Abstract
The outcomes of clinical trials using bone marrow stromal cell (BMSC) are variable; the degree of the expansion of BMSCs during clinical manufacturing may contribute to this variability since cell expansion is limited by senescence. Human BMSCs from aspirates of healthy subjects were subcultured serially until cell growth stopped. Phenotype and functional measurers of BMSCs from two subjects including senescence-associated beta-galactosidase staining and colony formation efficiency changed from an early to a senescence pattern at passage 6 or 7. Transcriptome analysis of 10 early and 15 late passage BMSC samples from 5 subjects revealed 2122 differentially expressed genes, which were associated with immune response, development, and cell proliferation pathways. Analysis of 57 serial BMSC samples from 7 donors revealed that the change from an early to senescent profile was variable among subjects and occurred prior to changes in phenotypes. BMSC age expressed as a percentage of maximum population doublings (PDs) was a good indicator for an early or senescence transcription signature but this measure of BMSC life span can only be calculated after expanding BMSCs to senescence. In order to find a more useful surrogate measure of BMSC age, we used a computational biology approach to identify a set of genes whose expression at each passage would predict elapsed age of BMSCs. A total of 155 genes were highly correlated with BMSC age. A least angle regression algorithm identified a set of 24 BMSC age-predictive genes. In conclusion, the onset of senescence-associated molecular changes was variable and preceded changes in other indicators of BMSC quality and senescence. The 24 BMSC age predictive genes will be useful in assessing the quality of clinical BMSC products.
Keywords: Bone marrow stromal cells, cellular therapy, gene expression profiling, Graft-versus-Host-Disease, regenerative medicine
Introduction
Bone marrow stromal cells (BMSCs) which are also known as mesenchymal stromal cells (MSCs) were first described by Friedenstein and colleagues (Friedenstein et al., 1966). They have anti-inflammatory and immunosuppressive properties and are being tested in a large number of clinical trials for many different applications (Ciccocioppo et al.; Dash et al., 2009; Hare et al., 2009; Joyce et al.; Karussis et al.; Le Blanc et al., 2004; Mazzini et al.; Pal et al., 2009; Saleem et al., 2000). In most cases a relatively small number of BMSCs are isolated from bone marrow aspirates or bone biopsies by plastic adherence and expanded by serial passage. The typical cell dose is 1 to 2 × 106 cells per kg of recipient weight or 75 to 200 × 106 cells. The production of those doses of BMSCs from 10 to 15 mL of aspirated marrow requires the expansion of BMSCs over 3 to 4 weeks. The expansion capability of BMSCs, however, is limited. Prolonged BMSCs culture leads to deterioration of their replication ability and eventually to senescence (Ksiazek, 2009; Wagner et al.). Those changes are associated with morphological changes, a reduction in proliferation ability, the loss of the ability to differentiate into bone, cartilage and adipose tissue (Banfi et al., 2000; Digirolamo et al., 1999; Sethe et al., 2006; Tanabe et al., 2008) and the down-regulation of stemness-related and DNA repair genes (Galderisi et al., 2009). Indiscriminate use of senescent BMSCs in clinical trials may lead to negative results and compromise the entire nascent field of BMSC therapy.
We investigated the effects of serial expansion on BMSCs isolated from marrow aspirates. BMSCs were analyzed for morphology, colony formation efficiency (CFE), immunophenotype, senescence associated beta-galactosidase (SA β-gal) staining and global gene expression profiles. We identified a BMSC transcription signature that was associated with senescence and found that the onset of the change from an early passage to a senescence transcription signature was variable among donors and preceded senescence-associated changes in phenotype and function. BMSC age, expressed as the percentage of life span completed and calculated using population doublings (PDs), was a good marker for an early or senescence transcription profile. Although this measure of BMSC age is valuable, the calculations can only be made retrospectively after expanding BMSCs to senescence. In order to find a surrogate measure of BMSC age, we used a computational biology approach to identify a set of genes whose expression at each passage would predict elapsed age of BMSCs. A least angle regression (LAR) method which uses a linear regression model to predict a continuous response was used to identify the least number of genes whose expression predicts BMSC age. We identified a set of 24 age-predictive genes, which will be useful in determining whether clinical lots of BMSCs have begun to show signs of senescence or not.
Material and Methods
Serial Culture of BMSCs
Marrow collection and BMSC culture were performed according to a Standard Operating Procedures (SOP) established in our lab (Sabatino et al, 2012). After obtaining informed consent, marrow was collected from the posterior iliac crest of 7 healthy donors. A total of 5 to 10 mL of marrow was collected in Bone Marrow Prep Syringes (Pharmacy Department, NIH, Bethesda, MD) and then washed with 2.5x volume of HBSS (Lonza, Walkersville, MD); no density gradient centrifuge was performed. A single cell suspension was made with BMSC culture media (BMSC CM) [alpha MEM with 2 mM glutamine (Lonza), supplemented with 20% lot-selected FBS (Hyclone, Thermo Fischer Scientific, Waltham, MA) and 10 μg/ml Gentamicin] and plated at a density of 2 × 105/cm2 in T-75 flasks (Corning Life Sciences, Corning, NY) and were incubated at 37°C in 5% CO2. Non-adherent cells were removed after 24 hours; the media was changed every three days until the colonies reach 70–80% confluence when the primary BMSCs were harvested. The primary BMSCs were washed with 10mL HBSS twice and lifted with 5mL TrypLE Express (Invitrogen, Life Technologies, Grand Island, NY), the cells were then centrifuged at 406 × g for 10 minutes and the cell number was counted and viability was assessed by Trypan Blue exclusion method. The cells harvested at this stage were designated as Passage 1, and serial passage numbers were designated thereafter. After passage 1, the BMSCs were seeded on plastic surface at a density of 3000 cells per cm2, cultured and harvested when they reach 70–80% confluence.
Passage 4 BMSCs were banked as clinical products and the lot release criteria were (1) ≥ 70% viability, as determined by Trypan Blue exclusion method, (2) No bacteria growth after 14 days of inoculation, as determined by Bactec Plus, aerobic and anaerobic, (3) Negative for mycoplasma, as determined by PCR, (4) < 5 EU/ml Endotoxin, as measured by the Limulus Amebocyte Lysate (LAL) assay, (5) ≥80% of the cells express CD105, CD73 and CD90, and ≥60% of the cells express CD146, as measured by flow cytometry, (6) ≤5% of the cells express CD45, CD14, CD34, CD11b. In addition, we set up in process controls during the production process; (1) at primary culture stage, multiple BMSC colonies must be visible and the confluence must be ≥ 70% on the day of harvest; (2) at the expansion stage, the confluence should be ≥ 70% and the viability ≥ 70% on the day of harvest
The number of BMSCs at every passage was manually counted and the PD for each passage was calculated using equation:
where N = population doublings, N H = cell harvest number, and N I = plating cell number, Cumulative PDs were calculated in relation to the number of cells at the first passage. Population doubling time (PDT) was calculated for each passage. These studies were approved by an NHLBI committee on the use of human subjects in research.
Flow Cytometry Analysis
BMSC surface markers were analyzed by flow cytometry. The cells were stained with: CD73 (CD73-PE, BD Bioscience, San Diego, CA), CD105 (CD105-APC, eBioscience, San Diego, CA), CD146 (CD146-PE, BD Bioscience), CD44 (CD44-APC, BD Bioscience), and isotype control. Data were collected on a FACSCalibur (BD Bioscience) and analyzed using FlowJo software (Tree Star, Inc, Ashland, OR).
Cell Cycle Analysis
3×104 BMSCs were fixed with 4.5mL of ice-cold 70% ethanol for one hour and then washed thoroughly with phosphate buffered saline (PBS). The fixed BMSCs were incubated with 0.5mL Propidium Iodide /Trinton-x-100/RNase A (all from Sigma Aldrich, St. Louis, MO) at room temperature for 30 minutes. Data were collected on the FACSCalibur and analyzed using FlowJo software.
SA β-gal Staining
BMSCs were stained using a Senescence Associated Beta-Gal Staining kit (Sigma Aldrich) following the manufacturer’s protocol. Briefly, BMSCs were washed with PBS, fixed with Fixation Buffer for 7 minutes at room temperature, washed with PBS, and then incubated with staining mixture at 37°C overnight. The senescent cells under 10 randomly selected high-magnitude microscope fields (100 X) were counted on the following day.
CFE Enumeration
BMSCs were plated at 10/cm2 in 6-well plates and cultured for 13 days without changing culture medium. The colonies were then fixed with methanol for 30 minutes and stained with saturated methyl-violet water solution for 20 minutes. Colonies were observed under low magnitude light microscope field (25 X). Colonies, initiated by the colony-forming units–fibroblasts (CFU-Fs) and containing 50 or more cells of fibroblastic morphology were counted, and colony-forming efficiency (CFE; number of BMSC colonies/plating BMSCs) was calculated.
Mixed Lymphocyte Reaction Inhibition
The immunosuppressive properties of BMSCs were compared using Mixed Lymphocyte Reaction (MLR) assay (SAIC-Frederic, Frederic, MD). Ficoll-separated peripheral blood mononuclear cells (PBMC) were plated in 96-well plates at 1×105 responders per well. Responders were co-cultured with 2500cGy irradiated stimulator PBMC at a concentration of 1×105 cells/well. BMSCs at passage 3, 6 and 9 were added at the following concentrations: 104, 4×104 and 105 cells/well. Culture plates were incubated for 6 days in a humidified 5% CO2 incubator at 37°C. On the day of harvest, 0.5μCi of 3H-thymidine (3H-TdR) was added to each well for 4 hours with lymphocyte proliferation measured using a liquid scintillation counter. The effect of BMSCs on MLR was calculated as the percentage of the suppression compared with the proliferative response of the positive control without BMSC, where the positive control was set to 0% suppression. The experiments were performed three times for each variable described.
Cytokine and Growth Factor Analysis of BMSC Culture Supernatant
The cytokine and growth factor concentrations in BMSC culture supernatant were evaluated using SearchLight Protein Array Analysis (Aushon Biosystems, Billerica, MA). Culture supernatant was collected, centrifuged for 10 minutes at 1400 rpm to remove cell debris and then stored at −80°C. The supernatants of BMSC from donors 09FC20 (Passage 3, 6, 9) and W10003 (Passage 3, 6, 9, 11) were evaluated for CXCL12 (SDF1), IL-6, IL-8, IL-10, TGFB1, SPP1 (osteopontin, OPN), ICAM1, TNFRSF11B (osteoprotegerin, OPG), BDNF, CSF1 (M-CSF), LIF, HGF, VEGF, CCL22 (MDC), SAA1, PTGS2 (COX-2) and TNFSF10 (TRAIL).
Microarray Gene Expression Analysis
Total RNA was extracted using miRNeasy Mini Kit (Qiagen, Hilden, Germany) and assessed using Nano Drop 2000 (Thermo Scientific, Wilmington, DE). Microarray expression experiments were performed on 43×3443K Whole Human Genome Microarray (Agilent technologies, Santa Clara, CA, USA) according to our protocols (Wang et al., 2012). Generally, 0.5 μg of BMSC RNA was labeled with Cyanine 5-CTP, and Universal Human Reference RNA (Stratagene, Santa Clara, CA, USA) was labeled with Cyanine3-CTP using a Quick Amp Labeling kit (Agilent), which uses T7 RNA polymerase to simultaneously amplifies target material and incorporates Cy 3 or Cy 5-labeled CTP into complimentary RNA (cRNA). After purification, 825 ng of labeled cRNA from BMSC and reference RNA was pooled, fragmented and then hybridized on 4×44K microarrays for 17 hours at 65°C. Images of the arrays were acquired using a microarray scanner G2505B (Agilent technologies) and image analysis was performed using feature extraction software version 9.5 (Agilent Technologies). The Agilent GE2-v5_95 protocol was applied using default settings. The data were submitted to Gene Expression Omnibus (GEO) under accession number GSE34303.
Data Processing and Statistical Analyses
Global gene expression analysis was performed according to a standard procedure (Wang et al., 2012). Raw data was uploaded into mAdb database (http://madb.nci.nih.gov/) and then imported into BRB-ArrayTools (Simon et al., 2007) (http://linus.nci.nih.gov/BRB-ArrayTools.html). Tests for differences between early, and late stages were conducted for individual genes using two-sided t tests, considering P values of <0.001 as significant, with adjustment for the donor effect. The Benjamini and Hochberg method was used to estimate the false discovery rate. The same tests were also used to identify the differentially expressed genes among early, middle and late passage BMSCs, considering P values of <0.0001 as significant, with adjustment for the donor effect. The Pearson correlation coefficients were used to identify genes that were highly correlated with the BMSC age. Least Angle Regression (LAR) method was used to develop a linear model for predicting the life span (BMSC age) based on gene expression measurements (Efron et al., 2004). The error of the prediction model was estimated using a leave-one-donor-out cross validated approach (Zhao and Simon). Clusters and Heatmaps were generated using MultiExperiment Viewer (MeV4.6.2, http://www.tm4.org/mev/) (Saeed et al., 2003). Pathway analysis was based on GeneGo database (www.genego.com) and Ingenuity Pathway Analysis Database (www.ingenuity.com).
Validation of the prediction model using publicly available datasets
To assess the efficacy of our BMSC “age” prediction model, we analyzed datasets that were deposited in Gene Expression Ominibus (GEO), in which microarray analysis was performed on MSCs that were cultured to replicative senescence using Affymetrix Human Genome U133 Plus 2.0 array (GSE7888 and GSE9593). The microarray data was imported into BRB ArrayTools and filtered to show the 24 predictive genes. One gene FLJ00254 protein was not found on the platform Affymetrix Human Genome U133 plus 2.0 array and its annotation has been discontinued, so it was excluded from analysis. For those genes that had multiple probes, only the probe has the highest intensity was selected for further analysis. After that, the percentage of the maximum lifespan of each sample was calculated using the formula Σicixi + 0.143 where ci and xi are the coefficient and gene expression for the i-th gene, respectively. The gene expression here is log-intensity for single-channel data.
Results
BMSC Growth
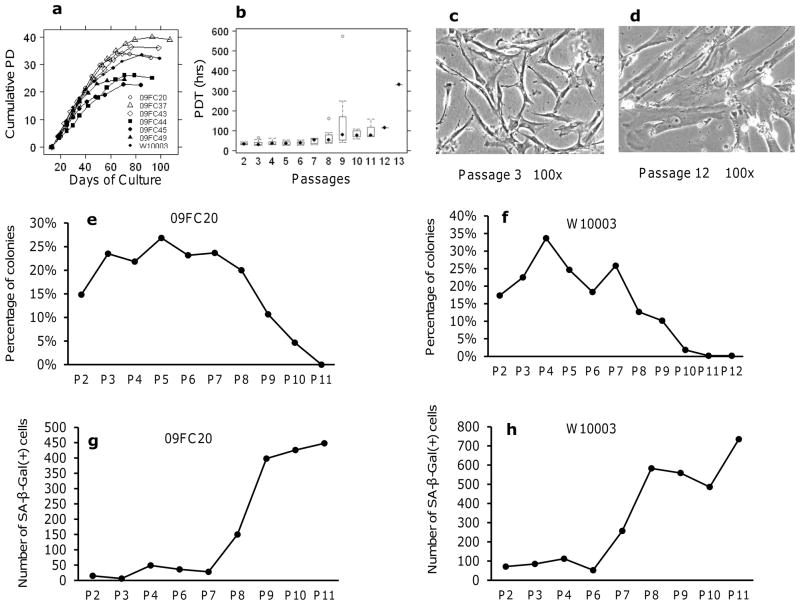
BMSCs used in this study were isolated and expanded from marrow aspirates collected from 7 consenting donors. The BMSCs were subcultured serially until replicative senescence. Growth kinetic analysis showed a rapid increase in proliferation in passage 2 and passage 3 cells, followed by a phase of slow growth and finally no proliferation. Growth began to slow after 5 to 7 passages and stopped after 8 to 12 passages (Fig. 1a). The BMSCs underwent a total of 25 to 40 population doublings (PDs) (Fig. 1a). Population Doubling Time (PDT) at each passage varied among individual donors, but steadily increased with passage number (Fig. 1b). The mean PDTs from passage 2 to passage 9 were 34.8±6.5 h or hours (passage 2), 37.4±13.7 h (passage 3), 38.5±11.3 h (passage 4), 36.7±10.2 h (passage 5), 39.0±9.6 h (passage 6), 46.9±13.3 h (passage 7), 69.9±44.1 h (passage 8) and 162.7±194.9 h (passage 9). BMSCs from 6 donors reached passage 10 with a mean PDT of 80.5 ±18.8 h. BMSCs from 3 donors reached passage 11 with a mean PDT of 101.1±48.5 h. Only one donor reached passages 12 and 13 and the PDTs were 113.10 h and 331.52 h respectively. BMSC morphology changed from thin and spindle-shaped among early passage cells (Fig. 1c) to large, flattened, and irregularly shaped among late passage cells (Fig. 1d).
Figure 1. Growth kinetics, morphology, CFE and senescence of BMSCs.
(a): The Cumulative Population Doublings (PD) for 7 donors. Each line represents BMSCs from one donor and each symbol represents one passage. Population doublings (PD) of each passage were added up to get the cumulative PD. (b): The mean Population Doubling Time (PDT) of multiple donors at each passage. (c) Morphology of BMSC at early passage. (d) Morphology of BMSC at late passage. The CFE assay was performed on BMSCs from donor 09FC20 (e) and donor W10003 (f), data were normalized by the number of plating BMSCs. The numbers of SA β-gal positive cells of BMSCs from 09FC20 (g) and W10003 (h) at each passage were shown. Passage (P) was indicated below x-axis.
Kinetics of Phenotype Changes Associated with BMSCs Senescence
To identify the onset of phenotype and functional changes associated with prolonged BMSC expansion, BMSCs from 2 donors (09FC20 and W10003) were thawed from a working cell bank and serially passaged until senescence. They were assessed for CFE, cell cycle, SA β-gal staining, cell surface markers, immunosuppressive effects and supernatant growth factors/cytokines levels.
In general, CFE was high initially (15–20%) and increased with passages until it reached a peak of 25–35% at passages 3 through 7 and then gradually fell until it reached 0 or 1 colony with less than 50 cells at senescence (Figs. 1e, f). SA β-Gal assay revealed that the number of senescent BMSCs was low and stable for early passages, passages 2 through 7 for 09FC20 and 2 through 6 for donor W10003. The number increased abruptly over the 2 following passages and increased more gradually over the later passages (Figs. 1g, h).
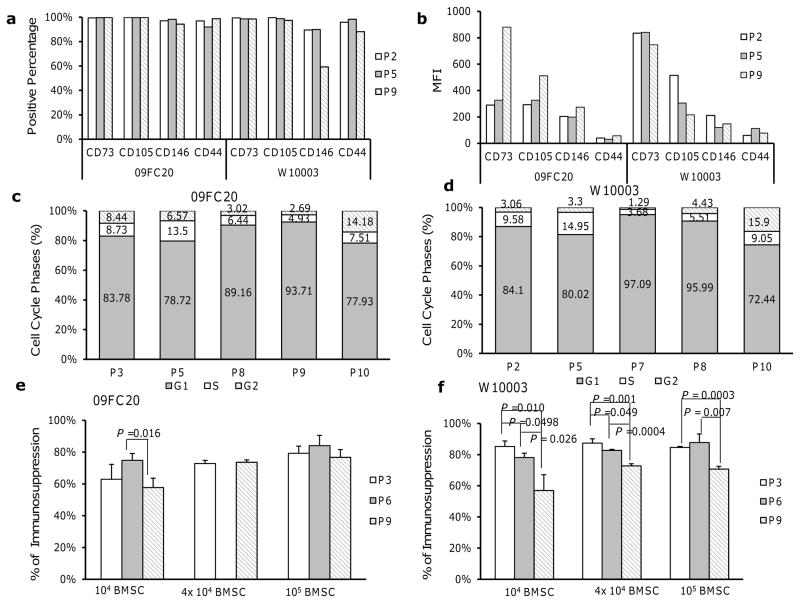
The percentage of CD73 and CD105-positive cells remained stable, while the percentage of CD146-postive cells decreased in late passage cells. The percentage of CD44-positive cells increased slightly in late passage 09FC20 cells, but decreased in late passage W10003 cells (Fig. 2a). The mean fluorescence intensity (MFI) of the BMSC surface markers CD73, CD105, CD146 and CD44 on donor 09FC20 cells increased with passage despite a higher isotype background at passage 9; while the MFI of these 4 surface markers decreased with passage of donor W10003 cells (Fig. 2b).
Figure 2. Surface marker, cell cycle phase and functional changes associates with BMSC expansion.
(a): The percentage of BMSCs expressing surface markers CD73, CD105, CD146 and CD44 from donor 09FC20 and W10003. (b): The mean fluorescence intensity (MFI) of surface markers for BMSCs from donor 09FC20 and W10003. (c): Results of cell cycle analysis on BMSCs from donor 09FC20. (d): Results of cell cycle analysis on BMSCs from donor W10003, passage (P), and cell cycle phage G1, S, G2 were indicated below x-axis. (e): The immunosuppressive effects of BMSCs from donor 09FC20. (f): The immunosuppressive effects of BMSCs from donor W10003. Number of BMSCs per well was indicated below the bars. Error bars indicated standard deviation. Statistical significances between passages 3, 6 and 9 were determined by two sample t-test, and P values were indicated above the bars.
Cell cycle analysis revealed that the proportion of cells in S phase increased from passages 2 or 3 to passage 5 and then decreased. The proportion of cells in G2 and S phases fell at passage 7, 8, and 9 and increased markedly at passage 10 (Figs. 2c and 2d). This finding was not surprising since an increased number of cells with 4C DNA content, probably G1 tetraploid cells, have been observed in premature senescence (Pitto et al., 2009).
We hypothesized that the immunosuppressive effects of BMSCs will be deteriorated by long-term culture and therefore examined the influence of BMSC expansion on their inhibition to lymphocyte proliferation using a mixed lymphocyte reaction (MLR) assay. As expected, BMSCs at passage 9 were markedly less suppressive than those at passage 3 and 6; while no significant differences were observed between passage 3 and passage 6 (Figs. 2e and 2f).
Cytokine/chemokine secretion was another key phenotype associated with senescence of BMSC. For example, X-ray irradiation induced the up-regulation of GRO, IL-8, IL-12 and MDC (CCL22) in human MSC supernatant (Wang and Jang, 2009); a modified secretory senescence (mSS) signature comprised of IL6, IL8, TGFB1, etc, revealed increasing senescence with passage of MSCs(Lafferty-Whyte et al.), 2010). We measured the concentrations of proteins whose genes were differentially expressed between early and late passage BMSCs (p<0.001), including CXCL12, VEGF, TNFSF10 (down-regulated in late passage cells), and IL-6, IL-8, TNFRSF11b, BDNF (up-regulated in late passage cells). Some genes whose expression changed with passages but with less statistical significance (p>0.001) were also measured. These genes included SPP1, CSF1, LIF, CCL22 and SAA1, which we think may be involved in the senescence of BMSCs, and TGFB1, HGF, and PTGS2 which are associated with the immune-modulatory functions of BMSCs. Among the proteins analyzed, only 4 were detected in culture medium, TGFB1 was of high level, while BDNF, CXCL12 and VEGF were at low levels. In the BMSC supernatant, IL-10, TNFSF10 and PTGS2 were too low to be detected. The concentrations of IL-6, IL-8, ICAM1 and TNFRSF11B were much higher in passage 9 supernatants than passage 3 and 6 supernatants (Table 1 and supplementary Material S1). The concentration of CSF1 and SPP1 increased drastically over passages and reach peak concentrations at passage 9 and passage 11. However, the levels of CXCL12, SAA1 and TGFB1 decreased greatly with passages; HGF, LIF were generally decreased but with donor variance; the other proteins BDNF and VEGF did not undergo significant change through the culture (Table 1 and Supplementary Material S1).
Table 1.
The concentrations (pg/ml) of proteins in the supernatant of BMSCs
| Factors | Culture Medium | Donor 09FC20 | Donor W1003 | |||||
|---|---|---|---|---|---|---|---|---|
| Passage 3 | Passage 6 | Passage 9 | Passage 3 | Passage 6 | Passage 9 | Passage 11 | ||
| IL-6 | - | 16421.68 | 10920.21 | 51574.52 | 23184.07 | 24462.14 | 40656.32 | 34727.54 |
| IL-8 | - | 630.94 | 480.49 | 6714.29 | 177.19 | 126.83 | 1207.35 | 4082.80 |
| BDNF | 25.0 | 308.76 | 902.57 | 935.42 | 346.28 | 552.37 | 468.25 | 413.18 |
| ICAM1 | - | 148.47 | 152.88 | 648.72 | 167.21 | 198.35 | 307.91 | 491.05 |
| CSF1 | - | 203.45 | 144.15 | 386.05 | 86.22 | 63.82 | 75.28 | 172.32 |
| TNFRSF11B | - | 47.25 | 275.99 | 1207.68 | 20.92 | 334.63 | 465.08 | 572.84 |
| SPP1 | - | 364.49 | 594.17 | 4002.43 | 2819.78 | 1047.16 | 2197.46 | 3637.69 |
| CXCL12 | 52.5 | 22712.19 | 18464.34 | 12244.89 | 12798.60 | 5418.17 | 3207.16 | 698.46 |
| TGFB1 | 17025.5 | 40134.34 | 36949.30 | 35784.64 | 29086.97 | 19178.13 | 20957.20 | 17920.62 |
| SAA1 | - | 26882.40 | 19814.58 | 18479.18 | 18063.59 | 11348.43 | 8784.44 | 9103.86 |
| VEGF | 3.2 | 16161.31 | 13102.05 | 22762.92 | 12757.75 | 5277.18 | 5839.16 | 6687.93 |
| LIF | - | 309.13 | 256.36 | 305.97 | 206.38 | 130.52 | 123.24 | 64.02 |
| HGF | - | 2692.63 | 5756.90 | 3744.90 | 42658.13 | 15782.62 | 9651.02 | 801.24 |
| CCL22 | - | 6.82 | 6.21 | 4.56 | 6.41 | 2.93 | 2.65 | 2.46 |
- not detected
Comparison of Early and Late Passage BMSCs from 5 Donors by Global Gene Expression Analysis
The CFE, PDT, cell cycle, SA-β-gal analysis suggested that BMSC are relatively stable for 6 or 7 passages. To further assess the nature of changes associated with senescence at a molecular level, we performed global transcriptome analysis on early and late passage BMSCs from 5 donors: 09FC37, 09FC43, 09FC44, 09FC45 and 09FC49. The early passage cells were from passages 2 and 3 and the late passage cells were from the three passages immediately prior to senescence. Unsupervised hierarchical clustering separated the samples into two clusters, one with all 10 early passage samples and one with all 15 late passage samples, passages 6 through 12 (Supplementary Material S2). We then identified 2122 differentially expressed genes between the two groups (2 sample t-test with blocking of donor effects, P-value <0.001, fold change > 1.5). Among these genes, 922 were up-regulated and 1200 down-regulated by early passage BMSCs compared to late passage cells (Supplementary Material S2). Ingenuity Pathway Analysis showed that genes up-regulated in early passage BMSCs were overrepresented in immune response pathways, development associated pathways, while the genes up-regulated in late passage BMSCs were over-represented in cell proliferation pathways, immune response pathways (Supplementary Material S2).
Kinetics of Molecular Changes Defined by Global Gene Expression Analysis of 7 Donors
Analysis of the early and late passage BMSCs from 5 donors revealed marked transcriptional differences but did not provide precise information as to the onset of the molecular changes associated with senescence, we therefore analyzed 57 BMSC samples from all 7 donors to clarify these changes.
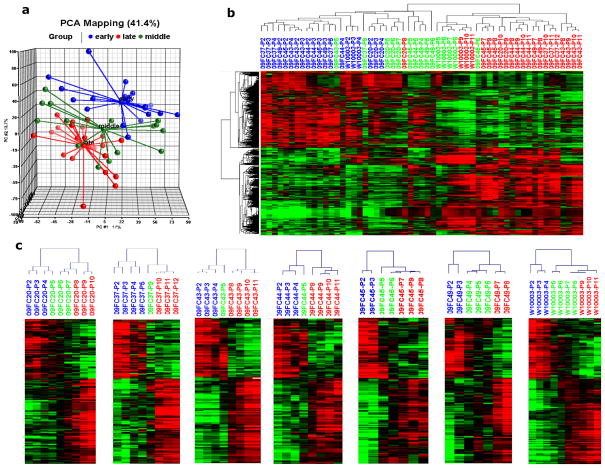
For further analysis we calculated BMSC “age” rather than passage number using a published method (Stenderup et al., 2003). The “age” of BMSCs at each passage was expressed as a percentage of the maximal life span, i.e. the population doubling (PD) at each passage was divided by the total number of PDs at senescence. The BMSC samples at each passage were then classified as early, middle and late stage based on their age. Passages whose accumulative PDs were less than 50% of maximum life span were considered early stage; those with 50% to 80% of maximum life span were middle stage; and those with accumulative PDs greater than 80% of maximum life span were late stage (Stenderup et al., 2003). Principal component analysis (PCA) using genes the expression of which in more than 20% of the BMSC samples was higher or lower than the median value by greater than 1.5-fold separated the samples into 3 groups; BMSCs at early stage were located primarily within a single group, BMSCs at late stage were primarily within another group, while BMSCs at middle stage were located in a group between them (Fig. 3a). The early stage group included BMSCs at passages 2 through 5, and late stage group included BMSCs from passages 8 through 11 (Fig. 3a). Interestingly, the BMSCs in middle stage group, representing a transition from early to late stage and with passage numbers ranging from 5 to 9, were quite heterogeneously distributed. For instance, the passage 9 BMSCs from donor 09FC37 and passage 7 cells of donor 09FC20 were close to early stage group, while passage 4 cells of donor 09FC49, passage 5 cells of donors 09FC44 and 09FC45 were close to late stage samples. These results revealed that there was variability among donor in the onset of molecular changes associated with senescence and, in general, the onset of molecule changes associated with senescence preceded the onset of phenotype and function changes.
Figure 3. Clustering of BMSCs from 7 donors.
(a): Principal Component Analysis (PCA) separated the BMSCs into three clusters, early passage cells (blue spheres), middle passage (green spheres) and late passage (red spheres). (b): Hierarchical clustering analysis using 1067 differentially expressed genes separated the 57 BMSC samples into two clusters. P stands for Passage, early stage cells were shown by blue font, middle stage by green and late stages by red. (C): Hierarchical clustering using the 155 age- correlated genes separated the individual samples from each donor into an earlier signature group and later signature group, P stands for Passage, blue font stands for early stage, green for middle stage and red for late stage.
For further analysis, we identified 1067 differentially expressed genes among early, middle and late stage BMSCs (group comparison analysis with donor effect blocked, and P value <0.0001 as significant). Mixed effects ANOVA models were also performed using stage as the fixed factor and donor as the random factor. With the mixed effects models, 1070 genes were found to be differentially expressed (p<0.0001) among stages, which is comparable to 1067 differentially expressed genes identified in class comparison among stages when blocking the donor effect. Hierarchical clustering analysis using the 1067 differentially expressed genes revealed that BMSCs were segmented into two major clusters (Fig. 3b); the first cluster contained all of the early stage samples plus most of the middle stage samples; while the second cluster included all late stage samples and the remaining middle stage samples (Fig. 3b). The middle age sample signature appeared to represent a transition from the early stage to the late stage. The early stage samples included passage 2 through 5 samples, but passage 4 and 5 samples were also present in the middle stage samples. These results show that BMSC age expressed as percentage of the maximal life span provides a good indication on whether BMSCs will have an early or late age molecular profile.
Those results showed that the expression of BMSC age as a percent of maximum life span is a useful marker of early and senescent BMSC transcription signatures. Unfortunately, it can only be calculated by growing the cells to senescence, a condition that is non-applicable to clinical manufacturing. Therefore we searched for genes whose expression could be used to predict BMSC age prospectively.
Identification of a set of BMSC age-predictive genes
Because of the relatively early onset of molecular changes associated with senescence in some donors, it would be worthwhile to identify markers that could be used to evaluate final clinical BMSC products and ensure they retain an early gene expression signature.
First, we calculated the Pearson correlation coefficients of each individual gene with the percentage of maximum life span, which stood for the BMSC age. This analysis identified 155 genes whose expression levels were highly correlated with the BMSC age (correlation efficient ≥ 0.7) (Supplementary Material S3). Then, we investigated the onset of the transition from an early to late life span signature for each of the 7 subjects using hierarchical clustering analysis using the 155 “age”-correlated genes. Hierarchical clustering separated the BMSC samples from each donor into two groups; an earlier gene expression signature group and a later signature group (Fig 3c.). For each of the 7 subjects all early stage BMSCs were in the earlier signature cluster and all late stage BMSCs were in the later signature cluster. Most of the middle stage BMSCs were clustered with late stage BMSCs, with single samples from only 2 donors clustering with early stage cells and samples from 1 donor clustering with both early and late signature groups. Specifically, earlier stage BMSCs included passage 2 and 3 for 09FC45 and 09FC49, passage 2, 3, and 4 for W10003, and passage 5 and less for 09FC20, 09FC37 and 09FC43, and 09FC44. However, some samples in the transition area, passages 6 and 7, were not available for 09FC37, 09FC43, and 09FC44; therefore the conclusions that can be made from these donors are somewhat limited. These results also show that the transition from an early to a late transcription signature is variable among donors.
GeneGo Process Network analysis of these 155 genes revealed that genes up-regulated at earlier passages were overrepresented by Notch Signaling, Wnt Signaling, TGF-β, GDF and Activin Signaling; and Development of Ossification and Bone Remodeling (Table 2); while genes up-regulated at late passages were overrepresented by Chromatin Modification; Blood Vessel Morphogenesis; Cell Cycle and IL-10 Anti-inflammatory Response (Table 2). Among these pathways, Wnt Signaling pathway is instrumental for the bone development; 3 genes, TGFB3, MAP2K6, and WISP1 were up-regulated by early passage BMSCs; Wnt-induced secreted protein 1 (WISP1) is a modulator of BMSC proliferation and osteogenic differentiation (Inkson et al., 2008), overexpression of WISP1 enhanced both in vitro osteogenic differentiation and osteogenesis in vivo (Ono et al.); it can also alleviate the biglycan (BGN) -inhibited proliferation of BMSC (Inkson et al., 2009).
Table 2.
GeneGo Process Network analysis of the 155 age – correlated genes
| Networks | Early passage up-regulated genes
|
Late passage up-regulated genes
|
||
|---|---|---|---|---|
| P-Value | Genes | P-Value | Genes | |
| Signal transduction_NOTCH signaling | 1.00E-04 | HEYL, TGFB3, MAP2K6, LFNG | 2.25E-02 | CCND1, FZD4, SMURF2, CCND1 |
|
| ||||
| Signal transduction_WNT signaling | 2.64E-04 | TGFB3, MAP2K6, WISP1 | 1.97E-01 | CCND1, FZD4 |
|
| ||||
| Transcription_Chromatin modification | 3.53E-01 | HIST1H4C, HIST1H4B | 2.70E-03 | HDAC9, HIST2H2AA3, HIST1H2BK, HIST1H2BE, HIST1H2BC |
|
| ||||
| Development_Blood vessel morphogenesis | 5.42E-01 | CXCL12 | 3.64E-03 | TEK, PLAT, EDN1, ADRB2, OXTR |
|
| ||||
| Development_EMT_Regulation of epithelial-to-mesenchymal transition | 6.93E-03 | MAP2K6, TGFB3, DES | 3.92E-03 | SMURF2, FZD4, EDN1, KRT14, KRT18 |
|
| ||||
| Cell cycle_G0–G1 | 2.14E-01 | CDKN2C | 4.16E-03 | CCND1, HDAC9 |
|
| ||||
| Inflammation_IL-10 anti-inflammatory response | 2.55E-01 | MAP2K6 | 7.34E-03 | CCND1, TLR4 |
|
| ||||
| Signal Transduction_TGF-beta, GDF and Activin signaling | 1.38E-02 | MAP2K6, RUNX2, TGFB3 | 5.09E-01 | CCND1 |
|
| ||||
| Development_Ossification and bone remodeling | 1.51E-02 | MAP2K6, RUNX2, TGFB3 | 5.21E-01 | FZD4 |
|
| ||||
| Cell adhesion_Platelet-endothelium-leucocyte interactions | 2.01E-02 | CD40, TGFB3 | 4.62E-02 | PLAT, EDN1, CD36 |
|
| ||||
| Reproduction_FSH-beta signaling pathway | 4.18E-01 | MAP2K6 | 4.62E-02 | CCND1, PSG1, PSG6 |
|
| ||||
| Signal Transduction_BMP and GDF signaling | 3.70E-02 | MAP2K6, RUNX2 | 3.46E-01 | SMURF2 |
|
| ||||
| Cytoskeleton_Intermediate filaments | 2.40E-01 | DES | 3.56E-02 | KRT14, KRT18 |
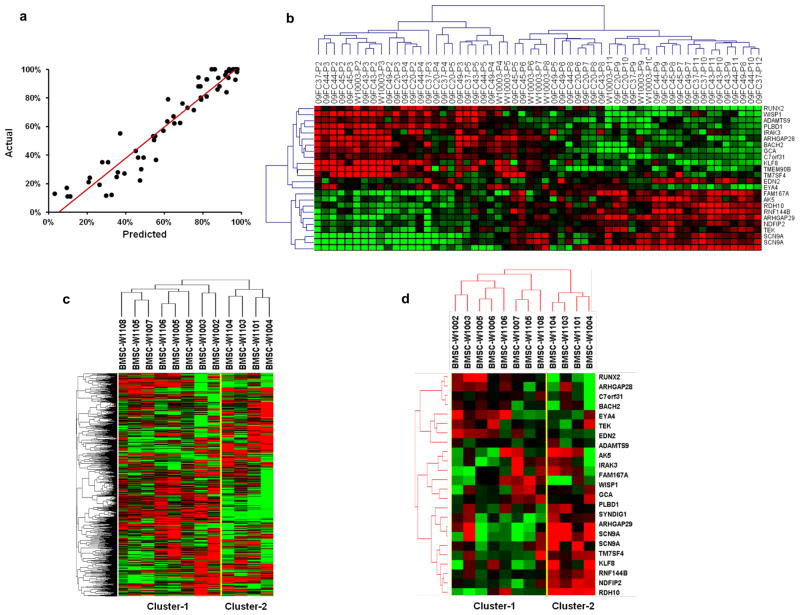
We then built a linear model for predicting a continuous response variable based on gene expression profile using Least Angle Regression/LASSO algorithm (Efron et al, 2004). This method avoids the over-fitting characteristic of least-squares linear regression when the number of variables (i.e., genes) is large compared to the number of cases (i.e., donors). In order to get an unbiased estimate of the true prediction error, we included leave-one-donor-out cross validation to estimate the squared prediction error for each model. In each round, the data of one donor was omitted. A linear model was built based on remaining data in a stepwise manner. The prediction error was recorded for the data withheld. This process was repeated for each donor. The model with the minimum estimated squared prediction error was selected as the final model. The figure 4a shows the scatter plot of the cross validated predicted response values versus the actual values, which indicated that the R square, a measure of the predicted life span vs. the actual life span, was 0.91.
Figure 4. Prediction model of BMSC age.
(a): Scatter-plot of the actual life span vs the predicted life span of each sample (black dots) from the leave-one-donor-out cross-validation. (b) Hierarchical clustering analysis using the 24 age-predictive genes separated the 57 samples into one cluster of 25 earlier passage BMSCs and one with 32 later passage BMSCs, the later cluster contained one sub-cluster with intermediate passage cells and one sub-cluster of late passages. (c): Unsupervised hierarchical clustering separated the 12 clinical BMSC lots into two clusters: cluster-1 with 8 lots and cluster-2 with 4 lots. (d): Hierarchical clustering using the 24 age-predictive genes also separated the 12 lots into two clusters, one with 8 lots (cluster-1) and the other one with 4 lots (cluster-2).
The prediction model identified a data set with 24 genes which were able to predict the life span of each BMSC sample at different passages. The names of the 24 genes and the prediction model were shown in Table 3, and the expression of the 24 genes was described in Supplementary Spreadsheet S4. Hierarchical clustering analysis using the 24 predictive genes separated the 57 BMSC samples into one cluster of 25 earlier passage BMSC samples and one with 32 later passage BMSCs (Fig. 4b). The later passage group contained two sub-clusters, one with 12 intermediate passage cells, passages 5 through 8, and a late passage group, passages 7 through 12 (Fig. 4c).
Table 3. Prediction model of BMSC age.
The predicted age of new samples can be calculated by the expression of these 24 genes and the formula: Σicixi + 0.143 where ci and xi are the coefficient and gene expression for the i-th gene, respectively. The gene expression is the (normalized) log ratios for dual-channel data.
| UG Cluster | Symbol | Map Location | Coefficient | |
|---|---|---|---|---|
| 1 | Hs.559718 | AK5 | 1p31 | 0.007 |
| 2 | Hs.439145 | SCN9A | 2q24 | 0.007 |
| 3 | Hs.535845 | RUNX2 | 6p21 | −0.003 |
| 4 | Hs.124638 | TMEM90B | 20p11.21 | −0.011 |
| 5 | Hs.656071 | ADAMTS9 | 3p14.1 | −0.001 |
| 6 | Hs.269764 | BACH2 | 6q15 | −0.014 |
| 7 | Hs.646614 | KLF8 | Xp11.21 | −0.001 |
| 8 | Hs.244940 | RDH10 | 8q21.11 | 0.038 |
| 9 | Hs.483238 | ARHGAP29 | 1p22.1 | −0.026 |
| 10 | Hs.1407 | EDN2 | 1p34 | 0.008 |
| 11 | Hs.131933 | PLBD1 | 12p13.1 | −0.068 |
| 12 | Hs.369265 | IRAK3 | 12q14.3 | −0.009 |
| 13 | Hs.148741 | RNF144B | 6p22.3 | 0.005 |
| 14 | Hs.525093 | NDFIP2 | 13q31.1 | 0.003 |
| 15 | Hs.124299 | FAM167A | 8p23-p22 | 0.009 |
| 16 | Hs.183114 | ARHGAP28 | 18p11.31 | 0.002 |
| 17 | Hs.596680 | EYA4 | 6q23 | −0.005 |
| 18 | Hs.652230 | TM7SF4 | 8q23 | −0.001 |
| 19 | Hs.89640 | TEK | 9p21 | 0.032 |
| 20 | Hs.377894 | GCA | 2q24.2 | −0.017 |
| 21 | Hs.439145 | SCN9A | 2q24 | 0.02 |
| 22 | Hs.681802 | FLJ00254 protein | 1 | 0.019 |
| 23 | Hs.492974 | WISP1 | 8q24.22 | −0.001 |
| 24 | Hs.122055 | C7orf31 | 7p15.3 | −0.006 |
Assessment of Clinical BMSCs’ Quality Using the Age-Predictive Gene Set
To determine if the 24 age-predictive genes might be useful for evaluating clinical BMSC products, we assessed 12 BMSC lots produced in our clinical processing laboratory using good manufacturing practices (Sabatino et al.) 2012). All lots were isolated from marrow aspirates of healthy donors and expanded by 4 serial passages. Unsupervised hierarchical clustering using 4895 genes with a standard deviation greater than 0.4 separated the 12 lots into two clusters: one with 8 lots (cluster-1) and one with 4 lots (cluster-2) (Fig. 4c), and clustering using the 24 age-predictive genes separated the 12 lots into the same two clusters (Fig. 4d). The median age of the 8 donors in cluster-1 was 23.3 years and ranged from 21.7 to 42.5 years, while the median age of the 4 donors in cluster-2 was 58.1 years and ranged from 21.3 to 67.9 years. All 8 lots in cluster-1 met lot release criteria, but only one lot in cluster-2 met lot release criteria. One of the three lots failing to meet lot release criteria, lot W1103 had higher CD34 positive percentage, which exceeded our release criteria of <5%. The other two lots W1004 and W1104, which were from the 2 oldest donors, 67 and 59 years of age, had slow proliferation during primary culture stage and failed to meet the standards of our manufacturing protocol(Sabatino et al.). Although lot W1101 in cluster-2 met the lot release criteria, it was from the third oldest donor, 57 years of age. Interestingly, the calculated percentage of maximal life span of samples in cluster-2 was much higher than those in cluster-1 (Table 4). These results showed that the age-predictive data set was able to identify slower growing and likely “older” BMSCs and may be useful in identifying poor quality BMSCs.
Table 4.
Calculated percentage of maximum life span of clinical BMSC products
| Donor | Age (years) | Calculated Percentage of Maximum Life Span | Met Lot Release Criteria | Cluster |
|---|---|---|---|---|
| BMSC-W1002 | 22.9 | 41.47% | Yes | Cluster-1 |
| BMSC-W1003 | 22.8 | 35.97% | Yes | Cluster-1 |
| BMSC-W1005 | 21.7 | 33.59% | Yes | Cluster-1 |
| BMSC-W1006 | 23.7 | 42.69% | Yes | Cluster-1 |
| BMSC-W1007 | 27.2 | 25.40% | Yes | Cluster-1 |
| BMSC-W1105 | 27.6 | 30.21% | Yes | Cluster-1 |
| BMSC-W1106 | 22.7 | 35.74% | Yes | Cluster-1 |
| BMSC-W1108 | 42.5 | 35.83% | Yes | Cluster-1 |
| BMSC-W1004 | 67.9 | 58.56% | No* | Cluster-2 |
| BMSC-W1101 | 56.8 | 47.65% | Yes | Cluster-2 |
| BMSC-W1103 | 21.3 | 45.40% | No$ | Cluster-2 |
| BMSC-W1104 | 59.4 | 52.68% | No* | Cluster-2 |
The percentage of maximum life span was calculated based on the prediction model described in Table 3.
Poor proliferation during primary culture
high CD34+ percentage
Validation of the prediction model using publicly available datasets
To assess the efficacy of our BMSC “age” prediction model, we analyzed datasets deposited in GSE7888 and GSE9593. The expression of the predictive genes was filtered out and the percentage of maximum lifespan was calculated using our formula. Due to the discrepancy of the platform used (Agilent microarrays in our study vs Affymetrix microarrays for the other studies), the calculated percentages for some samples were off scale. However, for the BMSCs from 9 subjects analyzed, 6 from GSE7888 and 3 from GSE9593, the calculated percentage of maximum lifespan increased with passage. When the cells entered the senescent stage, this percentage was much higher than for those at early passages (supplementary file 5 and 6). These results indicate that our prediction model is of universal interest for evaluating the in vitro senescence of BMSCs.
Discussion
While BMSCs are being used clinically to treat a wide variety of conditions, the results of these clinical trials have been variable. This may be due in part to differences in BMSC manufacturing methods. One variable among BMSC manufacturing methods is the number of passages used to produce the final product. Some laboratories use 3 or 4 passages while others use 5 or 6 passages (Horwitz et al., 2002; Koc et al., 2002; Koc et al., 2000; Le Blanc and Ringden, 2007; Le Blanc et al., 2003). It is known that prolonged culture of BMSCs is associated with the loss of proliferation potential and likely the loss of function (Banfi et al., 2000; Tanabe et al., 2008). We characterized changes that occurred in BMSCs with prolonged culture and found that molecular changes preceded phenotype and functional changes. The marked differences in molecular signatures between early and late passages occurred over multiple passages and the timing of these changes varied among individuals.
Conventional measurements of senescence such as SA β-gal staining, CFE, and PDTs suggested that BMSCs were relatively stable for several passages before showing signs of senescence. The proportion of cells staining with SA β-gal was very low until passage 6 or 7. CFE levels were somewhat variable, but did not fall below passage 2 levels until after passage 7 or 8. PDTs remained stable until passage 7.
The results of gene expression profiling were consistent with the results of SA β-gal staining and CFE assays in that passage 7 and greater BMSCs showed significant signs of senescence for all three assays. In contrast, gene expression analysis differed from the other two assays in that it consistently characterized only passages 2 through 5 as early passage BMSCs. Gene expression analysis generally placed passage 6 cells in a transition between early and late passages and passage 7 and later were generally in the late passage group. This suggests that the molecular changes associated with BMSC senescence begin at passage 4 or 5, but do not result in a senescence-associated phenotype until passage 6, 7 or later. While all functionally important BMSC properties may not be affected by senescence, the results of gene expression analysis suggest that if a critical BMSC function or property does change with cell passage, significant changes may occur before traditional markers of senescence are apparent.
When the molecular signatures of each donor’s BMSC samples were analyzed individually, we noticed that the BMSC samples were clustered from lowest to highest passages for all 7 donors. This is consistent with the results of Wagner and others (Schallmoser et al.; Wagner et al., 2008), who analyzed serial passages of BMSCs from three donors. These results suggest that some changes in BMSCs occur with each passage; however, the magnitudes of these changes with each passage are minor relative to the difference between the earliest and latest passage cells. This also provides further evidence that the transition from an early to late passage phenotype is gradual and all early passage cells may not have identical functional properties.
A number of important differences in gene expression occurred with prolonged BMSC passage. Stem cell related genes, including Wnt and Notch signaling genes, were down-regulated in late passage cells. In addition, genes involved with DNA replication, recombination and repair, cell cycle, chromatin modification and repair were up-regulated at late cell passages. This is consistent with the association of senescence with cell cycle arrest, resistance to apoptosis and chromatin reorganization (Schallmoser et al.). A number of immune response related genes were also down-regulated in late passage cells, suggesting that differences in anti-inflammatory and immune modulatory properties likely exist between early and late passage BMSCs. We found that the suppression of BMSCs on MLRs was reduced in late passage cells, but further functional studies are required to determine the effects of BMSC age on specific immune modulatory or anti-inflammatory effects.
We also characterized growth factors and cytokines in the BMSC culture supernatant, which revealed results consistent with a previously described senescence-associated secretory phenotype, including rising levels of IL-6, IL-8, ICAM-1 and TNFRSF11B (Coppe et al., 2008; Lafferty-Whyte et al.; Wang and Jang, 2009). Cytokine/chemokine secretion was another key phenotype associated with senescence of BMSC. For example, X-ray irradiation induced up-regulation of GRO, IL-8, IL-12 and MDC (CCL22) in human MSC supernatant (Wang and Jang, 2009). Senescence signaling gradually increases with passages when analyzed using all biomarkers, but there was a sharp increase in the senescence signaling in certain passages, specifically, at passage 6 in Damage Associated Senescence markers and at passage 7 in Modified Secretory Senescence markers (Lafferty-Whyte et al., 2010). When we calculated the percentage of maximum lifespan of BMSCs using our prediction model and the same dataset (GSE9593), we identified a very similar trend (supplementary file 6). Senescent human fibroblasts secrete numerous proteins, including IL-6 and IL8, and shed cell surface molecules ICAM1 and TNFRSF11B (Coppe et al., 2008); TNFRSF11B was expressed to a higher extent in senescent hMSC (Benisch et al.) 2012). Some proteins secreted by senescent cells can act in an autocrine manner to reinforce the senescence growth arrest or promote degenerative of hyperproliferative changes in neighboring cells. For instance, IL-6 prevents cell cycle progression once the senescence program is engaged (Kuilman et al., 2008) and IL-8 reinforces senescence via CXCR2 receptor (Acosta et al., 2008). We also observed the sharp increase of SPP1 (OPN) at late passage BMSCs. SPP1 serves as an attachment protein linking cells to the bone mineral and facilitates osteoclast migration to sites of resorption. In addition to its function as a structural protein, SPP is also involved in signaling transduction. For example, it is involved in the IL-12 and IFN- γ responses. It increases IL- 12 secretion of mouse macrophages, but suppressed the IL-10 response and thus it represents an essential early step in the pathway that leads to type 1 immunity (Ashkar et al., 2000). Interestingly, SPP1 was up-regulated with the age of subjects providing the BMSCs (Jiang et al.) 2011), and these data were in agreement with our data to some extent. Therefore it will be very interesting to investigate the roles of these cytokines in human BMSC. Genes involved in the Development of Ossification and Bone Remodeling were up-regulated in early passage BMSCs, which is consistent with the reports that late-passage BMSCs exhibited decreased bone formation capacity in both in vivo experiments (Stenderup et al., 2003) and in vitro differentiation assays (Siddappa et al., 2007).
Considerable variability was detected among BMSCs from the 7 subjects. The total number of passages and accumulative PDs of each donor’s BMSCs was highly variable. More important, the onset of BMSC transition from an earlier to a later transcription profile varied among the donors. This transition occurred after passage 5 in four donors, after passage 4 in one donor and after passage 3 in two donors. Because of this biological variability, it is important to develop biomarkers that can be used to assess clinical BMSC lots to determine if they show signs of changes associated with prolonged passage at the time of final harvest. Therefore, we searched for genes that predicted BMSC age based on the hypothesis that most critical BMSC functions will change with prolonged passage, and the changes in the expression of genes reflecting BMSC age can be used as a surrogate marker for quality control of clinical BMSCs products. However, global gene expression analysis yields notoriously large data sets that contain thousands of genes as potential markers and it is essential to refine these data sets using appropriate models. The LAR algorithm developed by Efron et al. (2004) is one such model. It attempts to avoid the over-fitting characteristic of least-squares linear regression when the number of variables (genes) is large compared to the number of cases (samples). This algorithm has been implemented into BRB-ArrayTools as a plug-in for building a linear model for predicting a continuous response variable based on gene expression measurements (Zhao and Simon). Using the LAR model, we identified a set of 24 genes predictive of BMSC age, which were as effective as the entire gene set for identifying clinical BMSC lots that grew slowly and likely were functionally “older” at the time of harvest. Furthermore, the age predictive data set recognized one BMSC lot that did not meet lot release criteria based on CD34+ cell percentage as being different or “older” than the BMSC lots meeting lot release criteria. That 24-gene set included functionally important genes such as WISP1, RUNX2, KLF8, TM7SF4, ADAMTS9, IRAK3 and EYA4 whose expression decreased with BMSC age, while the expression of TEK, ARHGAP29 and SCN9A was increased with age. The expression of RUNX2, a regulator of osteoblasts differentiation (Ducy et al., 1997), has been reported to be decreased in late passage BMSCs (Tanabe et al., 2008), but the other genes have not previously been found to be associated with BMSC age. WISP1 modulates BMSC proliferation and osteogenic differentiation (Inkson et al., 2008), TM7SF4 plays a role in osteoclastogenesis (Kukita et al., 2004), IRAK3 is a negative regulator of Toll-like receptor (TLR) signaling (Kobayashi et al., 2002), EYA4 is a transcription activator involved with the regulation of the innate immune response (Okabe et al., 2009), KLF8 is a transcription factor that involved with the regulation of epithelial to mesenchymal transition (EMT) (Wang et al., 2007), ARHGAP29 may be a tumor suppressor gene (Ripperger et al., 2007) and TEK is a receptor for angiopoietin-1 (Oskowitz et al.).
In conclusion, the onset of molecular changes associated with BMSC passage precede the onset of senescence-associated changes in commonly used indicators of BMSC quality such as SA β-gal staining, PDT and CFE. Those molecular changes occurred over multiple passages and the timing of the changes varied among individuals. A set of 24 age-predictive genes may be useful in assessing the quality of clinical BMSC products.
Supplementary Material
The concentrations of 14 proteins in the supernatant of BMSCs from 2 donors 09FC20 (red line) and W10003 (green line) were shown. The X-axis indicates BMSC passage (P) and the y-axis indicates the concentrations of proteins expressed as pg/ml.
Dataset GSE7888 obtained from serial passages of BMSCs was downloaded and imported into BRBArray-tools, the log-intensity of the predictive genes was acquired, and the percentage of maximum lifespan of BMSCs at each passage was calculated using our prediction model. Each plot displayed the results of one of six donors, which were labeled by the names used in the original database. In each plot, the x-axis indicates the passage number of individual samples and the y-axis indicates the calculated percentage of maximum lifespan, which denote the “age” of BMSC.
Dataset GSE9593 obtained from serial passages of BMSCs was downloaded and imported into BRBArray-tools, the log-intensity of the predictive genes was acquired, and the percentage of maximum lifespan of BMSCs at each passage was calculated using our prediction model. Each plot displayed the results of one of three donors, which were labeled by the names used in the original database. In each plot, the x-axis indicates the passage number of individual samples and the y-axis indicates the calculated percentage of maximum lifespan, which denote the “age” of BMSC.
Acknowledgments
The authors thank the staff of the Cell Processing Laboratory, Department of Transfusion Medicine, Clinical Center, NIH for their support of these studies. We also thank Dr. Francesco Marincola and Dr. Ena Wang (Department of Transfusion Medicine, Clinical Center, NIH) for useful discussions and comments on the manuscript. This work was funded by NIH Bone Marrow Stromal Cell Transplantation Center (BMSCTC) and the Department of Transfusion Medicine, Clinical Center NIH.
Footnotes
The authors declared no conflict of interest.
Contributor Information
Jiaqiang Ren, Email: renj@mail.nih.gov.
David F. Stroncek, Email: DStroncek@cc.nih.gov.
Yingdong Zhao, Email: zhaoy@ctep.nci.nih.gov.
Ping Jin, Email: Pjin@cc.nih.gov.
Luciano Castiello, Email: castiellol@cc.nih.gov.
Sara Civini, Email: civinis@cc.nih.gov.
Huan Wang, Email: Heidi.wang@nih.gov.
Ji Feng, Email: fenj2@cc.nih.gov.
Katherine Tran, Email: katherine.tran@nih.gov.
Sergei A. Kuznetsov, Email: skuznetsov@dir.nidcr.nih.gov.
Pamela G. Robey, Email: probey@dir.nidcr.nih.gov.
Marianna Sabatino, Email: sabatinom@cc.nih.gov.
References
- Acosta JC, O’Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, Fumagalli M, Da Costa M, Brown C, Popov N, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–1018. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- Ashkar S, Weber GF, Panoutsakopoulou V, Sanchirico ME, Jansson M, Zawaideh S, Rittling SR, Denhardt DT, Glimcher MJ, Cantor H. Eta-1 (osteopontin): an early component of type-1 (cell-mediated) immunity. Science. 2000;287:860–864. doi: 10.1126/science.287.5454.860. [DOI] [PubMed] [Google Scholar]
- Banfi A, Muraglia A, Dozin B, Mastrogiacomo M, Cancedda R, Quarto R. Proliferation kinetics and differentiation potential of ex vivo expanded human bone marrow stromal cells: Implications for their use in cell therapy. Exp Hematol. 2000;28:707–715. doi: 10.1016/s0301-472x(00)00160-0. [DOI] [PubMed] [Google Scholar]
- Benisch P, Schilling T, Klein-Hitpass L, Frey SP, Seefried L, Raaijmakers N, Krug M, Regensburger M, Zeck S, Schinke T, et al. The transcriptional profile of mesenchymal stem cell populations in primary osteoporosis is distinct and shows overexpression of osteogenic inhibitors. PLoS One. 7:e45142. doi: 10.1371/journal.pone.0045142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccocioppo R, Bernardo ME, Sgarella A, Maccario R, Avanzini MA, Ubezio C, Minelli A, Alvisi C, Vanoli A, Calliada F, et al. Autologous bone marrow-derived mesenchymal stromal cells in the treatment of fistulising Crohn’s disease. Gut. 60:788–798. doi: 10.1136/gut.2010.214841. [DOI] [PubMed] [Google Scholar]
- Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, Nelson PS, Desprez PY, Campisi J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash NR, Dash SN, Routray P, Mohapatra S, Mohapatra PC. Targeting nonhealing ulcers of lower extremity in human through autologous bone marrow-derived mesenchymal stem cells. Rejuvenation Res. 2009;12:359–366. doi: 10.1089/rej.2009.0872. [DOI] [PubMed] [Google Scholar]
- Digirolamo CM, Stokes D, Colter D, Phinney DG, Class R, Prockop DJ. Propagation and senescence of human marrow stromal cells in culture: a simple colony-forming assay identifies samples with the greatest potential to propagate and differentiate. Br J Haematol. 1999;107:275–281. doi: 10.1046/j.1365-2141.1999.01715.x. [DOI] [PubMed] [Google Scholar]
- Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- Efron B, Hastie T, Johnstone I, Tibshirani R. Least angle regression. Annals of Statistics. 2004;32:407–499. [Google Scholar]
- Friedenstein AJ, Piatetzky S, II, Petrakova KV. Osteogenesis in transplants of bone marrow cells. J Embryol Exp Morphol. 1966;16:381–390. [PubMed] [Google Scholar]
- Galderisi U, Helmbold H, Squillaro T, Alessio N, Komm N, Khadang B, Cipollaro M, Bohn W, Giordano A. In vitro senescence of rat mesenchymal stem cells is accompanied by downregulation of stemness-related and DNA damage repair genes. Stem Cells Dev. 2009;18:1033–1042. doi: 10.1089/scd.2008.0324. [DOI] [PubMed] [Google Scholar]
- Hare JM, Traverse JH, Henry TD, Dib N, Strumpf RK, Schulman SP, Gerstenblith G, DeMaria AN, Denktas AE, Gammon RS, et al. A randomized, double-blind, placebo-controlled, dose-escalation study of intravenous adult human mesenchymal stem cells (prochymal) after acute myocardial infarction. J Am Coll Cardiol. 2009;54:2277–2286. doi: 10.1016/j.jacc.2009.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz EM, Gordon PL, Koo WK, Marx JC, Neel MD, McNall RY, Muul L, Hofmann T. Isolated allogeneic bone marrow-derived mesenchymal cells engraft and stimulate growth in children with osteogenesis imperfecta: Implications for cell therapy of bone. Proc Natl Acad Sci U S A. 2002;99:8932–8937. doi: 10.1073/pnas.132252399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inkson CA, Ono M, Bi Y, Kuznetsov SA, Fisher LW, Young MF. The potential functional interaction of biglycan and WISP-1 in controlling differentiation and proliferation of osteogenic cells. Cells Tissues Organs. 2009;189:153–157. doi: 10.1159/000151377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inkson CA, Ono M, Kuznetsov SA, Fisher LW, Robey PG, Young MF. TGF-beta1 and WISP-1/CCN-4 can regulate each other’s activity to cooperatively control osteoblast function. J Cell Biochem. 2008;104:1865–1878. doi: 10.1002/jcb.21754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang SS, Chen CH, Tseng KY, Tsai FY, Wang MJ, Chang IS, Lin JL, Lin S. Gene expression profiling suggests a pathological role of human bone marrow-derived mesenchymal stem cells in aging-related skeletal diseases. Aging (Albany NY) 3:672–684. doi: 10.18632/aging.100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce N, Annett G, Wirthlin L, Olson S, Bauer G, Nolta JA. Mesenchymal stem cells for the treatment of neurodegenerative disease. Regen Med. 5:933–946. doi: 10.2217/rme.10.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karussis D, Karageorgiou C, Vaknin-Dembinsky A, Gowda-Kurkalli B, Gomori JM, Kassis I, Bulte JW, Petrou P, Ben-Hur T, Abramsky O, et al. Safety and immunological effects of mesenchymal stem cell transplantation in patients with multiple sclerosis and amyotrophic lateral sclerosis. Arch Neurol. 67:1187–1194. doi: 10.1001/archneurol.2010.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Hernandez LD, Galan JE, Janeway CA, Jr, Medzhitov R, Flavell RA. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell. 2002;110:191–202. doi: 10.1016/s0092-8674(02)00827-9. [DOI] [PubMed] [Google Scholar]
- Koc ON, Day J, Nieder M, Gerson SL, Lazarus HM, Krivit W. Allogeneic mesenchymal stem cell infusion for treatment of metachromatic leukodystrophy (MLD) and Hurler syndrome (MPS-IH) Bone Marrow Transplant. 2002;30:215–222. doi: 10.1038/sj.bmt.1703650. [DOI] [PubMed] [Google Scholar]
- Koc ON, Gerson SL, Cooper BW, Dyhouse SM, Haynesworth SE, Caplan AI, Lazarus HM. Rapid hematopoietic recovery after coinfusion of autologous-blood stem cells and culture-expanded marrow mesenchymal stem cells in advanced breast cancer patients receiving high-dose chemotherapy. J Clin Oncol. 2000;18:307–316. doi: 10.1200/JCO.2000.18.2.307. [DOI] [PubMed] [Google Scholar]
- Ksiazek K. A comprehensive review on mesenchymal stem cell growth and senescence. Rejuvenation Res. 2009;12:105–116. doi: 10.1089/rej.2009.0830. [DOI] [PubMed] [Google Scholar]
- Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, Aarden LA, Mooi WJ, Peeper DS. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- Kukita T, Wada N, Kukita A, Kakimoto T, Sandra F, Toh K, Nagata K, Iijima T, Horiuchi M, Matsusaki H, et al. RANKL-induced DC-STAMP is essential for osteoclastogenesis. J Exp Med. 2004;200:941–946. doi: 10.1084/jem.20040518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafferty-Whyte K, Bilsland A, Cairney CJ, Hanley L, Jamieson NB, Zaffaroni N, Oien KA, Burns S, Roffey J, Boyd SM, et al. Scoring of senescence signalling in multiple human tumour gene expression datasets, identification of a correlation between senescence score and drug toxicity in the NCI60 panel and a pro-inflammatory signature correlating with survival advantage in peritoneal mesothelioma. BMC Genomics. 11:532. doi: 10.1186/1471-2164-11-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Blanc K, Rasmusson I, Sundberg B, Gotherstrom C, Hassan M, Uzunel M, Ringden O. Treatment of severe acute graft-versus-host disease with third party haploidentical mesenchymal stem cells. Lancet. 2004;363:1439–1441. doi: 10.1016/S0140-6736(04)16104-7. [DOI] [PubMed] [Google Scholar]
- Le Blanc K, Ringden O. Immunomodulation by mesenchymal stem cells and clinical experience. J Intern Med. 2007;262:509–525. doi: 10.1111/j.1365-2796.2007.01844.x. [DOI] [PubMed] [Google Scholar]
- Le Blanc K, Tammik L, Sundberg B, Haynesworth SE, Ringden O. Mesenchymal stem cells inhibit and stimulate mixed lymphocyte cultures and mitogenic responses independently of the major histocompatibility complex. Scand J Immunol. 2003;57:11–20. doi: 10.1046/j.1365-3083.2003.01176.x. [DOI] [PubMed] [Google Scholar]
- Mazzini L, Ferrero I, Luparello V, Rustichelli D, Gunetti M, Mareschi K, Testa L, Stecco A, Tarletti R, Miglioretti M, et al. Mesenchymal stem cell transplantation in amyotrophic lateral sclerosis: A Phase I clinical trial. Exp Neurol. 223:229–237. doi: 10.1016/j.expneurol.2009.08.007. [DOI] [PubMed] [Google Scholar]
- Okabe Y, Sano T, Nagata S. Regulation of the innate immune response by threonine-phosphatase of Eyes absent. Nature. 2009;460:520–524. doi: 10.1038/nature08138. [DOI] [PubMed] [Google Scholar]
- Ono M, Inkson CA, Kilts TM, Young MF. WISP-1/CCN4 regulates osteogenesis by enhancing BMP-2 activity. J Bone Miner Res. 26:193–208. doi: 10.1002/jbmr.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskowitz A, McFerrin H, Gutschow M, Carter ML, Pochampally R. Serum-deprived human multipotent mesenchymal stromal cells (MSCs) are highly angiogenic. Stem Cell Res. 6:215–225. doi: 10.1016/j.scr.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal R, Venkataramana NK, Bansal A, Balaraju S, Jan M, Chandra R, Dixit A, Rauthan A, Murgod U, Totey S. Ex vivo-expanded autologous bone marrow-derived mesenchymal stromal cells in human spinal cord injury/paraplegia: a pilot clinical study. Cytotherapy. 2009;11:897–911. doi: 10.3109/14653240903253857. [DOI] [PubMed] [Google Scholar]
- Pitto L, Rizzo M, Simili M, Colligiani D, Evangelista M, Mercatanti A, Mariani L, Cremisi F, Rainaldi G. miR-290 acts as a physiological effector of senescence in mouse embryo fibroblasts. Physiol Genomics. 2009;39:210–218. doi: 10.1152/physiolgenomics.00085.2009. [DOI] [PubMed] [Google Scholar]
- Ripperger T, von Neuhoff N, Kamphues K, Emura M, Lehmann U, Tauscher M, Schraders M, Groenen P, Skawran B, Rudolph C, et al. Promoter methylation of PARG1, a novel candidate tumor suppressor gene in mantle-cell lymphomas. Haematologica. 2007;92:460–468. doi: 10.3324/haematol.10337. [DOI] [PubMed] [Google Scholar]
- Sabatino M, Ren J, David-Ocampo V, England L, McGann M, Tran M, Kuznetsov SA, Khuu H, Balakumaran A, Klein HG, et al. The establishment of a bank of stored clinical bone marrow stromal cell products. J Transl Med. 10:23. doi: 10.1186/1479-5876-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Saleem KS, Suzuki W, Tanaka K, Hashikawa T. Connections between anterior inferotemporal cortex and superior temporal sulcus regions in the macaque monkey. J Neurosci. 2000;20:5083–5101. doi: 10.1523/JNEUROSCI.20-13-05083.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schallmoser K, Bartmann C, Rohde E, Bork S, Guelly C, Obenauf AC, Reinisch A, Horn P, Ho AD, Strunk D, et al. Replicative senescence-associated gene expression changes in mesenchymal stromal cells are similar under different culture conditions. Haematologica. 95:867–874. doi: 10.3324/haematol.2009.011692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethe S, Scutt A, Stolzing A. Aging of mesenchymal stem cells. Ageing Res Rev. 2006;5:91–116. doi: 10.1016/j.arr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Siddappa R, Licht R, van Blitterswijk C, de Boer J. Donor variation and loss of multipotency during in vitro expansion of human mesenchymal stem cells for bone tissue engineering. J Orthop Res. 2007;25:1029–1041. doi: 10.1002/jor.20402. [DOI] [PubMed] [Google Scholar]
- Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-ArrayTools. Cancer Inform. 2007;3:11–17. [PMC free article] [PubMed] [Google Scholar]
- Stenderup K, Justesen J, Clausen C, Kassem M. Aging is associated with decreased maximal life span and accelerated senescence of bone marrow stromal cells. Bone. 2003;33:919–926. doi: 10.1016/j.bone.2003.07.005. [DOI] [PubMed] [Google Scholar]
- Tanabe S, Sato Y, Suzuki T, Suzuki K, Nagao T, Yamaguchi T. Gene expression profiling of human mesenchymal stem cells for identification of novel markers in early- and late-stage cell culture. J Biochem. 2008;144:399–408. doi: 10.1093/jb/mvn082. [DOI] [PubMed] [Google Scholar]
- Wagner W, Ho AD, Zenke M. Different facets of aging in human mesenchymal stem cells. Tissue Eng Part B Rev. 16:445–453. doi: 10.1089/ten.TEB.2009.0825. [DOI] [PubMed] [Google Scholar]
- Wagner W, Horn P, Castoldi M, Diehlmann A, Bork S, Saffrich R, Benes V, Blake J, Pfister S, Eckstein V, et al. Replicative senescence of mesenchymal stem cells: a continuous and organized process. PLoS One. 2008;3:e2213. doi: 10.1371/journal.pone.0002213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Jang DJ. Protein kinase CK2 regulates cytoskeletal reorganization during ionizing radiation-induced senescence of human mesenchymal stem cells. Cancer Res. 2009;69:8200–8207. doi: 10.1158/0008-5472.CAN-09-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Jin P, Sabatino M, Ren J, Civini S, Bogin V, Ichim TE, Stroncek DF. Comparison of endometrial regenerative cells and bone marrow stromal cells. Journal of translational medicine. 2012;10:207. doi: 10.1186/1479-5876-10-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zheng M, Liu G, Xia W, McKeown-Longo PJ, Hung MC, Zhao J. Kruppel-like factor 8 induces epithelial to mesenchymal transition and epithelial cell invasion. Cancer Res. 2007;67:7184–7193. doi: 10.1158/0008-5472.CAN-06-4729. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Simon R. Development and validation of predictive indices for a continuous outcome using gene expression profiles. Cancer Inform. 9:105–114. doi: 10.4137/cin.s3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The concentrations of 14 proteins in the supernatant of BMSCs from 2 donors 09FC20 (red line) and W10003 (green line) were shown. The X-axis indicates BMSC passage (P) and the y-axis indicates the concentrations of proteins expressed as pg/ml.
Dataset GSE7888 obtained from serial passages of BMSCs was downloaded and imported into BRBArray-tools, the log-intensity of the predictive genes was acquired, and the percentage of maximum lifespan of BMSCs at each passage was calculated using our prediction model. Each plot displayed the results of one of six donors, which were labeled by the names used in the original database. In each plot, the x-axis indicates the passage number of individual samples and the y-axis indicates the calculated percentage of maximum lifespan, which denote the “age” of BMSC.
Dataset GSE9593 obtained from serial passages of BMSCs was downloaded and imported into BRBArray-tools, the log-intensity of the predictive genes was acquired, and the percentage of maximum lifespan of BMSCs at each passage was calculated using our prediction model. Each plot displayed the results of one of three donors, which were labeled by the names used in the original database. In each plot, the x-axis indicates the passage number of individual samples and the y-axis indicates the calculated percentage of maximum lifespan, which denote the “age” of BMSC.