Abstract
Plasminogen activator inhibitor 1 (PAI-1) has been associated with metabolic disorders, through different mechanisms, which could involve changes in DNA methylation. This work aimed to assess the potential relationships of the cytosine methylation levels within SERPINE1 gene transcriptional regulatory region, which codes for PAI-1, in peripheral white blood cells with anthropometrical, metabolic and inflammatory features. Forty-six obese subjects with metabolic syndrome features followed Control or Metabolic Syndrome Reduction in Navarra (RESMENA) energy-restricted (−30%E) diets for 8 weeks. SERPINE1 transcriptional regulatory region methylation at baseline was analyzed by a microarray technical. Both dietary strategies reduced anthropometric and biochemical parameters. The Control group significantly reduced plasma PAI-1 concentrations but not the RESMENA group. Participants from both nutritional interventions with higher SERPINE1 methylation levels at baseline showed significantly major reductions in body weight, total fat mass, android fat mass, total cholesterol and triglycerides, as compared with those with lower initial SERPINE1 methylation levels. In conclusion, the DNA methylation levels of SERPINE1 transcriptional regulatory region were associated with some metabolic and anthropometric changes in obese subjects with metabolic syndrome under energy restriction, suggesting a complex epigenetic network in the regulation of this recognized pro-inflammatory marker. (www.clinicaltrials.gov; NCT01087086)
Keywords: DNA methylation, PAI-1, SERPINE1 gene, metabolic syndrome, energy restriction
Introduction
Plasminogen activator inhibitor-1 (PAI-1), encoded by the SERPINE1 gene, is the principal inhibitor of tissue plasminogen activator and urokinase, and hence is an inhibitor of fibrinolysis.(1) This serine protease is produced by the vascular endothelium, the liver, the monocytes/macrophagues, the platelets and the adipose tissue.(2) High plasma levels of PAI-1 have been associated with an increased risk of suffering cardiovascular diseases.(3) Furthermore, PAI-1 dependent mechanisms are also implicated in the pathogenesis of obesity, insulin resistance and type 2 diabetes.(4) The metabolic syndrome (MetS) encompasses a cluster of comorbidities linked to obesity, which increase cardiovascular risk,(5) where SERPINE1 overexpression may participate.(6) In fact, increased PAI-1 levels can be considered as a component of the syndrome.(6)
Since obesity onset and related alterations development involve multiple factors including inflammatory mediators,(7) the understanding of this disease remains as a complex challenge for the scientific community. In the last years epigenetic has emerged as a new framework of research studying gene expression changes not due to variations in the DNA nucleotide sequence but to other mechanisms.(8) On the other hand, dietary treatments based on calorie restriction are the first-choice implement for treating obesity(9) and, in turn, epigenetic has been proposed as an influential factor on the variable responses to a low-calorie strategy.(10,11)
Thus, taking into account this scenario the present study aimed to assess the potential relationships of the cytosine methylation levels of SERPINE1 gene transcriptional regulatory region in peripheral white blood cells (WBC) with anthropometrical, metabolic and inflammatory features in a subset of the Metabolic Syndrome Reduction in Navarra (RESMENA) project population after following an energy-restricted dietary program.
Material and Methods
Subjects and study protocol
The current analysis was conducted within the RESMENA project, a randomized controlled trial,(12) from which a subsample of 48 obese adults presenting MetS was selected. Two subjects did not overcome quality controls and therefore the sample size for the outcomes presented is this work was n = 46. Participants underwent two energy-restricted dietary patterns (Control diet or RESMENA diet) for 8 weeks, as described elsewhere.(13) The study was approved by the Ethics Committee of the University of Navarra (065/2009) and appropriately registered at www.clinicaltrials.gov; NCT01087086. Consequently, all the participants gave written informed consent for participation in agreement with the Declaration of Helsinki. This work was performed following the CONSORT 2010 guidelines.
Dietary intake assessment, anthropometry and blood pressure
Assessment and information about dietary intake (total energy intake), anthropometrical measurements (body weight, height, waist circumference and triceps skinfold thickness) were conducted according to previously described procedures.(13) Body mass index (BMI) was calculated by the ratio between weight (kg) and the squared height (m2) and body composition analyses by bioimpedance and Dual-energy X-ray Absorptiometry (DXA) were carried out following validated protocols as described elsewhere.(12) Systolic and diastolic blood pressures were measured following standardized World Health Organization criteria.(14)
Biological samples and metabolic and inflammatory markers
Venous blood samples were drawn after a 12-h overnight fast by venipuncture. The EDTA-plasma and serum samples as well as WBC were separated from whole blood by centrifugation at 3,500 rpm, 5°C, 15 min (Model 5804R, Eppendorf, Germany), and were frozen immediately at −80°C until assay (WBC in buffy-coat). Plasma concentrations of triglycerides, total cholesterol and high density lipoprotein–cholesterol (Wako Chemicals, GmbH, Nuiss, Germany) and glucose (Horiba ABX Diagnostics, Montpellier, France) were measured by specific colorimetric assays, using an automated analyzer system Pentra C-200 (HORIBA ABX, Madrid, Spain). Serum fasting insulin was measured by an enzyme immunoassay kit (Mercodia, Sweden). Insulin resistance was estimated by the HOMA-IR, through calculations as follows: HOMA-IR = [fasting glucose (mM) × fasting insulin (mU/l)]/22.5 as described elsewhere.(15) Plasma concentrations of PAI-1 (BioVendor, Germany), interleukin 6 (R&D Systems, Minneapolis, MN), tumor necrosis factor alpha (TNF-α) (R&D Systems) and high-sensitive C-reactive protein (Demeditec, Germany) were measured using enzyme immunoassay based kits by means of an automated analyzer system (Triturus, Grifols, Barcelona, Spain). In our laboratory, the inter- and intra-assay were <10% for all analytical determinations.
DNA isolation and DNA Methylation Study
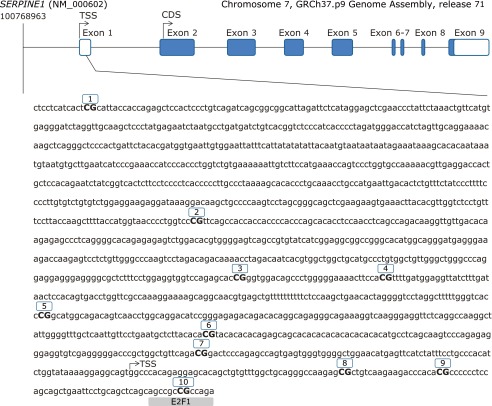
Genomic DNA from WBC was extracted using the Master Pure kit (Epicenter, Madison, WI), whose quality was assessed with PicoGreen dsDNA Quantitation Reagent (Invitrogen, Carlsbad, CA). A total of 500 ng of DNA was modified by using EZ-96 DNA Methylation Kit (Zymo Research Corporation, Irvine, CA) according to the manufacturer’s instructions, converting thus cytosine into uracil. Array-based specific DNA methylation analysis was performed with the Infinium Human Methylation 450K bead chip technology (Illumina, San Diego, CA). Bisulfite-treated genomic DNA was whole-genome amplified, hybridized to HumanMethylation450 BeadChips (Illumina) and scanned using the Illumina iScanSQ platform. The intensity of the images was extracted with the GenomeStudio Methylation Software Module (ver. 1.9.0, Illumina). Ten CpG sites of the SERPINE1 gene that codes for the PAI-1 protein were selected. CpG sites located in the transcriptional regulatory region (promoter, 5'-untranslated region and exon 1) are reported (Fig. 1). Reference names and characteristics of the selected Cytosine-phosphate-guanine (CpG) are shown in Table 1.
Fig. 1.
Genomic localization and nucleotide sequence of 10 CpGs sites covered by the Illumina probe for the study of DNA methylation levels of plasminogen activator inhibitor type 1 promoter (from −1408 to +113 bp). Transcription Start Site (TSS). CpG 10 has a putative consensus sequences for (E2F-1), found with MatInspector.
Table 1.
Information of the selected CpG sites for the SERPINE1 gene
| CpG ID1 | Illumina ID | CHR position2 | Reference3 |
|---|---|---|---|
| 1 | cg20773815 | 7:100768974 | c.−1396 |
| 2 | cg08053846 | 7:100769605 | c.−765 |
| 3 | cg24539923 | 7:100769903 | c.−467 |
| 4 | cg19722814 | 7:100769933 | c.−437 |
| 5 | cg25826546 | 7:100770060 | c.−310 |
| 6 | cg20438404 | 7:100770192 | c.−178 |
| 7 | cg08506775 | 7:100770286 | c.−84 |
| 8 | cg02273392 | 7:100770414 | c.+45 |
| 9 | cg15874872 | 7:100770434 | c.+65 |
| 10 | cg20583316 | 7:100770476 | c.+107 |
1Studied CpG identifier. 2Genome assembly: GRCh37, Ensemble release 71. 3It begins in the first nucleotide of exon 1.
Statistical analyses
Results are shown as mean ± SD. Variable distribution was determined by the Shapiro-Wilk test and no normal variables were logarithmically transformed. Statistical comparisons between groups were performed by the parametric Student t test. Pearson correlations were fitted to evaluate the potential correlations of SERPINE1 transcriptional regulatory region methylation with adiposity indicators, metabolic features and dietary factors. SERPINE1 CpG 10 site was selected for further statistical analyses, since in the preliminary assessment (Supplemental Table 1) showed a potential association with several adiposity indicators. All participants were assigned into two groups according the median of DNA SERPINE1 CpG 10 methylation (%): “lower DNA SERPINE1 methylation” (≤7%) and “higher DNA SERPINE1 methylation” (>7%) for comparisons. Statistical analyses were performed with SPSS 15.0 software (SPSS Inc., Chicago, IL) for Windows XP (Microsoft, Redmond, WA). P value <0.05 was considered as statistically significant.
Results
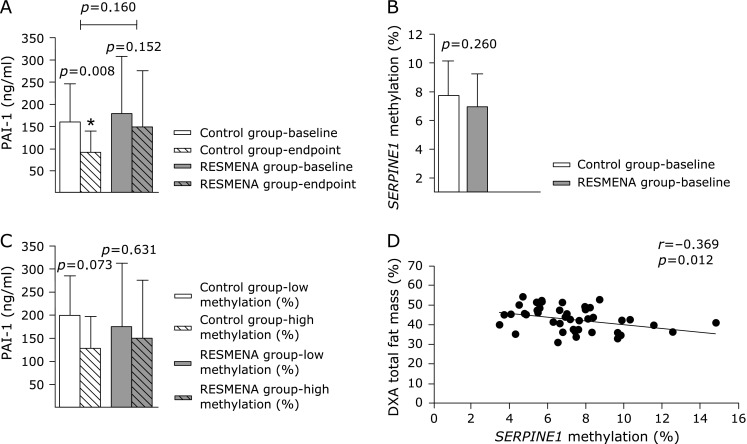
Statistical between-diet differences were not found (p>0.05) after the 8 weeks of dietary intervention, neither for anthropometric, nor for routine biochemical parameters (Table 2). PAI-1 concentration levels were significantly decreased within the Control group (p = 0.008) whereas this marker did not show statistical variations within the RESMENA group (Fig. 2A). There were not between-group differences concerning SERPINE1 methylation levels at baseline (p = 0.260, Fig. 2B). We did not find any interaction between the dietary group and SERPINE1 methylation levels (Fig. 2C). Therefore, the sample was merged and considered as a whole for subsequent analyses. Interestingly, a preliminary analysis showed significant associations between baseline SERPINE1 methylation levels and the changes in body weight (r = –0.360, p = 0.016), total fat mass (r = –0.354, p = 0.018) and android fat mass (r = –0.330, p = 0.029), which were not found for PAI-1 plasma concentrations. For further assessing the relationships between changes on anthropometrical variables, clinical and metabolic features, energy intake as well as pro-inflammatory markers concentrations and SERPINE1 methylation, the sample was categorized according to median DNA methylation baseline levels of SERPINE1 transcriptional regulatory region (Table 3). Differences were found neither on BMI, nor on waist circumference due to methylation level. However body weight and body composition-related variables changes were statistically different according to SERPINE1 transcriptional regulatory region methylation. In addition, total fat mass significantly correlated with DNA methylation levels of SERPINE1 CpG 10 site (Fig. 2D).
Table 2.
Changes in selected anthropometric and biochemical variables. Comparison between dietary treatments at the end of the intervention
| Characteristics | Control
(n = 19) |
RESMENA
(n = 27) |
p value difference | ||||
|---|---|---|---|---|---|---|---|
| Baseline | Final | p value | Baseline | Final | p value | ||
| Energy intake (kcal) | 2198 ± 428 | 1450 ± 347 | <0.001 | 2158 ± 522 | 1323 ± 305 | <0.001 | 0.601 |
| Body weight (kg) | 105.01 ± 17.44 | 98.04 ± 16.92 | <0.001 | 101.32 ± 18.18 | 94.53 ± 17.84 | <0.001 | 0.827 |
| BMI (kg/m2) | 36.60 ± 3.70 | 34.17 ± 3.71 | <0.001 | 36.19 ± 3.68 | 33.76 ± 3.91 | <0.001 | 1,000 |
| Waist circumference (cm) | 114.25 ± 10.20 | 107.82 ± 11.49 | <0.001 | 111.20 ± 13.48 | 104.35 ± 13.03 | <0.001 | 0.728 |
| Triceps skinfold (mm) | 28.00 ± 10.00 | 24.60 ± 9.40 | 0.006 | 31.60 ± 8.00 | 28.60 ± 7.60 | 0.001 | 0.764 |
| Bioimpedance Total Fat mass (%) | 37.83 ± 8.11 | 34.54 ± 7.55 | <0.001 | 40.33 ± 5.93 | 37.58 ± 7.24 | <0.001 | 0.558 |
| Bioimpedance Total Fat mass (kg) | 39.62 ± 10.34 | 33.71 ± 8.56 | <0.001 | 40.81 ± 9.39 | 35.58 ± 9.80 | <0.001 | 0.547 |
| DXA Total Fat mass (%) | 40.88 ± 6.17 | 38.92 ± 6.83 | 0.001 | 44.12 ± 5.75 | 41.85 ± 6.59 | <0.001 | 0.617 |
| DXA Trunk Fat mass (%) | 47.68 ± 5.57 | 45.26 ± 6.93 | 0.014 | 49.97 ± 5.13 | 46.83 ± 6.48 | <0.001 | 0.422 |
| DXA Android Fat mass (%) | 53.71 ± 6.44 | 51.05 ± 7.65 | 0.035 | 56.78 ± 5.53 | 53.00 ± 7.26 | <0.001 | 0.357 |
| DXA Total Fat mass (kg) | 42.65 ± 7.79 | 37.91 ± 8.48 | <0.001 | 44.36 ± 9.23 | 39.22 ± 9.56 | <0.001 | 0.604 |
| Glucose (mg/dl)* | 120.22 ± 24.61 | 107.99 ± 14.50 | 0.003 | 123.00 ± 39.09 | 104.05 ± 18.90 | 0.022 | 0.505 |
| Insulin (µU/ml)* | 17.71 ± 10.08 | 10.95 ± 8.71 | <0.001 | 14.51 ± 9.17 | 9.53 ± 6.65 | <0.001 | 0.345 |
| HOMA-IR* | 5.34 ± 3.48 | 3.10 ± 2.95 | <0.001 | 4.56 ± 3.37 | 2.60 ± 2.08 | 0.001 | 0.696 |
| Total cholesterol (mg/dl) | 217 ± 50 | 200 ± 43 | 0.042 | 220 ± 53 | 204 ± 48 | 0.043 | 0.919 |
| HDL-c (mg/dl) | 43 ± 10 | 39 ± 7 | 0.017 | 44 ± 12 | 41 ± 10 | 0.041 | 0.458 |
| Triglycerides (mg/dl) | 205 ± 103 | 150 ± 58 | 0.009 | 193 ± 123 | 143 ± 86 | 0.003 | 0.821 |
| SBP (mmHg) | 150 ± 14 | 139 ± 15 | 0.007 | 149 ± 20 | 136 ± 17 | 0.003 | 0.782 |
| DBP (mmHg) | 85 ± 9 | 80 ± 10 | 0.044 | 86 ± 8 | 78 ± 10 | <0.001 | 0.504 |
| CRP (mg/l) | 4.53 ± 7.31 | 2.63 ± 2.62 | 0.298 | 5.53 ± 9.99 | 4.51 ± 6.76 | 0.185 | 0.224 |
| IL-6 (pg/ml) | 2.48 ± 0.99 | 2.38 ± 1.19 | 0.536 | 2.74 ± 1.99 | 2.96 ± 1.73 | 0.419 | 0.740 |
| TNF-α (pg/ml) | 1.11 ± 1.86 | 0.96 ± 1.72 | 0.127 | 0.99 ± 0.94 | 0.94 ± 1.04 | 0.511 | 0.291 |
Data are mean ± SD. p values from Student t test. *Log transformed variables. BMI, body mass index; DXA, dual-energy x-ray absoptiometry; HOMA-IR, homeostasis model assessment-insulin resistance; HDL-c, high density lipoprotein cholesterol; SBP, systolic blood pressure; DBP, diastolic blood pressure; CRP, c-reactive protein; IL-6, interleukin-6; TNF-α, tumor necrosis factor-α.
Fig. 2.
Changes in PAI-1 plasma concentrations in Control and RESMENA groups from baseline to the endpoint of the intervention and differences between them (A); comparison between Control and RESMENA groups levels of SERPINE1 CpG 10 methylation (%) at baseline (B); comparison of PAI-1 plasma concentrations within Control and RESMENA groups according to SERPINE1 CpG 10 baseline methylation level (%) (C) and correlation analysis between methylation in the SERPINE1 CpG 10 and total fat mass by Dual-energy X-ray absorptiometry at baseline (D). PAI-1: plasminogen activator inhibitor 1.
Table 3.
Changes in selected characteristics of the participants, categorized by median of DNA SERPINE1 CpG 10 methylation (≤ or >7%) at baseline
| Characteristics | Lower DNA SERPINE1 methylation % (n = 23) | Higher DNA SERPINE1 methylation % (n = 23) | p value |
|---|---|---|---|
| Energy intake (kcal) | –717 ± 107 | –882 ± 121 | 0.311 |
| Body weight (kg) | –5.90 ± 2.53 | –7.82 ± 2.68 | 0.016 |
| BMI (kg/m2) | –2.22 ± 0.20 | –2.64 ± 0.20 | 0.143 |
| Waist circumference (cm) | –6.07 ± 0.69 | –7.28 ± 0.95 | 0.308 |
| Triceps skinfold (mm) | –2.01 ± 0.79 | –4.53 ± 1.08 | 0.065 |
| Bioimpedance Total Fat mass (%) | –1.84 ± 0.48 | –4.11 ± 0.68 | 0.009 |
| Bioimpedance Total Fat mass (kg) | –3.96 ± 0.46 | –7.06 ± 0.89 | 0.003 |
| DXA Total Fat mass (%) | –1.84 ± 0.42 | –2.45 ± 0.42 | 0.309 |
| DXA Trunk Fat mass (%) | –2.07 ± 0.68 | –3.62 ± 0.52 | 0.077 |
| DXA Android Fat mass (%) | –1.96 ± 0.81 | –4.68 ± 0.79 | 0.021 |
| DXA Total Fat mass (kg) | –4.28 ± 0.51 | –5.68 ± 0.51 | 0.060 |
| Glucose (mg/dl) * | –1.31 ± 0.54 | –1.03 ± 0.54 | 0.120 |
| Insulin (µU/ml) * | –0.53 ± 0.50 | –0.79 ± 0.33 | 0.066 |
| HOMA-IR* | –0.19 ± 0.45 | –0.19 ± 0.48 | 0.951 |
| Total cholesterol (mg/dl) * | –1.2 ± 0.6 | –1.4 ± 0.4 | 0.048 |
| HDL-c (mg/dl) | –3.6 ± 6.1 | –2.8 ± 6.8 | 0.688 |
| Triglycerides (mg/dl)* | –1.4 ± 0.5 | –1.8 ± 0.5 | 0.011 |
| SBP (mmHg) | –11 ± 4 | –15 ± 4 | 0.515 |
| DBP (mmHg) | –5 ± 2 | –8 ± 2 | 0.218 |
| CRP (mg/l)* | –0.10 ± 0.78 | –0.20 ± 0.71 | 0.746 |
| PAI-1 (ng/ml) | –38.72 ± 23.24 | –49.77 ± 15.48 | 0.692 |
| IL-6 (pg/ml)* | 0.43 ± 0.53 | 0.28 ± 0.62 | 0.550 |
| TNF-α (pg/ml) | –0.01 ± 0.08 | –0.14 ± 0.08 | 0.245 |
Data are mean ± SD. p values from Student t test. *Log transformed variables. BMI, body mass index; DXA, dual-energy x-ray absoptiometry; HOMA-IR, homeostasis model assessment-insulin resistance; HDL-c, high density lipoprotein cholesterol; SBP, systolic blood pressure; DBP, diastolic blood pressure; CRP, c-reactive protein; PAI-1, plasminogen activator inhibitor-1; IL-6, interleukin-6; TNF-α, tumor necrosis factor-α.
Participants allocated in the higher DNA SERPINE1 methylation group evidenced greater reductions on body weight, total fat mass and android fat mass (p<0.05, Table 3). Moreover, the same trend was found regarding other adiposity indicators such as triceps skinfold thickness (p = 0.065) or trunk fat mass (p = 0.077). Glucose metabolism parameters did not show significant differences concerning SERPINE1 methylation levels, although a trend toward signification was found for insulin (p = 0.066), showing a greater reduction the higher DNA SERPINE1 methylation group (Table 3). Interestingly, lipid profile indicators, such as total cholesterol and triglycerides, were greatly reduced in those subjects with high DNA methylation levels of SERPINE1 CpG 10 site (p = 0.048 and p = 0.011, respectively). There were no between-groups differences when assessing blood pressure, energy intake or the inflammatory markers plasma concentrations (Table 3).
Discussion
This investigation evidenced an interesting association of baseline methylation levels of SERPINE1 gene transcriptional regulatory region, which codes for PAI-1, with adiposity measurements after 8 weeks of energy-restricted dietary treatment in obese subjects presenting MetS features. Indeed, PAI-1 is well recognized as a proinflammatory and cardiovascular risk marker. Therefore, our objective was to extend the knowledge on signaling pathway mechanisms and hence we explored SERPINE1 methylation status. Interestingly, under the study conditions, SERPINE1 methylation levels better correlated with changes in some anthropometric measurements such as body weight as well as total and android fat mass indicative of obesity and closely related to latter cardiovascular damage,(16–18) which were not found for the PAI-1 values in plasma.
DNA methylation is an epigenetic event that may be involved in several human diseases by altering gene expression.(19) Thus, it has been widely demonstrated to be a potential contributing factor to cancer risk,(20) but some recent researches have shown that differential variability in methylation is also an important feature of obesity.(21) In fact, specific DNA regions have been reported to be differentially methylated depending on weight loss response in obese adolescents.(22) Moreover, some genes have been revealed to be hypermethylated in lean compared with obese subject.(19,23) Contrariwise, some studies reported no associations between BMI and global DNA methylation.(24,25) In any case, studies investigating the association between obesity and WBC DNA methylation are still limited.
Inflammation constitutes a mediating mechanism for obesity-linked comorbidities such as cardiovascular diseases, insulin resistance or type 2 diabetes.(26) Thus, inflammation indicators are gaining much research attention. This investigation focused on PAI-1, since PAI-1 high plasma levels have been associated with inflammation and increased risk of cardiovascular diseases.(3) Moreover, PAI-1 dependent mechanisms are implicated in the pathogenesis of obesity, insulin resistance and type 2 diabetes(4) and it has actually been proposed as constituent of the MetS definition,(6) which is the condition of the sample selected for this research. Other inflammatory markers were evaluated also reported as related to obesity.(27–29) However, we failed to find significant differences neither after the intervention nor between diets. PAI-1 was the only parameter that significantly reduced within the Control group, whereas not within the RESMENA group. This finding is in agreement with other studies that reported beneficial effects on fasting values of inflammation markers of the American Heart Association pattern in which the Control diet was based.(30,31)
The sample was categorized according to the median value of SERPINE1 methylation levels noting that those participants allocated in the high-methylation group evidenced a greater reduction of body-weight, total and android fat mass, triglycerides and total cholesterol, which represents an encouraging result since all of them are associated with obesity and derived comorbidities.(32,33) To the best of our knowledge, this is the first study evaluating SERPINE1 methylation level in relation to obesity in humans. However, the epigenetic control of this molecule has been previously studied, and CpG methylation in the 5'-flanking region of the PAI-1 gene appears not to be involved in the developments of gastric cancer.(34,35) Recently, a genome-wide methylation analysis on adipose tissue of pig breeds displaying distinct fat level found intrinsic methylation differences of some inflammatory markers, such as SERPINE1 gene, between various adipose depots depending on their localization.(35)
The findings of this research suggest a possible role for SERPINE1 methylation as predictive of the effects of a dietary treatment for obesity, which is in accordance with previous studies investigating other inflammatory markers. In this sense, TNF-α is an inflammatory and immune response mediator cytokine(36) and variability in this gene promoter methylation profile has been suggested to could predict the susceptibility to weight loss as well as to some comorbidities such as hypertension or type 2 diabetes.(10,37)
To understand the mechanisms by which DNA methylation levels in the analyzed region could be related to anthropometric and metabolic features, we searched for consensus response elements using MatInspector(38) in the entire SERPINE1 region, since the binding of several transcription factors is extremely sensitive to methylation of specific CpG dinucleotides in regulatory regions.(39) After the bioinformatics analysis, was identified that the CpG10 site matches a core-binding consensus motif for the E2F transcription factor 1. This transcriptional factor was described as a known transcriptional repressor of SERPINE1.(40) The fact that E2F1 can bind to this region depending if CpG10 site is methylated or unmethylated indicates a functionality of the transcriptional regulation by methylation.
On the other hand, SERPINE1 methylation levels were found to significantly correlate on a negative manner with the percentage of total fat mass measured by DXA at baseline. This negative association between the higher methylation percentage, which normally lead to lower mRNA levels,(41) and the lower values of obesity indicators suggest that a lower inflammatory status may result on beneficial effects for the obesity condition.
This work has the limitation that we have not measured expression levels, although previous reports show that CpG methylation of the SERPINE1 gene 5'-flanking region is inversely correlated with SERPINE1 mRNA levels in human cell lines.(41) Also, the determination of SERPINE1 methylation levels at the end of the intervention would be useful for giving support to the stated hypothesis. Moreover the outcomes would benefit of a higher sample size and a longer intervention period. The fact that different methods for assessing body composition, such as bioimpedance and DXA showed the same behavior concerning methylation is a strong point of this research.
In conclusion, this study reports for first time an association of methylation levels of SERPINE1 gene transcriptional regulatory region with body weight, total and android fat mass, triglycerides and cholesterol reduction within a hypocaloric intervention, suggesting that methylation changes may contribute to understand obesity management. Moreover, SERPINE1 regulatory region methylation could be a good predictive tool to the response to a nutritional energy-restricted intervention.
Acknowledgments
We wish to thank our physician Blanca E. Martinez de Morentin, our nurse Salome Perez and the technician Veronica Ciaurriz for excellent technical assistance as well as the volunteers who participated in this study. This work was supported by Health Department of the Government of Navarra (48/2009) and the Linea Especial about Nutrition, Obesity and Health (University of Navarra LE/97). Also CIBERobn and RETICS schemes from the isciii are gratefully credited. Government of Navarra provided a pre-doctoral research grant to P.L.L (233/2009). M.L.M. holds a Juan de la Cierva fellowship from Spanish Ministry of Economic and Competitiveness.
Abbreviations
- BMI
body mass index
- DXA
dual-energy X-ray absorptiometry
- MetS
metabolic syndrome
- PAI-1
plasminogen activator inhibitor 1
- RESMENA
metabolic syndrome reduction in Navarra
- TNF-α
tumor necrosis factor-α
- WBC
white blood cells
Conflict of Interest
No potential conflicts of interest were disclosed.
Supplementary Material
References
- 1.Declerck PJ, Gils A. Three decades of research on plasminogen activator inhibitor-1: a multifaceted serpin. Semin Thromb Hemost. 2013;39:356–364. doi: 10.1055/s-0033-1334487. [DOI] [PubMed] [Google Scholar]
- 2.Huang J, Sabater-Lleal M, Asselbergs FW, et al. Genome-wide association study for circulating levels of PAI-1 provides novel insights into its regulation. Blood. 2012;120:4873–4881. doi: 10.1182/blood-2012-06-436188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohler HP, Grant PJ. Plasminogen-activator inhibitor type 1 and coronary artery disease. N Engl J Med. 2000;342:1792–1801. doi: 10.1056/NEJM200006153422406. [DOI] [PubMed] [Google Scholar]
- 4.Festa A, Williams K, Tracy RP, Wagenknecht LE, Haffner SM. Progression of plasminogen activator inhibitor-1 and fibrinogen levels in relation to incident type 2 diabetes. Circulation. 2006;113:1753–1759. doi: 10.1161/CIRCULATIONAHA.106.616177. [DOI] [PubMed] [Google Scholar]
- 5.Eckel RH, Grundy SM, Zimmet PZ. The metabolic syndrome. Lancet. 2005;365:1415–1428. doi: 10.1016/S0140-6736(05)66378-7. [DOI] [PubMed] [Google Scholar]
- 6.Alessi MC, Juhan-Vague I. PAI-1 and the metabolic syndrome: links, causes, and consequences. Arterioscler Thromb Vasc Biol. 2006;26:2200–2207. doi: 10.1161/01.ATV.0000242905.41404.68. [DOI] [PubMed] [Google Scholar]
- 7.Martinez JA, Parra MD, Santos JL, Moreno-Aliaga MJ, Marti A, Martinez-Gonzalez MA. Genotype-dependent response to energy-restricted diets in obese subjects: towards personalized nutrition. Asia Pac J Clin Nutr. 2008;17:119–122. [PubMed] [Google Scholar]
- 8.Campión J, Milagro F, Martínez JA. Epigenetics and obesity. Prog Mol Biol Transl Sci. 2010;94:291–347. doi: 10.1016/B978-0-12-375003-7.00011-X. [DOI] [PubMed] [Google Scholar]
- 9.Abete I, Astrup A, Martinez JA, Thorsdottir I, Zulet MA. Obesity and the metabolic syndrome: role of different dietary macronutrient distribution patterns and specific nutritional components on weight loss and maintenance. Nutr Rev. 2010;68:214–231. doi: 10.1111/j.1753-4887.2010.00280.x. [DOI] [PubMed] [Google Scholar]
- 10.Cordero P, Campion J, Milagro FI, et al. Leptin and TNF-alpha promoter methylation levels measured by MSP could predict the response to a low-calorie diet. J Physiol Biochem. 2011;67:463–470. doi: 10.1007/s13105-011-0084-4. [DOI] [PubMed] [Google Scholar]
- 11.Campión J, Milagro FI, Martinez JA. Individuality and epigenetics in obesity. Obes Rev. 2009;10:383–392. doi: 10.1111/j.1467-789X.2009.00595.x. [DOI] [PubMed] [Google Scholar]
- 12.Zulet MA, Bondia-Pons I, Abete I, et al. The reduction of the metabolyc syndrome in Navarra-Spain (RESMENA-S) study: a multidisciplinary strategy based on chrononutrition and nutritional education, together with dietetic and psychological control. Nutr Hosp. 2011;26:16–26. [PubMed] [Google Scholar]
- 13.Lopez-Legarrea P, de la Iglesia R, Abete I, et al. Short-term role of the dietary total antioxidant capacity in two hypocaloric regimes on obese with metabolic syndrome symptoms: the RESMENA randomized controlled trial. Nutr Metab (Lond) 2013;10:22. doi: 10.1186/1743-7075-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitworth JA, World Health Organization, International Society of Hypertension Writing Group 2003 World Health Organization (WHO)/International Society of Hypertension (ISH) statement on management of hypertension. J Hypertens. 2003;21:1983–1992. doi: 10.1097/00004872-200311000-00002. [DOI] [PubMed] [Google Scholar]
- 15.Morimoto A, Tatsumi Y, Deura K, et al. Impact of impaired insulin secretion and insulin resistance on the incidence of type 2 diabetes mellitus in a Japanese population: the Saku study. Diabetologia. 2013;56:1671–1679. doi: 10.1007/s00125-013-2932-y. [DOI] [PubMed] [Google Scholar]
- 16.Lee MJ, Wu Y, Fried SK. Adipose tissue heterogeneity: implication of depot differences in adipose tissue for obesity complications. Mol Aspects Med. 2013;34:1–11. doi: 10.1016/j.mam.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hermsdorff HH, Barbosa KB, Volp AC, et al. Gender-specific relationships between plasma oxidized low-density lipoprotein cholesterol, total antioxidant capacity, and central adiposity indicators Eur J Prev Cardiol 2012; in press. [DOI] [PubMed] [Google Scholar]
- 18.Jahagirdar R, Hemchand KP, Chiplonkar SA, Khadilkar VV, Khadilkar AV. Relationship between body mass index, fat distribution and cardiometabolic risk factors in Indian children and adolescents. Pediatr Obes. 2012;7:E37–E41. doi: 10.1111/j.2047-6310.2012.00056.x. [DOI] [PubMed] [Google Scholar]
- 19.Zhang FF, Santella RM, Wolff M, Kappil MA, Markowitz SB, Morabia A. White blood cell global methylation and IL-6 promoter methylation in association with diet and lifestyle risk factors in a cancer-free population. Epigenetics. 2012;7:606–614. doi: 10.4161/epi.20236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 21.Xu X, Su S, Barnes VA, et al. A genome-wide methylation study on obesity: differential variability and differential methylation. Epigenetics. 2013;8:522–533. doi: 10.4161/epi.24506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moleres A, Campión J, Milagro FI, et al. Differential DNA methylation patterns between high and low responders to a weight loss intervention in overweight or obese adolescents: the EVASYON study. Faseb J. 2013;27:2504–2512. doi: 10.1096/fj.12-215566. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Zhu H, Snieder H, et al. Obesity related methylation changes in DNA of peripheral blood leukocytes. BMC Med. 2010;8:87. doi: 10.1186/1741-7015-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang FF, Cardarelli R, Carroll J, et al. Physical activity and global genomic DNA methylation in a cancer-free population. Epigenetics. 2011;6:293–299. doi: 10.4161/epi.6.3.14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu ZZ, Hou L, Bollati V, et al. Predictors of global methylation levels in blood DNA of healthy subjects: a combined analysis. Int J Epidemiol. 2012;41:126–139. doi: 10.1093/ije/dyq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bondia-Pons I, Ryan L, Martinez JA. Oxidative stress and inflammation interactions in human obesity. J Physiol Biochem. 2012;68:701–711. doi: 10.1007/s13105-012-0154-2. [DOI] [PubMed] [Google Scholar]
- 27.Richard C, Couture P, Desroches S, Lamarche B. Effect of the mediterranean diet with and without weight loss on markers of inflammation in men with metabolic syndrome. Obesity (Silver Spring) 2013;21:51–57. doi: 10.1002/oby.20239. [DOI] [PubMed] [Google Scholar]
- 28.Moreira PF, Dalboni MA, Cendoroglo M, Santos GM, Cendoroglo MS. Postprandial interleukin-6 response in elderly with abdominal obesity and metabolic syndrome. J Nutr Health Aging. 2013;17:206–210. doi: 10.1007/s12603-012-0400-x. [DOI] [PubMed] [Google Scholar]
- 29.Hermsdorff HH, Puchau B, Zulet MA, Martínez JA. Association of body fat distribution with proinflammatory gene expression in peripheral blood mononuclear cells from young adult subjects. Omics. 2010;14:297–307. doi: 10.1089/omi.2009.0125. [DOI] [PubMed] [Google Scholar]
- 30.Bhupathiraju SN, Lichtenstein AH, Dawson-Hughes B, Tucker KL. Adherence index based on the AHA 2006 diet and lifestyle recommendations is associated with select cardiovascular disease risk factors in older Puerto Ricans. J Nutr. 2011;141:460–469. doi: 10.3945/jn.110.133603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krauss RM, Eckel RH, Howard B, et al. AHA Dietary Guidelines: revision 2000: a statement for healthcare professionals from the Nutrition Committee of the American Heart Association. Stroke. 2000;31:2751–2766. doi: 10.1161/01.str.31.11.2751. [DOI] [PubMed] [Google Scholar]
- 32.Jago R, Mendoza JA, Chen T, Baranowski T. Longitudinal associations between BMI, waist circumference, and cardiometabolic risk in US youth: Monitoring implications. Obesity (Silver Spring) 2013;21:E271–E279. doi: 10.1002/oby.20080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hernández-Mijares A, Bañuls C, Gómez-Balaguer M, Bergoglio M, Victor VM, Rocha M. Influence of obesity on atherogenic dyslipidemia in women with polycystic ovary syndrome. Eur J Clin Invest. 2013;43:549–556. doi: 10.1111/eci.12080. [DOI] [PubMed] [Google Scholar]
- 34.Liu J, Li X, Yu N, et al. Genetic instability and CpG methylation in the 5'-flanking region of the PAI-1 gene in Chinese patients with gastric cancer. Genet Mol Res. 2012;11:2899–2908. doi: 10.4238/2012.May.18.11. [DOI] [PubMed] [Google Scholar]
- 35.Li M, Wu H, Wang T, et al. Co-methylated genes in different adipose depots of pig are associated with metabolic, inflammatory and immune processes. Int J Biol Sci. 2012;8:831–837. doi: 10.7150/ijbs.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wieser V, Moschen AR, Tilg H. Inflammation, cytokines and insulin resistance: a clinical perspective. Arch Immunol Ther Exp (Warsz) 2013;61:119–125. doi: 10.1007/s00005-012-0210-1. [DOI] [PubMed] [Google Scholar]
- 37.Campión J, Milagro FI, Goyenechea E, Martínez JA. TNF-alpha promoter methylation as a predictive biomarker for weight-loss response. Obesity (Silver Spring) 2009;17:1293–1297. doi: 10.1038/oby.2008.679. [DOI] [PubMed] [Google Scholar]
- 38.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burnett E, Christensen J, Tattersall P. A consensus DNA recognition motif for two KDWK transcription factors identifies flexible-length, CpG-methylation sensitive cognate binding sites in the majority of human promoters. J Mol Biol. 2001;314:1029–1039. doi: 10.1006/jmbi.2000.5198. [DOI] [PubMed] [Google Scholar]
- 40.Venugopal J, Hanashiro K, Nagamine Y. Regulation of PAI-1 gene expression during adipogenesis. J Cell Biochem. 2007;101:369–380. doi: 10.1002/jcb.21173. [DOI] [PubMed] [Google Scholar]
- 41.Gao S, Skeldal S, Krogdahl A, Sørensen JA, Andreasen PA. CpG methylation of the PAI-1 gene 5'-flanking region is inversely correlated with PAI-1 mRNA levels in human cell lines. Thromb Haemost. 2005;94:651–660. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.