Abstract
Oncorhynchus mykiss have a diverse array of life history types, and understanding the relationship among types is important for management of the species. Patterns of gene flow between sympatric freshwater resident O. mykiss, commonly known as rainbow trout, and anadromous O. mykiss, commonly known as steelhead, populations are complex and poorly understood. In this study, we attempt to determine the occurrence and pathways of gene flow and the degree of genetic similarity between sympatric resident and anadromous O. mykiss in three river systems, and investigate whether resident O. mykiss are producing anadromous offspring in these rivers, two of which have complete barriers to upstream migration. We found that the population structure of the O. mykiss in these rivers appears to be influenced more by the presence of a barrier to upstream migration than by life history type. The sex ratio of resident O. mykiss located above a barrier, and smolts captured in screw traps was significantly skewed in favor of females, whereas the reverse was true below the barriers, suggesting that male resident O. mykiss readily migrate downstream over the barrier, and that precocious male maturation may be occurring in the anadromous populations. Through paternity analyses, we also provide direct confirmation that resident O. mykiss can produce offspring that become anadromous. Most (89%) of the resident O. mykiss that produced anadromous offspring were males. Our results add to the growing body of evidence that shows that gene flow does readily occur between sympatric resident and anadromous O. mykiss life history types, and indicates that resident O. mykiss populations may be a potential repository of genes for the anadromous life history type.
Introduction
Salmonids show extensive within-species life history variation in a variety of traits, such as age and size at maturity, seasonal spawn timing, and migration and mating behavior. Such diversity is believed to increase resilience to unfavorable environmental variables, and buffers population fluctuations over time [1]. Many salmonid species have both an anadromous life history type, which migrates from freshwater to saltwater, and then returns to freshwater to spawn, and a resident life history type, which remains in fresh water for its entire life. Anadromy allows for individual fish to take advantage of the greater productivity in the ocean compared to freshwater [2], resulting in greater growth opportunity that should then lead to greater reproductive output [3]. It comes with a cost, however, as anadromous individuals may experience greater predation and greater physiological stress and energy expenditure that is involved with migrating between fresh and salt water. The propensity to migrate to salt water may be dependent on numerous factors such as population density, condition of individuals, and sex [3]. The persistence and frequency of both anadromous and resident types, often in sympatry, suggests that neither one has a consistently greater fitness. This could be due to fluctuations in the variables that determine which type is favored, resulting in frequent changes over time in which life history type is favored [4].
The salmonid species Oncorhynchus mykiss can occur as either a freshwater resident type, commonly known as rainbow trout, or an anadromous type, known as steelhead, both of which frequently occur in sympatry [5]. Resident O. mykiss spend their entire lives in freshwater streams or lakes, whereas anadromous O. mykiss migrate from their place of birth in freshwater to the marine environment where they live for a variable number of years before maturing and returning to freshwater (usually the same stream in which they were born) to spawn. The relationship between resident O. mykiss and anadromous O. mykiss populations where they occur in the same river, particularly in regard to gene flow between the two types, is important to discern for proper management of the populations. If there is significant gene flow between the two life history types in the same river, then they both may need to be included in the same conservation unit for purposes of managing that population [6].
Patterns of gene flow between resident and anadromous O. mykiss populations are no doubt complex and may differ among river systems. A few locations have shown significant genetic differences between sympatric resident and anadromous O. mykiss populations [7,8], but most have not [8-14]. This implies that significant gene flow can occur between sympatric resident and anadromous O. mykiss populations, despite the fact that the propensity to migrate to saltwater appears to be under genetic control [15-17] and is strongly, but not completely, influenced by parental life history type [18]. Otolith microchemistry analysis has confirmed that female resident O. mykiss can produce anadromous offspring, and that female anadromous O. mykiss can produce resident offspring [7,19-21]. In addition, the propensity to smolt and tolerate saltwater can be highly heritable in O. mykiss [15]. However, no direct links have been shown between paternal life history and offspring life history in natural populations. Speculation and indirect evidence suggests that resident O. mykiss males spawn readily with anadromous O. mykiss females [5,22-25]. However, such matings have not been confirmed empirically. In a small southeast Alaska population, all possible mating combinations between resident O. mykiss and anadromous O. mykiss produced both life history types, but the resident O. mykiss were from a landlocked population that had originally been derived from O. mykiss residing in the below-barrier, anadromous portion of the stream, and all offspring were raised in a captive rearing environment [15]. The occurrence and frequency of the paternal contribution to “life history switching” (one form producing the other) in natural populations of O. mykiss are unknown.
The possibility that gene flow occurs between sympatric resident and anadromous O. mykiss populations leads to several considerations. Resident O. mykiss may provide a reservoir of genetic material for the anadromous O. mykiss population if they are similar and interbreed regularly without compromising fitness. Such a situation would be critically important for anadromous O. mykiss populations for which there are conservation concerns. Resident O. mykiss could also help to maintain a larger effective population size in an O. mykiss population [26], another important consideration for populations with conservation concerns. On the other hand, introgression of resident O. mykiss genes could have a detrimental effect on anadromous O. mykiss populations by reducing the proportion of individuals that migrate to sea and their fitness in the marine environment [27], to the extent there is genetic control over anadromy. Natural or manmade barriers to migration (e.g., waterfalls, dams) add complexity to the relationship between sympatric life history types within a river system [9,28-31]. Barriers may promote divergence between above-barrier, resident populations and a below-barrier population containing fish of one or both life history types. One-way migration of a limited number of fish downstream over the barrier may provide a means for gene flow into the below-barrier population from the above-barrier population [32].
In this study, we 1) determine the occurrence and pathways of gene flow and degree of genetic similarity between sympatric resident and anadromous O. mykiss in three river systems, and 2) investigate whether resident O. mykiss are producing anadromous offspring in these rivers, two of which have barriers to upstream migration.
Methods
Sample collection
All necessary scientific collection permits for this study were obtained from the Washington Department of Fish and Wildlife by the National Oceanic and Atmospheric Administration (NOAA). Oncorhynchus mykiss is listed as a threatened species in Puget Sound under the U.S Endangered Species Act (ESA), and all ESA consultation requirements were met.
O. mykiss samples were collected from three rivers that flow into Hood Canal in Washington State – the Duckabush, Hamma Hamma, and South Fork Skokomish (hereafter, referred to as Skokomish) rivers (Figure 1). The majority of O. mykiss used for this study were sampled non-lethally by temporarily anesthetizing the fish using tricaine mesylate (MS-222), removing a small portion of fin tissue using scissors, and storing the tissue in 95% ethanol. Fish for which otolith analyses were required were killed by subjecting them to a lethal dose of MS-222. They were stored frozen at -80°C until their otoliths were removed. Resident O. mykiss and parr of an unknown life history type were collected via hook-and-line angling during the summer months (late July through early September, Table 1). In Hood Canal, O. mykiss adopting an anadromous life history (i.e., smolts) out migrate during springtime (April - June) from freshwater to marine waters, predominately at 2 years of age, and at an average size of 170 mm. Any fish greater than 200 mm remaining in the river during summertime have substantial opportunity for further growth prior to the next spring migration window, and are therefore almost certainly adopting a resident life history, and are above the minimum threshold size for maturation [5]. Based on this information, we categorized fish in our study as residents if their length was equal to or greater than 200 mm. The Duckabush River and the Hamma Hamma River each contain a natural falls (at river kilometer 12.1 and 3.8, respectively) that presents a barrier to further upstream migration by returning anadromous O. mykiss. Resident O. mykiss from these rivers were collected from both above and below the barriers. Fish that were moving downstream were also captured in the lower reaches of the rivers in rotary screw traps during April and May and were categorized as smolts, and thus were adopting an anadromous life history, if they were greater than 125 mm. This threshold size for smoltification is consistent with other populations [33]. At this size fish show silvering and loss of parr marks, development of dark margins on the fins, loosening of the scales and body elongation [34]. Ideally, we would also have collected adult anadromous O. mykiss from these rivers; however, that was not possible for these locations. The samples used were collected as part of an ongoing genetic monitoring study of anadromous O. mykiss supplementation in Hood Canal. The Duckabush and Skokomish River samples were all collected before any current supplementation efforts would have affected the samples, whereas the Hamma Hamma River samples were collected after the river had been supplemented with captively reared smolts and adults, which originated from the Hamma Hamma River, beginning in 2000 [32,35].
Figure 1. Map of the study area.
The three rivers where O. mykiss samples were collected.
Table 1. The location, life stage (and for resident O. mykiss, if the fish was collected above or below a barrier to upstream migration), years sampled, and number genotyped of O. mykiss analyzed.
| Location | Life Stage | Years Sampled | N |
|---|---|---|---|
| Duckabush River | Resident O. mykiss above | 2006 - 2008 | 89 |
| Resident O. mykiss below | 2006-2008, 2010, 2011 | 119 | |
| Parr | 2006, 2007 | 39 | |
| Smolts | 2007 - 2011 | 172 | |
| Hamma Hamma River | Resident O. mykiss above | 2002-2005, 2008 | 136 |
| Resident O. mykiss below | 2006-2008 | 85 | |
| Parr | 2006 - 2008 | 29 | |
| Smolts | 2007 - 2011 | 215 | |
| Skokomish River | Resident O. mykiss | 2006 - 2008, 2010, 2011 | 111 |
| Parr | 2006 - 2008 | 83 | |
| Smolts | 2006 - 2011 | 385 |
Genotyping
We genotyped all samples for 15 microsatellite DNA loci – Ocl1 [36], Oke4 [37], Oki23 [38], Ogo4 [39], Omy1001, Omy1011 [40], Omy77 [41], Omy325 [42], Omy7iNRA (K. Gharbi, University of Glasgow, Glasgow, United Kingdom, unpublished data), Oneu14 [43], Ots100 [44], Ots3, Ots4 [45], Ssa289 [46], Ssa407, and Ssa408 [47]. Genomic DNA was isolated using Promega Wizard DNA Purification Kits (Promega Corp.), and then amplified for each locus using polymerase chain reactions (PCR). The resulting PCR products were sized using an Applied Biosystems 3100 genetic analyzer. Genotypes for each individual were determined using GeneScan and Genotyper software (Applied Biosystems Inc.).
Within river population structure
Genetic diversity among and within rivers was measured by conducting AMOVA analyses [48] with the program Arlequin [49]. Significance was tested using 1,000 random permutations.
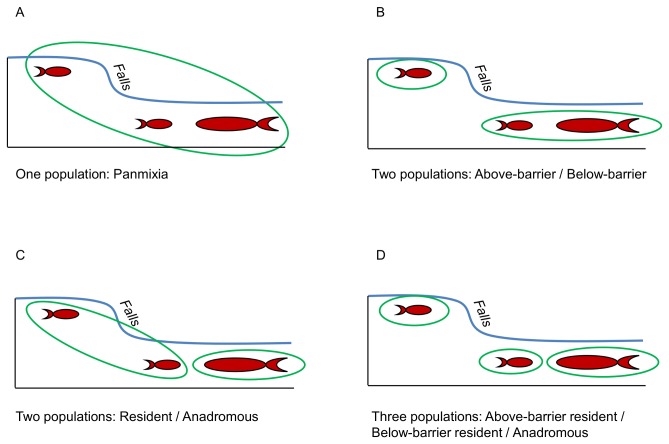
Several possible configurations of population structure within each river were tested to determine which one more closely defines the true configuration. Our samples could be sub-divided based upon life history type (resident O. mykiss vs. anadromous O. mykiss) as defined above, and for the Duckabush and Hamma Hamma rivers, based upon location above or below a barrier to upstream migration. We hypothesized that one of four different configurations of population sub-division could be present, and divided samples within each river into groups accordingly for analyses (Figure 2). We used three methods to search for the population structure configuration that showed the most differentiation among sample groups, thus representing the greatest departure from panmixia. These results allowed us to make inferences about the number of distinct O. mykiss populations in each river.
Figure 2. Pictorial representation of the hypothetical population structures tested for resident and anadromous O. mykiss samples.
The small and large fish icons represent resident and anadromous O. mykiss, respectively, the falls represent a barrier to upstream migration, and the green ovals encompass the life-history type and location of fish considered as a single population for each scenario. For the Skokomish River, which does not contain a barrier to migration, only the one population, panmixia (A), and two population, resident / anadromous (C) scenarios were considered.
First, genetic diversity among sample groups was measured by calculating pairwise F ST values and corresponding 95% confidence intervals in the program FSTAT [50]. The F ST values were plotted to compare their values among the different sample groups. Secondly, we performed leave-one-out tests in GENECLASS2 [51] using the method of Rannala and Mountain [52]. This test removes an individual from the sample groups and assigns it to a group as if it were a fish of unknown origin. After this is completed for every individual in the sample set, the assignments are compared to the known origin of each fish to measure the accuracy of the assignments. As gene flow among populations decreases, the percentage of correct assignments is expected to increase, as has been shown through simulations [53]. The binomial probability of the number of correct assignments observed was calculated to determine if they were greater than the number expected by chance [53]. Thirdly, we ran the program STRUCTURE [54], varying the number of populations (K) in each river from 1 - 5. STRUCTURE uses a Bayesian clustering analysis to infer the number of populations that exist in a sample without defining the populations a priori. We used a burn in length of 100,000 iterations followed by 200,000 iterations, using the correlated allele frequencies model. We ran 20 simulations for each value of K to calculate a mean log probability value (L(K)) for each K value. We then used the method of Evanno et al. [55] to calculate the value ΔK, the rate of change of L(K) between successive K values, which is a better estimator of the number of clusters in a group compared to L(K) alone. The greatest ΔK value was considered to be the true value of K.
Parentage analysis
Parentage analyses were performed to determine whether any of the resident O. mykiss sampled above or below the barriers produced parr or smolts. Individuals having identical genotypes were searched for using GenAlEx [56] so we could exclude any parr-resident O. mykiss parentage matches that represented the same individual sampled at two different times, rather than a true parent-offspring match. CERVUS [57] was used to calculate the average number of alleles per locus and the average non-exclusion probability for the first parent. The average non-exclusion probability is the probability that a candidate parent that is not the true parent of an offspring will not be excluded as a potential parent for that offspring.
Parentage assignments of resident O. mykiss as candidate parents and parr and smolts as candidate offspring were made using the program CERVUS [57]. Because O. mykiss are iteroparous [5], the samples collected were considered as candidate parents for parr and smolt samples collected in years both before and after the resident O. mykiss samples were collected. We only included individuals in the parentage analyses that were genotyped for at least 12 of the 15 loci. Parental matches were accepted only if there were no allele mismatches between a candidate parent and offspring, and if the maximum likelihood confidence value for the pair was > 0.95. These were very conservative measures that resulted in fewer parent-offspring matches than if an exclusionary method or a maximum likelihood method alone was used. However, these stringent criteria serve to increase the certainty of our reported parent-offspring matches. We chose these criteria because of the potentially high number of unsampled parents in our study, as we did not have samples from any adult anadromous O. mykiss, and we did not know what proportion of the resident O. mykiss population we had sampled. Confidence values of parentage assignments are determined in CERVUS using the results of parentage simulations. For these calculations we used simulation parameters of 100,000 offspring and 500 candidate parents, and assumed that we sampled only 10% of the total number of potential parents. Although we did not have an estimate of the size of the resident O. mykiss populations in these rivers, once again, we believe these were very conservative parameters aimed at increasing the confidence in our results. Additional simulations were run assuming we had sampled 50% and 75% of the total number of potential parents, to assure that the parameters we had chosen were not severely biasing our results.
Genetic sex identification
All resident O. mykiss were genotyped for the locus OmyY1 [58] to determine their genotypic sex, which then would allow us to examine if geneflow from the resident population into the anadromous population is sex-specific. We also genotyped all smolts captured in the screw traps for OmyY1 so that the sex ratio of fish from different locations and different life histories could be compared. OmyY1 is a male-specific marker that can be used to determine the genotypic sex of O. mykiss when it is amplified in combination with microsatellite loci (M. Campbell, Idaho Department of Fish and Game, Eagle, Idaho, personal communication). We combined OmyY1 primers along with those for Ogo4, Omy7, and Ots4 into a single PCR reaction. Genomic DNA from each resident O. mykiss identified as a parent was amplified and genotyped for OmyY1 at least twice to confirm the results. The sex ratio of O. mykiss collected in each location was tested for deviance from an expected 1:1 ratio by calculating the observed ratio’s binomial probability.
Otolith analysis
Evidence for anadromous female x resident male matings was investigated first by determining maternal life history of each parr using otolith microchemistry analysis. Reliable determinations of maternal life history have been documented for these populations, including the same parr genetically analyzed in this study (see Berejikian et al. [59] for methods and parr maternity for each population). Only parr that were identified as having an anadromous O. mykiss mother from the otolith analyses were then analyzed to determine their paternity by the parentage analysis described above. In this manner, we were able to conclude if anadromous females were only spawning with anadromous males, or if some resident males were also spawning with the anadromous females. Due to the need to lethally sample fish to obtain their otoliths and the conservation concerns pertaining to these populations, we did not sample any smolts for otolith analyses.
Results
Genotyping results
Samples from all three rivers were highly polymorphic as the mean number of alleles per locus was 16.0 for the Duckabush River, 16.4 for the Hamma Hamma River, and 14.9 for the Skokomish River. The average non-exclusion probability of the first parent for all populations was < 0.00009. Four individuals (one resident O. mykiss, two smolts, one parr) were found to have genotypes matching other individuals, and were subsequently dropped from any further analyses, as they were most likely individuals that had been sampled at two different times.
Within river population structure
The AMOVA analyses revealed that more variation existed among the different rivers (3.3%), then among differing samples within each river (2.6%) . Both of these values are significantly greater than zero (P < 0.001).
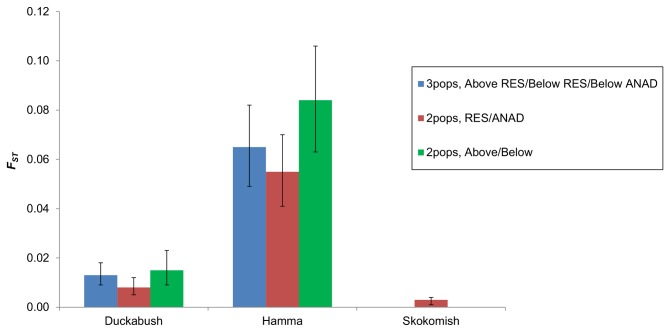
The pairwise F ST values showed some differences among the different population configurations within each river, but none of them were significantly different from one another as all of the 95% confidence intervals overlapped (Figure 3). The two population, above / below configuration had the greatest F ST values for both the Duckabush and Hamma Hamma river samples. For the Skokomish River, which lacks a barrier to migration, the two population, resident / anadromous O. mykiss configuration had a very low F ST value (0.003) that was just barely above zero (lower 95% confidence interval = 0.001). This value is also lower than similar comparisons (below resident O. mykiss vs. below anadromous O. mykiss; not shown on figure) in the Duckabush River (F ST = 0.005) and the Hamma Hamma River (F ST = 0.016).
Figure 3. Pairwise F ST estimates for resident (RES) and anadromous (ANAD) O. mykiss sampled from three rivers.
Samples are grouped in various population structure configurations depending on their location above or below a barrier to upstream migration, and life history type.
Results from the leave-one-out tests suggest that for all population structure configurations tested, the number of correct assignments was significantly greater than random expectations (P < 0.001; Figure 4). For the Duckabush and Hamma Hamma rivers, the two populations, above / below configuration had the greatest number of correct assignments, with 84.7% and 90.5% correct, respectively. The only configuration tested for the Skokomish River (two populations, resident O. mykiss / anadromous O. mykiss), had a correct assignment value of only 72.6%.
Figure 4. Leave-one-out assignment tests for resident (RES) and anadromous (ANAD) O. mykiss sampled from three rivers.
Samples are tested for the percent of correct assignments to population of origin given different population structure configurations depending on sample’s location above or below a barrier to upstream migration, and life history type.
The STRUCTURE analyses found that for the Duckabush and Hamma Hamma rivers, the greatest ΔK was obtained when K = 2 (Figure 5). The values of ΔK for the Hamma Hamma River are considerably greater than those of the other rivers, suggesting stronger differentiation between the above and below Hamma Hamma River populations than those in the Duckabush River. Initially, it appears that the highest ΔK for the Skokomish River is when K = 3, a value not even considered in our population configurations. However, ΔK cannot find the best K if the true value of K = 1 and will return illogical results [55]. In addition, the L(K) values for the Skokomish River do not follow the typical pattern of continually increasing values, most likely due to the fact that substantial gene flow between populations can reduce the reliability of estimating the true value of K from STRUCTURE results [53]. Thus, we did not consider the STRUCTURE estimate to be accurate, as our other two methods provided strong evidence that O. mykiss in the Skokomish River consist of a single panmictic population.
Figure 5. Most likely number of O. mykiss populations (K) in three rivers determined from STRUCTURE analyses.
The most likely value of K is the one with the greatest rate of change (ΔK) between successive L(K) values.
Parentage results
A total of 19 (2.5%) of the 772 smolts examined were identified as offspring of resident O. mykiss (Table 2). It is important to note that these proportions should be considered the minimum proportion of smolts that had a resident parent, as we most likely did not sample every resident in each stream. Thus, other smolts could have been the offspring of unsampled residents. Parentage results using simulation data where we assumed we had sampled 50% or 75% or the total number of potential parents did identify a small number of additional offspring-parent matches (N = 1 & 1, Duckabush River; N = 3 & 6, Hamma Hamma River; N = 1 & 3, Skokomish River), which showed that we were not severely biasing our results with our chosen parameters. Of the resident O. mykiss identified as parents of smolts, 17 (89.5%) were males and only 2 (10.5%) were females. Three male resident O. mykiss were identified as the parent of two smolts each, and one male was identified as the parent of both a smolt and a parr. All other parents identified matched just one smolt or parr each. No smolts could be identified as coming from a resident x resident mating, as none of them were assigned to both a male and female resident O. mykiss. Once again, this result does not preclude that possibility, as it is highly likely that there were resident O. mykiss present in these populations that we did not sample. None of the resident O. mykiss that were collected above the barrier to upstream migration in the Hamma Hamma River were identified as parents of any smolts. However, one smolt from the Duckabush River was identified as an offspring of a female resident O. mykiss collected above the barrier.
Table 2. The number and percentage of smolts in samples from each river that had a resident O. mykiss parent.
| River | Male Parent | Female Parent | Percent of Sample |
|---|---|---|---|
| Duckabush | 9 | 2 | 6.4% |
| Hamma Hamma | 6 | 0 | 2.8% |
| Skokomish | 2 | 0 | 0.5% |
Four of the 150 parr examined were matched to a resident O. mykiss parent (2 in the Duckabush River; 1 each in the Hamma Hamma and Skokomish rivers). All of these parents were identified as males by the genotypic sex analysis. Because we know from otolith analyses that all of these parr had an anadromous maternal origin, they must have been produced by a male resident O. mykiss spawning with a female anadromous O. mykiss.
Sex ratio
The sex ratio of all populations of residents and smolts deviated significantly from the expected 1:1 ratio (Table 3). For the above-barrier resident populations in the Duckabush and Hamma Hamma rivers, we found significantly more females than males, as was also the case for outmigrating smolts, caught in the screw traps, from each of the three rivers. Conversely, in the below-barrier resident populations in the Duckabush and Hamma Hamma rivers, and in the Skokomish River resident O. mykiss population, males made up a significantly greater proportion of each population.
Table 3. Total number of female and male O. mykiss found in each sampling location, the female : male ratio, and the probability that the observed sex ratio is 1:1.
| Location | N total | N females | N males | Ratio | P |
|---|---|---|---|---|---|
| Duckabush River, Above-barrier residents | 89 | 60 | 29 | 2.1 | 0.001 |
| Duckabush River, Below-barrier residents | 119 | 44 | 75 | 0.6 | 0.003 |
| Duckabush River, Screw trap smolts | 135 | 94 | 41 | 2.3 | 0.000 |
| Hamma Hamma River, Above-barrier residents | 136 | 93 | 43 | 2.2 | 0.000 |
| Hamma Hamma River, Below-barrier residents | 85 | 29 | 56 | 0.5 | 0.002 |
| Hamma Hamma River, Screw trap smolts | 208 | 130 | 78 | 1.7 | 0.000 |
| Skokomish River, In river residents | 111 | 30 | 81 | 0.4 | 0.000 |
| Skokomish River, Screw trap smolts | 291 | 169 | 122 | 1.4 | 0.003 |
Discussion
The population structure of the O. mykiss in these rivers appears to be influenced more by the presence of a barrier to upstream migration than by the life history type of the fish. Our results show that in areas within a river where resident and anadromous O. mykiss are able to mix freely, there is less genetic differentiation between the life history types than there is between fish sampled above and below the barrier (i.e., Figure 2B). However, there does appear to be enough gene flow between fish above and below the barriers to prevent complete reproductive isolation. Although the barriers present in the Duckabush and Hamma Hamma rivers presumably block gene flow from below-barrier O. mykiss into the above-barrier population, gene flow is not necessarily blocked in the opposite direction. Results from previous studies have implied that resident O. mykiss are capable of surviving a descent over waterfalls [18,31], including those in the Hamma Hamma River [32]. Thus, the potential does exist for gene flow between the above-barrier and below-barrier populations, albeit in a single direction. Our results suggest that gene flow between the above-barrier and below-barrier populations has been more substantial in the Duckabush River, as evidenced by its much lower AMOVA values between the above-barrier and below-barrier groups compared to the Hamma Hamma River. However, past stocking practices of hatchery raised O. mykiss has also likely affected the population structure in the Hamma Hamma River. Non-native resident O. mykiss, derived from the McCloud River, California, were regularly stocked into the Hamma Hamma River, above its barrier to upstream migration, from the mid-1970s through 1996 [59]. If the introduced fish were reproductively successful and introgressed into the gene pool of the native population, an increased level of genetic differentiation between life history types and locations would have occurred, as non-native genes from the hatchery stock were introduced only into the above-barrier resident O. mykiss population. This is evident when our results from the Hamma Hamma River are compared to the other populations, which have not received any reported releases of non-native O. mykiss. The Hamma Hamma River samples had much greater pairwise FST values for all of the population configurations tested, and the ΔK values from the STRUCTURE analyses were several magnitudes greater than they were for either the Duckabush or Skokomish River samples. We would expect that over time, the non-native genes introduced above the barrier will become introgressed into the below-barrier population, but the speed at which this occurs depends upon the level of gene flow between locations via fish migrating downstream over the barrier, and the reproductive success of the hatchery stock relative to the native fish.
The sex ratios we observed in the above-barrier resident O. mykiss samples suggest that migration over the barriers may be sex biased. The above-barrier samples were highly female-skewed, whereas the below-barrier samples were male-skewed, indicating that males migrate downstream over the barriers at a greater frequency then females. This is especially important given our finding that gene flow from the resident populations into the anadromous populations occurs primarily via resident males spawning with anadromous females. Sex-biased migration of resident males over the barriers into lower reaches of the river would increase the number of resident males in the steelhead spawning areas, and thus, increase the opportunities for resident male and anadromous female matings. Resident O. mykiss are known to be highly migratory in freshwater systems [11,60], but in contrast to our results, Olsen et al. [11] found no evidence of sex-biased dispersal. However, in that study, dispersal was measured between locations within a river that were not separated by a barrier to migration, as in our study. Sex-specific density dependent factors above the barrier, such as competition among males for mates, could drive some males to venture over the barrier in search of areas with less competition. There could also be other explanations for our results, such as higher mortality in males in the above-barrier locations accounting for the female-skewed ratio in those locations, or precocious male maturation in anadromous O. mykiss, accounting for the male-skewed ratios in below-barrier populations (discussed below).
The F ST values we calculated were significantly greater than zero when considering the two populations (resident and anadromous), indicating some degree of reproductive isolation between life history types, especially in the Hamma Hamma River. The assignment tests for the configuration that considered both a resident and anadromous population were significantly more accurate than what would be expected by chance if only a single panmictic population existed. Reproductive isolation between life history types may be promoted by size assortative mating, evident in some salmonids [61,62], or partial spatial [7] or temporal [12] isolation in spawning. However, even a low mating frequency between life history types could result in substantial gene flow between them, thus partially counteracting divergence caused by genetic drift or selection [63].
Our results provide direct confirmation through paternity analyses that resident O. mykiss produce offspring that become anadromous. This was especially true in the Duckabush River, where we found the greatest proportion of smolts (6.4%) that had a resident O. mykiss as one of their parents. This could be related to the relatively low numbers of returning anadromous O. mykiss that have been observed there. For example, the annual average number of anadromous O. mykiss redds observed from 2008-2010 was 11.7 for the Duckabush River, 64.3 for the Hamma Hamma River, and 249.7 for the Skokomish River [64]. If there are few anadromous male O. mykiss on the spawning grounds, some resident males may be spawning with anadromous females with little to no competition from anadromous males. Our results suggest this is occurring, as we found four parr that were the offspring of matings between male resident and female anadromous O. mykiss. This could create what Araki et al. [24] called “genetic compensation between life history forms,” and speculated that it could be a means of stabilizing the effective size of an O. mykiss population during times of low anadromous O. mykiss abundance. A study by Berejikian et al. [59] found much higher proportions of below-barrier, O. mykiss parr with resident mothers in the Duckabush (42.0%) and Hamma Hamma (58.7%) rivers compared to the Skokomish River (5.6%). They attributed the greater percentage of offspring with resident O. mykiss mothers in the Duckabush and Hamma Hamma rivers to the presence of large-scale habitat features and the presence of an above-barrier resident population for each of those rivers. This suggests that resident O. mykiss comprise a significant proportion of the O. mykiss populations in those two rivers, providing ample opportunities for matings between resident males and anadromous females.
Further gene flow between life history types in these rivers may be occurring due to precocious male maturation of anadromous O. mykiss offspring. Precocious male maturation occurs when male offspring of anadromous O. mykiss mature in freshwater without ever migrating to salt water, essentially adopting a resident O. mykiss life history. As discussed earlier, the male-skewed sex ratio in the below-barrier resident O. mykiss locations may be at least partially attributable to sex-biased migration over the barrier. However, no barrier exists in the South Fork Skokomish River, so the male-skewed sex ratio we observed there is likely due to precocious male maturation. This can also be clearly seen in the sex ratios of the smolt samples captured in screw traps (Table 3). Those results show that for all three rivers, significantly more females are outmigrating to marine waters then are males. McMillan et al. [12] also found a male-dominated population of wild resident O. mykiss in a Washington state river that did not have a barrier to migration, and Rundio et al. [65] found a male-skewed sex ratio in a California resident O. mykiss population that they attributed to differential rates of anadromy between sexes. We presume that precocious males would be more likely to spawn with resident O. mykiss due to their similar life history characteristics, thus providing a means of gene flow from the anadromous O. mykiss population into the resident O. mykiss population.
Conservation of life history diversity is important to the long term persistence of a population, as a population with life history diversity may be able to better withstand catastrophic events that might cause the extinction of a singular life history type [1,63]. Our evidence of significant gene flow between O. mykiss life history types is important to conservation and management issues related to this species [66]. Two of the more important issues considered are 1) in which rivers should sympatric resident and anadromous O. mykiss be managed as a single population versus multiple populations, and 2) do resident O. mykiss represent a repository of genes for a given river that can be used to restore the anadromous life history type if it has been lost (due to migration barriers, for example; [29,67])? Our results add to the growing body of evidence that shows that in many rivers gene flow does readily occur between resident and anadromous O. mykiss life history types. The sympatric resident and anadromous O. mykiss present in each of the locations we sampled do not constitute reproductively isolated populations, and appear to have a low level of gene flow between them. This is important because when a particular life history type is not entirely under genetic control, the loss of one life history type would not necessarily be permanent if that type can arise from the surviving type [68].
As for the second question, the barriers in our study area created more reproductive isolation than life history type did, but there was still enough gene flow present to prevent complete reproductive isolation. In the Duckabush River, we identified a smolt as having a mother who originated from above the anadromous barrier in that system, indicating that for that location the resident O. mykiss isolated above the barrier are capable of producing offspring that will adopt an anadromous life history. Our results, added to the results of previous studies [7,19-21,25,69], clearly show that resident O. mykiss can produce offspring that become anadromous, thereby suggesting that resident O. mykiss populations may represent a repository of genes for an anadromous population. However, the rate at which residents produce anadromous migrants, and the survival of those migrants, will need to be determined to help us understand the net contribution that resident O. mykiss can make to anadromous O. mykiss productivity.
Acknowledgments
The authors wish to thank the numerous personnel from the Hood Canal Salmon Enhancement Group, Long Live the Kings, NOAA Fisheries, the Skokomish Tribal Nation, and the Washington Department of Fish and Wildlife who helped with data and sample collection, and the Robbins family for providing access to the Hamma Hamma River. Thanks also to Jeff Hard, Krista Nichols, Linda Park and David Teel who provided valuable reviews of earlier versions of this manuscript.
Funding Statement
Primary funding for this work was provided by the National Oceanic and Atmospheric Administration. Additional support was provided by the Hood Canal Salmon Enhancement Group, Long Live the Kings, the Skokomish Tribal Nation, the U.S. Fish and Wildlife Service, and the Washington Department of Fish and Wildlife. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Greene CM, Hall JE, Guilbault KR, Quinn TP (2010) Improved viability of populations with diverse life-history portfolios. Biol Lett 6: 382-386. doi: 10.1098/rsbl.2009.0780. PubMed: 20007162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gross MR, Coleman RM, McDowall RM (1988) Aquatic productivity and the evolution of diadromous fish migration. Science 239: 1291-1293. doi: 10.1126/science.239.4845.1291. PubMed: 17833216. [DOI] [PubMed] [Google Scholar]
- 3. Hendry AP, Bohlin T, Jonsson B, Berg OK (2004) To sea or not to sea? Anadromy versus non-anadromy in salmonids. In: Hendry AP, Stearns SC. Evolution illuminated: salmon and their relatives. New York: Oxford University Press; pp. 92-125. [Google Scholar]
- 4. Seamons TR, Bentzen P, Quinn TP (2007) DNA parentage analysis reveals inter-annual variation in selection: results from 19 consecutive brood years in steelhead trout. Evol Ecol Res 9: 409-431. [Google Scholar]
- 5. Behnke RJ (2002) Trout and salmon of North America. New York: The Free Press. [Google Scholar]
- 6. Busby PJ, Wainwright TC, Bryant GJ, Lierheimer LJ, Waples RS et al. (1996) Status review of West Coast steelhead from Washington, Idaho. Oregon, and California: National Oceanic and Atmospheric Administration; Technical Memorandum NMFS-NWFSC-27 [Google Scholar]
- 7. Zimmerman CE, Reeves GH (2000) Population structure of sympatric anadromous and nonanadromous Oncorhynchus mykiss: evidence from spawning surveys and otolith microchemistry. Can J Fish Aquat Sci 57: 2152-2162. doi: 10.1139/f00-192. [DOI] [Google Scholar]
- 8. Narum SR, Contor C, Talbot A, Powell MS (2004) Genetic divergence of sympatric resident and anadromous forms of Oncorhynchus mykiss in the Walla Walla River, U.S.A. J Fish Biol 65: 471-488. doi: 10.1111/j.0022-1112.2004.00461.x. [DOI] [Google Scholar]
- 9. Currens KP, Schreck CB, Li HW (1990) Allozyme and morphological divergence of rainbow trout (Oncorhynchus mykiss) above and below waterfalls in the Deschutes River, Oregon. Copeia: 1990:730-746. [Google Scholar]
- 10. Docker MF, Heath DD (2003) Genetic comparison between sympatric anadromous steelhead and freshwater resident rainbow trout in British Columbia, Canada. Conserv Genet 4: 227-231. doi: 10.1023/A:1023355114612. [DOI] [Google Scholar]
- 11. Olsen JB, Wuttig K, Fleming D, Kretschmer EJ, Wenburg JK (2006) Evidence of partial anadromy and resident-form dispersal bias on a fine scale in populations of Oncorhynchus mykiss . Conserv Genet 7: 613-619. doi: 10.1007/s10592-005-9099-0. [DOI] [Google Scholar]
- 12. McMillan JR, Katz SL, Pess GR (2007) Observational evidence of spatial and temporal structure in a sympatric anadromous (winter steelhead) and resident rainbow trout mating system on the Olympic Peninsula, Washington. Trans Am Fish Soc 136:736-748
- 13. McPhee MV, Utter F, Stanford JA, Kuzishchin KV, Savvaitova KA et al. (2007) Population structure and partial anadromy in Oncorhynchus mykiss from Kamchatka: relevance for conservation strategies around the Pacific Rim. Ecol Freshw Fish 16: 539-547. doi: 10.1111/j.1600-0633.2007.00248.x. [DOI] [Google Scholar]
- 14. Heath DD, Bettles CM, Jamieson S, Stasiak I, Docker MF (2008) Genetic differentiation among sympatric migratory and resident life history forms of rainbow trout in British Columbia. Trans Am Fish Soc 137: 1268-1277. doi: 10.1577/T05-278.1. [DOI] [Google Scholar]
- 15. Thrower FP, Hard JJ, Joyce JE (2004) Genetic architecture of growth and early life-history transitions in anadromous and derived freshwater populations of steelhead. J Fish Biol 65: 286-307. doi: 10.1111/j.0022-1112.2004.00551.x. [DOI] [Google Scholar]
- 16. Nichols KM, Edo AF, Wheeler PA, Thorgaard (2008) The genetic basis of smoltification-related traits in Oncorhynchus mykiss . Genetics 179: 1559-1575. doi: 10.1534/genetics.107.084251. PubMed: 18562654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hecht BC, Campbell NR, Holecek DE, Narum SR (2012) Genome-wide association reveals genetic basis for the propensity to migrate in wild populations of rainbow and steelhead trout. Mol Ecol, 22: 3061–76. doi: 10.1111/mec.12082. PubMed: 23106605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hayes SA, Hanson CV, Pearse DE, Bond MH, Garza JC et al. (2012) Should I stay or should I go? The influence of genetic origin on emigration behavior and physiology of resident and anadromous juvenile Oncorhynchus mykiss . N J Fish Manag 32: 772-780. doi: 10.1080/02755947.2012.686953. [DOI] [Google Scholar]
- 19. Donohoe CJ, Adams PB, Royer CF (2008) Influence of water chemistry and migratory distance on ability to distinguish progeny of sympatric resident and anadromous rainbow trout (Oncorhynchus mykiss). Can J Fish Aquat Sci 65: 1060-1075. doi: 10.1139/F08-029. [DOI] [Google Scholar]
- 20. Zimmerman CE, Edwards GW, Perry K (2009) Maternal origin and migratory history of steelhead and rainbow trout captured in rivers of the Central Valley, California. Trans Am Fish Soc 138: 280-291. doi: 10.1577/T08-044.1. [DOI] [Google Scholar]
- 21. Thibault I, Hedger RD, Dodson JJ, Shiao J-C, Iizuka Y et al. (2010) Anadromy and the dispersal of an invasive fish species (Oncorhynchus mykiss) in Eastern Quebec, as revealed by otolith microchemistry. Ecol Fresh. Fish 19: 348-360. [Google Scholar]
- 22. Seamons TR, Bentzen P, Quinn TP (2004) The mating system of steelhead, Oncorhynchus mykiss, inferred by molecular analysis of parents and progeny. Envir Biol. Fish 69: 333-344. doi: 10.1023/B:EBFI.0000022893.88086.8f. [DOI] [Google Scholar]
- 23. Kuligowski DR, Ford MJ, Berejikian BA (2005) Breeding structure of steelhead inferred from patterns of genetic relatedness among nests. Trans Am Fish Soc 134: 1202-1212. doi: 10.1577/T04-187.1. [DOI] [Google Scholar]
- 24. Araki H, Ardren WR, Olsen E, Cooper B, Blouin MS (2006) Reproductive success of captive-bred steelhead trout in the wild: evaluation of three hatchery programs in the Hood River. Conserv Biol 21: 181-190. PubMed: 17298524. [DOI] [PubMed] [Google Scholar]
- 25. Christie MR, Marine ML, Blouin MS (2011) Who are the missing parents? Grandparentage analysis identifies multiple sources of gene flow into a wild population. Mol Ecol 20: 1263-1276. doi: 10.1111/j.1365-294X.2010.04994.x. PubMed: 21244538. [DOI] [PubMed] [Google Scholar]
- 26. Araki H, Waples RS, Ardren WR, Cooper B, Blouin MS (2007) Effective population size of steelhead trout: influence of variance in reproductive success, hatchery programs, and genetic compensation between life-history forms. Mol Ecol 16: 953-966. doi: 10.1111/j.1365-294X.2006.03206.x. PubMed: 17305853. [DOI] [PubMed] [Google Scholar]
- 27. Thrower FP, Hard JJ (2009) Effects of a single event of close inbreeding on growth and survival in steelhead. Conserv Genet 10: 1299-1307. doi: 10.1007/s10592-008-9709-8. [DOI] [Google Scholar]
- 28. Thrower F, Guthrie C III, Nielsen J, Joyce J (2004b) A comparison of genetic variation between an anadromous steelhead, Oncorhynchus mykiss, population and seven derived populations sequestered in freshwater for 70 years. Environ Biol Fishes 69: 111-125. doi: 10.1023/B:EBFI.0000022880.52256.92. [DOI] [Google Scholar]
- 29. Winans GA, McHenry ML, Baker J, Elz A, Goodbla A et al. (2008) Genetic inventory of anadromous Pacific salmonids of the Elwha River prior to dam removal. Nw Sci 82: 128-141. [Google Scholar]
- 30. Clemento AJ, Anderson EC, Boughton D, Girman D, Garza JC (2009) Population genetic structure and ancestry of Oncorhynchus mykiss populations above and below dams in south-central California. Conserv Genet 10: 1321-1336. doi: 10.1007/s10592-008-9712-0. [DOI] [Google Scholar]
- 31. Pearse DE, Hayes SA, Bond MH, Hanson CV, Anderson EC et al. (2009) Over the falls? Rapid evolution of ecotypic differentiation in steelhead/rainbow trout (Oncorhynchus mykiss). J Hered 100: 515-525. doi: 10.1093/jhered/esp040. PubMed: 19561050. [DOI] [PubMed] [Google Scholar]
- 32. Van Doornik DM, Berejikian BA, Campbell LA, Volk EC (2010) The effect of a supplementation program on the genetic and life history characteristics of an Oncorhynchus mykiss population. Can J Fish Aquat Sci 67: 1449-1458. doi: 10.1139/F10-073. [DOI] [Google Scholar]
- 33. Berejikian BA, Larsen DA, Swanson P, Moore ME, Tatara CP et al. (2012) Development of natural growth regimes for hatchery-reared steelhead to reduce residualism, fitness loss, and negative ecological interactions. Envir Biol. Fish 94: 29-44. doi: 10.1007/s10641-011-9788-0. [DOI] [Google Scholar]
- 34. Moore M, Berejikian BA, Tezak EP (2012) Variation in the early marine survival and behavior of natural and hatchery-reared Hood Canal steelhead. PLOS ONE 7(11): e49645. doi: 10.1371/journal.pone.0049645. PubMed: 23185393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Berejikian BA, Johnson T, Endicott RS, Lee-Waltermire J (2008) Increases in steelhead (Oncorhynchus mykiss) redd abundance resulting from two conservation hatchery strategies in the Hamma Hamma River, Washington. Can J Fish Aquat Sci 65:754-764
- 36. Condrey MJ, Bentzen P (1998) Characterization of coastal cutthroat trout (Oncorhynchus clarki clarki) microsatellites and their conservation in other salmonids. Mol Ecol 7: 787–789. PubMed: 9640655. [PubMed] [Google Scholar]
- 37. Buchholz W, Miller SJ, Spearman WJ (1999) Summary of PCR primers for salmonid genetic studies. Alaska: United States Fish and Wildlife Service; Fisheries Progress Report 99-1 [Google Scholar]
- 38. Smith CR, Koop BF, Nelson RJ (1998) Isolation and characterization of coho salmon (Oncorhynchus kisutch) microsatellites and their use in other salmonids. Mol Ecol 7: 1614-1616. PubMed: 9819912. [PubMed] [Google Scholar]
- 39. Olsen JB, Bentzen P, Seeb JE (1998) Characterization of seven microsatellite loci derived from pink salmon. Mol Ecol 7: 1087-1089. PubMed: 9711869. [PubMed] [Google Scholar]
- 40. Spies IB, Brasier DJ, O' Reilly PTL, Seamons TR, Bentzen P (2005) Development and characterization of novel tetra-, tri-, and dinucleotide microsatellite markers in rainbow trout (Oncorhynchus mykiss). Mol Ecol Notes 5: 278-281. doi: 10.1111/j.1471-8286.2005.00900.x. [DOI] [Google Scholar]
- 41. Morris DB, Richard KR, Wright JM (1996) Microsatellites from rainbow trout (Oncorhynchus mykiss) and their use for genetic study of salmonids. Can J Fish Aquat Sci 53: 120-126. doi: 10.1139/f95-161. [DOI] [Google Scholar]
- 42. O’Connell M, Danzmann RG, Cornuet J-M, Wright JM, Ferguson MM (1997) Differentiation of rainbow trout (Oncorhynchus mykiss) populations in Lake Ontario and the evaluation of the stepwise mutation and infinite allele mutation models using microsatellite variability. Can J Fish Aquat Sci 54: 1391-1399. doi: 10.1139/f97-043. [DOI] [Google Scholar]
- 43. Scribner KT, Gust JR, Fields RL (1996) Isolation and characterization of novel microsatellite loci: cross-species amplification and population genetic applications. Can J Fish Aquat Sci 53: 685-693. doi: 10.1139/f95-229. [DOI] [Google Scholar]
- 44. Nelson RJ, Beacham TD (1999) Isolation and cross-species amplification of microsatellite loci useful for study of Pacific salmon. Anim Genet 30: 228-229. doi: 10.1046/j.1365-2052.1999.00404-4.x. PubMed: 10442992. [DOI] [PubMed] [Google Scholar]
- 45. Banks MA, Blouin MS, Baldwin BA, Rashbrook VK, Fitzgerald HA et al. (1999) Isolation and inheritance of novel microsatellites in Chinook salmon (Oncorhynchus tschawytscha). J Hered 90: 281-288. doi: 10.1093/jhered/90.2.281. [DOI] [Google Scholar]
- 46. McConnell S, Hamilton L, Morris D, Cook D, Paquet D et al. (1995) Isolation of salmonid microsatellite loci and their application to the population genetics of Canadian east coast stocks of Atlantic salmon. Aquaculture 137: 19-30. doi: 10.1016/0044-8486(95)01111-0. [DOI] [Google Scholar]
- 47. Cairney M, Taggart JB, Høyheim B (2000) Characterization of microsatellite and minisatellite loci in Atlantic salmon (Salmo salar L.) and cross-species amplification in other salmonids. Mol Ecol 9: 2175-2178. doi: 10.1046/j.1365-294X.2000.105312.x. PubMed: 11123640. [DOI] [PubMed] [Google Scholar]
- 48. Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 131: 479-491. PubMed: 1644282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinform Online 1: 47-50. [PMC free article] [PubMed] [Google Scholar]
- 50. Goudet J (2002) FSTAT, version 2.9.3.2. Available: www2.unil.ch/popgen/softwares/fstat.htm. Accessed 19 October 2011.
- 51. Piry S, Alapetite A, Cornuet J-M, Paetkau D, Baudouin L et al. (2004) GeneClass2: A software for genetic assignment and first-generation migrant detection. J Hered 95: 536-539. doi: 10.1093/jhered/esh074. PubMed: 15475402. [DOI] [PubMed] [Google Scholar]
- 52. Rannala B, Mountain JL (1997) Detecting immigration by using multilocus genotypes. Proc Natl Acad Sci U S A 94: 9197-9201. doi: 10.1073/pnas.94.17.9197. PubMed: 9256459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Waples RS, Gaggiotti O (2006) What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Mol Ecol 15: 1419-1439. doi: 10.1111/j.1365-294X.2006.02890.x. PubMed: 16629801. [DOI] [PubMed] [Google Scholar]
- 54. Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155: 945-959. PubMed: 10835412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611-2620. doi: 10.1111/j.1365-294X.2005.02553.x. PubMed: 15969739. [DOI] [PubMed] [Google Scholar]
- 56. Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Resour 6: 288-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kalinowski ST, Taper ML, Marshall TC (2007) Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16: 1099-1106. doi: 10.1111/j.1365-294X.2007.03089.x. PubMed: 17305863. [DOI] [PubMed] [Google Scholar]
- 58. Brunelli JP, Wertzler KJ, Sundin K, Thorgaard GH (2008) Y-specific sequences and polymorphisms in rainbow trout and Chinook salmon. Genome 51: 739-748. doi: 10.1139/G08-060. PubMed: 18772952. [DOI] [PubMed] [Google Scholar]
- 59. Berejikian BA, Campbell LA, Moore ME (2013) Large-scale freshwater habitat features influence the degree of anadromy in eight Hood Canal Oncorhynchus mykiss populations. Can J Fish Aquat Sci 70: 756-765. doi: 10.1139/cjfas-2012-0491. [DOI] [Google Scholar]
- 60. Meka JM, Knudsen EE, Douglas DC, Benter RB (2003) Variable migratory patterns of different adult rainbow trout life history types in a southwest Alaska watershed. Trans Am Fish Soc 132: 717-732. doi: 10.1577/T01-166. [DOI] [Google Scholar]
- 61. Foote CJ, Larkin PA (1988) The role of male choice in the assortative mating of sockeye salmon and kokanee, the anadromous and nonanadromous forms of Oncorhynchus nerka . Behaviour 106: 43-62. doi: 10.1163/156853988X00089. [DOI] [Google Scholar]
- 62. Maekawa K, Nakano S, Yamamoto S (1994) Spawning behavior and size-assortative mating of Japanese charr in an artificial lake-inlet stream system. Envir Biol. Fish 39: 109-117. doi: 10.1007/BF00004927. [DOI] [Google Scholar]
- 63. Allendorf FW, Luikart G (2007) Conservation and the genetics of populations. Malden, MA: Blackwell Publishing. [Google Scholar]
- 64.WDFW (Washington Department of Fish and Wildlife) (2012) Spawning Ground Survey Database. Available: www.wdfw.wa.gov/conservation/fisheries/sgs/.
- 65. Rundio DE, Williams TH, Pearse DE, Lindley ST (2012) Male-biased sex ratio of nonanadromous Oncorhynchus mykiss in a partially migratory population in California. Ecol Freshw Fish 21: 293-299. doi: 10.1111/j.1600-0633.2011.00547.x. [DOI] [Google Scholar]
- 66. Good TP, Waples RS, Adams P, Bjorkstedt EP, Boughton D et al. (2005) Updated status of federally listed ESUs of West Coast salmon and steelhead. National Oceanic and Atmospheric Administration Technical Memorandum NMFS-NWFSC-66.
- 67. Holecek DE, Scarnecchia DL, Miller SE (2012) Smoltification in an impounded, adfluvial redband trout population upstream from an impassable dam: does it persist? Trans Am Fish Soc 141: 68-75. doi: 10.1080/00028487.2011.651550. [DOI] [Google Scholar]
- 68. Beechie T, Buhle E, Ruckelshaus M, Fullerton A, Holsinger L (2006) Hydrologic regime and the conservation of salmon life history diversity. Biol Conserv 130: 560-572. doi: 10.1016/j.biocon.2006.01.019. [DOI] [Google Scholar]
- 69. Pascual M, Bentzen P, Rossi CR, Mackey G, Kinnison MT et al. (2001) First documented case of anadromy in a population of introduced rainbow trout in Patagonia, Argentina. Trans Am Fish Soc 130:53-67