Abstract
Uptake, transport, and stabilization of xanthophylls in the human retina are important components of a complex multistep process that culminates in a nonuniform distribution of these important nutrients in the retina. The process is far from understood; here, we consider the potential role of interphotoreceptor retinoid-binding protein (IRBP) in this process. IRBP is thought to facilitate the exchange of 11-cis-retinal, 11-cis-retinol, and all-trans-retinol between the retinal pigment epithelium (RPE), photoreceptors and Müller cells in the visual cycle. Structural and biochemical studies suggest that IRBP has a variety of nonequivalent ligand binding sites that function in this process. IRBP is multifunctional, being able to bind a variety of physiologically significant molecules including fatty acids in the subretinal space. This wide range of binding activities is of particular interest because it is unknown whether the lutein and zeaxanthin found in the macula originate from the choroidal or retinal circulations. If from the choroidal circulation, then IRBP is a likely mediator for their transport across the interphotoreceptor matrix. In this report, we explore the binding interactions of retinoids, fatty acids, and carotenoids with IRBP using surface plasmon resonance (SPR)-based biosensors. IRBP showed similar affinity toward retinoids and carotenoids (1–2 µM), while fatty acids had approximately 10 times less affinity. These results suggest that further studies should be carried out to evaluate whether IRBP has a physiologically relevant role in binding lutein and zeaxanthin in the interphotoreceptor matrix.
Keywords: Carotenoids, Interphotoreceptor matrix, Interphotoreceptor retinoidbinding protein, Macular pigments, Retinoids, SPR
Introduction
Although there are more than 16 carotenoids in human serum, only lutein, zeaxanthin and meso-zeaxanthin (a metabolite of lutein) are present in the macular region of the retina [1, 2]. This remarkable selectivity is achieved by a complex multi-step transport process. Dietary carotenoids released from ingested food matrix are first incorporated into lipid micelles before absorption by duodenal mucosal cells [3]. Carotenoids are subsequently taken up into the mucosal cells by an apical membrane transporter, scavenger receptor class B type I (SR-BI) protein [4, 5]. In the intestinal mucosal cells, these pigments are incorporated into the chylomicrons towards the lymphatic, or portal circulations [3, 6]. In the liver, carotene remnants from the chylomicrons are transported by low-density lipoprotein (LDL) and very low-density lipoproteins (VLDL), and the xanthophylls are mainly carried by high-density lipoprotein (HDL) to the target tissues [1, 3, 6, 7]. Ultimately, the specific carotenoid-binding proteins (GSTP1 for zeaxanthin and meso-zeaxanthin; StARD3 for lutein) mediate the stable accumulation of these pigments in the macula [8, 9].
It is not clear whether the lutein and zeaxanthin present in the macular region originate from the choroidal or from the retinal circulation, but clues can be provided from the retinoids, well characterized relatives of the carotenoids that also selectively accumulate in the mammalian retina. It is well known in retinoid metabolism that coordinated biochemical activity between the choroidal circulation, retinal pigment epithelium (RPE), and photoreceptor cells is involved in the transport of retinoids in and out of the retina [10]. The photoreceptors receive a constant supply of retinoids from the choroidal circulation via the RPE and the interphotoreceptor matrix (IPM). Serum retinol-binding protein (RBP4) delivers retinol from the choroidal circulation to the basal RPE surface via a STRA6 receptor in a manner similar to the reported delivery of lutein and zeaxanthin bound to HDL via the SR-B1 receptor on the RPE [11, 12]. Within the RPE, various retinoids are bound to specific intracellular proteins such as cellular retinol-binding protein (CRBP) and cellular retinal-binding protein (CRALBP), and there is likewise a substantial amount of carotenoids in the RPE/choroid [13], presumably bound to binding proteins as well.
As the most abundant protein component of the IPM, interphotoreceptor retinoid-binding protein (IRBP) could have a role in the trafficking of carotenoids between the RPE and retina or a role in sequestering them within the subretinal space. IRBP is a 140 kDa extracellular glycolipoprotein thought to facilitate the exchange of visual cycle retinoids between the RPE, rods, cones, and Müller glia. IRBP appears to be important for targeting and protecting all-trans-retinol, 11-cis-retinol, and 11-cis-retinal as they are exchanged between these cells during the visual cycle [14–19]. IRBP is secreted into the IPM by both rod and cone cells [20, 21]. The binding of retinoids to IRBP helps to protect them from isomerization and oxidative damage [22]. IRBP is widely distributed among vertebrates, and apart from the IPM, IRBP is expressed in the pineal gland and also is present vitreous [16, 23]. IRBP is able to bind other hydrophobic molecules such as fatty acids in a non-selective manner [24]. If the carotenoids present in the macular region come from the choroidal circulation, we can hypothesize that IRBP is a previously unrecognized carotenoid transporter across the IPM.
As a first step to evaluate this hypothesis, we compared the IRBP binding affinities of carotenoids relative to retinoids and fatty acids. A surface plasmon resonance (SPR) biosensor platform was used for the biophysical characterization of these interactions to provide analytical consistency because previously used fluorescence methods are not usable with non-fluorescent carotenoids, and radiolabeled carotenoids are not readily available.
Experimental
Reagents
Amine-coupling reagents (N-hydroxysuccinimide (NHS), 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride (EDC), and 1 M sodium ethanolamine hydrochloride, pH 8.5) were purchased from Biacore AB (Uppsala, Sweden). All chemicals used were of the highest purity available and were purchased from Sigma Chemicals (Saint Louis, Missouri) unless otherwise stated. Zeaxanthin was a gift from Zeavision (Chesterfield, Missouri), meso-zeaxanthin was a gift from DSM Nutritional Products, Ltd. (Basel, Switzerland), lutein was a gift from Kemin Health (Des Moines, Iowa), astaxanthin was a gift from Cardax Pharmaceuticals (Honolulu, Hawaii), and fatty acids were purchased from Cayman Chemicals (Ann Arbor, Michigan).
Instrumentation
SPR analyses were conducted at 25°C using a fully automated SensiQ Pioneer optical biosensor equipped with carboxyl modified sensor chips (SensiQ Technologies, Inc., Oklahoma City, OK). Using this platform, a protein surface and an unmodified reference surface were prepared for simultaneous analysis. Immediately after chip docking, each channel was primed with water. Prior to each interaction analysis, the biosensor detector response was normalized by an automated procedure, and the instrument was primed five times with freshly degassed running buffer. Data were collected at 2.5 Hz.
Protein Immobilization and Analyte Interactions
IRBP was extracted from dark adapted detached bovine retinas and purified to homogeneity by a combination of Con-A affinity, ion exchange, and size-exclusion chromatography. The concentration of the purified protein was determined by absorbance spectroscopy and amino acid analysis. We chose to use bovine IRBP, as it is an easily obtainable native form of IRBP with correct post-translational modification folding. The over all identity of bovine and human IRBP is 84% [25], and ligand-binding characteristics are similar [26]. Although the bovine retina does not have a macula, xanthophyll levels in the bovine retina are comparable to human peripheral retina [27, 28].
Bovine IRBP was immobilized at 10 µg/mL in 10 mM sodium acetate (pH 4.5) on the sensor chip surface using standard amine-coupling to obtain a density of 10–12 kRU. Each of the five carotenoids was dissolved in DMSO to achieve a high concentration, and 10 mM PBS (pH 7.4) with 0.01% Tween-20 and 3% DMSO was used as the running buffer. Retinoids were prepared in a manner similar to the carotenoids. For fatty acid analysis, HEPES buffer (pH 7.4) with 0.01% Tween-20 and 3% ethanol was used as the running buffer. Typically, the analyte concentration series spanned 0.01–15 µM. Five of these blanks were analyzed at the beginning of the analysis, and the remaining blanks were interspersed throughout the analysis for double-referencing purposes. Each of the analytes was run in triplicate. The analyte concentration series was injected as two-fold dilutions (seven dilutions) in running buffer using the FastStep™ gradient injection mode. The details of the FastStep™ SPR experimental method can be found elsewhere [29, 30]. For affinity and stoichiometry determinations, SPR response data (sensorgrams) were zeroed at the beginning of each injection and double referenced. Kinetic rate constants were extracted by globally fitting the data using the Qdat™ analysis software (Biologic Software, Australia).
Results and Discussion
IRBP is believed to play an important role in the visual cycle and in retinal physiology by binding and protecting retinoids and fatty acids within the IPM [23]. There is controversy regarding the number and affinity of IRBPs various ligand binding sites. For example, the reported number of sites in bovine IRBP for retinol and fatty acids has ranged from one to three [22, 31, 32]. These studies utilized either intrinsic fluorescence of the ligand (retinoids), fluorescent analogs (fatty acids), or radioligand binding assays to determine ligand binding stoichiometry and affinity. Fluorescence-based assays would be difficult to apply to lutein and zeaxanthin because these carotenoids have almost no intrinsic fluorescence, and synthesis of fluorescent analogs would be likely to alter their interaction with the ligand binding site. Radioligand methods are cumbersome, expensive, and difficult to obtain kinetic data, and radioactive carotenoids are not readily available. We therefore chose surface plasmon resonance (SPR) to study the interaction of IRBP with its ligands based on our successful studies of the specific carotenoid-binding proteins of the human macula, GSTP1 (zeaxanthin and meso-zeaxanthin) and StARD3 (lutein) [8, 9]. Furthermore, SPR techniques have recently been used to study retinoid-binding proteins [33, 34]. One of the major advantages of the SPR approach is that we are able to monitor the interactions in real-time, and all classes of analytes (retinoids, fatty acids, and carotenoids) can be studied using a single immobilized protein chip; thus, the sample-to-sample preparation variability is very low, and all affinity values are obtained from the same surface. Since SPR is a label-free method, we can use the native analytes as opposed to fluorescent analogs [35, 36]. This can eliminate many artifacts that may arise from these analog molecules and assist in mimicking physiological conditions. The double referencing method with blank buffer used in the SPR assays avoids potential artifacts from non-specific binding [37].
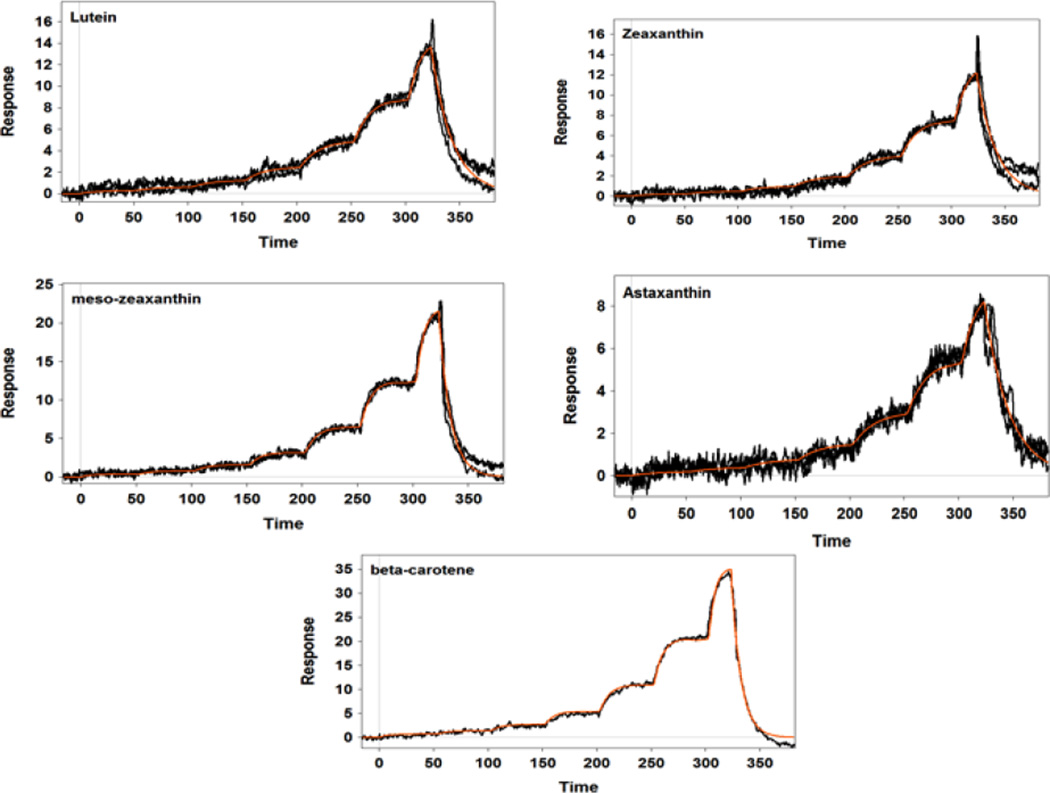
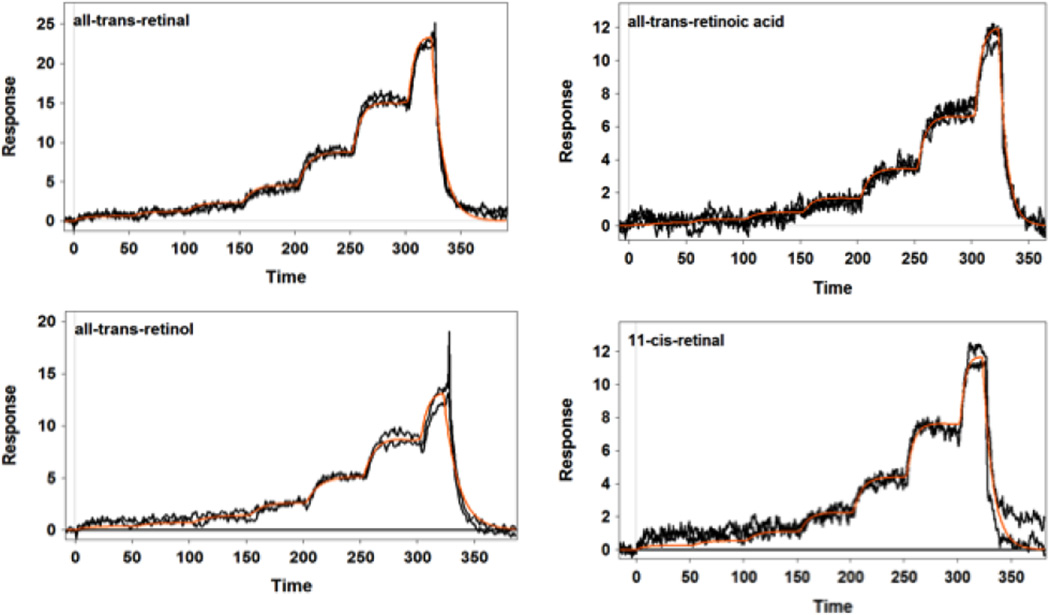
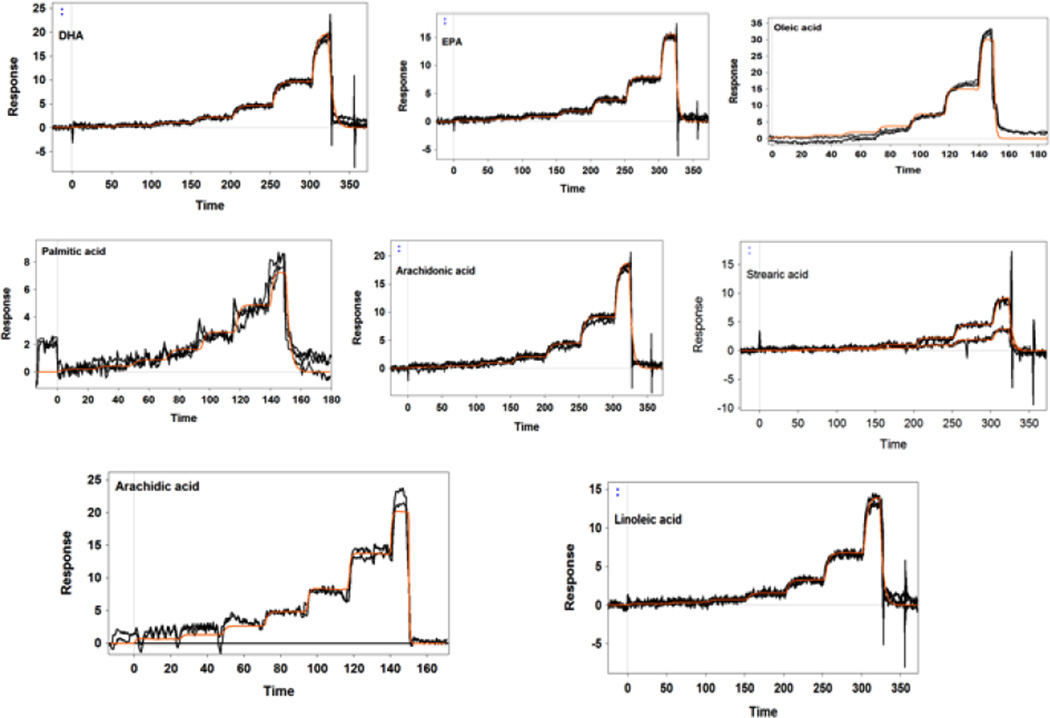
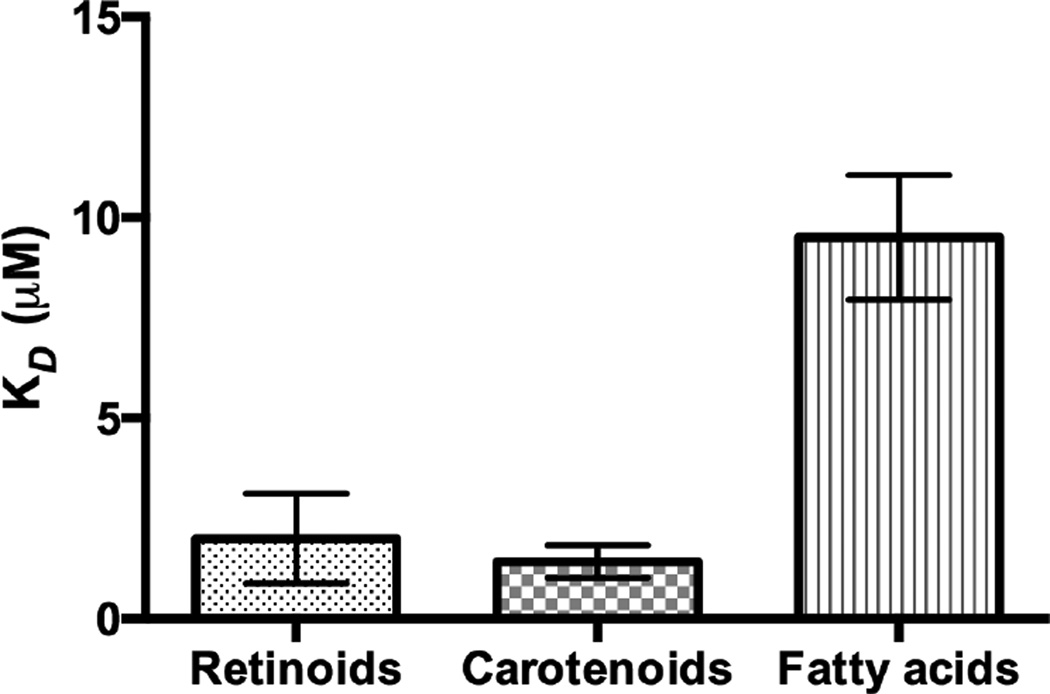
Our SPR studies revealed that IRBP was able to bind all tested retinoids, fatty acids, and carotenoids with moderate affinity and relatively low selectivity. Figure 1 displays the SPR sensorgrams of the interaction of IRBP with carotenoids. As shown in Table 1, lutein and β-carotene had relatively tight binding with a KD close to 1 µM. Zeaxanthin, meso-zeaxanthin, and astaxanthin had weaker affinities in the 2 µM range. These KD values are close to those of human serum albumin and β-lactoglobulin that we have reported previously for carotenoids [30]. Figures 2 and 3 show the SPR sensorgrams of the interaction of retinoids and fatty acids with IRBP, respectively. As tabulated in Table 1, 11-cis-retinal had an affinity close to 1 µM followed by all-trans-retinol, retinal, and retinoic acids, while fatty acids in general exhibited relatively lower binding affinity compared to both carotenoids and retinoids. The average affinities of retinoids, carotenoids, and fatty acids are graphically represented in Figure 4. As shown in Table 1, ligand-binding stoichiometry slightly over 3:1 was observed for retinoids and carotenoids whereas for fatty acids it was close to 4:1. This observation is consistent with the idea that different classes of ligand-binding sites exist and that homologous sites in each module are not equivalent. Structural evidence for a restrictive hydrophobic cavity and a shallow hydrophobic cleft have been obtained from structural and site directed mutagenesis studies [38–40]. Ongoing studies will determine the site(s) responsible for carotenoid binding site, and whether this site is unique or shared by IRBP’s other physiological ligands.
Figure 1.
SPR sensorgram responses of the interaction of IRBP with carotenoids. Carotenoids were tested at 10 µM concentrations with a 7-step serial dilution using FastStepTM injections. The orange line shows the model fit.
Table 1.
Ligand binding parameters determined at 25°C .
| Analyte (n=3) |
ka (M−1s−1) | kd (s−1) | KD (µM) | Stoichiometry (No. of sites) |
|---|---|---|---|---|
| Astaxanthin | 2.68 ± 0.9 ×104 | 0.04 ± 0.5 | 1.64 ± 0.06 | 3.4 |
| β-carotene | 1.22 ± 0.2 ×105 | 0.11 ± 0.8 | 0.92 ± 0.10 | 3.1 |
| Lutein | 4.90 ± 0.9 ×104 | 0.05 ± 0.4 | 1.06 ± 0.20 | 3.4 |
| meso-zeaxanthin | 5.16 ± 0.8 ×104 | 0.09 ± 0.6 | 1.85 ± 0.30 | 3.2 |
| Zeaxanthin | 3.30 ± 0.1 ×104 | 0.05 ± 0.5 | 1.64 ± 0.50 | 3.3 |
| all-trans-retinol | 6.20 ± 0.1 ×104 | 0.08 ± 0.8 | 1.31 ± 0.30 | 3.5 |
| all-trans-retinal | 4.47 ± 0.3 ×104 | 0.09 ± 0.4 | 2.18 ± 0.20 | 3.2 |
| 11-cis-retinal | 1.10 ± 0.1 ×105 | 0.11 ± 0.9 | 1.00 ± 0.20 | 3.1 |
| all -trans -retinoic acid | 4.00 ± 0.1 ×104 | 0.14 ± 0.2 | 3.50 ± 0.10 | 3.4 |
| Arachidonic acid | 2.40 ± 0.3 ×104 | 0.24 ± 0.3 | 10.0± 0.10 | 3.8 |
| Arachidic acid | 2.80 ± 0.1 ×105 | 2.8 ± 0.10 | 10.2 ± 0.70 | 3.7 |
| Docosahexaenoic acid (DHA) | 2.00 ± 0.2 ×104 | 0.2 ± 0.20 | 10.0 ± 0.10 | 3.4 |
| Eicosapentaenoic acid (EPA) | 3.00 ± 0.2 ×104 | 0.28 ± 0.4 | 9.40 ± 0.70 | 3.4 |
| Linoleic acid | 2.30 ± 0.4 ×104 | 0.26 ± 0.4 | 11.0 ± 0.20 | 3.7 |
| Oleic acid | 1.50 ± 0.2 ×105 | 1.10 ± 0.3 | 7.30 ± 0.70 | 3.6 |
| Palmitic acid | 3.60 ± 0.2 ×105 | 0.40 ± 0.1 | 11.1 ± 0.70 | 3.7 |
| Stearic acid | 3.20 ± 0.4 ×104 | 0.23 ± 0.6 | 7.03 ± 0.10 | 3.7 |
Errors represent the residuals from the model fit.
KD Equilibrium dissociation constant
ka Association rate constant
kd Dissociation rate constant
Figure 2.
SPR profiles of retinoid binding to IRBP. IRBP was tested at 5 µM concentrations with a 7-step two-fold dilution using FastStep™ injections. Orange line shows the model fit.
Figure 3.
SPR responses of fatty acid binding to IRBP. Fatty acids were tested at 20 µM concentrations with a 7-step two-fold dilution series using FastStep™ injections. Orange line shows the model fit.
Figure 4.
Comparison of the affinities of retinoids, carotenoids, and fatty acids. Error bars denote the standard deviation for each analyte group.
The interaction affinities that we observed indicate that IRBP is a non-selective hydrophobic binder of retinoids, fatty acids, and carotenoids. This non-selective nature is important because of the strategic location of IRBP in the eye where shuttling, buffering, and protection of a variety of small hydrophobic molecules is needed. This is not to say that IRBP does not have ligand-binding sites tailored for the above classes of physiological ligands, however. Future studies using site directed mutagenesis guided by structural data from X-ray crystallography studies will be done to evaluate specific ligand-binding domains within IRBP.
IRBP could protect retinoids and xanthophylls from breakdown, and it also could protect the membranes lining the IPM from any disruptive membranolytic effects of these molecules. In normal conditions, the retina is very close to the RPE, making it difficult to quantify the concentrations of molecules present in the IPM. Future studies will determine the amount of carotenoids that can be extracted from the IPM and whether these are bound to IRBP. In pathological conditions such as rhegmatogenous retinal detachment, the space between the RPE and the retina fills with sub-retinal fluid (SRF) which can be analyzed. Several reports confirm the presence of retinoids, carotenoids, and IRBP in SRF [41, 42].
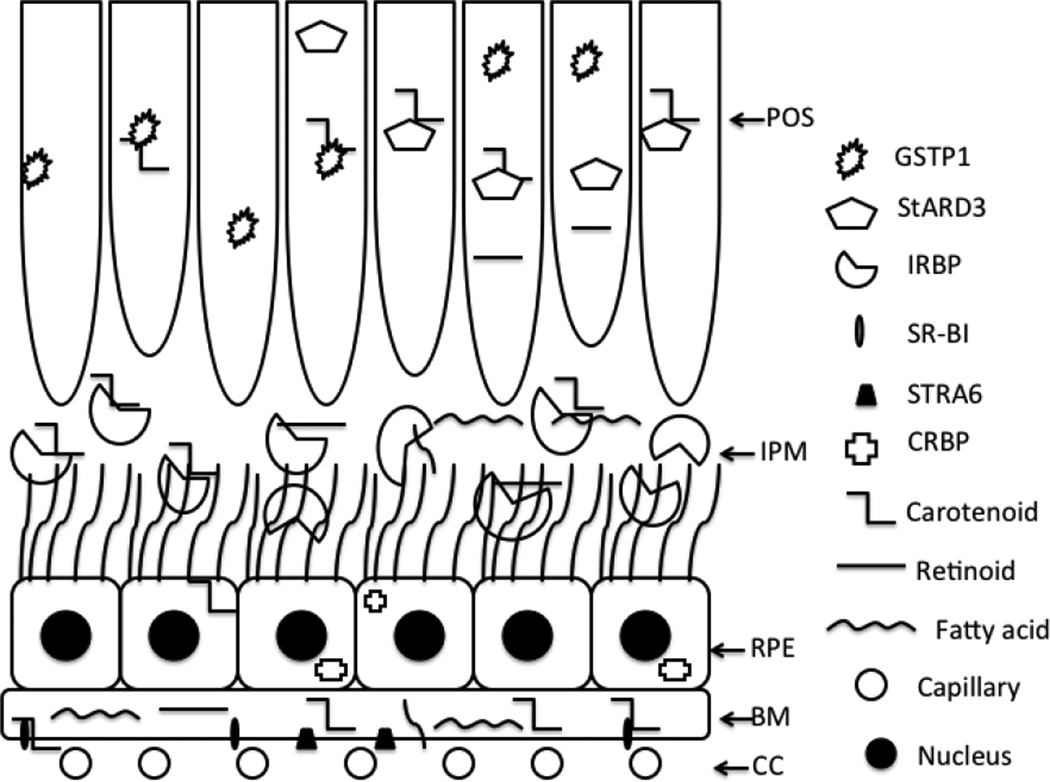
Figure 5 shows a schematic diagram of the flow of pigments between the RPE and photoreceptors in the mammalian visual cycle. In a physiological system, there are scenarios where a single protein can bind more than one hydrophobic ligand non-covalently [11]. Our results suggest that IRBP is such a protein. Since IRBP is present in high concentration in the IPM, and its affinities toward pigments are relatively strong, we hypothesize that IRBP acts as a key transporter protein in a reaction cascade between the RPE and the retina.
Figure 5.
Possible transport of pigments between RPE and retinal cells in the mammalian visual cycle. Choriocapillaris (CC); Bruch’s membrane (BM); retinal pigment epithelium (RPE); interphotoreceptor matrix (IPM); photoreceptor outer segment (POS).
While studying the interaction of fatty acids and IRBP, Putilina et al (1997) concluded that binding sites of IRBP primarily involve the C-10 area of the fatty acid analog [31]. Since the interaction is based on a specific short portion of these fatty acids, their binding sites on IRBP could accommodate a variety of fatty acids with different chain length [31]. Noy et al (1996) found that binding of fatty acids to IRBP could change the binding affinity of retinoids with IRBP [43]. They proposed that binding of a fatty acids such as docosahexaenoic acid (DHA) could act as a “switch” that will promote IRBP to release retinoids at the proper target, based on the concentration gradient of DHA that occurs between photoreceptor cells and RPE [23]. Further studies are needed to see whether carotenoid binding to IRBP is regulated in a similar manner.
Conclusion
Although IRBP may function as a key extracellular transporter of multiple hydrophobic molecules important in visual function, study of its ligand-binding properties has been limited to cumbersome radio-ligand assays and fluorescence spectroscopy. The latter requires that the ligand is fluorescent and able to quench protein tryptophans. Therefore, binding studies of visual cycle retinoids to IRBP have relied primarily on all-trans-retinol, due to its favorable fluorescent properties. Nevertheless, such assays sometimes require unrealistic assumptions regarding the equivalency of the quantum yield at different binding sites.
Here, we show that SPR can be used to conveniently study non-fluorescent ligands such as the carotenoids to obtain rate equilibrium constants of their interactions with IRBP. Our SPR studies revealed that IRBP is able to bind a variety of carotenoids at an affinity comparable to the retinoids and with much stronger affinity than any fatty acid tested. Although we cannot rule out direct delivery of lutein and zeaxanthin to the macula via the retinal circulation, IRBP’s high concentration in the IPM and its ability bind a variety of carotenoids with micromolar affinity lend support to the hypothesis that IRBP serves as a physiologically relevant carrier of carotenoids from the RPE to the retina across the IPM. Future studies will continue to evaluate this interesting new function for IRBP.
Highlights.
-
➢
Interphotoreceptor retinoid-binding protein (IRBP) binds a variety of ligands.
-
➢
A surface plasmon resonance biosensor was employed to study these interactions.
-
➢
IRBP is able to bind carotenoids and retinoids with similar affinities.
-
➢
IRBP has 10 times less affinity for ocular fatty acids.
-
➢
IRBP could serve as a carrier of carotenoids across the interphotoreceptor matrix (IPM).
Acknowledgements
This work was supported by NIH grant R01 EY11600 (P.S.B.), Lowy Medical Research Foundation (P.S.B.), core instrumentation grant EY14800 (R. Marc/P.S.B), Merit Review Award I01BX007080 from the Biomedical Laboratory Research & Development Service of the Veterans Affairs Office of Research and Development (F.G.-F.), NIH R01 EY09412 (D. Ghosh/F.G.-F), R24 EY 016662 core instrumentation grant, and Unrestricted Research Grants from Research to Prevent Blindness to the Department of Ophthalmology at State University of New York at Buffalo, and to the Moran Eye Center, University of Utah.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Li B, Vachali P, Bernstein PS. Photochem Photobiol. 2010;9:1418–1425. doi: 10.1039/c0pp00126k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdel-Aal E-S, Akhtar H, Zaheer K, Ali R. Nutrients. 2013;5:1169–1185. doi: 10.3390/nu5041169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parker RS. FASEB J. 1996;10:542–551. [PubMed] [Google Scholar]
- 4.During A, Doraiswamy S, Harrison EH. J Lipid Res. 2008;49:1715–1724. doi: 10.1194/jlr.M700580-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kiefer C, Sumser E, Wernet MF, Von Lintig J. Proc Natl Acad Sci U S A. 2002;99:10581–10586. doi: 10.1073/pnas.162182899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrison EH. Biochim Biophys Acta. 2012;1821:70–77. doi: 10.1016/j.bbalip.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loane E, Nolan JM, O'Donovan O, Bhosale P, Bernstein PS, Beatty S. Surv Ophthalmol. 2008;53:68–81. doi: 10.1016/j.survophthal.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 8.Li B, Vachali P, Frederick JM, Bernstein PS. Biochemistry. 2011;50:2541–2549. doi: 10.1021/bi101906y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhosale P, Larson AJ, Frederick JM, Southwick K, Thulin CD, Bernstein PS. J Biol Chem. 2004;279:49447–49454. doi: 10.1074/jbc.M405334200. [DOI] [PubMed] [Google Scholar]
- 10.Bok D. Eye (Lond) 1990;4(Pt 2):326–332. doi: 10.1038/eye.1990.44. [DOI] [PubMed] [Google Scholar]
- 11.Kawaguchi R, Yu J, Honda J, Hu J, Whitelegge J, Ping P, Wiita P, Bok D, Sun H. Science. 2007;315:820–825. doi: 10.1126/science.1136244. [DOI] [PubMed] [Google Scholar]
- 12.Sun H, Kawaguchi R. Int Rev Cell Mol Biol. 2011;288:1–41. doi: 10.1016/B978-0-12-386041-5.00001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernstein PS, Khachik F, Carvalho LS, Muir GJ, Zhao DY, Katz NB. Exp Eye Res. 2001;72:215–223. doi: 10.1006/exer.2000.0954. [DOI] [PubMed] [Google Scholar]
- 14.Adler AJ, Spencer SA. Experimental Eye Research. 1991;53:337–346. doi: 10.1016/0014-4835(91)90239-b. [DOI] [PubMed] [Google Scholar]
- 15.Chader GJ. Invest Ophthalmol Vis Sci. 1989;30:7–22. [PubMed] [Google Scholar]
- 16.Gonzalez-Fernandez F. J Ophthalmic Vis Res. 2012;7:100–104. [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez-Fernandez F, Ghosh D. Exp Eye Res. 2008;86:169–170. doi: 10.1016/j.exer.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez-Fernandez F. Vision Res. 2003;43:3021–3036. doi: 10.1016/j.visres.2003.09.019. [DOI] [PubMed] [Google Scholar]
- 19.McBee JK, Palczewski K, Baehr W, Pepperberg DR. Prog Retin Eye Res. 2001;20:469–529. doi: 10.1016/s1350-9462(01)00002-7. [DOI] [PubMed] [Google Scholar]
- 20.Hollyfield JG, Fliesler SJ, Rayborn ME, Fong SL, Landers RA, Bridges CD. Invest Ophthalmol Vis Sci. 1985;26:58–67. [PubMed] [Google Scholar]
- 21.Gonzalez-Fernandez F, Kittredge KL, Rayborn ME, Hollyfield JG, Landers RA, Saha M, Grainger RM. J Cell Sci. 1993;105(Pt 1):7–21. doi: 10.1242/jcs.105.1.7. [DOI] [PubMed] [Google Scholar]
- 22.Crouch RK, Hazard ES, Lind T, Wiggert B, Chader G, Corson DW. Photochem Photobiol. 1992;56:251–255. doi: 10.1111/j.1751-1097.1992.tb02154.x. [DOI] [PubMed] [Google Scholar]
- 23.Noy N. Biochem J. 2000;348(Pt 3):481–495. [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y, Saari JC, Noy N. Biochemistry. 1993;32:11311–11318. doi: 10.1021/bi00093a007. [DOI] [PubMed] [Google Scholar]
- 25.Si JS, Borst DE, Redmond TM, Nickerson JM. Gene. 1989;80:99–108. doi: 10.1016/0378-1119(89)90254-0. [DOI] [PubMed] [Google Scholar]
- 26.Lin ZY, Si JS, Nickerson JM. Invest Ophthalmol Vis Sci. 1994;35:3599–3612. [PubMed] [Google Scholar]
- 27.Dachtler M, Kohler K, Albert K. J Chromatogr B Biomed Sci Appl. 1998;720:211–216. doi: 10.1016/s0378-4347(98)00431-9. [DOI] [PubMed] [Google Scholar]
- 28.Bhosale P, Serban B, Bernstein PS. Arch Biochem Biophys. 2009;483:175–181. doi: 10.1016/j.abb.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rich RL, Quinn JG, Morton T, Stepp JD, Myszka DG. Analytical Biochemistry. 2010;407:270–277. doi: 10.1016/j.ab.2010.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vachali P, Li B, Nelson K, Bernstein PS. Arch Biochem Biophys. 2012;519:32–37. doi: 10.1016/j.abb.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Putilina T, Sittenfeld D, Chader GJ, Wiggert B. Biochemistry. 1993;32:3797–3803. doi: 10.1021/bi00065a036. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y, Noy N. Biochemistry. 1994;33:10658–10665. doi: 10.1021/bi00201a013. [DOI] [PubMed] [Google Scholar]
- 33.Campos-Sandoval JA, Redondo C, Kinsella GK, Pal A, Jones G, Eyre GS, Hirst SC, Findlay JB. J Med Chem. 2011;54:4378–4387. doi: 10.1021/jm200256g. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y, Kerimo A, Khan S, Wei LN. Mol Endocrinol. 2002;16:2528–2537. doi: 10.1210/me.2002-0124. [DOI] [PubMed] [Google Scholar]
- 35.Piliarik M, Vaisocherova H, Homola J. Methods Mol Biol. 2009;503:65–88. doi: 10.1007/978-1-60327-567-5_5. [DOI] [PubMed] [Google Scholar]
- 36.Schuck P. Curr Opin Biotechnol. 1997;8:498–502. doi: 10.1016/s0958-1669(97)80074-2. [DOI] [PubMed] [Google Scholar]
- 37.Myszka DG. Journal of Molecular Recognition. 1999;12:279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 38.Gonzalez-Fernandez F, Baer CA, Ghosh D. BMC Biochem. 2007;8:15. doi: 10.1186/1471-2091-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonzalez-Fernandez F, Bevilacqua T, Lee KI, Chandrashekar R, Hsu L, Garlipp MA, Griswold JB, Crouch RK, Ghosh D. Invest Ophthalmol Vis Sci. 2009;50:5577–5586. doi: 10.1167/iovs.08-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loew A, Gonzalez-Fernandez F. Structure. 2002;10:43–49. doi: 10.1016/s0969-2126(01)00698-0. [DOI] [PubMed] [Google Scholar]
- 41.Bridges CD, Price J, Landers RA, Fong SL, Liou GI, Hong BS, Tsin AT. Invest Ophthalmol Vis Sci. 1986;27:1027–1030. [PubMed] [Google Scholar]
- 42.Chan C, Leung I, Lam KW, Tso MO. Curr Eye Res. 1998;17:890–895. doi: 10.1076/ceyr.17.9.890.5141. [DOI] [PubMed] [Google Scholar]
- 43.Chen Y, Houghton LA, Brenna JT, Noy N. J Biol Chem. 1996;271:20507–20515. doi: 10.1074/jbc.271.34.20507. [DOI] [PubMed] [Google Scholar]