Abstract
Cysteine S-nitrosylation is a post-translational modification regulating protein function and nitric oxide signaling. Herein the selectivity, reproducibility, and sensitivity of a mass spectrometry-based proteomic method for the identification of endogenous S-nitrosylated proteins is outlined. The method enriches for either S-nitrosylated proteins or peptides through covalent binding of the cysteine sulfur with phenylmercury at pH=6.0. Phenylmercury reacts selectively and efficiently with S-nitrosocysteine since no reactivity can be documented for disulfides, sulfinic or sulfonic acids, S-glutathionylated, S-alkylated or S- sulfhydrylated cysteine residues. A specificity of 97 ± 1 % for the identification of S-nitrosocysteine peptides in mouse liver tissue is achieved by the inclusion of negative controls. The method enables the detection of 36 S-nitrosocysteine peptides starting with 5 pmoles S-nitrosocysteine/mg of total tissue protein. Both the percentage of protein molecules modified as well as the occupancy by S-nitrosylation can be determined. Overall, selective, sensitive and reproducible enrichment of S-nitrosylated proteins and peptides is achieved by the use of phenylmercury. The inclusion of appropriate negative controls secures the precise identification of endogenous S-nitrosylated sites and proteins in biological samples.
Keywords: cysteine modification, mass spectrometry, nitric oxide, protein S-nitrosylation
Introduction
Protein S-nitrosylation, the covalent addition of a nitric oxide equivalent to a reduced thiol, is considered the major non-soluble guanylate cyclase-regulated mechanism for the biological functions of nitric oxide in vivo[1–6]. Experimental data indicated that S-nitrosylation regulates enzymatic activity, alters the topology of modified proteins and influences protein-protein interactions in vivo [7–12]. Despite these important findings that highlight the biological significance of protein S-nitrosylation, critical aspects such as the mechanism(s) of S-nitrosylation in vivo, the factors that govern its selectivity, the dependency of the modification on different isoforms of nitric oxide synthases (NOS), as well as the physiological function of this protein modification require further exploration. To gain insights into these areas of investigation the global identification of endogenous S-nitrosoproteome(s) would be exceptionally valuable. Over the last decade the identification of S-nitrosylated proteins has been achieved primarily by the use of the original and several variations of the biotin switch method [13–16]. Other methods based on chemical enrichment have been also employed (for recent reviews ref [17–19]). We used the principles of the Saville reaction [20] to develop a mass spectrometry-based proteomic method for the precise identification of protein S-nitrosocysteine sites in vivo [21]. As with every method, it is imperative that the selectivity, specificity, reproducibility and the quantitative capacity are explored in detail. In this article we report that the phenylmercury-assisted capture provides selective, sensitive and reproducible enrichment for S-nitrosylated proteins and peptides present in complex biological samples. This chemical enrichment coupled with mass spectrometric identification enables the precise mapping of endogenous S-nitrosoproteomes.
Materials and methods
Chemicals and reagents
Bovine insulin solution (10 mg/mL), rabbit glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) and mouse monoclonal anti-GAPDH antibody were purchased from Sigma-Aldrich (St Louis, MO). All chemicals used were of analytical grade.
Glyceraldehyde 3-phosphate dehydrogenase treatment
Rabbit glyceraldehyde-3-phosphate dehydrogenase at concentration of 5µg/µl (140 µM) was exposed to 10 equivalents of S-nitrosoglutathione (GSNO), oxidized glutathione (GSSG), hydrogen peroxide (H2O2), N-ethlymaleimide (NEM) and sodium hydrosulfide (NaHS) for 30 min at room temperature in the dark. The excess reagents were removed by micro bio-spin chromatography columns (Biorad, Hercules, CA) according to manufacturer instructions. Protein concentration was determined by BCA assay and samples were stored at −80°C until use.
Evaluation of displacement capacity of phenylmercury resin
Liver homogenates were exposed to 2 µM GSNO for 30 min in triplicate (N = 3). The homogenate was divided equally between two tubes and blocked with S-methyl methanethiosulfonate (MMTS). Then, one sample was mixed for 1 hr at room temperature with 2ml activated phenylmercury resin whereas the second left untreated. The phenylmercury resin exposed sample was centrifuged at 1000 × g for 5 min and the supernatant was collected. Total protein was normalized between samples and protein S-nitrosocysteine content was quantified by reductive chemistries coupled to ozone-based chemiluminescence as previously described [22]. The limit of detection for this experiment was 3 pmoles of S-nitrosocysteine per mg of total protein.
In solution digestion of modified GAPDH and mass spectrometric analysis
Mapping of modified cysteine residues on GAPDH as a result of treatment with NEM, GSNO, GSSG, NaHS and H2O2 respectively was facilitated by in solution digestion of GAPDH preparations (0.5 µg/µl in 50 mM NaHCO3) with trypsin at ratio of 1:100 enzyme to protein for 6 h at 37°C. Five µl of each tryptic digest (40ng/µl) were analyzed by mass spectrometry as previously described [21].
Phenylmercury resin assisted capture
Phenylmercury resin assisted capture was performed as previously described [21]with a minor modification. Sixty µg of protein mixture (1:1 ratio of GAPDH and insulin) in 250mM Hepes, 1mM DTPA, 0.1mM neocupreine, 2.5% SDS, pH 7.7 reacted with 35 mM MMTS for 30 min at 50 °C. The blocking buffer was exchanged to 250 mM MES, 1 mM DTPA pH 6.0 by micro bio-spin chromatography columns. Sodium docecyl sulfate was added to protein preparations at concentration of 1% and protein suspensions were loaded onto activated organic mercury columns for 1 h at room temperature. The unbound fraction was collected, columns were extensively washed according to our standard protocol [23]and bound proteins were eluted into 4 ml β-mercaptoethanol. Both bound and unbound fractions were concentrated using 3kDa cut off microconcentrators (Millipore, Billerica, MA). For western blot analysis against GAPDH, 1 µg of unbound and 20% of the bound fraction respectively were used. Densitometric analysis was employed to quantify the amount of GAPDH present in bound and unbound fractions based on the fluorescence intensity of a series of pure GAPDH loaded onto the same gel. For bovine insulin 1 µg of unbound fraction and 50% of bound fraction were separated on SDS protein gel. To preserve disulfide bonds on insulin, inclusion of β-mercaptoethanol in the LDS (lithium docedyl sulfate) loading buffer as well as boiling of the samples was omitted. Proteins were resolved using MES SDS running buffer instead of MOPS SDS running buffer. The gel was stained with silver using SilverQuest kit (Invitrogen, Carlsbad, CA) according to the manufacturer instructions. For the identification of the sites of modification, proteins were processed for phenylmercury assisted capture followed by on column trypsin digestion and performic acid elution as previously described [21].
Results and discussion
Chemical selectivity and efficiency of phenylmercury for S-nitrosocysteine
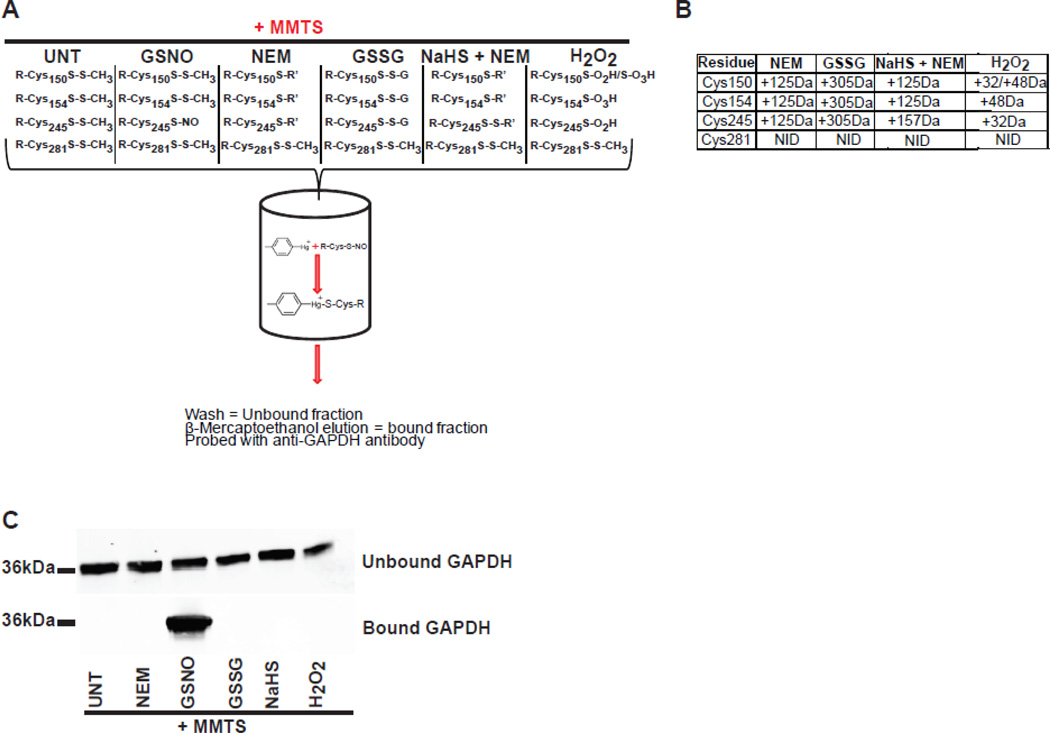
The method relies on the chemical reactivity of phenylmercury towards S-nitrosocysteine which was originally described by Saville [20]. The reaction of phenylmercury with S-nitrosocysteine displaces nitric oxide forming a covalent bond between the mercury and the sulfur atom. The phenylmercury resin is synthesized by conjugating ρ-amino-phenylmercuric acetate to N-hydroxysuccinimide- activated Affi-Gel 10 agarose beads. The reactive group, phenylmercury, is activated by base hydrolysis of the acetate group at pH 8.8. Phenylmercury has been used previously for the specific capture of proteins with reduced cysteine residues [24]. Therefore it is essential that reduced cysteine residues are blocked effectively. This is achieved by S-methylsulfonylation of reduced cysteine residues with MMTS. We previously reported that the efficiency of S-methylsulfonylation is 99.95% in biological samples and that S-methylsulfonylated cysteine residues do not react with phenylmercury [25]. However, in biological samples, other modifications of cysteine residues that do not react with MMTS are present and are potentially reactive towards phenylmercury. Known post- translational modifications of cysteine residues include oxidation to disulfide, oxidation to sulfinic or sulfonic acids, S-glutathionylation, S-alkylation, and S- sulfhydrylation [17]. Therefore the selectivity of phenylmercury for the detection of S-nitrosocysteine was tested with model proteins. First the reactivity of phenylmercury with bovine insulin was tested. Insulin was selected because of the presence of three well-characterized disulfide bonds and the absence of reduced cysteine residues in the structure of the protein. Phenylmercury does not react with insulin as evident by; i) the absence of insulin in silver stained protein gel of the bound fraction eluted from the phenylmercury resin and ii) the nonexistence of insulin-derived cysteine containing peptides after mass spectrometric analysis of trypsin digested bound fraction (data not shown). Next, the reactivity of the phenylmercury with purified rabbit glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was tested. GAPDH was selected since four reduced cysteine residues at positions 150, 154, 245 and 281 are present in the crystal structure of the protein (Uniprot accession number P46046) and previous studies have determined that some of these residues particularly residues 150 and 245 are modified by S-nitrosylation in vivo or in cellular model systems [26,27]. Using well-described chemical modifiers we generated GAPDH with altered cysteine residues (Figure 1A). The site-specific modified GAPDH proteins were characterized by mass spectrometry. Reacting GAPDH with N-ethylmaleimide generated alkylated cysteine residues at Cys150, Cys154 and Cys245 (Figure 1B). Upon incubation of GAPDH with oxidized glutathione (GSSG), S-glutathionylated adducts on cysteine residues Cys150, Cys154 and Cys245 were detected (Figure 1B). Sodium hydrosulfate (NaHS) was used to generate S-sulfhydrylated GAPDH. S-Sulfhydrylated thiol (S-SH) is prone to oxidation making its detection by mass spectrometry challenging. Therefore, to prevent further oxidation of S-sulfhydrylated GAPDH the protein was treated with N-ethylmaleimide which alkylates S-sulfhydrylated thiols [28]. S-Sulfhydrylated cysteine residues were detected by mass spectrometry having an additional mass of 157 Da corresponding to the S-N-ethylmaleimide adducts. Cysteine 245 was detected with an additional mass of 157 Da whereas cysteine residues Cys150 and Cys154 were detected with additional mass of 125 Da (Figure 1B) indicating that under these experimental conditions only cysteine 245 is modified by S-sulfhydrylation. Hydrogen peroxide treatment of GAPDH generated sulfinic and sulfonic acids on residues Cys150, sulfonic acid on Cys154 and sulfinic acid on Cys245 (Figure 1B). Finally, treatment with S-nitrosoglutathione (GSNO) resulted in S-nitrosylated GAPDH on cysteine residue 245 (Figure 2B and 2C). These differently modified GAPDH preparations were used to test their reactivity with phenylmercury resin. Only S-nitrosylated GAPDH reacted with phenylmercury as documented by: i) the presence of GAPDH in the bound fraction (Figure 1C) and ii) by mass spectrometric detection of VPTPNVSVVDLTC245R peptide (Figure 2B and 2C). By employing the same methodologies, none of the differently modified cysteine residues were detected indicating the selectivity of the phenylmercury for S-nitrosocysteine.
Figure 1. Phenylmercury resin selectively enriches for S-nitrosylated proteins.
(A) Schematic representation of phenylmercury assisted capture of S-nitrosylated proteins. GAPDH preparations react with MMTS which S-methylsulfonate reduce thiols and prevents their subsequent reaction with phenylmercury. MMTS-blocked GAPDH preparations are loaded onto activated phenylmercury-resin-containing columns and one hour later the resin is washed extensively to remove the unbound proteins. Bound proteins are released by β-mercaptoethanol. (B) Mapping of cysteine modifications on GAPDH after different chemical treatments. GAPDH, treated with different chemical agents was digested in solution with trypsin and the resulting peptides were identified by LC-MS/MS. These peptides were identified having a mass indicative of the predicted mass shift expected for the chemical treatments. Cysteine containing peptides with an additional mass of 29 Da, corresponding to nitric oxide adduct on reduced cysteine, were not identified due to the labile nature of S-nitrosocysteine bond. NID; none identified. (C) Representative western blot using antibodies against GAPDH in bound and unbound fractions collected after phenylmercury resin-assisted capture. Note the presence of immunoreactivity in the bound fraction corresponding to GAPDH treated with GSNO indicating that phenylmercury resin selectively enriches for S-nitrosocysteine.
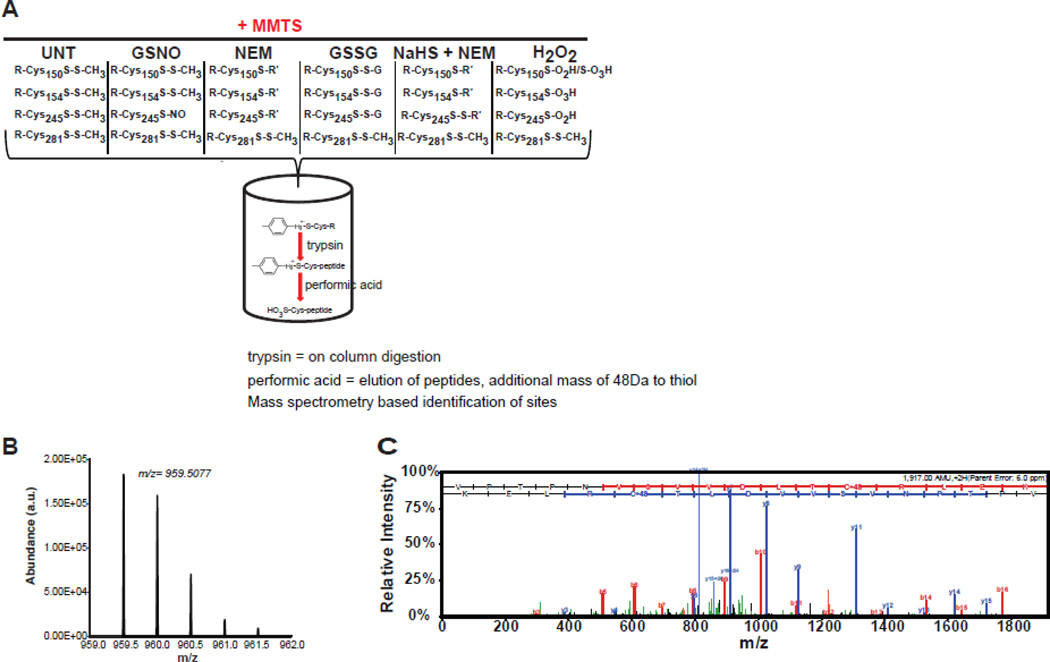
Figure 2. Phenylmercury resin selectively enriches for S-nitrosocysteine.
(A) Schematic representation of phenylmercury resin-assisted identification of S-nitrosocysteine peptides. After covalent attachment of S-nitrosylated proteins and extensive washes bound proteins are subjected to on-column trypsin digestion followed by extensive washes to remove unbound peptides. Cysteine containing peptides are released from the resin using performic acid which cleaves the mercury-thiol bond and oxidizes thiol to sulfonic acid (+48 Da). The sites of modification are identified by mass spectrometric analysis of eluted peptides. (B) Representative MS spectra of doubly-charged sulfonic acid-containing tryptic peptide VPTPNVSVVDLTC245R from GAPDH acquired in the bound fraction after mercury assisted capture of GSNO-treated GAPDH. (C) MS/MS spectra confirmed the sequence and site of sulfonic acid-containing peptide from GAPDH. MS/MS spectra passed automatic and manual filter criteria and were identified with high SEQUEST cross correlation (Xc) score. The experiments presented to this figure were repeated once more with identical results.
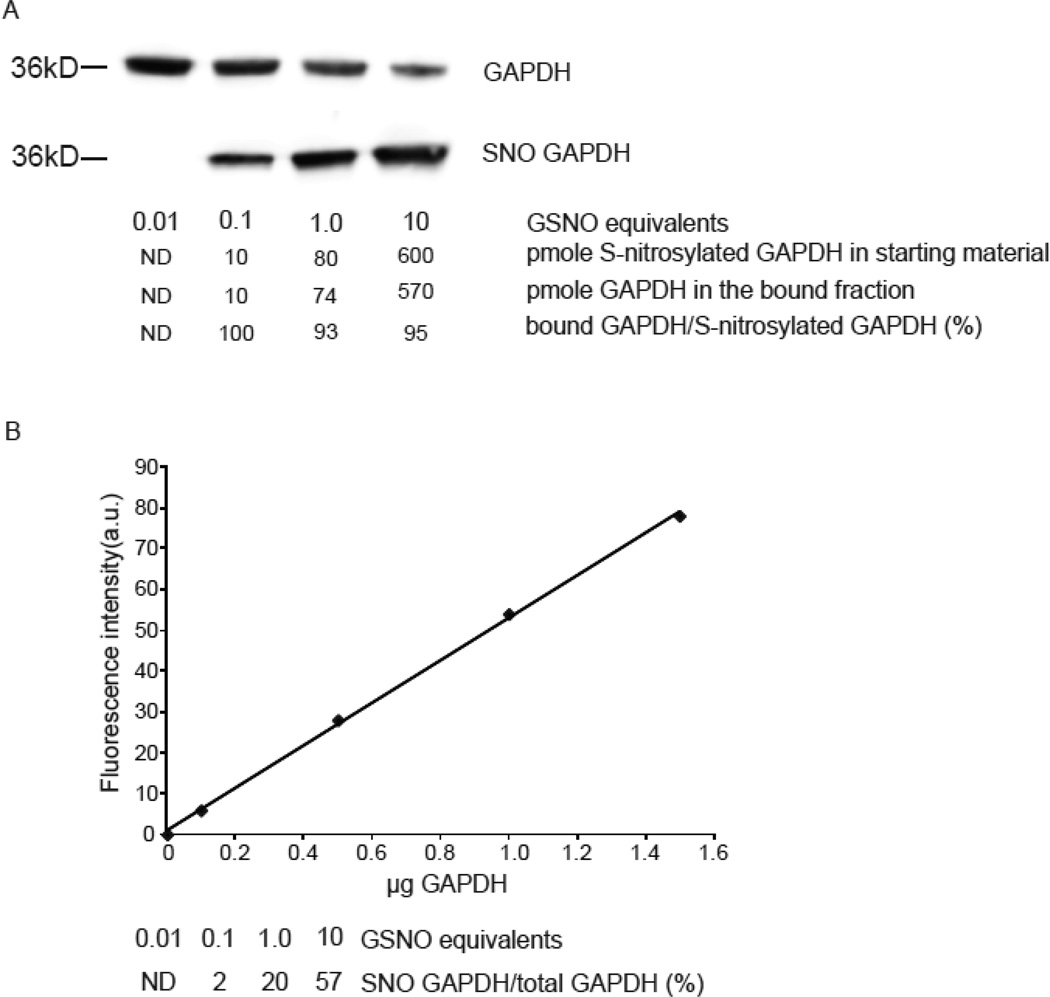
The binding efficiency of phenylmercury resin for S-nitrosocysteine was evaluated by generating GAPDH with various levels of S-nitrosylation (Figure 3A). These preparations of GAPDH were reacted with phenylmercury and the bound and unbound fractions were quantified by western blot using a standard curve constructed with purified GAPDH. Based on the densitometric analysis, it was determined that 2, 20 and 57 percent of the molecules of GAPDH were captured by phenylmercury resin (Figure 3B). Using molecular mass of 36 kDa for GAPDH the amount of bound protein was converted to pmole of bound GAPDH. These values were compared with the levels of S-nitrosylated GAPDH that were independently measured in the protein used for input using reduction and chemiluminescence detection of S-nitrosylated GAPDH. It was found that across the three samples, bound GAPDH represented 96 ± 4 % of S-nitrosylated GAPDH that was present in the original preparations. Therefore the phenylmercury resin enriches for proteins with S-nitrosocysteine with high efficiency even with a starting S-nitrosocysteine concentration as low as 0.2 pmole per µg of protein.
Figure 3. Phenylmercury resin enriches quantitatively for S-nitrosocysteine.
(A) Representative western blot with anti-GAPDH antibodies in bound and unbound fractions collected after phenylmercury resin-assisted capture. GAPDH (140 µM) was exposed to the indicated equivalents of GSNO and 50 µg from each preparation was used for phenylmercury-assisted capture. To avoid signal saturation, 1 µg from the unbound fraction was loaded to the gels. For the same reason, the bound fractions were loaded as follows; 100%, 100%, 20% and 10% of the volume of bound fraction corresponded to GAPDH treated with 0.01, 0.1, 1 and 10 equivalents of GSNO respectively. The levels of S-nitrosocysteine per µg of protein were 0.2, 1.6 and 12 pmole as quantified by reduction of S-NO bond followed by chemiluminescence-based detection of liberated NO equivalents. (B) GAPDH was used to titrate antibody binding and construct this standard curve. Densitometric analysis was employed to determine the fraction of GAPDH present in the bound and unbound fractions. The experiment repeated once more with similar results.
In addition, the efficiency of phenylmercury resin to displace cysteine-bound nitric oxide in complex mixtures was evaluated by reduction coupled to ozone-based chemiluminescence detection of S-nitrosocysteine. Incubation of GSNO-exposed, MMTS-blocked mouse liver homogenate with phenylmercury resin resulted in a 94.4 ± 1.4 % decrease in protein S-nitrosocysteine levels. Collectively the data indicate that phenylmercury displaces nitric oxide from S-nitrosocysteine leading to selective and efficient capture of S-nitrosylated proteins and peptides.
Application of the mercury resin assisted capture of S-nitrosylated proteins in complex biological samples
The application phenylmercury resin-assisted capture for the identification of S-nitrosylated proteins in complex biological mixtures requires three basic steps [21]. Initially, the reduced cysteine residues in the biological sample are S-methylsulfonated by reacting with MMTS. Blocked-protein homogenates react with phenylmercury, which displaces nitric oxide and forms a covalent bond with sulfur immobilizing proteins on the resin. Bound proteins are released intact using reduction of the mercury-sulfur bond with β-mercaptoethanol. The bound fraction can be then separated on regular SDS-PAGE gels followed by immunodetection of specific proteins provided that antibodies are available. Alternatively for the global identification of proteins in the bound fraction, the proteins are separated a short distance (2 cm) by SDS-PAGE followed by in-gel trypsin digestion and LC-MS/MS-based identification of the resulting peptides. Additional approaches can be used for reducing the complexity of the bound fraction and facilitate the identification of low abundance proteins. For example, a typical workflow includes the separation of the tryptic peptides to ten fractions by ion-exchange chromatography, followed by separation of each of the ten fractions by reverse-phase chromatography and MS/MS detection. Proteins are considered for further analysis only if at least two peptides that pass our previously described stringent criteria are detected [29].
However, we cannot rely on the positive identification of these proteins as being S-nitrosylated without knowing the site of modification. Therefore, we simultaneously perform peptide capture as follows. After intact proteins are bound to the phenylmercury resin, the proteins are digested on column with trypsin followed by extensive washes of the resin. The bound peptides are then eluted from the resin by performic acid oxidation. Mild performic acid releases the bound peptides and more importantly oxidizes cysteine sulfur to sulfonic acid (+48Da). Under electrospray ionization, sulfonic acid-containing peptides are stable and yield typical y- and b-ion peptide fragments pairs from collision-induced dissociation (CID) as would be observed for cysteine carbamidomethyl-containing peptides. At times the presence of sulfonic acid may also generate increased confidence in peptide database search scores [30,31]. However, for tryptic peptides with more than one cysteine residues the use of performic acid for elution does not permit the discrimination of which residue was modified by S-nitrosylation. This situation was encountered during the analysis of endogenous S-nitrosocysteine proteomes and it appears to be a rare occurrence as less than 1% peptides identified in the liver S-nitrosocysteine proteome had more than one cysteine residues [21]. The post mass spectrometry data analysis includes the generation of DTA files by extracting the MS/MS spectra from RAW files (Figure 4). DTA files are submitted to Sorcerer-SEQUEST for performing the database search against a Uniprot reference database. We perform database searches including cysteine sulfonic acid as static modification since we have determined that performic acid treatment oxidizes 100% of cysteine sulfur to sulfonic acid. Potential sequence-to-spectrum peptide assignments generated by Sorcerer-SEQUEST are loaded into Scaffold to validate protein identifications and perform manual inspection of MS/MS spectra as well as to compare protein identifications across experimental conditions. While this secures the precise identification of the S-nitrosocysteine residues, it does not control for inclusion of false positive identifications that may result from incomplete blocking of reduced cysteine residues or non-specific interactions with the resin during enrichment. To secure the specificity of the identification each sample analyzed is paired with a negative control that is processed in parallel with the sample. Negative controls are generated either by chemical treatment (dithiothreitol (DTT), Cu/ascorbate and HgCl2) or by UV-light illumination. Displacement of nitric oxide from S-nitrosocysteine residues by pretreatment of biological samples with UV, DTT, Cu-ascorbate, or HgCl2 decreased protein S-nitrosocysteine levels by greater than 99% (quantified by ozone-chemiluminescence) and also eliminated reaction with the phenylmercury compounds [21]. All cysteine containing peptides that were identified in the negative controls were considered as false positive and were removed from the final analysis. The number of peptides identified in the negative samples was used for the calculation of the false identification rate (FIR) of the method. Using wild type mouse liver homogenates we calculated 3 ± 1% FIR across seven biological replicates. Besides liver, we calculated the false identification rate in wild type mouse heart, kidney, lung and thymus using three biological replicates. In these organs the FIR did not exceed 3%. This FIR is in agreement and compare favorably with the greater than 90% specificity reported for immobilized metal affinity chromatography used for the enrichment of phosphopeptides from mouse liver [32]. As an additional measure of method specificity we considered the non-cysteine peptides that were identified during each run. Across the five mouse organs these peptides represented lower than 2% of the total peptides identified. Based on these results we conclude that the specificity of the method is equal or greater than 95% for complex biological samples.
Figure 4. Workflow of LC-MS/MS post-acquisition data analysis.
RAW files generated by LC-MS/MS analysis from protein (left) and peptide (right) capture experiments are filtered according to the criteria presented here in order to extract the MS/MS spectra. DTA files, comprise the MS/MS spectra, are searched by Sorcerer-SEQUEST against a forward-reverse reference database. Potential sequence-to-spectrum peptide assignments generated by Sorcerer-SEQUEST are loaded into Scaffold (Proteome Software, Portland, OR) to validate protein identifications and perform manual inspection of MS/MS spectra as well as to compare protein identifications across experimental conditions. Proteins are considered for further analysis if at least two peptides passing the criteria for acceptance are identified. Peptides are considered for further analysis if meet all the criteria presented in this figure.
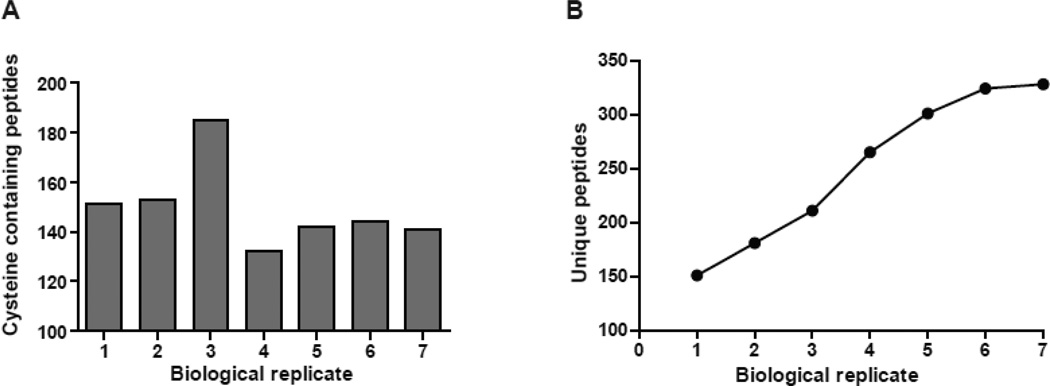
The technical reproducibility of the method was determining as follows; the same liver homogenate was analyzed through phenylmercury assisted protein and peptide capture three times. The technical reproducibility for each triplicate was calculated by dividing the number of proteins/sites identified in that sample by the number of proteins/sites identified across all three replicates. The technical reproducibility of the method is defined as the average ± standard deviation (SD) of the values calculated for each triplicate. According to this formula technical reproducibility of 80 ± 3% and 81 ± 2% for proteins and peptides respectively was calculated. This technical reproducibility compares well with reported variance of mass spectrometry based methods for the identification of protein acetylation and glycation {{226 Choudhary,C. 2009};[33]. In addition, we determined the minimum number of biological replicates that is required to cover at least 95% of the endogenous S-nitrosoproteome (Figure 5). In a typical experiment starting with 3 mg of liver homogenate, corresponding to 150 pmole S-nitrosocysteine resulted in the identification of nearly 150 cysteine-containing peptides per sample (Figure 5A). Under these conditions the analysis of seven different biological replicates resulted in the acquisition of 328 S-nitrosocysteine peptides from a wild type mouse liver (Figure 5B). In subsequent experiments we increased the protein concentration of the starting material to 30 mg and were able to identify the same number of cysteine containing peptides using only three biological replicates. As long as there are no limitations in tissue availability, higher starting protein concentrations will reduce the number of biological replicates required to achieve comprehensive coverage of the S-nitrosocysteine proteome under the current workflow.
Figure 5. Biological reproducibility of mercury assisted capture.
Wild-type mouse liver homogenates were processed for protein and peptide capture through phenylmercury assisted capture. Cysteine-containing peptides were matched to their corresponding proteins. (A) Number of cysteine containing peptides that were matched in seven different homogenates. (B) Number of unique cysteine-containing peptides identified across seven liver homogenates. Note that the number of unique peptides did not increase significantly (<5%) between the sixth and seventh biological replicate indicating that under these experimental conditions six biological replicates are required to acquire the endogenous S-nitrosoproteome.
The sensitivity and selectivity of the method in complex biological mixtures was further evaluated by the use of liver homogenates from mice lacking the endothelial nitric oxide synthase (eNOS−/− mice). The absence of eNOS from the liver results in diminished generation of nitric oxide, which in turn affects protein S-nitrosylation. Indeed, the liver homogenates from eNOS−/− mice had only 5 ± 5 pmoles S-nitrosocysteine per mg of protein, 10 fold lower as compared to wild-type mice. This resulted in a significant reduction in the number of peptides identified by the method as only 36 S-nitrosocysteine peptides were detected across three biological replicates from eNOS null mouse liver [15,21]. Collectively, these experiments show that the reaction of protein S-nitrosocysteine with the phenylmercury is specific, efficient, and through the inclusion of negative controls, achieved selective identification of endogenous nitrosocysteine peptides, the ultimate qualifier for the unambiguous assignment of S-nitrosylated proteins. Since our primary goal is to broaden the discovery and increase the depth of the in vivo S-nitrosocysteine proteome while minimizing bias due to biological variance, we devise the following work flow: (a) From all biological replicates, sulfonic acid-containing peptides, identified by SEQUEST database searches, are pooled (from this list peptides present in the corresponding UV-pretreated samples are removed). Although protein identification can be accomplished from a single peptide we have elected to increase the confidence by matching the single peptide with the modified cysteine to proteins identified by the same method but independently of the peptides. (b) Therefore, all proteins identified by protein capture were pooled (those also identified in the UV-pretreated samples were removed). By matching sulfonic acid-containing peptides with corresponding protein identifications, we precisely pinpoint the site of modification and independently match the modified peptides with protein identification providing additional confidence for the correct identification of the modified proteins.
The method was applied to multiple organs from wild type mouse to precisely identify 1011 S-nitrosocysteine sites in 468 proteins [27]. The data showed that the method is capable of covering a reasonable dynamic range in terms of molecular mass and distribution in different tissue organelles including the nucleus [27]. Ontological and pathway analysis coupled with site-directed mutagenesis and structure-function analysis uncover a potential role of S-nitrosylation in the regulation of fatty acid β-oxidation in the mitochondria. The method has been also applied to tissues of mice lacking S-nitrosoglutathione reductase (GSNOR−/−), the enzyme that metabolizes GSNO in vivo [34], revealing a potential role of S-nitrosylation in the regulation of apoptotic cell death in the thymus [35].
Quantification of the occupancy of cysteine residues by S-nitrosylation in vivo
The method can be utilized for label free quantification of endogenous S-nitrosylation sites by mass spectrometry. For this purpose a reference proteome which comprised all phenylmercury-reactive cysteine residues present in the biological sample was generated by omitting the blocking step with MMTS. For the identification of S-nitrosylation sites MMTS-blocked homogenate is processed in parallel as described above. The cysteine containing peptides that are identified in both samples are extracted from the base peak chromatogram and their current intensities are divided. The ratio of ion current intensity multiplied by 100 represents the occupancy by S-nitrosylation for this site. We applied this approach to determine the occupancy by S-nitrosylation for GAPDH in wild type mouse liver homogenates. Two cysteine-containing peptides for GAPDH were identified in both blocked and unblocked homogenates (Table 1). These peptides were extracted from the base peak chromatograms and their current intensities were compared. The occupancy by S-nitrosylation for the peptide that contains Cys150 and Cys154 is 6 ± 3% whereas the peptide that contains Cys245 is occupied by 1.0 ± 0.4 % under physiological conditions. S-nitrosylation of Cys150 has been reported to have implications for the protein-protein interactions and cellular localization of GAPDH [26]. These protein-protein interactions and translocation of the S-nitrosylated GAPDH to the nucleus have been found to influence signaling events and determine the fate of neurons [36]. The biological implications of S-nitrosylation at Cys245 have not been examined. We speculate that poly-S-nitrosylation, in accordance with the known effects of other modifications such as lysine acetylation [37], may promote conformational changes which in turn influence the protein interactions, stability and trafficking of GAPDH.
Table 1. Occupancy by S-nitrosylation of GAPDH in vivo.
Liver homogenates were processed for phenylmercury assisted capture according to the standard protocol (blocked) or by omitting the blocking step with MMTS (unblocked). In the blocked homogenate are identified S-nitrosylated cysteine residues only whereas in the unblocked homogenate are identified all phenylmercury-reactive cysteine residues, i.e. S-nitrosylated plus reduced. Cysteine containing peptides that were identified for GAPDH in both samples were extracted from the base peak chromatogram and their intensities were divided. The ratio of blocked/unblocked multiplied by 100 represents the occupancy by S-nitrosylation for each cysteine containing peptide.
| Peptide sequence | Intensity blocked(× E+06) | Intensity unblocked (× E+08) | Occupancy % |
|---|---|---|---|
| IVSNASC150TTNC154LAPLAK | 7.6 ± 1.2 | 1.3 ± 0.1 | 6 ± 3 |
| VPTPNVSVVDLTC245R | 9.7 ± 1.4 | 9.7 ± 0.5 | 1 ± 0.4 |
Significance.
The current study describes a selective, sensitive and reproducible method for the acquisition of endogenously S-nitrosylated proteins and peptides. The acquisition of endogenous S-nitrosoproteomes provides robust data that is necessary for the investigating mechanism(s) of S-nitrosylation in vivo, the factors that govern its selectivity, the dependency of the modification on different isoforms of nitric oxide synthases (NOS), as well as the physiological functions of this protein modification.
Highlights.
-
➢
Selective and efficient enrichment for S-nitrosocysteine proteins and peptides by phenylmercury
-
➢
Site specific identification of S-nitrosylated peptides by mass spectrometry
-
➢
Work-flow, negative controls and implementation of the method in complex biological samples
Acknowledgements
This work was supported by the National Institutes of Health Grants AG13966, HL054926 and National Institute of Environmental Health Sciences Center of Excellence in Environmental Toxicology Grant ES013508. KR was supported by National Institutes of Health Training Grant T32AG000255. HI is the Gisela and Dennis Alter Research Professor of Pediatrics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ignarro LJ, Barry BK, Gruetter DY, Edwards JC, Ohlstein EH, Gruetter CA, Baricos WH. Guanylate cyclase activation of nitroprusside and nitrosoguanidine is related to formation of S-nitrosothiol intermediates. Biochem Biophys Res Commun. 1980;94:93–100. doi: 10.1016/s0006-291x(80)80192-6. [DOI] [PubMed] [Google Scholar]
- 2.Stamler JS, Simon DI, Osborne JA, Mullins ME, Jaraki O, Michel T, Singel DJ, Loscalzo J. S-nitrosylation of proteins with nitric oxide: synthesis and characterization of biologically active compounds. Proc Natl Acad Sci U S A. 1992;89:444–448. doi: 10.1073/pnas.89.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seth D, Stamler JS. The SNO-proteome: causation and classifications. Curr Opin Chem Biol. 2011;15:129–136. doi: 10.1016/j.cbpa.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith BC, Marletta MA. Mechanisms of S-nitrosothiol formation and selectivity in nitric oxide signaling. Curr Opin Chem Biol. 2012;16:498–506. doi: 10.1016/j.cbpa.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakamura T, Lipton SA. Redox modulation by S-nitrosylation contributes to protein misfolding, mitochondrial dynamics, and neuronal synaptic damage in neurodegenerative diseases. Cell Death Differ. 2011;18:1478–1486. doi: 10.1038/cdd.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shahani N, Sawa A. Protein S-nitrosylation: role for nitric oxide signaling in neuronal death. Biochim Biophys Acta. 2012;1820:736–742. doi: 10.1016/j.bbagen.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benhar M, Forrester MT, Hess DT, Stamler JS. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 2008;320:1050–1054. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitchell DA, Marletta MA. Thioredoxin catalyzes the S-nitrosation of the caspase-3 active site cysteine. Nat Chem Biol. 2005;1:154–158. doi: 10.1038/nchembio720. [DOI] [PubMed] [Google Scholar]
- 9.Kornberg MD, Sen N, Hara MR, Juluri KR, Nguyen JV, Snowman AM, Law L, Hester LD, Snyder SH. GAPDH mediates nitrosylation of nuclear proteins. Nat Cell Biol. 2010;12:1094–1100. doi: 10.1038/ncb2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsushita K, Morrell CN, Cambien B, Yang SX, Yamakuchi M, Bao C, Hara MR, Quick RA, Cao W, O'Rourke B, Lowenstein JM, Pevsner J, Wagner DD, Lowenstein CJ. Nitric oxide regulates exocytosis by S-nitrosylation of N-ethylmaleimide-sensitive factor. Cell. 2003;115:139–150. doi: 10.1016/s0092-8674(03)00803-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho DH, Nakamura T, Fang J, Cieplak P, Godzik A, Gu Z, Lipton SA. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mannick JB, Schonhoff C, Papeta N, Ghafourifar P, Szibor M, Fang K, Gaston B. S-Nitrosylation of mitochondrial caspases. J Cell Biol. 2001;154:1111–1116. doi: 10.1083/jcb.200104008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 14.Lopez-Sanchez LM, Muntane J, de la Mata M, Rodriguez-Ariza A. Unraveling the S-nitrosoproteome: tools and strategies. Proteomics. 2009;9:808–818. doi: 10.1002/pmic.200800546. [DOI] [PubMed] [Google Scholar]
- 15.Raju K, Doulias PT, Tenopoulou M, Greene JL, Ischiropoulos H. Strategies and tools to explore protein S-nitrosylation. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbagen.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy E, Kohr M, Sun J, Nguyen T, Steenbergen C. S-nitrosylation: a radical way to protect the heart. J Mol Cell Cardiol. 2012;52:568–577. doi: 10.1016/j.yjmcc.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murray CI, Van Eyk JE. Chasing cysteine oxidative modifications: proteomic tools for characterizing cysteine redox status. Circ Cardiovasc Genet. 2012;5:591. doi: 10.1161/CIRCGENETICS.111.961425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kettenhofen NJ, Broniowska KA, Keszler A, Zhang Y, Hogg N. Proteomic methods for analysis of S-nitrosation. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;851:152–159. doi: 10.1016/j.jchromb.2007.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giustarini D, Milzani A, Dalle-Donne I, Rossi R. Detection of S-nitrosothiols in biological fluids: a comparison among the most widely applied methodologies. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;851:124–139. doi: 10.1016/j.jchromb.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 20.Saville B. A Scheme for the Colorimetric Determination of Microgram Amounts of Thiols. Analyst. 1958;83:670–672. [Google Scholar]
- 21.Doulias PT, Greene JL, Greco TM, Tenopoulou M, Seeholzer SH, Dunbrack RL, Ischiropoulos H. Structural profiling of endogenous S-nitrosocysteine residues reveals unique features that accommodate diverse mechanisms for protein S-nitrosylation. Proc Natl Acad Sci U S A. 2010;107:16958–16963. doi: 10.1073/pnas.1008036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang K, Ragsdale NV, Carey RM, MacDonald T, Gaston B. Reductive assays for S-nitrosothiols: implications for measurements in biological systems. Biochem Biophys Res Commun. 1998;252:535–540. doi: 10.1006/bbrc.1998.9688. [DOI] [PubMed] [Google Scholar]
- 23.Doulias PT, Raju K, Greene JL, Tenopoulou M, Ischiropoulos H. Mass spectrometry-based identification of S-nitrosocysteine in vivo using organic mercury assisted enrichment. Methods. 2012 doi: 10.1016/j.ymeth.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mcdonagh J, Waggoner WG, Hamilton EG, Hindenbach B, Mcdonagh RP. Affinity chromatography of human plasma and platelet factor XIII on organomercurial agarose. Biochim Biophys Acta. 1976;446:345–357. doi: 10.1016/0005-2795(76)90002-7. [DOI] [PubMed] [Google Scholar]
- 25.Greco TM, Hodara R, Parastatidis I, Heijnen HF, Dennehy MK, Liebler DC, Ischiropoulos H. Identification of S-nitrosylation motifs by site-specific mapping of the S-nitrosocysteine proteome in human vascular smooth muscle cells. Proc Natl Acad Sci U S A. 2006;103:7420–7425. doi: 10.1073/pnas.0600729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, Ferris CD, Hayward SD, Snyder SH, Sawa A. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 27.Doulias PT, Tenopoulou M, Greene JL, Raju K, Ischiropoulos H. Nitric oxide regulates mitochondrial Fatty Acid metabolism through reversible protein s-nitrosylation. Sci Signal. 2013;6:rs1. doi: 10.1126/scisignal.2003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sen N, Paul BD, Gadalla MM, Mustafa AK, Sen T, Xu R, Kim S, Snyder SH. Hydrogen sulfide-linked sulfhydration of NF-kappaB mediates its antiapoptotic actions. Mol Cell. 2012;45:13–24. doi: 10.1016/j.molcel.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keene SD, Greco TM, Parastatidis I, Lee SH, Hughes EG, Balice-Gordon RJ, Speicher DW, Ischiropoulos H. Mass spectrometric and computational analysis of cytokine-induced alterations in the astrocyte secretome. Proteomics. 2009;9:768–782. doi: 10.1002/pmic.200800385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matthiesen R, Bauw G, Welinder KG. Use of performic acid oxidation to expand the mass distribution of tryptic peptides. Anal Chem. 2004;76:6848–6852. doi: 10.1021/ac049032l. [DOI] [PubMed] [Google Scholar]
- 31.Pesavento JJ, Garcia BA, Streeky JA, Kelleher NL, Mizzen CA. Mild performic acid oxidation enhances chromatographic and top down mass spectrometric analyses of histones. Mol Cell Proteomics. 2007;6:1510–1526. doi: 10.1074/mcp.M600404-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Feng S, Ye M, Zhou H, Jiang X, Jiang X, Zou H, Gong B. Immobilized zirconium ion affinity chromatography for specific enrichment of phosphopeptides in phosphoproteome analysis. Mol Cell Proteomics. 2007;6:1656–1665. doi: 10.1074/mcp.T600071-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Q, Tang N, Brock JW, Mottaz HM, Ames JM, Baynes JW, Smith RD, Metz TO. Enrichment and analysis of nonenzymatically glycated peptides: boronate affinity chromatography coupled with electron-transfer dissociation mass spectrometry. J Proteome Res. 2007;6:2323–2330. doi: 10.1021/pr070112q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu L, Yan Y, Zeng M, Zhang J, Hanes MA, Ahearn G, McMahon TJ, Dickfeld T, Marshall HE, Que LG, Stamler JS. Essential roles of S-nitrosothiols in vascular homeostasis and endotoxic shock. Cell. 2004;116:617–628. doi: 10.1016/s0092-8674(04)00131-x. [DOI] [PubMed] [Google Scholar]
- 35.Yang Z, Wang ZE, Doulias PT, Wei W, Ischiropoulos H, Locksley RM, Liu L. Lymphocyte development requires S-nitrosoglutathione reductase. J Immunol. 2010;185:6664–6669. doi: 10.4049/jimmunol.1000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sen N, Hara MR, Kornberg MD, Cascio MB, Bae BI, Shahani N, Thomas B, Dawson TM, Dawson VL, Snyder SH, Sawa A. Nitric oxide-induced nuclear GAPDH activates p300/CBP and mediates apoptosis. Nat Cell Biol. 2008;10:866–873. doi: 10.1038/ncb1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM, Jacobson MP, Verdin E. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 2010;12:654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]