 1O2 stress, but not under organic peroxide stress. In this study we show that both RSP_1090 and RSP_1091 are required for full resistance towards 1O2. Moreover, we revealed that the DegS and RseP homologs RSP_3242 and RSP_2710 contribute to 1O2 resistance and promote ChrR proteolysis. The RpoE signaling pathway in R. sphaeroides is therefore highly similar to that of Escherichia coli, although very different anti-sigma factors control RpoE activity. Based on the acquired results, the current model for RpoE activation in response to 1O2 exposure in R. sphaeroides was extended.
Introduction
Light and oxygen in combination with a photosensitizer lead to the formation of toxic singlet oxygen (1O2). The photosensitizer absorbs light and transfers energy to molecular oxygen, causing a spin conversion of an electron, thereby forming the highly reactive 1O2 [1]. Excess of 1O2 is toxic for the cell, as it can react with macromolecules like proteins, lipids and nucleic acids [2,3]. The cell needs to respond to this so called photooxidative stress to prevent cellular damages which consequently would lead to cell death.
Facultative photosynthetic α-proteobacteria like Rhodobacter sphaeroides induce the formation of the photosynthetic apparatus when the oxygen tension in the environment decreases. The synthesized bacteriochlorophyll molecules and their precursors can act as potent cellular photosensitizers. Nevertheless, even when photosynthetic pigments are highly abundant in the cell, R. sphaeroides grows well in the presence of light and oxygen. The presence of carotenoids protects against 1O2 caused damages and in addition, R. sphaeroides mounts a molecular response to 1O2 exposure, which is independent of carotenoids [4,5]. This response partly depends on the alternative group IV sigma factor RpoE. RpoE is inactivated by forming a stable complex with its cognate anti-sigma factor ChrR in a 1:1 stoichiometry [6,7]. When R. sphaeroides cells are exposed to 1O2, the RpoE:ChrR complex dissociates, RpoE binds to the RNA polymerase and induces the expression of target genes [4,6]. When the crystal structure of the RpoE:ChrR complex was solved it was shown that the zinc containing anti-sigma domain (ASD) of ChrR is necessary for the interaction with RpoE [7]. The ASD is conserved in many bacterial anti-sigma factors [7]. A second zinc containing ChrR domain, the cupin like domain (CLD), is necessary for activation of RpoE by 1O2. It was proposed that amino acid side chains or a ligand in the ChrR-CLD are targets of unknown chemical modification by 1O2 that lead to dissociation of the RpoE:ChrR complex [7]. The CLD could also play a role in promoting an association of the RpoE:ChrR complex with the photosynthetic membrane, the main source of 1O2 generation [8].
In bacteria one mechanism of sigma factor activation is the proteolysis of the cognate anti-sigma factor. In the Gram negative bacterium Escherichia coli, the alternative sigma factor σE (also known as RpoE) is inactivated by the binding of its cognate anti-sigma factor RseA, which is membrane localized. Under cell envelope stress conditions, RseA is stepwise proteolyzed, thus RpoE is released and can bind to the RNA polymerase [9]. Interestingly, the N-terminal ASD of ChrR and RseA are similar in structure, but not in amino acid sequence [7,10].
Homologs of the RpoE:ChrR complex can be found in many α-, β- and γ-proteobacteria [11]. In the α-proteobacterium Caulobacter crescentus RpoE activity is not only induced by 1O2, but also by exposure to organic peroxide (tert-butyl-hydroperoxide, tBOOH), cadmium and UV-A irradiation [12]. Specific amino acid residues in the anti-sigma factor ChrR may be required for the specific response to either 1O2, organic peroxide and UV-A irradiation or cadmium [12].
The R. sphaeroides RpoE regulon is well defined, but the exact mechanism of RpoE:ChrR dissociation is still unknown. Recent work reported that the anti-sigma factor ChrR is degraded in the presence of 1O2 and tBOOH [13,14], but the proteases involved in ChrR proteolysis are yet unknown. This motivated us to search for factors that are involved in RpoE activation under photooxidative stress. A Tn5 mutagenesis of the R. sphaeroides wild type revealed that insertion of Tn5 into the RSP_1090 generated a strain highly sensitive to 1O2. Consequently, we investigated the impact of genes encoded in the RSP_1091-1087 operon in the photooxidative stress response and showed that RSP_1090 affects the stability of ChrR. In E. coli the proteases DegS and RseP are involved in proteolysis of the RpoE anti-sigma factor RseA. Because the Tn5 mutagenesis did not reveal 1O2 sensitive protease-mutants in R. sphaeroides and the RSP_1090 product has no homology to proteases, the DegS and RseP homologs RSP_3242 and RSP_2710 were deleted in R. sphaeroides in order to elucidate if these proteases are involved in ChrR degradation and RpoE activation. Our results support a function of these proteases in singlet oxygen-dependent proteolysis of ChrR. Therefore, central factors involved in RpoE activation are shared between R. sphaeroides and E. coli despite the limited similarities of the anti-sigma factors ChrR and RseA and the different signals leading to RpoE activation.
Results
Insertion of Tn5 in the gene RSP_1090 leads to decreased RpoE activity and increased sensitivity towards 1O2
We performed a Tn5 mutagenesis in the R. sphaeroides 2.4.1 wild type harboring the reporter plasmid pPHUphrAlacZ to identify unknown factors triggering RpoE activation. The plasmid harbors the lacZ gene preceded by the RpoE-inducible phrA promoter [15]. We screened for those Tn5 mutants which showed decreased or even no β-galactosidase activity upon exposure to 1O2. Additionally, mutants of interest should not have an insertion of Tn5 in the reporter plasmid pPHUphrAlacZ and in the rpoE gene including the rpoE upstream regulatory region, respectively, and should be more sensitive to 1O2 than the wild type. Several mutants were found that carried the transposon in the rpoE locus and the reporter plasmid, respectively. After screening around 18.000 Tn5 mutants, we finally found one mutant which passed the selection criteria. Vectorette PCR [16] identified the Tn5 insertion into the gene RSP_1090, which encodes a protein of unknown function (Figure 1), that was previously annotated to encode a protein involved in cyclopropane fatty acid synthesis [17]. RSP_1090 is part of the putative RSP_1091-1087 operon located upstream of rpoEchrR and belongs to the recently defined RpoE regulon [13,17].
Figure 1. Genetic organization of the RSP_1091-1087 and rpoEchrR operons on the R. sphaeroides chromosome 1.
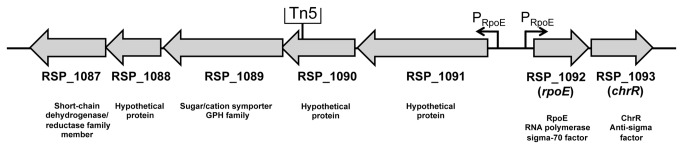
The insertion site of Tn5 which resulted in reduced RpoE activity is indicated. The Tn5 inserted 683 bp downstream of the start codon of the RSP_1090 gene. RSP_1090 located in a putative operon with RSP_1091, RSP_1089, RSP_1088 and RSP_1087. Both operons are preceded by an RpoE dependent promoter. Annotated protein functions are depicted below the locus tag numbers.
Deletion of RSP_1090 leads to decreased RpoE activity under 1O2 stress, but not under organic peroxide stress
The deletion of the RSP_1090 gene in the R. sphaeroides wild type was performed by the insertion of a kanamycin resistance cassette without transcriptional terminator to avoid polar effects on the transcription of downstream genes.
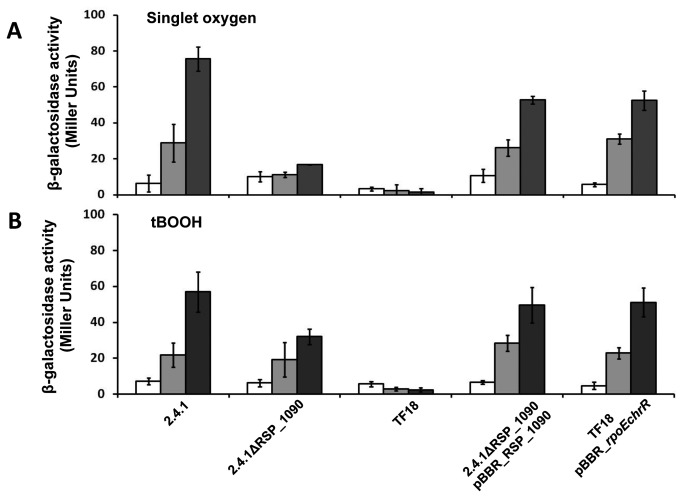
To analyze the role of RSP_1090 in the RpoE response we monitored RpoE activity via the expression of a phrA-lacZ fusion in response to 1O2 and tBOOH (Figure 2). In the wild type strain β-galactosidase activity increased strongly after 1O2 exposure (Figure 2A). In contrast to the wild type, a minor increase in β-galactosidase activity was found for 2.4.1ΔRSP_1090. We did not observe any increase in β-galactosidase activity for strain TF18 which lacks rpoE and chrR. Strain 2.4.1ΔRSP_1090 was complemented with pBBR2.4.1_RSP_1090, harboring RSP_1090 flanked by the RpoE promoter located upstream of the putative RSP_1091-1087 operon. Strain TF18 was complemented with a copy of the rpoEchrR operon by using the same vector. Both strains showed higher β-galactosidase activities compared to the deletion strains, but did not match the wild type after 3 h of stress exposure (Figure 2A). For the experiments performed with organic peroxide (Figure 2B), the β-galactosidase activities were similar to those observed for the 1O2 stress experiment, except for the RSP_1090 deletion strain. RpoE activity was induced in 2.4.1ΔRSP_1090 after organic peroxide exposure, but compared to the wild type the observed activities were significantly lower after 3 h of tBOOH exposure (Figure 2B). The finding that the RpoE activity is strongly impaired in the absence of RSP_1090 under 1O2 is in agreement with recent studies [14].
Figure 2. RpoE activity is negatively affected in strain 2.4.1ΔRSP_1090 especially under 1O2 stress.
β-galactosidase activity of the R. sphaeroides wild type 2.4.1, strain 2.4.1ΔRSP_1090 and TF18 harboring the reporter plasmid pPHUphrAlacZ. Complemented mutant strains were also included. Cells were grown aerobically in the dark to an OD660nm of 0.4 and were exposed to high light (800 W m-2) and 50 nM methylene blue (A) or to 360 µM of tBOOH (B) for 0 min, 60 min and 180 min. The data represent the mean of three independent experiments. Error bars indicate the standard deviation.
Both, RSP_1091 and RSP_1090 are required for full defense against 1O2
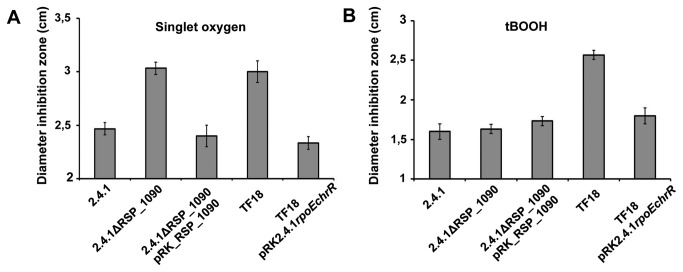
The sensitivity of strain 2.4.1ΔRSP_1090 to 1O2 and organic peroxide was tested by inhibition zone assays (Figure 3). The mutant was more sensitive to 1O2 compared to the wild type as indicated by larger inhibition zones and similar in sensitivity to the rpoEchrR deletion strain TF18 (Figure 3A). In contrast, strain 2.4.1ΔRSP_1090 was as sensitive to organic peroxide as the wild type, but strain TF18 showed a higher sensitivity (Figure 3B).
Figure 3. The RSP_1090 deletion strain is more sensitive to 1O2 than the wild type.
Inhibition of growth of the R. sphaeroides wild type 2.4.1, strain 2.4.1Δ1090 and TF18(rpoEchrR -) by 1O2 (A) and organic peroxide (B). The data represent the mean of three independent experiments. Error bars indicate the standard deviation.
Both complemented mutant strains, 2.4.1ΔRSP_1090pRK_RSP_1090 and TF18pRK2.4.1rpoEchrR, showed a similar sensitivity to 1O2 and to organic peroxide as the wild type (Figure 3 A and B). We also tested the sensitivity of the strains 2.4.1ΔRSP_1090, TF18 and the wild type harboring the empty vector pRK415 and we did not observe any difference in sensitivity compared to the respective strains lacking pRK415 (Table S1).
To address the question which genes of the RSP_1091-1087 operon are required for full activation of RpoE and to exclude polar effects of the RSP_1090 deletion strain, we deleted the entire RSP_1091-1087 operon in the R. sphaeroides wild type by the insertion of a kanamycin resistance cassette. Complementation of the mutant was then performed by reintroducing either the entire operon, RSP_1091, RSP1090 or a combination of RSP1091 and RSP1090 in trans on pRK415. Significantly larger inhibition zone assays showed that the RSP1091-1087 mutant was more sensitive to 1O2 compared to the wild type (Figure 4). Reintroduction of RSP_1091-1087 on pRK415 fully restored the wild type phenotype. It was not possible to restore the wild type phenotype by reintroducing either RSP1090 or RSP1091 on a low copy plasmid, because inhibition zones were similar to those observed for strain 2.4.1ΔRSP_1091-1087 (Figure 4). Only a combination of both genes restored the wild type phenotype. Therefore, RSP1091 and RSP1090 are both required for defense against 1O2 stress and full activation of the RpoE-dependent 1O2 stress response.
Figure 4. Inhibition by 1O2 in 2.4.1ΔRSP_1091-1087 complementation strains.
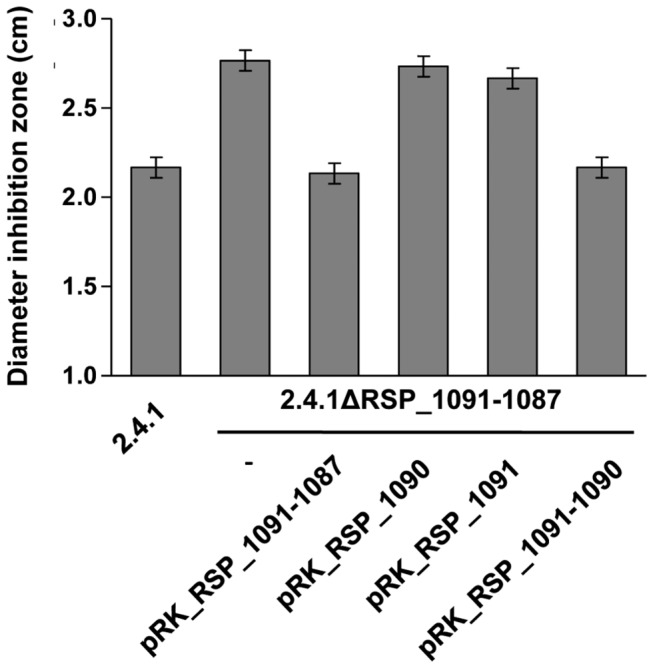
Inhibition of growth by 1O2 of the R. sphaeroides wild type 2.4.1, strain 2.4.1ΔRSP_1091-1087 and complementation with RSP_1091-1087, RSP_1090, RSP1091 and RSP_1091-1090 in pRK415. The data represent the mean of three independent experiments. Error bars indicate the standard deviation.
The RSP_1091-1090 locus is well conserved in the α-proteobacteria
Homologs of RSP_1090 proteins were identified by using the BLAST option on the integrated microbial genome (IMG) website. In the Bacteria a number of 337 genomes contained RSP_1090 with an upstream encoded homolog of RSP_1091, in most cases in proximity to rpoE and chrR homologs. RSP_1090 and RSP1091 were annotated in R. sphaeroides to be related to putative cylcopropane/cyclopropene fatty acid synthesis proteins. However, this annotation appears not to be justified due to the weak homologies to verified cylcopropane/cyclopropene fatty acid synthesis proteins. RSP_1091 and RSP_1090 were found in many α-proteobacteria, whereas conservation of RSP_1089, RSP_1088 and RSP_1087 is restricted to species belonging to the Rhodobacteraceae as e.g. Roseobacter denitrificans OCh114 (Figure S1). However, the RSP_1089-1087 homologs are absent in more distantly related Rhodobacteraceae as Oceanicola granulosus HTCC2516 (Figure S1), which underlines a genetic context specific to bacteria closely related to Rhodobacter. Also Caulobacter sp. K31 harbors RSP_1091 and RSP_1090 homologs encoded in close distance to rpoE and chrR homologs (Figure S1), but in other C. crescentus strains those homologs were not located together with rpoE and chrR homologs. In Rhizobium etli CIAT 652 no RSP_1091 and RSP_1090 homologs were located close to a chrR homolog, but rpoE was missing (Figure S1). In summary, the RSP_1091 and RSP_1090 homologs are well conserved in the α-proteobacteria and are frequently encoded adjacent to rpoE and chrR homologous genes.
The ChrR protein is rapidly degraded under singlet oxygen stress in the presence of RSP_1090
For R. sphaeroides we analyzed the levels of ChrR and RpoE in the R. sphaeroides wild type under 1O2 stress and non-stress conditions (Figure 5A). Polyclonal antibodies raised against the His6-tagged version of ChrR and RpoE were applied to detect changes in the levels of both proteins. ChrR and RpoE were detected in the absence of 1O2 (0 min, Figure 5A) and the level of both proteins increased within 60 min of 1O2 exposure. In strain TF18, neither protein was detectable (Figure 5A). Because proteolysis of the cognate anti-sigma factor is one known mechanism for sigma factor activation in Gram negative and Gram positive bacteria [18], we tested the stability of ChrR and RpoE after translation inhibition using chloramphenicol (Figure 5B). If RpoE activity is regulated by ChrR proteolysis, ChrR stability should be negatively affected under 1O2 stress conditions. In the presence of 1O2 and chloramphenicol two bands specific for ChrR were detected (Figure 5B). Without exposure to 1O2 ChrR was stable for at least 20 min in the R. sphaeroides wild type, but only a faint signal was detected after 60 min. In the presence of 1O2, the signal for the lower ChrR band was strongly decreased within 5 min, but the upper band was detectable even after 60 min. In contrast, RpoE was rather stable for 20 min under both conditions and detected in lower amounts after 60 min of 1O2 stress conditions (Figure 5B). As RpoE activity and resistance to 1O2 are decreased in the absence of RSP_1090, we tested for ChrR proteolysis in the RSP_1090 deletion strain. ChrR was more stable in the RSP_1090 mutant under 1O2 stress conditions (Figure 5C) compared to the wild type strain (Figure 5B). Its half-life in the RSP_1090 deletion strain was similar to the wild type under non-stress conditions (Figure 5B and C). Our results therefore verify that ChrR stability is decreased upon exposure of Rhodobacter to 1O2 stress and that degradation of ChrR demands RSP_1090 [13,14].
Figure 5. Protein levels and stabilities of RpoE and ChrR under non-stress and 1O2 stress conditions.
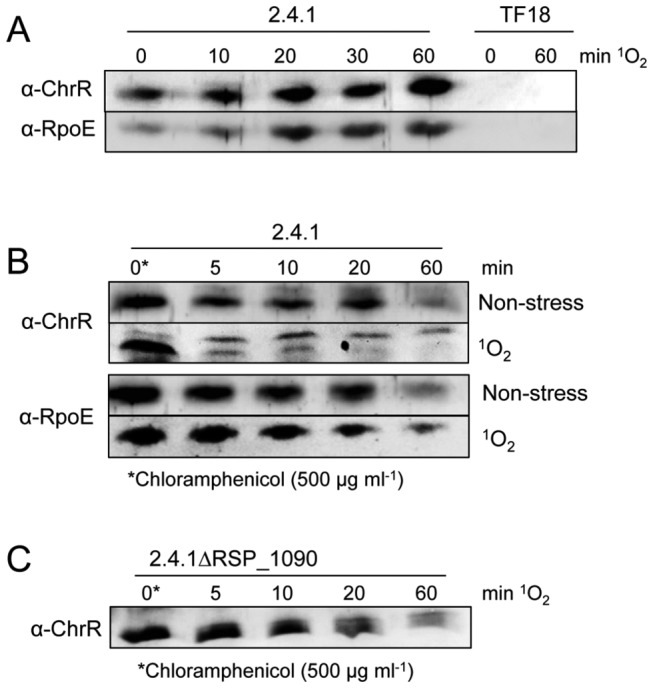
For Western blotting 240 µg of total protein were used. Loading of equal amounts of proteins was confirmed by Ponceau staining (not shown). Antibodies (α-RpoE and α-ChrR) were raised against the recombinant His6-tagged RpoE and ChrR proteins, respectively. 1O2 stress was induced at time point 0 min (OD660nm 0.4). (A) Levels of RpoE and ChrR in the R. sphaeroides wild type and the TF18 strain at different time points of 1O2 exposure (high light 800 W m-2; 50 nM methylene blue). (B) Stability of RpoE and ChrR in the R. sphaeroides wild type under non-stress (50 nM methylene blue; dark) and 1O2 stress conditions (high light 800 W m-2; 50 nM methylene blue). (C) Stability of ChrR in the presence of 1O2 in the RSP_1090 deletion mutant. To check ChrR stability under stress conditions, translation was inhibited by adding chloramphenicol (500 µg ml-1) after cultures were exposed for 60 min to 1O2 (time point 0 min). For non-stress conditions chloramphenicol was added 1 hour after OD660nm 0.4 (time point 0 min), while cultures were further incubated in the dark under aerobic conditions. The wild type control is depicted in Figure 5B. Western blots were developed using α-RpoE and α-ChrR, respectively, and anti-rabbit IgG conjugated with alkaline phosphatase.
RSP_1090 dependent ChrR degradation does not require de novo synthesis of proteases
ChrR is rapidly degraded under 1O2 exposure, but it remained unclear if the involved protease/proteases are already synthesized prior to stress exposure and therefore proteolytic activity is increased upon stress exposure or if de novo synthesis of involved factors is required. To address this question, we analyzed ChrR stability in the wild type strain by adding chloramphenicol 10 minutes before stress induction (Figure 6). ChrR levels were similar before and 10 min after addition of chloramphenicol in the absence of 1O2. ChrR was then rapidly degraded under 1O2 exposure and the signal of the lower ChrR band was abolished after 5 min of stress exposure (Figure 6). Required factors leading to ChrR degradation are therefore present before stress exposure and not synthesized de novo. To test for contribution of RSP_1090 for the activation of ChrR proteolysis, we repeated the above described experiment with the RSP1090 mutant. In contrast to the wild type, ChrR was detectable in 2.4.1∆RSP_1090 (Figure 6) even after 60 min of stress. The results indicate that the respective protease/proteases involved in ChrR proteolysis are present under non-stress conditions and that ChrR proteolysis is activated upon 1O2 stress exposure. This process depends on RSP1090, which as well does not require de novo protein synthesis.
Figure 6. Protein stabilities of ChrR in the R. sphaeroides wild type 2.4.1 and strain 2.4.1ΔRSP_1090.
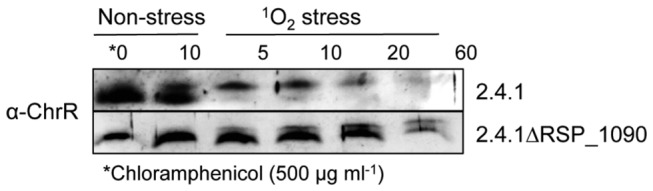
For Western blotting 240 µg of total protein were used. Loading of equal amounts of proteins was confirmed by Ponceau staining (not shown). Stability of ChrR in the wild type and strain 2.4.1ΔRSP_1090, with chloramphenicol treatment 10 min before induction of 1O2 stress (high light 800 W m-2; 50 nM methylene blue).
DegS and RseA type proteases are involved in RpoE activation in R. sphaeroides
In E. coli RpoE is involved in regulation of the membrane stress response [9]. Its activity is controlled by the membrane bound anti-sigma factor RseA, which undergoes regulated proteolysis. First the trypsin-like serine endoprotease DegS cleaves the periplasmic domain of RseA, then the transmembrane domain is cleaved by the zinc-metallo protease RseP. Finally cytoplasmic proteases degrade the part of RseA, which is still bound to RpoE [19] and RpoE can consequently activate its target genes. The R. sphaeroides protein RSP_3242 shares 37 % identity with the E. coli DegS protein and the RSP_2710 protein shares 31 % identity with RseP. To test whether these proteases have a similar function in RpoE-dependent signaling in R. sphaeroides as in E. coli we constructed strains lacking the respective genes. In addition we constructed a strain lacking the RSP_1096/1097 genes, which are in close neighborhood to the rpoE-chrR operon on the chromosome and encode a putative zinc-metallo protease. All strains showed similar growth behavior as the wild type (data not shown).
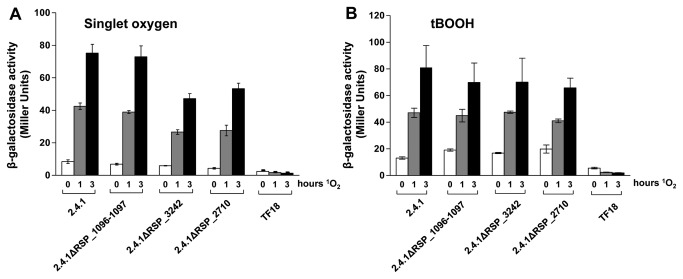
Less efficient proteolytic degradation of ChrR should result in lower activity of RpoE and consequently in lower resistance to 1O2. Therefore we tested the sensitivity of all three mutants against this substance. Strain 2.4.1ΔRSP_1096-1097 showed similar sensitivity in inhibition zone assays as the parental wild type strain, indicating no major role of the deleted genes in RpoE signaling (Figure 7). In contrast, the strains 2.4.1ΔRSP_3242 and 2.4.1ΔRSP_2710 showed significantly increased sensitivity against 1O2 when compared to the wild type. Their sensitivity was, however, lower than that of strain TF18.
Figure 7. Deletion of the degS and rseP homologous genes RSP_3242 and RSP_2710 affects sensitivity to 1O2.
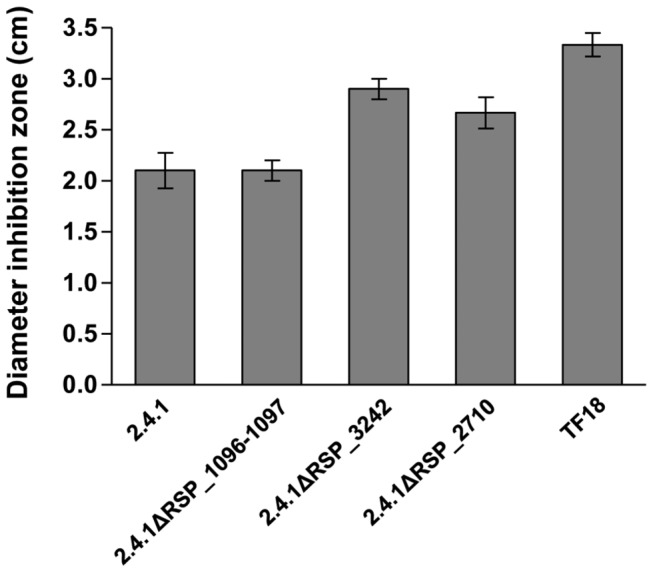
Inhibition of growth of the R. sphaeroides wild type 2.4.1, strains 2.4.1ΔRSP_1096/1097, 2.4.1ΔRSP_3242, 2.4.1ΔRSP_2710 and TF18(rpoEchrR -) by 1O2. The data represent the mean of three independent experiments. Error bars indicate the standard deviation.
To further elucidate the role of the three proteases in RpoE-dependent signaling the expression of an phrA-lacZ reporter gene in response to 1O2 and organic peroxide was analyzed. While the increase of β-galactosidase activity after the 1O2 exposure was nearly identical in the wild type and strain 2.4.1ΔRSP_1096-1097, the increase of β-galactosidase activity was clearly reduced in strains 2.4.1ΔRSP_3242 and 2.4.1ΔRSP_2710 (Figure 8A). As a negative control strain TF18 was used, which is lacking rpoEchrR and therefore exhibits no or only basal β-galactosidase activity. In contrast to 1O2, organic peroxide exposure did not lead to significantly decreased expression of the phrA-lacZ reporter gene in the protease mutant strains (Figure 8B).
Figure 8. RpoE activity is negatively affected in strains 2.4.1ΔRSP_3242 and 2.4.1ΔRSP_2710.
β-galactosidase activity of the R. sphaeroides wild type 2.4.1 and strains 2.4.1ΔRSP_1096/1097, 2.4.1ΔRSP_3242, 2.4.1ΔRSP_2710 and TF18 harboring the reporter plasmid pPHUphrAlacZ. Cells were grown aerobically in the dark to an OD660nm of 0.4 and were exposed to high light (800 W m-2) and 50 nM methylene blue (A) or to 360 µM tBOOH (B). The data represent the mean of three independent experiments. Error bars indicate the standard deviation.
Our results suggest a role of RSP_3242 and RSP_2710 in ChrR degradation. Therefore we directly tested the turn-over of ChrR in these two mutant strains.
DegS and RseA type proteases are involved in ChrR degradation in R. sphaeroides
We compared the decay of ChrR in the presence of 1O2 in the mutants lacking RSP_3242 or RSP_2710 to the decay in wild type cells. In all strains we observed two ChrR specific bands. While the lower band was maximal before addition of chloramphenicol and showed decreased abundance after its addition, the upper band strongly increased directly after addition of chloramphenicol and disappeared at later time points.
The half-life of the lower band clearly increased in the RSP_3242 and RSP_2710 mutants compared to the wild type (Figure 9). In the wild type strain the upper ChrR band had a much longer half-life than the lower band. The half-life of the upper band strongly increased in the RSP_2710 mutant. These data support a contribution of the DegS and RseA homologs in proteolytic degradation of the ChrR anti sigma factor.
Figure 9. Deletion of DegS and RseA like proteases increases ChrR stability.
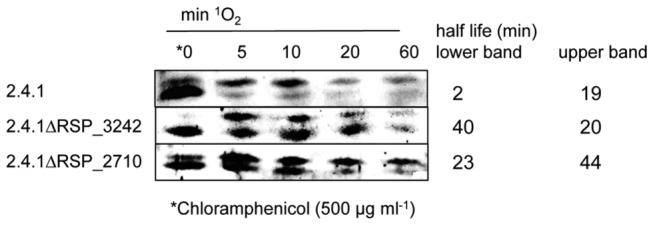
Stability of ChrR in the R. sphaeroides wild type and strains 2.4.1ΔRSP_3242, 2.4.1ΔRSP_2710 under 1O2 stress conditions (high light 800 W m-2; 50 nM methylene blue). To check ChrR stability under stress conditions, translation was inhibited by adding chloramphenicol (500 µg ml-1) after cultures were exposed for 60 min to 1O2 (time point 0 min). Western blots were developed using α-ChrR and anti-rabbit IgG conjugated with alkaline phosphatase.
Discussion
RpoE activation under 1O2 stress is regulated by proteolysis of ChrR and requires RSP_1090 and RSP1091
In this study we demonstrate that under 1O2 stress RpoE is activated by rapid proteolysis of the anti-sigma factor ChrR and show that ChrR proteolysis under 1O2 stress is dependent on RSP_1090. Our result is in line with the recent finding that an in frame deletion of RSP_1091-1090 leads to decreased activation of rpoE expression in the presence of 1O2 and that ChrR proteolysis demands RSP_1090 [13,14]. Nam et al. [14] reported that isolation of an in frame deletion of only RSP_1090 was not possible and suggested that the corresponding protein may be needed for viability in the absence of 1O2. The isolation of an RSP_1090 Tn5 mutant and construction of a knock out of RSP_1090 in this study demonstrate that the gene is not essential and failure to obtain a mutant may rather be due to technical reasons. The finding that the RSP_1090 deletion strain is negatively impaired in response to 1O2, but not to organic peroxide, strongly indicates that 1O2 and organic peroxide act independently on RpoE activation.
Moreover, we demonstrate that RSP_1090 triggered ChrR proteolysis under 1O2 does not require de novo synthesis of proteases. The required proteases are already present before stress exposure and proteolysis of ChrR is quickly activated upon stress exposure. This allows a rapid activation of the RpoE response and subsequent stress adaptation. We demonstrate that both genes RSP_1091 and RSP_1090 are required for full resistance to 1O2 and that RSP_1091-1090 homologs and their genetic location are highly conserved in other genomes. This finding supports that a combination of both genes is required for full activation of RpoE dependent defense mechanisms. As in R. sphaeroides, in many other genomes the RSP_1090 and RSP_1091 homologs are encoded next to sigma factor encoding genes, often co-localized with an anti-sigma factor encoding gene. The activity of those sigma factors may be controlled as well by RSP_1090 and RSP_1091 dependent proteolysis of the cognate anti-sigma factor, which most likely represents a highly conserved mechanism.
Organic peroxide stress activates RpoE via ChrR proteolysis, but does not depend on RSP_1091 and RSP_1090
Exposure of R. sphaeroides to organic peroxide leads to RpoE activation and ChrR proteolysis [14]. Here we further demonstrate that RpoE activation in the presence of organic peroxide does not require RSP_1090 or the DegS and RseP homologs, indicating that the response to 1O2 and organic peroxide is mediated via different signal chains to RpoE. It was recently shown that organic peroxide and 1O2 promote the dissociation of the RpoE:ChrR complex and that dissociation involves the ChrR C-terminal domain, which contains two conserved cysteine residues [20]. Oxidants such as organic peroxide are known to affect proteins via e.g. the modification of cysteine residues [21]. Organic peroxide might promote RpoE:ChrR dissociation by oxidation of one or both of the two cysteines within the ChrR C-terminal domain, as hypothesized recently [14]. Free ChrR could be targeted by proteases different from DegS and RseP. How organic peroxide eventually leads to RpoE activation and the identification of the involved protease remains to be elucidated.
Activation of the RpoE response in R. sphaeroides shows homology to the RpoE/RseA system in E. coli
As RSP_1090 and RSP_1091 do not show any homology to known proteases, we assume that proteolysis is not directly linked to the RSP_1090-1091 gene products. In E. coli RpoE is activated upon cell envelope stress by the stepwise proteolysis of the cognate anti-sigma factor RseA, due to the proteolytic activity of DegS and RseP [9]. In R. sphaeroides homologs to DegS and RseP exist. As the anti-sigma factor domains (ASD) of RseA and ChrR exhibit structure homology [7], the DegS and RseP homologs RSP_3242 and RSP_2710 were possible candidates for ChrR proteolysis in R. sphaeroides. In fact, our results show the involvement of RSP_3242 and RSP_2710 in ChrR proteolysis upon 1O2 stress exposure. Sigma factor activation by proteolysis of the cognate anti-sigma factor is a common mechanism within bacterial species [18] and was recently shown for RpoE/ChrR [14].
Besides RseA, RpoE activity in E. coli is negatively regulated by RseB, a protein which directly binds to the periplasmic domain of RseA [22-24]. Binding of RseB to RseA prevents RseP from degrading intact RseA, ensuring that RseA proteolysis is only initiated when DegS is activated upon stress [24]. A further signal is required that inhibits RseB, as RseB binding to RseA prevents cleavage by activated DegS [9,25,26]. A recent study provides evidence that intermediates in LPS transport and assembly are the second signal for RpoE activation, in this context LPS antagonizes RseA-RseB binding [27].
A RseB homolog was not found in the R. sphaeroides genome, but an RseB like action of RSP_1091 and RSP_1090 in R. sphaeroides is conceivable. RSP_1091 exhibits a putative transmembrane domain and could therefore be membrane-localized. Similar to RseA, ChrR could be membrane-localized as it exhibits a putative N-terminal transmembrane domain [28]. Membrane localization of the RpoE:ChrR complex is supported by Western blot experiments in which RpoE and ChrR were both detected in soluble (periplasmic/cytoplasmic) and insoluble (membrane) protein fractions. In non-stressed and 1O2 stressed wild type cultures RpoE and ChrR were more abundant in the insoluble fractions (data not shown). According to DegS and RseP, the proteases RSP_3242 and RSP_2710 carry at least one putative membrane-spanning segment. The subcellular localization of RSP_1091, RSP_1090 and ChrR and a possible interaction between these proteins will be investigated in future studies.
Interestingly, RSP_1091 is predicted to bind FAD or NAD. The N-terminus of the R. sphaeroides AppA protein was found to bind FAD non-covalently and was later termed BLUF (blue light using FAD). The BLUF domain was shown to function as a novel photoreceptor [29,30]. RSP_1091 could be involved in light- or redox-dependent sensing of 1O2 and might transmit the signal to RSP_1090, RSP_3242 or RSP_2710 to trigger ChrR proteolysis. The light- or redox- dependent activation of RpoE would be one of at least two possible mechanisms of RpoE activation, as RpoE activation by organic peroxide is light independent. Further studies on the localization of the involved factors and the function of RSP_1091 and RSP_1090 are in progress to unravel the detailed mechanism of 1O2 dependent RpoE activation.
Singlet oxygen signal transduction in R. sphaeroides
How 1O2 is sensed and recognized by the cells is far from being unraveled, but this study provides important insights into the conversion of the 1O2 signal to a transcriptional response in R. sphaeroides. In the previous model of RpoE activation [13] proteases were not included. Our results and another recent study [14] provide the experimental evidence that RSP_1091 and RSP_1090 are required for RpoE activation. It is important to note that the RSP_1091-1087 operon is under RpoE control [4] and induced by 1O2 exposure [31], suggesting a regulatory feedback loop. RSP_1091 and RSP_1090 are expressed under non-stress conditions, but the mRNA levels increase at least 4 fold after 1O2 exposure [31]. Once RpoE is activated upon stress induction, the expression of the rpoEchrR operon itself and the RSP_1091-1087 operon is induced by RpoE. When we analyzed RpoE and ChrR protein levels and stabilities under 1O2 stress (Figure 5 A and B), we observed increasing RpoE and ChrR levels over time in equal amounts, but interestingly ChrR stability was highly decreased under 1O2 stress in contrast to RpoE stability. This finding indicates that the ChrR turnover rate is much higher under 1O2 stress compared to RpoE. Therefore, RSP_1091 and RSP_1090 seem to be crucial to further enhance ChrR proteolysis to keep RpoE activity high during stress exposure.
The proteases RSP_2710 and RSP_3242, which are involved in proteolytic degradation of ChrR are not induced by 1O2 and not controlled by RpoE [31]. Envelope stress in E. coli that leads to dissociation of the anti-sigma factor RseA from RpoE is initiated by misfolding and assembly of outer membrane proteins in the periplasm [32]. In detail, the DegS protease is activated by unassembled porin monomers [10]. Several periplasmic or membrane stress factors lead to activation of DegS and subsequent release of RpoE [9]. Therefore, the mechanism of converting a stress signal as 1O2 formation into a cascade that leads to activation of RpoE may be highly similar in R. sphaeroides compared to what is known in E. coli, despite the cognate anti-sigma factors are not homologous.
Conclusions
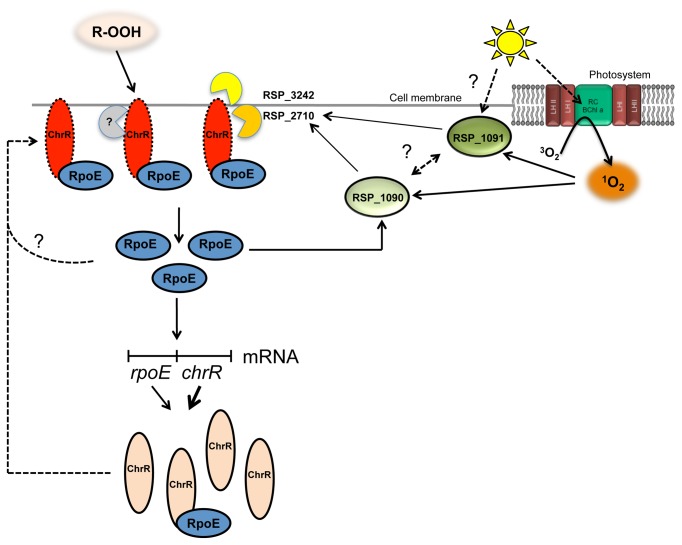
Our current model (Figure 10) shows the activation of RpoE in the response to photooxidative stress. Under non-stress conditions the ChrR proteolysis rate is low, but ensures a basal level and activity of RpoE in the cell. Upon exposure to 1O2, as well as organic peroxide stress, the anti-sigma factor ChrR is rapidly degraded. The 1O2 induced proteolysis requires RSP_1091 and RSP_1090 and involves at least two proteases, the DegS and RseP homologs RSP_3242 and RSP_2710. ChrR proteolysis leads to RpoE release, RpoE binds to the RNA-polymerase and induces target gene expression, including the rpoEchrR and the RSP_1091-1087 operon. Increased levels of RSP_1091 and RSP_1090 promote ongoing ChrR proteolysis to maintain high RpoE activity, which displays a positive regulatory loop. Activation of RpoE in the presence of organic peroxide does not require the DegS and RseP homologs RSP_3242 and RSP_2710 for ChrR proteolysis, but so far unknown proteases. In this study further components of the cascade involved in 1O2 signaling were identified, but the direct link between RSP_1091 and RSP_1090 and the proteases RSP_2710 and RSP_3242 in ChrR proteolysis and subsequent RpoE activation remain to be elucidated.
Figure 10. Current model of RpoE activation by ChrR proteolysis in R. sphaeroides under 1O2 stress.
The model displays the mechanism of RpoE activation in the response to 1O2. The localization of ChrR at the membrane is speculative. Solid black arrows indicate positive effects. Dashed arrows indicate hypothetical effects. A detailed explanation is given in the conclusion part.
Material and Methods
Bacterial strains and growth conditions
R. sphaeroides strains were grown at 32°C in minimal salt medium containing malate as carbon source [33]. Aerobic growth conditions with a concentration of 160 to 180 µM of dissolved oxygen were established by gassing cultures with air in flat glass bottles or by continuous shaking of Erlenmeyer flasks at 140 rpm with a culture volume of 20%. In semiaerobic cultures a volume of 80% in Erlenmeyer flasks and shaking at 140 rpm lead to a dissolved oxygen concentration of approximately 25 µM. When necessary kanamycin (25 µg ml-1), tetracycline (1.5 µg ml-1), trimethoprim (50 µg ml-1) or gentamycine (10 µg ml-1) was added to liquid and solid growth media (1.6% agar). Antibiotics were omitted from pre-cultures, cultures and agar plates used for R. sphaeroides strains during stress experiments and inhibition zone assays (see below). E. coli strains were grown aerobically at 37°C in LB medium under continuous shaking at 180 rpm or on solid growth media.
Sensitivity to 1O2 and organic peroxide
Measurement of sensitivity to 1O2 was performed as described before [34]. The measurement of sensitivity to organic peroxide was performed similarly to 1O2 experiments. Instead of methylene blue 5 µl of 700 mM tBOOH were added to the filter disks and the agar plates were incubated 48 hours in the dark.
Tn5-mutagenesis of R. sphaeroides 2.4.1pPHUphrAlacZ
The R. sphaeroides wild type harboring plasmid pPHUphrAlacZ was grown under semiaerobic conditions in the presence of 1.5 µg ml-1 tetracycline. The E. coli strain S17-1pSUP202 was grown aerobically in LB medium containing 20 µg ml-1 tetracycline. For Tn5-mutagenesis 1 ml of exponential-phase R. sphaeroides culture was mixed with 200 µl exponential-phase E. coli culture. The cells were centrifuged at 5.000 rpm for 5 min at room temperature. The supernatant was removed and the cells were washed in 1 ml of malate minimal medium. After a second centrifugation the supernatant was discarded and the cells resuspended in 200 µl of malate minimal medium. The cell suspension was transferred onto nitrocellulose membranes (Whatman, Dassel, Germany) placed on PY agar plates. After 5 hours of incubation at 32°C the filter was transferred into a fresh 1.5 ml tube containing 1 ml of malate minimal medium. After vortexing, the whole suspension was diluted in malate minimal medium and 50 µl aliquots were plated on malate minimal agar plates containing 2 µM of Rose Bengal, tetracycline (1.5 µg ml-1) and kanamycin (25 µg ml-1). The plates were incubated at 32°C for 3 days in the dark.
Screening for Tn5-mutants with decreased β-galactosidase activity under 1O2 stress
Agar plates containing R. sphaeroides colonies were placed under a daylight fluorescent tube (20 W m-2) to induce RpoE activity. After 2 hours the plates were sprayed with X-Gal (20 mg ml-1) and incubated for 6 hours at room temperature. Such colonies were further investigated, which were not or only slightly colored blue. To test for Tn5 insertion into plasmid pPHUphrAlacZ, and consequently leading to a false negative result, pPHUphrAlacZ was isolated from Tn5 mutants and electroporated into E. coli JM109. The E. coli cells were plated on LB agar plates containing 20 µg ml-1 tetracycline (pPHUphrAlacZ) and 25µg ml-1 kanamycin (Tn5). Cells growing on kanamycin had a Tn5 insertion in the plasmid pPHUphrAlacZ and respective Tn5 mutants were excluded.
Identification of Tn5 insertion sites by vectorette PCR
In principle, the Vectorette PCR was performed as described previously [16]. For the synthesis of the Vectorette units two imperfect complementary DNA oligonucleotides (Vectorette oligonucleotide_1 and Vectorette oligonucleotide_2, Table S2) were incubated for 5 min at 65°C. After addition of 5 µl 25 mM MgCl2 solution the oligonucleotides were cooled down slowly to room temperature. After digestion of chromosomal DNA with the blunt end restriction enzymes PvuII, FspI and DpnI (New England Biolabs), the digested DNA was purified with phenol/chloroform/isoamylalcohol (AppliChem, Darmstadt, Germany) and precipitated with ethanol and sodium acetate. The Vectorette units were ligated with the digested DNA, ligation was performed for 12 hours at 15°C. The ligated DNA was used as template in a PCR reaction using Taq DNA polymerase (Qiagen) and the primers 224new and Tnp_out_new (Table S2). After PCR the samples were separated on a 1% agarose gel and DNA fragments were gel extracted with the Gel extraction kit QIAEX III (Qiagen). The purified DNA fragment was cloned into the pDrive vector (Qiagen) and sequenced with the primer Tnp_out_new.
β-galactosidase activity assay
This was carried out as described previously [35].
Photooxidative stress conditions
Photooxidative stress conditions were performed as described earlier [5], except the final concentration of methylene blue. In brief, cultures were grown under semiaerobic conditions over night to obtain pigmented cells. Cultures were diluted to an OD660nm of 0.2 and allowed to double once under aerobic growth conditions in darkened flat glass bottles. High light conditions were generated by illumination with 800 W m-2 white light. For photooxidative stress 1O2 producing methylene blue was added to liquid cultures at a final concentration of 50 nM prior to illumination.
Construction of R. sphaeroides deletion mutants
R. sphaeroides deletions strains 2.4.1ΔRSP_1090, 2.4.1ΔRSP_1091-1087, 2.4.1ΔRSP_1096-1097, 2.4.1ΔRSP_3242 and 2.4.1ΔRSP_2710 were generated by transferring the respective suicide plasmid pPHU2.4.1RSP_1090::Km, pPHU2.4.1RSP_1091-1087::Km, pPHU2.4.1ΔRSP_1096-1097::Km, pPHU2.4.1ΔRSP_3242::Km and pPHU2.4.1ΔRSP_2710::Km (Table 1 and 2) into R. sphaeroides 2.4.1. Knockout candidates were screened for insertion of the kanamycin cassette into the chromosome by homologous recombination. For construction of pPHU2.4.1RSP_1090::Km parts of the gene RSP_1090 together with upstream and downstream regions were amplified by PCR using the oligonucleotides 2.4.1RSP1090_knockout-up_EcoRI, 2.4.1RSP1090_knockout-up_PstI, 2.4.1RSP1090_knockout-down_PstI and 2.4.1RSP1090_knockout-down_SphI (Table S2). Using the same strategy, parts of RSP_1091-1087 operon together with RSP_1091 upstream and RSP_1087 downstream regions were amplified by PCR using the oligonucleotides 2.4.1RSP_1091_knockout-up_EcoRI, 2.4.1RSP_1091_knockout-up_PstI, 2.4.1RSP_1087_knockout-down_PstI and 2.4.1RSP1087_knockout-down_SphI (Table S2). For construction of pPHU2.4.1RSP_1096-1097::Km RSP_1096-1097 together with upstream and downstream regions were amplified by PCR using the oligonucleotides 2.4.1RSP1096/97_knockout-up_EcoRI, 2.4.1RSP1096/97_knockout-up_PstI, 2.4.1RSP1096/97_knockout-down_PstI and 2.4.1 RSP1096/97_knockout-down_SphI (Table S2). For deletion of RSP_3242, part of RSP_3242 together with upstream and downstream regions were amplified by PCR using the oligonucleotides 2.4.1RSP3242_knockout-up_EcoRI, 2.4.1RSP3242_knockout-up_PstI, 2.4.1RSP3242_knockout-down_PstI and 2.4.1RSP3242_knockout-down_SphI (Table S2). RSP_2710 together with upstream and downstream regions were amplified by PCR using the oligonucleotides 2.4.1RSP2710_knockout-up_EcoRI, 2.4.1RSP2710_knockout-up_PstI, 2.4.1RSP2710_knockout-down_PstI and 2.4.1 RSP2710_knockout-down_SphI (Table S2). The obtained PCR fragments were cloned into pPHU281 [35] using the appropriate restriction endonucleases. Then the kanamycin cassette obtained from plasmid pUC4K [36] was inserted into the PstI restriction site to obtain the plasmids pPHU2.4.1RSP_1090::Km, pPHU2.4.1RSP_1091-1087::Km, pPHU2.4.1Δ1096-1097::Km, pPHU2.4.1ΔRSP_3242::Km and pPHU2.4.1ΔRSP_2710::Km. The plasmids were transferred into E. coli strain S17-1 [37] and mobilized into R. sphaeroides strains by biparental conjugation. Conjugants were selected on malate minimal medium agar plates containing 25 µg ml-1 kanamycin. PCR analyses of chromosomal DNA isolated from kanamycin resistant and tetracycline sensitive conjugants were carried out to confirm the double crossover event of the kanamycin cassette into the R. sphaeroides chromosome.
Table 1. Strains.
| Strains | Description | Source/Reference |
|---|---|---|
| E. coli | ||
| S17-1 | recA pro; hsdR; RP4- 2- Tc::Mu-Km::tn7; tra +; Kmr; Spr | [37] |
| JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi (lac–proAB) | New England Biolabs |
| M15(pREP4) | E. coli strain M15 containing pREP4 plasmid encoding lac repressor in trans, Kmr | Qiagen |
| M15(pREP4)pQE30_2.4.1rpoE | E. coli M15 harboring pQE30_2.4.1rpoE, Kmr, Apr | This study |
| M15(pREP4)pQE30_2.4.1chrR | E. coli M15 harboring pQE30_2.4.1chrR, Kmr, Apr | This study |
| R. sphaeroides | ||
| 2.4.1 | Wild type | [40] |
| 2.4.1pPHUphrAlacZ | 2.4.1 harboring pPHUphrAlacZ, Tcr | [15] |
| 2.4.1pRK415 | Wild type harboring pRK415, Tcr | This study |
| 2.4.1ΔRSP_1090 | 2.4.1RSP_1090::Kmr cassette, Kmr | This study |
| 2.4.1ΔRSP_1090pRK415 | 2.4.1ΔRSP_1090 harboring pRK415, Kmr, Tcr | This study |
| 2.4.1ΔRSP_1091-1087 | 2.4.1RSP_1091-1087::Kmr cassette, Kmr | This study |
| 2.4.1ΔRSP_1096-1097 | 2.4.1RSP_1096-1097::Kmr cassette, Kmr | This study |
| 2.4.1ΔRSP_2710 | 2.4.1RSP_2710::Kmr cassette, Kmr | This study |
| 2.4.1ΔRSP_3242 | 2.4.1RSP_3242::Kmr cassette, Kmr | This study |
| 2.4.1ΔRSP_1090pRKRSP_1090 | 2.4.1ΔRSP_1090 harboring pRKRSP_1090, Kmr, Tcr | This study |
| 2.4.1ΔRSP1090pPHUphrAlacZ | 2.4.1ΔRSP_1090 harboring pPHUphrAlacZ, Kmr,Tcr | This study |
| 2.4.1ΔRSP1090pPHUphrAlacZpBBR_RSP_1090 | 2.4.1ΔRSP_1090 pPHUphrAlacZ harboring pBBR_RSP_1090, Kmr,Tcr, Gmr | This study |
| 2.4.1ΔRSP_1091-1087pRKRSP_1090 | 2.4.1ΔRSP_1091-1087 harboring pRKRSP_1090, Kmr, Tcr | This study |
| 2.4.1ΔRSP_1091-1087pRKRSP_1091 | 2.4.1ΔRSP_1091-1087 harboring pRKRSP_1091, Kmr, Tcr | This study |
| 2.4.1ΔRSP_1091-1087pRKRSP_1091-1090 | 2.4.1ΔRSP_1091-1087 harboring pRKRSP_1091-1090, Kmr, Tcr | This study |
| 2.4.1ΔRSP_1091-1087pRKRSP_1091-1087 | 2.4.1ΔRSP_1091-1087 harboring pRKRSP_1091-1087, Kmr, Tcr | This study |
| TF18 | rpoEchrR mutation in 2.4.1, Tpr | [41] |
| TF18pRK415 | TF18 harboring pRK415, Tpr, Tcr | This study |
| TF18pPHUphrAlacZ | TF18 harboring pPHUphrAlacZ, Tpr, Tcr | [15] |
| TF18pPHUphrAlacZ pBBR_2.4.1rpoEchrR | TF18 pPHUphrAlacZ harboring pBBR_2.4.1rpoEchrR Tpr, Tcr, Gmr | This study |
| 2.4.1ΔRSP_1096-1097pPHUphrAlacZ | 2.4.1ΔRSP_1096-1097 harboring pPHUphrAlacZ, Tpr, Tcr | This study |
| 2.4.1ΔRSP_2710pPHUphrAlacZ | 2.4.1ΔRSP_2710harboring pPHUphrAlacZ, Tpr, Tcr | This study |
| 2.4.1ΔRSP_3242pPHUphrAlacZ | 2.4.1ΔRSP_3242 harboring pPHUphrAlacZ, Tpr, Tcr | This study |
| TF18pRK2.4.1rpoEchrR | TF18 harboring pRK2.4.1rpoEchrR, Tpr, Tcr | [42] |
Table 2. Plasmids.
| Plasmids | Description | Source/Reference |
|---|---|---|
| pPHU281 | Tcr, suicide vector for R. sphaeroides, Tcr | [35] |
| pUC4K | Kmr, source of Kmr cassette | [36] |
| pSUP202 | Tcr, Kmr, Suicide vector used for Tn5 mutagenesis | [37] |
| pRK415 | Tcr | [43] |
| pBBR1MCS-5 | Gmr | [44] |
| pPHUphrAlacZ | pPHU234 with phrA upstream-region, Tcr | [15] |
| pPHU2.4.1RSP_1090::Kmr | pPHU281 with Kmr cassette, flanked by the up- and downstream region of RSP_1090 | This study |
| pPHU2.4.1RSP_RSP_1091-1087::Kmr | pPHU281 with Kmr cassette, flanked by the upstream region of RSP_1091 | This study |
| and downstream region of RSP_1087 | ||
| pPHU2.4.1RSP_1096-1097::Kmr | pPHU281 with Kmr cassette, flanked by the up- and downstream region of RSP_1096-1097 | This study |
| pPHU2.4.1RSP_2710::Kmr | pPHU281 with Kmr cassette, flanked by the up- and downstream region of RSP_2710 | This study |
| pPHU2.4.1RSP_3242::Kmr | pPHU281 with Kmr cassette, flanked by the up- and downstream region of RSP_3242 | This study |
| pRKRSP_1090 | pRK415 harboring a 0.8 kb fragment containing RSP_1090 flanked by the 64 bp upstream region of RSP_1091 and 7 bp downstream region of RSP_1090 | This study |
| pRK2.4.1rpoEchrR | pRK415 harboring a 1.6 kb fragment containing 2.4.1 rpoEchrR flanked by the 241 bp upstream and 158 bp downstream regions | [42] |
| pRK2.4.1RSP_1091 | pRK415 harboring a 1.4 kb fragment containing RSP_1091 flanked by the 97 bp upstream region of RSP_1090 and 24 bp downstream region of RSP_1091 | This study |
| pRK2.4.1RSP_1091-1090 | pRK415 harboring a 2.1 kb fragment containing entire sequence of RSP_1091 and RSP_1090 flanked by the 97 bp upstream region of RSP_1091 and 19 bp downstream of RSP_1090 | This study |
| pRK2.4.1RSP_1091-1087 | pRK415 harboring a 4.6 kb fragment containing entire sequence of RSP_1091-1087 flanked by 99 bp upstream of RSP_1091 and 78 bp downstream of RSP_1087 | This study |
| pBBR2.4.1RSP_1090 | pBBR1MCS-5 harboring a 0.8 kb fragment containing RSP_1090 flanked by the 64 bp upstream region of RSP_1091 and 7 bp downstream region of RSP_1090 | This study |
| pBBR_2.4.1rpoEchrR | pBBR1MCS-5 harboring a 1.6 kb fragment containing 2.4.1 rpoEchrR flanked by the 241 bp upstream and 158 bp downstream regions | This study |
| pQE30_2.4.1rpoE | pQE30 harboring the entire rpoE gene lacking the first codon | This study |
| pQE30_2.4.1chrR | pQE30 harboring the entire chrR gene lacking the first codon | This study |
| pQE30 | Apr, vector used for protein overexpression in E. coli M15(REP4) | Qiagen |
| pDrive cloning vector | Apr; Kmr | Qiagen |
Complementation of the R. sphaeroides RSP_1090 and RSP_1091-1087 deletion mutants
For complementation of strain 2.4.1ΔRSP_1090 and 2.4.1ΔRSP_1091-1087 with RSP_1090 a 821 bp PCR fragment, containing the entire gene along with 64 bp of the upstream region of RSP_1091, including the RpoE dependent RSP_1091 promoter, and 7 bp downstream of the last RSP_1090 codon, was amplified using the oligonucleotides 2.4.1RSP1090com_up_SigE1091 and 2.4.1RSP1090com_down (Table S2). The obtained PCR fragment was cloned into the pDrive vector (Qiagen). Digestion of the pDrive vector, containing the insert, with PstI and XbaI followed by cloning with the same restriction sites into plasmid pRK415 yielded plasmid pRK_RSP_1090. This plasmid was subsequently transformed into E. coli S17-1 and conjugated with strain 2.4.1ΔRSP_1090 to obtain the complemented strain 2.4.1ΔRSP_1090pRK_RSP_1090. Cloning of the PCR fragment with the same restriction enzymes into the vector pBBR1MCS-5 yielded plasmid pBBR2.4.1_RSP_1090.
The same strategy was applied for complementation of 2.4.1ΔRSP_1091-1087 with RSP1091, RSP_1090, RSP_1091-1090 and RSP_1091-1087. For complementation with RSP_1091, a 1.4 kb PCR fragment, containing the entire sequence of RSP_1091, along with 97 bp of the upstream region of RSP_1091 and 24 bp downstream of the last RSP_1091 codon, was amplified using the oligonucleotides 2.4.1RSP1091com_up_KpnI and 2.4.1RSP1091com_dn_XbaI (Table S2). For complementation with RSP_1091-1090, a 2.1 kb PCR fragment, containing the entire sequence of both the genes, RSP_1090 and RSP1091, along with 97 bp of the upstream region of RSP_1091 and 19 bp downstream of the last RSP_1090 codon, was amplified using the oligonucleotides 2.4.1RSP1091com_up_KpnI and 2.4.1RSP1090com_dn_XbaI (Table S2). Finally, for complementation with RSP_1091-1087 a 4.6 kb PCR fragment, containing the entire region of RSP_1091-1087 along with 99 bp of the upstream region of RSP_1091 and 78 bp downstream of the RSP_1087, was amplified using the oligonucleotides 2.4.1RSP1091-87com_up and 2.4.1RSP1091-87com_down (Table S2). The obtained PCR fragments were cloned into the pDrive vector (Qiagen). Digestion of the pDrive vector with KpnI and XbaI was followed by cloning with the same restriction sites into plasmid pRK415 yielding the strains 2.4.1ΔRSP_1091-1087pRK2.4.1RSP_1091, 2.4.1ΔRSP_1091-1087pRK2.4.1RSP_1091-1090, and 2.4.1ΔRSP_1091-1087pRK2.4.1RSP_1091-1087.
Construction of strains 2.4.1ΔRSP_1096-1097pPHUphrAlacZ, 2.4.1ΔRSP_2710pPHUphrAlacZ, 2.4.1ΔRSP_3242pPHUphrAlacZ, and TF18pPHUphrAlacZ
For construction of the strains 2.4.1ΔRSP_1096-1097pPHUphrAlacZ, 2.4.1ΔRSP_2710pPHUphrAlacZ, 2.4.1ΔRSP_3242pPHUphrAlacZ and TF18pPHUphrAlacZ the plasmid pPHUphrAlacZ was transferred to strains 2.4.1ΔRSP_1096-1097, 2.4.1ΔRSP_2710, 2.4.1ΔRSP_3242 and TF18 by biparental conjugation.
Construction of strains 2.4.1ΔRSP_1090pPHUphrAlacZ, 2.4.1ΔRSP_1090pPHUphrAlacZpBBR2.4.1_RSP_1090 and TF18pPHUphrAlacZpBBR_2.4.1rpoEchrR
For construction of strain 2.4.1ΔRSP_1090pPHUphrAlacZ the plasmid pPHUphrAlacZ was transferred to strain 2.4.1ΔRSP_1090 by biparental conjugation. Transfer of plasmid pBBR2.4.1_RSP_1090 to strain 2.4.1ΔRSP_1090pPHUphrAlacZ yielded strain 2.4.1ΔRSP_1090pPHUphrAlacZpBBR2.4.1_RSP_1090. For construction of plasmid pBBR2.4.1rpoEchrR the plasmid pRK2.4.1rpoEchrR [38] was digested with EcoRI. The obtained 2.4.1rpoEchrR fragment was cloned into pBBR1MCS-5 and transformed in E. coli S17-1 and conjugated with strain TF18pPHUphrAlacZ yielding TF18pPHUphrAlacZpBBR2.4.1rpoEchrR.
Construction and purification of recombinant proteins and production of antibodies
The rpoE (RSP_1092) and the chrR (RSP_1093) gene of R. sphaeroides 2.4.1 were PCR amplified, from the second to the last codon, using the oligonucleotides rpoE-A-4, rpoE-B542, chrR-A-4 and chrR-B-638. The PCR products were cloned into the pQE30 vector (Qiagen). Overexpression in E. coli M15 (pREP4) cells was performed as described earlier [39]. Purification using nickel-nitriloacetic-acid (Ni-NTA) agarose was performed under denaturing conditions in accordance with the manufacturer’s instructions (Qiagen). For production of polyclonal antibodies raised against His6-RpoE, 750 µg of recombinant protein were separated by SDS-PAGE and stained with ice cold 3 M potassium chloride solution. Protein bands were cut out of the gel, production of antibodies in rabbits was performed by BioGenes, Berlin. Antibodies were purified by using CNBr-activated sepharose (GE Healthcare, Munich) coupled with His6-RpoE. For production of polyclonal antibodies in rabbits raised against His6-ChrR, purified recombinant protein was sent to Davids Biotechnologie in Regensburg. Antibodies were purified by using affinity purification via western blotting. The His6-ChrR protein bands were excised from the membrane and incubated with serum for three hours. After washing the membrane with 1xTBS, the bound antibody was eluted with acidic glycine buffer and immediately neutralized with 1M Tris (pH 8.0).
Western blot experiments
For Western blot experiments R. sphaeroides cultures were grown under non-stress or 1O2 stress conditions as described above. To determine stability of RpoE and ChrR chloramphenicol was added in a final concentration of 500 µg ml-1. Aliquots were taken at different time points, rapidly cooled and ice-cold trichloroacetic acid (10% final concentration) was added and incubated on ice for one hour. For precipitation of the protein samples were centrifuged at 13.000 rpm for 10 minutes. The supernatant was aspirated and the pellet washed twice with ice-cold 100% acetone. After evaporation of residual acetone, the cell pellet was suspended in 1 fold tris buffer saline (1xTBS) with 0.05% tween-20. Equal amounts of total protein (300 µg) were separated on a 12% PAA-SDS gel and transferred to a nitrocellulose membrane (Whatman). Proteins were stained and fixed with Ponceau S (Sigma Aldrich), destained with sodium hydroxide and the membrane was blocked at room temperature for 1 hour in blocking buffer (1xTBS) containing 5% (w/v) of milk powder (Roth). After blocking, the purified primary antibodies, α-RpoE or α-ChrR, diluted 1:5.000 in blocking buffer, were added to the membrane and incubated for 3 hours. After washing the membrane 3 times for 5 min in 1xTBS buffer, the secondary antibody (anti-rabbit IgG conjugated with peroxidases, produced in goat, Sigma Aldrich) was added (diluted 1:15.000 in blocking buffer) and the membrane further incubated for 2 hours at room temperature. The membrane was washed 3 times with 1xTBS for 5 minutes. The washing step was repeated 2 times. Western blots were developed using the lumi-light western blotting substrate 1 and 2 (Roche).
Supporting Information
The RSP_1091-1090 locus is well conserved in α-proteobacteria. Gene neighborhood of the R. sphaeroides 2.4.1 gene RSP_1090 in selected genomes of the α-proteobacteria. Homologs of RSP_1090 were searched by using the BLAST option on the integrated microbial genome (IMG) website. The genes encoding the retrieved homologs of RSP_1090 were subsequently analyzed with respect to the homology of proteins encoded by adjacent genes. In bacteria a number of 337 genomes contained RSP_1090 with an upstream located homolog of RSP_1091. Amino acid identities to R. sphaeroides 2.4.1 proteins are indicated.
(PDF)
Inhibition zone diameters. Sensitivity against 1O2 and organic peroxide (tBOOH) was tested for the R. sphaeroides wild type, strain 2.4.1ΔRSP_1090 and strain TF18. For strains harbouring plasmid pRK415 and the constructs pRKRSP_1090 and pRK_2.4.1rpoEchrR, also values for 1O2 and tBOOH inhibition zones were determined. The generation of 1O2 was achieved by applying 5µl of 10 mM methylene blue solution on filter discs placed on agar plates in the light. In the same manner 700 mM tBOOH was used, agar plates were incubated in the dark. In all cases the mean and standard deviation for three replicates are depicted. Mean values of three experiments are given, SD: standard deviation.
(DOC)
Oligonucleotides used throughout this study.
(DOC)
Acknowledgments
We thank Angelika Balzer for her help in constructing the protease deletion mutants and Silvia Caballero Ortíz for experimental assistance.
Funding Statement
This project was funded by the Deutsche Forschungsgemeinschaft grant Kl 563/ 20-2. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ogilby PR (2010) Singlet oxygen: there is indeed something new under the sun. Chem Soc Rev 39: 3181-3209. doi: 10.1039/b926014p. PubMed: 20571680. [DOI] [PubMed] [Google Scholar]
- 2. Davies MJ (2004) Reactive species formed on proteins exposed to singlet oxygen. Photochem Photobiol Sci 3: 17-25. doi: 10.1039/b307576c. PubMed: 14743273. [DOI] [PubMed] [Google Scholar]
- 3. Wilkinson F, Helman WP, Ross AB (1995) Rate constants for the dacay and reactions of the lowest electronically excited singlet-state of molecular-oxygen in solution - An expanded and revised compilation. J Phys Chem Ref Data 24: 663-1021. doi: 10.1063/1.555965. [DOI] [Google Scholar]
- 4. Anthony JR, Warczak KL, Donohue TJ (2005) A transcriptional response to singlet oxygen, a toxic byproduct of photosynthesis. Proc Natl Acad Sci U_S_A 102: 6502-6507. doi: 10.1073/pnas.0502225102. PubMed: 15855269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Glaeser J, Klug G (2005) Photo-oxidative stress in Rhodobacter sphaeroides: Protective role of carotenoids and expression of selected genes. Microbiology 151: 1927-1938. doi: 10.1099/mic.0.27789-0. PubMed: 15942000. [DOI] [PubMed] [Google Scholar]
- 6. Anthony JR, Newman JD, Donohue TJ (2004) Interactions between the Rhodobacter sphaeroides ECF sigma factor, σE, and its anti-sigma factor, ChrR. J Mol Biol 341: 345-360. doi: 10.1016/j.jmb.2004.06.018. PubMed: 15276828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Campbell EA, Greenwell R, Anthony JR, Wang S, Lim L et al. (2007) A conserved structural module regulates transcriptional responses to diverse stress signals in bacteria. Mol Cell 27: 793-805. doi: 10.1016/j.molcel.2007.07.009. PubMed: 17803943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cogdell RJ, Howard TD, Bittl R, Schlodder E, Geisenheimer I et al. (2000) How carotenoids protect bacterial photosynthesis. Philos Trans R Soc Lond B 355: 1345-1349. doi: 10.1098/rstb.2000.0696. PubMed: 11127989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chaba R, Grigorova IL, Flynn JM, Baker TA, Gross CA (2007) Design principles of the proteolytic cascade governing the sigma(E)-mediated envelope stress response in Escherichia coli: keys to graded, buffered, and rapid signal transduction. Genes Dev 21: 124-136. doi: 10.1101/gad.1496707. PubMed: 17210793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Campbell EA, Tupy JL, Gruber TM, Wang S, Sharp MM et al. (2003) Crystal structure of Escherichia coli sigma(E) with the cytoplasmic domain of its anti-sigma RseA. Mol Cell 11: 1067-1078. doi: 10.1016/S1097-2765(03)00148-5. PubMed: 12718891. [DOI] [PubMed] [Google Scholar]
- 11. Dufour YS, Landick R, Donohue TJ (2008) Organization and evolution of the biological response to singlet oxygen stress. J Mol Biol 383: 713-730. doi: 10.1016/j.jmb.2008.08.017. PubMed: 18723027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lourenço RF, Gomes SL (2009) The transcriptional response to cadmium, organic hydroperoxide, singlet oxygen and UV-A mediated by the sigma(E)-ChrR system in Caulobacter crescentus . Mol Microbiol 72: 1159-1170. doi: 10.1111/j.1365-2958.2009.06714.x. PubMed: 19400803. [DOI] [PubMed] [Google Scholar]
- 13. Glaeser J, Nuss AM, Berghoff BA, Klug G (2011) Singlet oxygen stress in microorganisms. Adv Microb Physiol 58: 141-173. doi: 10.1016/B978-0-12-381043-4.00004-0. PubMed: 21722793. [DOI] [PubMed] [Google Scholar]
- 14. Nam TW, Ziegelhoffer EC, Lemke RAS, Donohue TJ (2013) Proteins needed to activate a transcriptional response to the reactive oxygen species singlet oxygen. mBio 4: e00541–e00512. PubMed: 23300250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hendrischk A-K, Braatsch S, Glaeser J, Klug G (2007) The phrA gene of Rhodobacter sphaeroides encodes a photolyase and is regulated by singlet oxygen and peroxide in a σE-dependent manner. Microbiology 153: 1842-1851. doi: 10.1099/mic.0.2006/004390-0. PubMed: 17526841. [DOI] [PubMed] [Google Scholar]
- 16. Riley J, Butler R, Ogilvie D, Finniear R, Jenner D et al. (1990) A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res 18: 2887-2890. doi: 10.1093/nar/18.10.2887. PubMed: 2161516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ziegelhoffer EC, Donohue TJ (2009) Bacterial responses to photo-oxidative stress. Nat Rev Microbiol 7: 856- 863. PubMed: 19881522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ho TD, Ellermeier CD (2012) Extra cytoplasmic function sigma factor activation. Curr Opin Microbiol 15: 182-188. doi: 10.1016/j.mib.2012.01.001. PubMed: 22381678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Flynn JM, Levchenko I, Sauer RT, Baker TA (2004) Modulating substrate choice: the SspB adaptor delivers a regulator of the extracytoplasmic-stress response to the AAA plus protease ClpXP for degradation. Genes Dev 18: 2292-2301. doi: 10.1101/gad.1240104. PubMed: 15371343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Greenwell R, Nam TW, Donohue TJ (2011) Features of Rhodobacter sphaeroides ChrR required for stimuli to promote the dissociation of sigma(E)/ChrR complexes. J Mol Biol 407: 477-491. doi: 10.1016/j.jmb.2011.01.055. PubMed: 21295582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Imlay JA (2003) Pathways of oxidative damage. Annu Rev Microbiol 57: 395-418. doi: 10.1146/annurev.micro.57.030502.090938. PubMed: 14527285. [DOI] [PubMed] [Google Scholar]
- 22. De Las Peñas A, Connolly L, Gross CA (1997) The sigmaE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of sigmaE. Mol Microbiol 24: 373-385. doi: 10.1046/j.1365-2958.1997.3611718.x. PubMed: 9159523. [DOI] [PubMed] [Google Scholar]
- 23. Alba BM, Gross CA (2004) Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol Microbiol 52: 613-619. doi: 10.1111/j.1365-2958.2003.03982.x. PubMed: 15101969. [DOI] [PubMed] [Google Scholar]
- 24. Grigorova IL, Chaba R, Zhong HJ, Alba BM, Rhodius V, et al. (2004) Fine-tuning of the Escherichia coli sigma(E) envelope stress response relies on multiple mechanisms to inhibit signal-independent proteolysis of the transmembrane anti-sigma factor, RseA. Genes Dev 8: 2686-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cezairliyan BO, Sauer RT (2007) Inhibition of regulated proteolysis by RseB. Proc Natl Acad Sci U_S_A 104: 3771-3776. doi: 10.1073/pnas.0611567104. PubMed: 17360428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim DY, Jin KS, Kwon E, Ree M, Kim KK (2007) Crystal structure of RseB and a model of its binding mode to RseA. Proc Natl Acad Sci U_S_A 104: 8779-8784. doi: 10.1073/pnas.0703117104. PubMed: 17496148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lima S, Guo MS, Chaba R, Gross CA, Sauer RT (2013) Dual molecular signals mediate the bacterial response to outer-membrane stress. Science 340: 837-841. doi: 10.1126/science.1235358. PubMed: 23687042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cserzö M, Wallin E, Simon I, von Heijne G, Elofsson A (1997) Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng 10: 673-676. doi: 10.1093/protein/10.6.673. PubMed: 9278280. [DOI] [PubMed] [Google Scholar]
- 29. Braatsch S, Gomelsky M, Kuphal S, Klug G (2002) A single flavoprotein, AppA, integrates both redox and light signals in Rhodobacter sphaeroides . Mol Microbiol 45: 827-836. doi: 10.1046/j.1365-2958.2002.03058.x. PubMed: 12139627. [DOI] [PubMed] [Google Scholar]
- 30. Gomelsky M, Klug G (2002) BLUF: a novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem Sci 27: 497-500. doi: 10.1016/S0968-0004(02)02181-3. PubMed: 12368079. [DOI] [PubMed] [Google Scholar]
- 31. Berghoff BA, Konzer A, Mank NN, Looso M, Rische T et al. (2013) Integrative omics approach discovers dynamic and regulatory features of bacterial stress responses. PLOS Genet 9: e1003576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Raivio TL, Silhavy TJ (2001) Periplasmic stress and ECF sigma factors. Annu Rev Microbiol 55: 591-624. doi: 10.1146/annurev.micro.55.1.591. PubMed: 11544368. [DOI] [PubMed] [Google Scholar]
- 33. Drews G (1983) Mikrobiologisches Praktikum. Heidelberg: Springer Verlag. [Google Scholar]
- 34. Nuss AM, Glaeser J, Klug G (2009) RpoHII Activates oxidative-stress defense systems and is controlled by RpoE in the dinglet oxygen-fependent response in Rhodobacter sphaeroides . J Bacteriol 191: 220-230. doi: 10.1128/JB.00925-08. PubMed: 18978062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hübner P, Willison JC, Vignais PM, Bickle TA (1991) Expression of regulatory nif genes in Rhodobacter capsulatus . J Bacteriol 173: 2993-2999. PubMed: 1902215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vieira J, Messing J (1982) The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19: 259-268. doi: 10.1016/0378-1119(82)90015-4. PubMed: 6295879. [DOI] [PubMed] [Google Scholar]
- 37. Simon R, O'Connell M, Labes M, Pühler A (1986) Plasmid vectors for the genetic analysis and manipulation of Rhizobia and other Gram-negative bacteria. Methods Enzymol 118: 640-659. doi: 10.1016/0076-6879(86)18106-7. PubMed: 3005803. [DOI] [PubMed] [Google Scholar]
- 38. Berghoff BA, Glaeser J, Nuss AM, Zobawa M, Lottspeich F et al. (2011) Anoxygenic photosynthesis and photooxidative stress: A particular challenge for Roseobacter . Environ Microbiol 13: 775-791. doi: 10.1111/j.1462-2920.2010.02381.x. PubMed: 21108722. [DOI] [PubMed] [Google Scholar]
- 39. Anthony JR, Green HA, Donohue TJ (2003) Purification of Rhodobacter sphaeroides RNA polymerase and its sigma factors. Methods Enzymol 370: 54-65. doi: 10.1016/S0076-6879(03)70005-6. PubMed: 14712633. [DOI] [PubMed] [Google Scholar]
- 40. van Niel CB (1944) The culture, general physiology, morphology, and classification of the non-sulfur purple and brown bacteria. Bacteriol Rev 8: 1-118. PubMed: 16350090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schilke BA, Donohue TJ (1995) ChrR positively regulates transcription of the Rhodobacter sphaeroides cytochrome c2 gene. J Bacteriol 177: 1929-1937. PubMed: 7721683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Berghoff BA, Glaeser J, Sharma CM, Zobawa M, Lottspeich F et al. (2011) Contribution of Hfq to photooxidative stress resistance and global regulation in Rhodobacter sphaeroides . Mol Microbiol 80: 1479-1495. doi: 10.1111/j.1365-2958.2011.07658.x. PubMed: 21535243. [DOI] [PubMed] [Google Scholar]
- 43. Keen NT, Tamaki S, Kobayashi D, Trollinger D (1988) Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70: 191-197. doi: 10.1016/0378-1119(88)90117-5. PubMed: 2853689. [DOI] [PubMed] [Google Scholar]
- 44. Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA et al. (1995) Four new derivatives of the broad-host-range cloning verctor pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166: 175-176. doi: 10.1016/0378-1119(95)00584-1. PubMed: 8529885. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The RSP_1091-1090 locus is well conserved in α-proteobacteria. Gene neighborhood of the R. sphaeroides 2.4.1 gene RSP_1090 in selected genomes of the α-proteobacteria. Homologs of RSP_1090 were searched by using the BLAST option on the integrated microbial genome (IMG) website. The genes encoding the retrieved homologs of RSP_1090 were subsequently analyzed with respect to the homology of proteins encoded by adjacent genes. In bacteria a number of 337 genomes contained RSP_1090 with an upstream located homolog of RSP_1091. Amino acid identities to R. sphaeroides 2.4.1 proteins are indicated.
(PDF)
Inhibition zone diameters. Sensitivity against 1O2 and organic peroxide (tBOOH) was tested for the R. sphaeroides wild type, strain 2.4.1ΔRSP_1090 and strain TF18. For strains harbouring plasmid pRK415 and the constructs pRKRSP_1090 and pRK_2.4.1rpoEchrR, also values for 1O2 and tBOOH inhibition zones were determined. The generation of 1O2 was achieved by applying 5µl of 10 mM methylene blue solution on filter discs placed on agar plates in the light. In the same manner 700 mM tBOOH was used, agar plates were incubated in the dark. In all cases the mean and standard deviation for three replicates are depicted. Mean values of three experiments are given, SD: standard deviation.
(DOC)
Oligonucleotides used throughout this study.
(DOC)