Abstract
Background
Loss of PTEN has been shown to be associated with aggressive behavior of prostate cancer. It is less clear that loss of PTEN also increases the risk of cancer mortality. We investigated the association between PTEN expression and prostate cancer mortality, and the potential effect modification by IGF1R, a direct activator of the PI3K pathway.
Methods
Protein expression in tumor were evaluated using tumor tissues obtained from 805 participants of the Physicians’ Health and the Health Professionals Follow-up studies who were diagnosed with prostate cancer and underwent radical prostatectomy. Proportional hazard models were used to assess PTEN expression, and its interaction with IGF1R, in relation to lethal prostate cancer (cancer-specific death or distant metastases).
Results
Low PTEN expression was associated with an increase risk of lethal prostate cancer (HR = 1.7, 95% CI: 0.98-3.2, P for trend = 0.04). The association was attenuated after adjustment for Gleason grade, tumor stage, and PSA at diagnosis. A significant negative interaction between PTEN and IGF1R was found (P for interaction = 0.03). Either reduction in PTEN or increase in IGF1R expression was sufficient to worsen prognosis. Models including PTEN and IGF1R expression offer additional predicting power to prostate cancer survival, comparing to those only including demographic and clinical factors.
Conclusions
Low PTEN protein expression significantly increases the risk of lethal prostate cancer, particularly when the IGF1R expression remains at normal level.
Impact
PTEN and IGF1R expression in tumor are promising candidates for independent prognostic factors to predict lethal prostate cancer.
Keywords: Prostate cancer, PTEN, Tumor biomarker, Survival, Interaction
INTRODUCTION
The use of prostate-specific antigen (PSA) screening for prostate cancer has likely contributed to reductions in cancer-specific mortality (1, 2), while at the same time over-treatment is a concern for many PSA test-detected cancers (3, 4). The reason for such a controversy lies in the heterogeneous nature of prostate cancer, of which the clinical behaviors vary greatly (5-8). While many tumors grow slowly and would not pose a threat if left untreated, some clinically organ-confined tumors progress rapidly and develop metastasis even after local therapy. Therefore, it is of urgent need and great clinical importance to identify key factors or molecular events that can determine or predict the behaviors of the tumor.
One such marker for prostate cancer is phosphatase and tensin homolog deleted on chromosome 10 (PTEN), a tumor suppressor that negatively regulates the PI3K/Akt pathway (9, 10). Loss or inactivation of PTEN, usually leading to constitutive activation of the PI3K/Akt pathway, has been implicated in a number of human malignancies, including prostate cancer (11-13). In animal models, prostate-specific PTEN knockout can lead to tumorigenesis, progression, and metastasis of prostate cancer (14-16). In epidemiologic and clinical studies, loss or reduction of PTEN in prostate tumors has been shown to be associated with higher grade (17-20), more advanced stages (18, 20), greater angiogenesis (21), faster progression to androgen independence (22), and increased risk of recurrent disease (22-25). The role of PTEN loss in association with prostate cancer-specific mortality, however, is less clear. Both positive and null findings have been reported (18, 19, 26). Homozygous deletions or mutations of PTEN gene, resulting in total loss of PTEN activity, occur at relatively low frequencies of 10%-15% in primary prostate cancer (11, 21). However, a significantly higher proportion of tumors have reduced PTEN protein level (17, 20, 27), and such reductions may be sufficient to influence prostate cancer survival (24, 26).
One of the activators of the PI3K/Akt pathway is the insulin-like growth factor 1 (IGF1) (28, 29). Initiation of downstream signaling by IGF1 is predominantly mediated by the IGF1 receptor (IGF1R), which is ubiquitously expressed across different tissues, and promotes cell proliferation, differentiation, and inhibits apoptosis. Overexpression or constitutive activation of IGF1R is shown to promote tumor formation in mouse models (30, 31). Several clinical studies reported high IGF1R expression has been detected in human tumor specimens and the expression level correlates with metastasis (32-34). Epidemiologic studies on IGF1 signaling in relation to risk of prostate cancer have nearly exclusively focused on circulating IGF1 and IGF binding proteins (35-46). Although some recent findings have been null, the majority of the studies support a positive association between IGF1 and prostate cancer risk.
We hypothesize that reduction or loss of PTEN expression in prostate tumor contributes to cancer progression. Since the PI3K/PTEN/Akt pathway is downstream of IGF1/IGF1R, the effect of PTEN loss may be modified by IGF1 signaling. To test our hypotheses, we examined tumor protein expression of PTEN and IGF1R from specimens of 805 men who were diagnosed with prostate cancer and underwent radical prostatectomy as therapy, and followed them prospectively for cancer-specific mortality or development of distant metastasis. We evaluated whether PTEN protein expression in tumor is related to the risk of lethal prostate cancer, defined by cancer-specific mortality or distant metastasis. We also investigated IGF1R expression in tumor as a potential effect modifier for the association between PTEN expression and lethal prostate cancer risk. The research was approved by the Institutional Review Board at the Harvard School of Public Health and the Partners Health Care.
MATERIALS AND METHODS
Study Population
The men with prostate cancer in this study were participants in the prospective Physicians’ Health Study (PHS) and Health Professionals Follow-up Study (HPFS). The PHS was a randomized trial among 29,067 US male physicians ages 40-84 at randomization in the primary prevention of cancer and cardiovascular disease. Participants are sent annual questionnaires to ascertain disease endpoints as well as lifestyle, dietary and medical covariate data as part of the main trial. The HPFS is an ongoing prospective cohort study of 51,529 U.S. male health professionals who were aged 40 to 75 years at baseline in 1986. The participants have been sent a questionnaire every 2 years since 1986 to update their information on lifestyle factors, medical history, and disease outcomes.
The PHS and HPFS Prostate Tumor Cohort is a sample of participants for whom we had retrieved archival formalin-fixed, paraffin embedded prostatectomy (95%) and TURP (5%) tumor specimens from these men. The current analysis is based among 804 men (329 from PHS and 475 from HPFS) for whom the first batch of molecular assessment of PTEN expression was completed.
Ascertainment of Outcome
In both the PHS and HPFS cohorts, cases of prostate cancer were identified by self-report, then confirmed by review of medical records and pathology reports. Clinical information, such as tumor stage and PSA at diagnosis, was acquired through a standardized review of medical records. The cohorts were followed prospectively for cancer and all-cause mortality. Deaths were ascertained through repeated mailings, telephone calls to non-respondents, and searches of the National Death Index. All causes of death were confirmed by extensive review of death certificates and medical records. Follow-up for cancer was >96% complete and for mortality >98% complete. Participants who had prostate cancer were separately followed. Detailed information on treatment and development of metastasis was obtained via additional questionnaires and collection of medical records.
Immunohistochemistry
Angiogenesis
Protein expression of endothelial cell marker CD34 was ascertained on 5 micron sections using the anti-CD34 mouse monoclonal antibody (QBE ND 10, BioGenex, CA, diluted 1:200) and peroxidase blocking reagent (Dual Endogenous Enzyme Block, DakoCytomation, CA). Detection was accomplished by a peroxidase labeled polymer conjugated to anti-mouse IgG (DakoCytomation, CA), liquid 3,3′-diaminobenzidine (DAB)+ Substrate Chromogen System (K3468), and DakoCytomation EnVision+Dual Link System (DakoCytomation, CA). Immunohistochemistry was performed in an OptiMax automated cell Staining System (BioGenex Laboratories, CA). Slides were counterstained with hematoxylin (Sigma-Aldrich, MO). The size and architecture of microvessels were quantified by Semi-automated image analysis using Image ProPlus 4.5 software (Media Cybernetics, MD). Vessel size was determined as the average area comprised by a vessel (μm2). The irregularity of the vessel lumen was calculated by perimeter2 / 4*Π*area, with a value of 1.0 indicating a perfect circle and values greater than 1.0 indicating increasing irregularity.
Tissue Microarrays
To increase efficiency of immunohistochemical staining, we constructed high-density tumor tissue microarrays (TMA). Our pathologists (ML, MF, SF, RF) reviewed hematoxylin and eosin slides to confirm prostate cancer, and to provide uniform Gleason grading (47). TMAs were constructed by sampling 0.6 mm paraffin embedded tissue cores (at least 3 cores per subject) and embedding the cores on a recipient array block.
PTEN
Cytoplasmic expression of PTEN in tumor cells was ascertained using the Anti-PTEN rabbit polyclonal antibody (Zymed, San Francisco, CA; diluted 1:200) and was quantified using automated image analysis. Briefly, 4 micron sections of each TMA were mounted on a glass slide, deparaffinized and microwaved for antigen retrieval in citrate-based buffer. Hematoxylin was used as a counterstain and diaminobenzadine (DAB) for the immunohistochemical staining. The stained slides were then scanned using the Ariol Pathology and Imaging System (Applied Imaging, San Jose, CA), and PTEN expression was quantified using the Ariol Immunohistochemical Module. Following image acquisition of the stained slide, a custom mask was made for each core to limit analysis to tumor cells. The IHC module in Ariol outputs information on the staining intensity for a given core (range 0-255, mean and mode intensity across core), the area of DAB staining and blue counter-stain which allows calculation of percent of tumor tissue staining positive for PTEN (0-100%).
IGF1R and IR
Immunohistochemical staining was conducted on 5 micron sections of each TMA using the Anti-Insulin Receptor, B subunit, Rabbit immunoaffinity purified IgG (Upstate Cell Signaling Solutions, Lake Placid, NY) and the IGF-1R beta R rabbit polyclonal antibody (Santa Cruz Biotechnology Inc, Santa Cruz, CA) with enzyme labeled biotin streptavidin system and solvent resistant DAB Map kit (Ventana autostainer model Discover XT™, Ventana Medical System, Tuscan, AZ). Nonspecific reactivity was assessed by omission of the primary antibody. The specificity of staining for IR was confirmed by using placenta as a positive control. The slide was scanned with the BLISS system (Bacus Lab, North Lombard, IL) and scored manually by two pathologists who were blinded to clinical outcomes. For both receptors, intensity was scored from 0 to +3 – 0 (no staining by any tumor cells, 1 (faint or focal staining), 2 (moderate intensity in a convincing number of cells), and 3 (intense staining by a sufficient number of cells).
Apoptosis
The TUNEL assay was used on 5 micron sections to identify the percentage of tumor cells undergoing apoptosis. The procedure was carried out using the Apoptag Peroxidase In situ kit (Chemicon International, Temecula, CA) according to the manufacturer’s instructions. The entire area of each tumor core was evaluated. Apoptosis was quantified as the percentage of positively-stained area over the whole tumor area.
Ki-67
The expression of Ki-67, a cell proliferation marker, was assessed on 5 micron sections using a rabbit polyclonal antibody (Vector Labs, Burlingame, CA; diluted 1:1,500). After immunohistochemical staining, the tumor areas of each core were selected for quantitative image analysis using the Ariol instrument SL-50 (Applied Imaging, San Jose, CA). Cell proliferation was quantified as the percentage of Ki-67-positive nuclei over all tumor nuclei.
Statistical Analysis
In the analyses presented here, we used the baseline questionnaire data when the participants entered the original HPFS and PHS studies as baseline for demographic measures.
For each individual, we calculated the minimum, mean, and maximum values for percent area staining and staining intensity of PTEN across the TMA cores for an individual. These values were qualitatively similar; therefore we only presented the analyses using the mean values. We consider the percent area staining, reflecting the percent of tumor tissue with PTEN expression, as the most relevant measure to evaluate the potential role of PTEN in cancer progression. Additionally, we also generated a combined staining score, by multiplying the mean percent staining and the mean staining intensity, to reflect total PTEN expression in the tumor. These two measures were used as continuous variables in the analyses. They were also divided into quartiles according to the cohort distribution. In addition, we created an arbitrary cutoff point for each of these two measures by combining the bottom three quartiles as the low expression group. Less than 32% is considered as low percent staining, and less than 32.25 as low score.
We compared clinical characteristics of prostate cancer cases including age at diagnosis, baseline body mass index (BMI), tumor stage, Gleason grade, and PSA at diagnosis, as well as cellular biomarkers for characteristics of angiogenesis (microvessel size and irregularity), apoptosis, and cell proliferation, across quartiles of PTEN expression. We calculated a p-for-trend value from ANOVA for age at diagnosis, baseline BMI, angiogenic characteristics, apoptosis, and cell proliferation across PTEN quartiles. Using the Mantel-Haenszel statistic, we tested whether there is a linear trend for tumor stage, Gleason grade, or PSA at diagnosis across quartiles of PTEN expression.
We employed time-to-event analyses to evaluate the association of PTEN expression on development of lethal prostate cancer (cancer-specific death or development of distant metastasis). Cases were diagnosed with prostate cancer between 1983-2005 in PHS and 1986-2002 in HPFS. Person-time for each individual was calculated from the date of cancer diagnosis to development of metastases, death or end of follow-up (July 1, 2010 for the HPFS and March 1, 2010 for the PHS). Hazard ratios (HR) and 95% confidence intervals (CI) were estimated from the Cox proportional hazard regression models. We adjusted for age at diagnosis (5-year categories), diagnosis era (pre-PSA era: before 1989, peri-PSA era: 1989-1993, PSA era: after 1993), baseline BMI (kg/m2, continuous), and baseline smoking status (current, non-current). In additional models, we examined PTEN expression and lethal prostate cancer independent of tumor stage (T1/T2, T3, T4), Gleason score (6 or less, 7-10), and PSA at diagnosis (ng/ml; <4.1, 4.1-9.9, 10 or higher, or missing). The trend tests across quartiles of percent staining and the multiplicative score were performed by assigning median values of these categories and entering these values as continuous terms in the Cox model.
To investigate whether the associations between PTEN expression and lethal prostate cancer risk differ between the HPFS and PHS cohorts, we conducted survival analyses in the HPFS and PHS separately, and calculated the Q statistics, which follows an approximate χ2 distribution with 1 degree of freedom. Due to the absence of heterogeneity, the two cohorts were merged for all analyses.
We assessed whether the association between of PTEN expression and the development of lethal prostate cancer differed by IGF1R expression in a subset of individuals (N=651), who have both the PTEN and IGF1R expression measured. IGF1R expression was measured by a multiplicative score, similar to that of PTEN expression. The top tertile of the IGF1R multiplicative score was arbitrarily defined as high IGF1R expression. We performed stratified survival analyses by the IGF1R multiplicative score to evaluate the associations of percent PTEN staining and PTEN multiplicative staining score and lethal prostate cancer risk. We also generated interaction terms between PTEN expression measures and the IGF1R multiplicative score, and fitted multivariate Cox models with main effect measures and covariates. The significance of the interaction was determined using Wald test to test the beta-coefficients of the cross-product terms.
We evaluated the ability of PTEN and IGF1R expression to predict 10-year survival after diagnosis of prostate cancer using logistic regression, comparing the base model containing only clinical and demographic covariates to the model also containing PTEN and IGF1R expression and their interaction. Area under curve (AUC), net reclassification improvement (NRI), and integrated discrimination index (IDI) were calculated for these two nested models.
All statistical analyses were two-sided and a P-value of less than 0.05 was considered statistically significant. We conducted all analyses using the SAS software (SAS Institute, Inc., Version 9.1, Cary, NC).
RESULTS
The baseline characteristics of the HPFS and PHS Tumor Cohorts are presented in Table 1. These two cohorts were demographically and clinically similar. In HPFS, 475 men who were diagnosed with prostate cancer between 1986 and 2002 have been followed prospectively for clinical progression and mortality, with a median follow-up of 13.2 years. The average age at cancer diagnosis was 65.6 years. A total of 38 prostate cancer-specific death and 10 cases of distant metastatic prostate cancer were documented. In PHS, 330 men were diagnosed with prostate cancer between 1983-2005. The mean age at diagnosis was 66.4 years. During a median follow-up of 10.9 years, 25 cancer death and 3 distant metastases occurred. For both cohorts, the majority of the cancers were diagnosed after the clinical introduction of PSA test as prostate cancer screening. The proportion of current smokers was relatively low at baseline in both cohorts, compared to the general population.
Table 1.
Clinical and demographic characteristics of the HPFS (1986-2010) and PHS (1983-2010) Prostate Tumor Cohort
| HPFS (N=475) | PHS (N=330) | |
|---|---|---|
| Continuous | ||
|
| ||
| Age at baseline (yr), mean (sd) | 57.7 (7.4) | 56.3 (7.6) |
| Age at diagnosis (yr), mean (sd) | 65.6 (6.3) | 66.4 (6.2) |
| Follow-up (month), median (range) | 158.0 (1.0-281.0) | 130.9 (1.0-277.8) |
| Baseline BMI (kg/m2), median (range) | 24.8 (18.6-61.5) | 24.4 (18.3-35.9) |
| Mean percent area for PTEN+, mean (sd) | 0.25 (0.22) | 0.20 (0.18) |
| Mean multiplicative score for PTEN+, mean (sd) | 26.0 (24.4) | 19.9 (20.1) |
|
| ||
| Categorical | N (%) | N (%) |
|
| ||
| Outcomes | ||
| Prostate cancer death | 38 (8.0) | 25 (7.6) |
| Distant metastasis | 10 (2.1) | 3 (0.9) |
| Tumor stage | ||
| pT1b-pT2N0M0 | 323 (68.0) | 254 (77.0) |
| pT3N0M0 | 132 (27.8) | 63 (19.1) |
| pT4/N1/M1 | 20 (4.2) | 13 (3.9) |
| Gleason grade | ||
| 2-6 | 95 (20.0) | 88 (26.8) |
| 7 (3+4) | 116 (24.4) | 67 (20.3) |
| 7 (4+3) | 190 (40.0) | 120 (36.4) |
| 8-10 | 74 (15.6) | 54 (16.4) |
| (Missing 1 record) | ||
| Smoking status | ||
| Never | 238 (50.1) | 164 (49.7) |
| Past | 191 (40.2) | 120 (36.4) |
| Current | 46 (9.7) | 24 (7.3) |
| Missing | 0 (0) | 22 (6.7) |
| PSA at diagnosis | ||
| < 4 | 48 (10.1) | 31 (9.4) |
| 4-10 | 214 (45.1) | 184 (55.8) |
| >10 | 136 (28.6) | 67 (20.3) |
| Missing | 77 (16.2) | 48 (14.6) |
| Year of diagnosis | ||
| Before 1990 (Pre-PSA era) | 35 (7.4) | 25 (7.6) |
| 1990-1993 (Peri-PSA era) | 155 (32.6) | 63 (19.1) |
| After 1993 (PSA era) | 285 (60.0) | 242 (73.3) |
The characteristics of prostate cancer cases were compared across quartiles of PTEN expression, as shown in Table 2. No significant differences in age at cancer diagnosis, baseline BMI, or PSA at diagnosis were observed across quartiles of percent area for PTEN staining. Tumors with lower percent area for PTEN staining were more likely to be advanced stage (P for trend = 0.06) and higher Gleason grade (P for trend = 0.049). The results were similar when PTEN expression was stratified according to the multiplicative staining score (data not shown). Higher expression of PTEN appeared to be associated with more apoptosis in tumor cells (P for trend = 0.002). Such a trend was not present for either characteristics of angiogenesis including microvessel size and irregularity or cell proliferation.
Table 2.
Characteristics of prostate cancer cases across quartiles of PTEN expression in the HPFS (1986-2010) and PHS (1983-2010) Prostate Tumor Cohort
| N§ | Overall | Mean percent area of PTEN staining | P for Trend | ||||
|---|---|---|---|---|---|---|---|
| Q1 | Q2 | Q3 | Q4 | ||||
| Mean percent area of PTEN+, mean (sd) | 805 | 0.23 (0.21) | 0.04 (0.02) | 0.11 (0.03) | 0.23 (0.05) | 0.53 (0.17) | |
| Age at diagnosis (yr), mean (sd) | 805 | 65.9 (6.3) | 65.9 (6.4) | 65.3 (6.1) | 66.2 (6.2) | 66.5 (6.3) | 0.2† |
| Baseline BMI (kg/m2), mean (sd) | 805 | 24.9 (3.0) | 24.8 (2.6) | 25.0 (3.9) | 24.9 (2.8) | 24.9 (2.4) | 0.8† |
| Tumor stage, N (%) | 805 | ||||||
| pT1b-pT2N0M0 | 577 (71.7) | 128 (67.0) | 143 (68.4) | 154 (76.6) | 152 (74.5) | ||
| pT3N0M0 | 195 (24.7) | 54 (28.3) | 57 (27.3) | 41 (20.4) | 43 (21.1) | ||
| pT4/N1/M1 | 33 (4.1) | 9 (4.7) | 9 (4.3) | 6 (3.0) | 9 (4.4) | 0.06‡ | |
| Gleason grade, N (%) | 804 | ||||||
| 2-6 | 183 (22.8) | 36 (19.0) | 46 (22.0) | 45 (22.4) | 56 (27.5) | ||
| 7 | 493 (61.3) | 117 (61.6) | 131 (62.7) | 129 (64.2) | 116 (56.9) | ||
| 8-10 | 128 (15.9) | 37 (19.5) | 32 (15.3) | 27 (13.4) | 32 (15.7) | 0.049‡ | |
| PSA at diagnosis, N (%) | 680 | ||||||
| < 4 | 79 (11.6) | 18 (11.1) | 16 (9.0) | 24 (14.6) | 21 (11.9) | ||
| 4-10 | 398 (58.5) | 94 (58.0) | 110 (62.2) | 92 (56.1) | 102 (57.6) | ||
| >10 | 203 (29.9) | 50 (30.9) | 51 (28.8) | 48 (29.3) | 54 (30.5) | 0.7‡ | |
| Angiogenesis - CD34 staining | 351 | ||||||
| Vessel area∥ (μm2), mean (sd) | 549.2 (265.6) | 502.0 (246.9) | 556.4 (248.6) | 596.4 (319.7) | 530.4 (233.0) | 0.3† | |
| Irregularity of vessel lumen¶, mean (sd) | 3.87 (1.00) | 3.88 (0.99) | 3.67 (0.93) | 3.89 (1.06) | 4.01 (1.00) | 0.3† | |
| Apoptosis - TUNEL assay | |||||||
| Percentage of stained area, mean (sd) | 669 | 2.2 (4.5) | 1.6 (3.6) | 2.1 (4.2) | 1.8 (3.4) | 3.3 (6.3) | 0.002† |
| Cell proliferation - Ki-67 staining | |||||||
| Percentage of positive nuclei, mean (sd) | 773 | 0.73 (2.6) | 0.81 (2.8) | 0.48 (0.99) | 0.59 (1.4) | 1.04 (3.9) | 0.07† |
Missing data if N is not equal to 805
Smaller vessels are more angiogenic
Irregular vessels are more angiogenic, a score of 1.0 indicates a perfect circle
P for trend was calculated using ANOVA
P for trend was calculated from the Mantel-Haenszel statistic
We evaluated the association between PTEN expression in tumor and lethal prostate cancer (cancer death/distant metastasis); the results are shown in Table 3. Using the percent of tumor tissue with positive staining as a continuous variable, every 10% decrease in staining conferred a 20% increase in risk of lethal disease, after adjusting for age at diagnosis, diagnosis era, baseline BMI and smoking status. Men with the lowest percent staining (bottom quartile) were at markedly higher risk for lethal cancer (HR = 2.4, 95% CI: 1.2-4.7), compared to those with the highest percent staining (top quartile) (p for trend = 0.04). Low percent of tumor tissue stained positive for PTEN protein (< 32%) was associated with a borderline significant HR of 1.7 (95% CI: 0.98-3.2). However, when we further adjusted for clinical characteristics of the tumor, such as tumor stage, Gleason grade and PSA at diagnosis, percent of tumor tissue with PTEN expression no longer predicted lethal prostate cancer. The multiplicative score for PTEN staining, as a measure for total PTEN protein in tumor, yielded very similar findings.
Table 3.
Hazard ratios (HR) and 95% Confidence Interval (CI) of PTEN expression to predict time to lethal prostate cancer* in the HPFS (1986-2010) and PHS (1983-2010) Prostate Tumor Cohort
| PTEN expression | No. of lethal cases | HR† (95% CI) | HR‡ (95% CI) | HR§ (95% CI) |
|---|---|---|---|---|
| Percent Staining | ||||
| Per 10% decrease | 76 | 1.2 (1.0, 1.3) | 1.2 (1.0, 1.3) | 1.1 (0.93, 1.2) |
| Q1 - Lower | 24 | 2.4 (1.2, 4.7) | 2.4 (1.2, 4.7) | 1.6 (0.82, 3.3) |
| Q2 | 15 | 1.2 (0.59, 2.7) | 1.2 (0.56, 2.6) | 1.0 (0.44, 2.1) |
| Q3 | 22 | 1.7 (0.87, 3.5) | 1.7 (0.87, 3.5) | 1.6 (0.77, 3.2) |
| Q4 - Higher | 15 | 1 | 1 | 1 |
| Test for trend | P = 0.04 | P = 0.04 | P = 0.33 | |
| Low percent (<32%) | 61 | 1.8 (0.98, 3.2) | 1.7 (0.98, 3.2) | 1.4 (0.76, 2.6) |
| Multiplicative score | ||||
| Per SD decrease | 76 | 1.4 (1.1, 1.8) | 1.4 (1.1, 1.8) | 1.2 (0.88, 1.6) |
| Q1 - Lower | 24 | 2.6 (1.3, 5.2) | 2.6 (1.3, 5.2) | 1.8 (0.85, 3.6) |
| Q2 | 14 | 1.4 (0.62, 3.0) | 1.3 (0.6, 3.0) | 1.1 (0.48, 2.5) |
| Q3 | 25 | 2.3 (1.1, 4.7) | 2.3 (1.1, 4.7) | 2.0 (0.97, 4.3) |
| Q4 - Higher | 13 | 1 | 1 | 1 |
| Test for trend | P = 0.03 | P = 0.03 | P = 0.27 | |
| Low score (<32.25) | 63 | 2.1 (1.1, 3.9) | 2.1 (1.1, 3.9) | 1.6 (0.86, 3.1) |
Lethal prostate cancer is defined as fatal prostate cancer or distant metastasis
Hazard ratios adjusted for age at diagnosis and diagnosis era
Hazard ratios adjusted for age at diagnosis, diagnosis era, baseline BMI and smoking status
Hazard ratios adjusted for age at diagnosis, diagnosis era, baseline BMI, smoking status, Gleason grade, and PSA at diagnosis
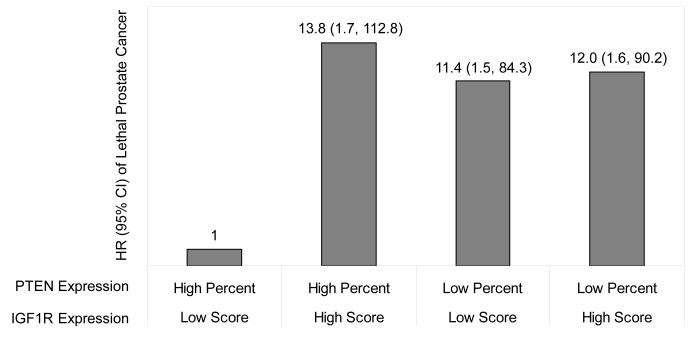
To examine the potential interaction between PTEN and IGF1R expression, we stratified the study population by IGF1R staining score. As shown in Table 4, the associations between measures of PTEN expression and development of lethal prostate cancer varied according to the level of IGF1R expression. When the expression of IGF1R protein is low in the tumor, low PTEN expression was significantly predictive of lethal disease, adjusted for both demographic and clinical parameters. A statistically significant trend in risk of lethal prostate cancer was present across the quartiles of PTEN expression (P = 0.01 for percent and P = 0.04 for score). Low percent and low score for PTEN staining were both associated a more-than-10-fold increase in the multivariate HR. However, when the expression of IGF1R protein is high in the tumor, no association between measures of PTEN expression and lethal prostate cancer was observed. A strong negative interaction was observed between PTEN and IGF1R expression (P = 0.03). In additional multivariate survival analyses presented in Figure 1, we used men with low score for IGF1R staining and high percent of tumor with PTEN staining as the reference group. High score for IGF1R staining alone was associated with an HR of 13.8 (95% CI: 1.7-112.8). Similarly, low percent of tumor with PTEN staining alone was associated with an HR of 11.4 (95% CI: 1.5-84.3). However, combining these two conditions did not confer additional risk for lethal prostate cancer (HR = 12.0, 95% CI: 1.6-90.2).
Table 4.
HR and 95% CI of PTEN expression to predict time to lethal prostate cancer* in the HPFS (1986-2010) and PHS (1983-2010) Prostate Tumor Cohort, stratified by IGF1R expression
| PTEN expression | Low score of IGF1R expression | High score of IGF1R expression | Pfor interaction | ||
|---|---|---|---|---|---|
| No. of lethal cases | HR† (95% CI) | No. of lethal cases | HR† (95% CI) | ||
| Percent Staining | |||||
| Per 10% decrease | 31 | 1.3 (0.96, 1.7) | 33 | 0.96 (0.77, 1.2) | 0.14‡ |
| Q1 - Lower | 12 | 17.6 (1.7, 184) | 10 | 0.77 (0.26, 2.2) | |
| Q2 | 5 | 10.4 (0.94, 115) | 6 | 0.49 (0.14, 1.8) | |
| Q3 | 12 | 13.3 (1.3, 134) | 9 | 1.1 (0.37, 3.6) | |
| Q4 - Higher | 2 | 1 | 8 | 1 | |
| Test for trend | P = 0.01 | P = 0.44 | |||
| Low percent (<32%) | 29 | 13.8 (1.5, 130) | 25 | 0.78 (0.31, 2.0) | 0.03‡ |
| Multiplicative score | |||||
| Per SD decrease | 31 | 1.7 (0.92, 3.2) | 33 | 0.94 (0.60, 1.5) | 0.15‡ |
| Q1 - Lower | 12 | 12.7 (1.3, 127) | 10 | 0.98 (0.30, 3.1) | |
| Q2 | 4 | 7.6 (0.66, 87) | 6 | 0.78 (0.20, 3.1) | |
| Q3 | 13 | 12.4 (1.2, 132) | 11 | 1.9 (0.57, 6.3) | |
| Q4 - Higher | 2 | 1 | 6 | 1 | |
| Test for trend | P = 0.04 | P = 0.77 | |||
| Low score (<32.25) | 29 | 11.3 (1.2, 109) | 27 | 1.2 (0.41, 3.6) | 0.09‡ |
Lethal prostate cancer is defined as fatal prostate cancer or distant metastasis
Hazard ratios adjusted for age at diagnosis, diagnosis era, baseline BMI, smoking status, Gleason grade, and PSA at diagnosis
Wald test was used to test the beta-coefficients of the cross-product terms between PTEN and IGF1R expression
Figure 1.
HR† and 95% CI of PTEN and IGF1R expression to predict time to lethal prostate cancer* in the HPFS (1986-2010) and PHS (1983-2010) Prostate Tumor Cohort
* Lethal prostate cancer is defined as fatal prostate cancer or distant metastasis.
† Hazard ratios adjusted for age at diagnosis, diagnosis era, baseline BMI, smoking status, Gleason grade, and PSA at diagnosis
To assess the ability of PTEN and IGF1R expression to predict 10-year survival after diagnosis of prostate cancer, we compared the base logistic regression model that included demographic factors such as smoking status and BMI, as well as prognostic factors including patient age, tumor stage, Gleason score, and PSA at diagnosis, to the model also including percent for PTEN staining, score for IGF1R staining, and their interaction. Adding PTEN and IGF1R expression to the base model increased AUC from 0.837 to 0.864, and improved NRI (NRI = 0.191, p = 0.004) and IDI (IDI = 0.0129, p = 0.0007) significantly. These results suggest that inclusion of PTEN and IGF1R expression and their interaction in the base model offer further predicting power for prostate cancer prognosis.
DISCUSSION
To our knowledge, our study is one of the largest studies to evaluate the role of PTEN protein expression level in relation to the risk of lethal prostate cancer. We observed a significant positive association between reduction in PTEN protein expression and development of lethal disease, although the association was attenuated after adjustment for tumor stage, Gleason grade, and PSA levels. We are also the first to report that the association of PTEN tumor expression with clinical outcome is strongly modified by the level of IGF1R expression in the tumor. These two factors negatively interact with each other: Unfavorable changes in protein expression of either biomarker alone are associated with increased risk, but combined unfavorable changes in both do not confer additional harm. PTEN and IGF1R expression in tumor predict prostate cancer survival independently of clinical prognostic factors.
Loss of PTEN expression has been shown to be correlated with indicators of poorer prognosis, such as high grade (17-20), advanced stage (18, 20), and the number of microvessels, a marker of angiogenesis (21). Several studies also reported that PTEN inactivation is associated with biochemical or clinical recurrence of prostate cancer (22-25). Three recent studies examined the effect of PTEN loss in relation to cause-specific death, which is the most relevant outcome for prostate cancer (18, 19, 26). Reid et al. performed fluorescence in situ hybridization (FISH) assays in TMAs to detect PTEN loss in 308 patients with conservatively managed prostate cancer and reported that loss of PTEN alone was not associated with prostate cancer survival (18). Pourmand et al. used tissue microdissection and polymerase chain reaction/single-strand conformation polymorphism methods to analyze PTEN mutations in 51 prostate tumor specimens obtained from radical prostatectomy or transurethral resection (19). Patients with PTEN mutations were found to have lower survival rates (P = 0.001). When Cox regression models were employed, with adjustment for Gleason grade, age and PSA at diagnosis, PTEN mutations were no longer associated with death by prostate cancer (P = 0.60). In the study by Sircar et al. (26), TMAs were constructed from transurethral resection samples of 59 patients with hormone refractory prostate cancer. The four-color FISH strategy was employed to evaluate genomic deletion of PTEN. The number of PTEN gene deletions (undeleted, heterozygous, and homozygous) was significantly correlated with prostate cancer-specific mortality (r = 0.332, P = 0.04). Yet presence versus absence of any PTEN deletion, by itself, was not correlated with cancer death (r = 0.189, P = 0.3). These findings also imply that the change in PTEN protein level is more relevant in association with prostate cancer survival.
The role of circulating IGF1 and its binding proteins such as IGFBP3 in prostate cancer has been extensively investigated (35-46). Evidence that high levels of circulating IGF1 significantly increase the risk of total prostate cancer became less than convincing with some recent reports of null findings (37, 41, 43). However, several studies, including the PHS and HPFS, have shown that IGF1 is more strongly associated with low-grade prostate cancer (35, 40, 45). A recent pooled analysis of individual patient data from 12 prospective studies yielded a significantly elevated risk for prostate cancer, comparing the highest vs. the lowest quintiles of the serum IGF-1 concentration (46). This increased risk became more pronounced when the analysis was restricted to low-grade prostate tumors, suggesting poorly-differentiated tumors, which are more autonomous, are less sensitive to the stimulus of IGF1. Such reduced dependence of IGF1 may be the result of alterations in expression of the downstream signaling pathway molecules, including those in the PI3K/Akt pathway. As described previously, loss of PTEN is strongly associated with high grade prostate tumor. We also found that lower PTEN levels were significantly associated with higher Gleason grade. Thus our finding that higher levels of IGF1R expression in tumor was related to lethal prostate cancer when PTEN level was high whereas lower PTEN was related to lethal outcome regardless IGF1R further support the notion that poorly-differentiated tumors are less sensitive to IGF1/IGF1R.
In addition to IGF1R, IGF1 also binds to and activate insulin receptor (IR) at a weaker affinity (29). However, no interaction between PTEN and IR expression was present in our study population (data not shown), suggesting the interaction between PTEN and IGF1R is specific to the IGF1/PI3K/Akt signaling cascade and the involvement of insulin signaling may be limited.
There are potential limitations in our study to be considered. First, the PHS and HPFS Tumor Cohort consists of physicians and health professionals, are mainly Caucasian, and have relatively high socio-economic status. Thus it is not a random sample of the general population. This may limit the generalizability of the results, though the validity of the study is not undermined. We may be able to gain insights into the underlying biological mechanism through which reduced PTEN expression may promote prostate cancer progression. Secondly, we have relatively few deaths or distant metastases from prostate cancer considering our large sample size. However, despite the small number of outcomes, we had sufficient power to detect the interaction between PTEN and IGF1R expression. Thirdly, the tumor specimens were obtained only when men underwent prostatectomy or TURP as curative treatment for their cancer. These are largely localized, organ-confined tumors. Those who were diagnosed with more advanced cancers may be ineligible for surgical intervention and have undergone hormonal, radiation therapy, or chemotherapy. Since loss or reduction of PTEN is generally regarded as a late event in prostatic tumorigenesis, and is associated with more malignant tumors and worse prognosis, it is likely that our analysis underestimates the role of PTEN expression in relation to prostate cancer mortality. The use of tumor blocks may generate certain heterogeneity for these tumors were diagnosed at different times, and fixed and processed in different pathological labs. Last, the presence of missing data such as PSA at diagnosis and baseline smoking status can lead to residual confounding by these factors. Uncontrolled confounding by unknown or unmeasured factors can not be ruled out either.
The prospective study design with long-term and complete follow-up adds to the strength of our analysis. Clinical characteristics of the cancers were systematically extracted from medical records in a standardized format. The study pathologists reviewed the records and provided uniform Gleason grades for all cases. These approaches should contribute to reduction in measurement errors. Additionally, we used cause-specific death and distant metastasis as the outcome, which are the most clinically relevant endpoints for prostate cancer.
In conclusion, low levels of PTEN protein are associated with an increased risk of lethal prostate cancer. This association is strongly modified by IGF1R expression in the tumor. Our results reinforce the idea that the PI3K/PTEN/Akt pathway plays a critical role in prostate cancer survival. PTEN and IGF1R are eligible independent predictors for prognosis. Their interaction may shine light on the complex picture of IGF axis in prostate tumorigenesis and cancer progression.
ACKNOWLEDGEMENTS
We are grateful to the participants and staff in the Physicians’ Health Study for their ongoing participation and support. We would also like to thank the participants and staff of the Health Professionals Follow-up Study, for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. We would like to thank Luba Bondarenko, Li Moy, and Julia Fleet for tumor block collection, Chungdak Li for construction of the tumor tissue microarrays, and Edward Stack for generating images on IHC staining of PTEN.
GRANT SUPPORT This project was funded by NIH R01 CA133891 (Giovannucci), CA136578 (Mucci), CA141298 (Stampfer), US Army Prostate Cancer Program W81XWH-05-1-0562 (Mucci). The Physicians’ Health Study is supported by grants CA-34933, CA-40360, and CA-091793 from the National Cancer Institute and grants HL-26490 and HL-34595 from the National Heart, Lung, and Blood Institute, Bethesda, MA. The Health Professionals Follow-up study is supported by NIH UM1 CA167552. Drs. Martin and Mucci are supported by the Prostate Cancer Foundation.
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST No potential conflicts of interest were disclosed.
REFERENCES
- 1.Andriole GL, Crawford ED, Grubb RL, 3rd, Buys SS, Chia D, Church TR, et al. Mortality results from a randomized prostate-cancer screening trial. N Engl J Med. 2009;360:1310–9. doi: 10.1056/NEJMoa0810696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schroder FH, Hugosson J, Roobol MJ, Tammela TL, Ciatto S, Nelen V, et al. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360:1320–8. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 3.Lee TH, Kantoff PW. McNaughton-Collins MF. Screening for prostate cancer. N Engl J Med. 2009;360:e18. doi: 10.1056/NEJMp0901825. [DOI] [PubMed] [Google Scholar]
- 4.Draisma G, Etzioni R, Tsodikov A, Mariotto A, Wever E, Gulati R, et al. Lead time and overdiagnosis in prostate-specific antigen screening: importance of methods and context. J Natl Cancer Inst. 2009;101:374–83. doi: 10.1093/jnci/djp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albertsen PC, Hanley JA, Fine J. 20-year outcomes following conservative management of clinically localized prostate cancer. JAMA. 2005;293:2095–101. doi: 10.1001/jama.293.17.2095. [DOI] [PubMed] [Google Scholar]
- 6.Porter CR, Kodama K, Gibbons RP, Correa R, Jr., Chun FK, Perrotte P, et al. 25-year prostate cancer control and survival outcomes: a 40-year radical prostatectomy single institution series. J Urol. 2006;176:569–74. doi: 10.1016/j.juro.2006.03.094. [DOI] [PubMed] [Google Scholar]
- 7.Johansson JE, Andren O, Andersson SO, Dickman PW, Holmberg L, Magnuson A, et al. Natural history of early, localized prostate cancer. JAMA. 2004;291:2713–9. doi: 10.1001/jama.291.22.2713. [DOI] [PubMed] [Google Scholar]
- 8.Bill-Axelson A, Holmberg L, Ruutu M, Haggman M, Andersson SO, Bratell S, et al. Radical prostatectomy versus watchful waiting in early prostate cancer. N Engl J Med. 2005;352:1977–84. doi: 10.1056/NEJMoa043739. [DOI] [PubMed] [Google Scholar]
- 9.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–8. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–7. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 11.Ali IU, Schriml LM, Dean M. Mutational spectra of PTEN/MMAC1 gene: a tumor suppressor with lipid phosphatase activity. J Natl Cancer Inst. 1999;91:1922–32. doi: 10.1093/jnci/91.22.1922. [DOI] [PubMed] [Google Scholar]
- 12.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 13.Osaki M, Oshimura M, Ito H. PI3K-Akt pathway: its functions and alterations in human cancer. Apoptosis. 2004;9:667–76. doi: 10.1023/B:APPT.0000045801.15585.dd. [DOI] [PubMed] [Google Scholar]
- 14.Ma X, Ziel-van der Made AC, Autar B, van der Korput HA, Vermeij M, van Duijn P, et al. Targeted biallelic inactivation of Pten in the mouse prostate leads to prostate cancer accompanied by increased epithelial cell proliferation but not by reduced apoptosis. Cancer Res. 2005;65:5730–9. doi: 10.1158/0008-5472.CAN-04-4519. [DOI] [PubMed] [Google Scholar]
- 15.Backman SA, Ghazarian D, So K, Sanchez O, Wagner KU, Hennighausen L, et al. Early onset of neoplasia in the prostate and skin of mice with tissue-specific deletion of Pten. Proc Natl Acad Sci U S A. 2004;101:1725–30. doi: 10.1073/pnas.0308217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S, Gao J, Lei Q, Rozengurt N, Pritchard C, Jiao J, et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 2003;4:209–21. doi: 10.1016/s1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- 17.Dreher T, Zentgraf H, Abel U, Kappeler A, Michel MS, Bleyl U, et al. Reduction of PTEN and p27kip1 expression correlates with tumor grade in prostate cancer. Analysis in radical prostatectomy specimens and needle biopsies. Virchows Arch. 2004;444:509–17. doi: 10.1007/s00428-004-1004-6. [DOI] [PubMed] [Google Scholar]
- 18.Reid AH, Attard G, Ambroisine L, Fisher G, Kovacs G, Brewer D, et al. Molecular characterisation of ERG, ETV1 and PTEN gene loci identifies patients at low and high risk of death from prostate cancer. Br J Cancer. 2010;102:678–84. doi: 10.1038/sj.bjc.6605554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pourmand G, Ziaee AA, Abedi AR, Mehrsai A, Alavi HA, Ahmadi A, et al. Role of PTEN gene in progression of prostate cancer. Urol J. 2007;4:95–100. [PubMed] [Google Scholar]
- 20.McMenamin ME, Soung P, Perera S, Kaplan I, Loda M, Sellers WR. Loss of PTEN expression in paraffin-embedded primary prostate cancer correlates with high Gleason score and advanced stage. Cancer Res. 1999;59:4291–6. [PubMed] [Google Scholar]
- 21.Giri D, Ittmann M. Inactivation of the PTEN tumor suppressor gene is associated with increased angiogenesis in clinically localized prostate carcinoma. Hum Pathol. 1999;30:419–24. doi: 10.1016/s0046-8177(99)90117-x. [DOI] [PubMed] [Google Scholar]
- 22.McCall P, Witton CJ, Grimsley S, Nielsen KV, Edwards J. Is PTEN loss associated with clinical outcome measures in human prostate cancer? Br J Cancer. 2008;99:1296–301. doi: 10.1038/sj.bjc.6604680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halvorsen OJ, Haukaas SA, Akslen LA. Combined loss of PTEN and p27 expression is associated with tumor cell proliferation by Ki-67 and increased risk of recurrent disease in localized prostate cancer. Clin Cancer Res. 2003;9:1474–9. [PubMed] [Google Scholar]
- 24.Yoshimoto M, Cunha IW, Coudry RA, Fonseca FP, Torres CH, Soares FA, et al. FISH analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor clinical outcome. Br J Cancer. 2007;97:678–85. doi: 10.1038/sj.bjc.6603924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koumakpayi IH, Le Page C, Mes-Masson AM, Saad F. Hierarchical clustering of immunohistochemical analysis of the activated ErbB/PI3K/Akt/NF-kappaB signalling pathway and prognostic significance in prostate cancer. Br J Cancer. 2010;102:1163–73. doi: 10.1038/sj.bjc.6605571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sircar K, Yoshimoto M, Monzon FA, Koumakpayi IH, Katz RL, Khanna A, et al. PTEN genomic deletion is associated with p-Akt and AR signalling in poorer outcome, hormone refractory prostate cancer. J Pathol. 2009;218:505–13. doi: 10.1002/path.2559. [DOI] [PubMed] [Google Scholar]
- 27.Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–24. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larsson O, Girnita A, Girnita L. Role of insulin-like growth factor 1 receptor signalling in cancer. Br J Cancer. 2005;92:2097–101. doi: 10.1038/sj.bjc.6602627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hartog H, Wesseling J, Boezen HM, van der Graaf WT. The insulin-like growth factor 1 receptor in cancer: old focus, new future. Eur J Cancer. 2007;43:1895–904. doi: 10.1016/j.ejca.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 30.Lopez T, Hanahan D. Elevated levels of IGF-1 receptor convey invasive and metastatic capability in a mouse model of pancreatic islet tumorigenesis. Cancer Cell. 2002;1:339–53. doi: 10.1016/s1535-6108(02)00055-7. [DOI] [PubMed] [Google Scholar]
- 31.Carboni JM, Lee AV, Hadsell DL, Rowley BR, Lee FY, Bol DK, et al. Tumor development by transgenic expression of a constitutively active insulin-like growth factor I receptor. Cancer Res. 2005;65:3781–7. doi: 10.1158/0008-5472.CAN-04-4602. [DOI] [PubMed] [Google Scholar]
- 32.Xie Y, Skytting B, Nilsson G, Brodin B, Larsson O. Expression of insulin-like growth factor-1 receptor in synovial sarcoma: association with an aggressive phenotype. Cancer Res. 1999;59:3588–91. [PubMed] [Google Scholar]
- 33.Belfiore A, Pandini G, Vella V, Squatrito S, Vigneri R. Insulin/IGF-I hybrid receptors play a major role in IGF-I signaling in thyroid cancer. Biochimie. 1999;81:403–7. doi: 10.1016/s0300-9084(99)80088-1. [DOI] [PubMed] [Google Scholar]
- 34.All-Ericsson C, Girnita L, Seregard S, Bartolazzi A, Jager MJ, Larsson O. Insulin-like growth factor-1 receptor in uveal melanoma: a predictor for metastatic disease and a potential therapeutic target. Invest Ophthalmol Vis Sci. 2002;43:1–8. [PubMed] [Google Scholar]
- 35.Nimptsch K, Platz EA, Pollak MN, Kenfield SA, Stampfer MJ, Willett WC, et al. Plasma insulin-like growth factor 1 is positively associated with low-grade prostate cancer in the Health Professionals Follow-up Study 1993-2004. Int J Cancer. 2011;128:660–7. doi: 10.1002/ijc.25381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Platz EA, Pollak MN, Leitzmann MF, Stampfer MJ, Willett WC, Giovannucci E. Plasma insulin-like growth factor-1 and binding protein-3 and subsequent risk of prostate cancer in the PSA era. Cancer Causes Control. 2005;16:255–62. doi: 10.1007/s10552-004-3484-8. [DOI] [PubMed] [Google Scholar]
- 37.Severi G, Morris HA, MacInnis RJ, English DR, Tilley WD, Hopper JL, et al. Circulating insulin-like growth factor-I and binding protein-3 and risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:1137–41. doi: 10.1158/1055-9965.EPI-05-0823. [DOI] [PubMed] [Google Scholar]
- 38.Stattin P, Rinaldi S, Biessy C, Stenman UH, Hallmans G, Kaaks R. High levels of circulating insulin-like growth factor-I increase prostate cancer risk: a prospective study in a population-based nonscreened cohort. J Clin Oncol. 2004;22:3104–12. doi: 10.1200/JCO.2004.10.105. [DOI] [PubMed] [Google Scholar]
- 39.Harman SM, Metter EJ, Blackman MR, Landis PK, Carter HB. Baltimore Longitudinal Study on A. Serum levels of insulin-like growth factor I (IGF-I), IGF-II, IGF-binding protein-3, and prostate-specific antigen as predictors of clinical prostate cancer. J Clin Endocrinol Metab. 2000;85:4258–65. doi: 10.1210/jcem.85.11.6990. [DOI] [PubMed] [Google Scholar]
- 40.Li L, Yu H, Schumacher F, Casey G, Witte JS. Relation of serum insulin-like growth factor-I (IGF-I) and IGF binding protein-3 to risk of prostate cancer (United States) Cancer Causes Control. 2003;14:721–6. doi: 10.1023/a:1026383824791. [DOI] [PubMed] [Google Scholar]
- 41.Mikami K, Ozasa K, Nakao M, Miki T, Hayashi K, Watanabe Y, et al. Prostate cancer risk in relation to insulin-like growth factor (IGF)-I and IGF-binding protein-3: A nested case-control study in large scale cohort study in Japan. Asian Pac J Cancer Prev. 2009;10(Suppl):57–61. [PubMed] [Google Scholar]
- 42.Mucci LA, Stark JR, Pollak MN, Li H, Kurth T, Stampfer MJ, et al. Plasma levels of acid-labile subunit, free insulin-like growth factor-I, and prostate cancer risk: a prospective study. Cancer Epidemiol Biomarkers Prev. 2010;19:484–91. doi: 10.1158/1055-9965.EPI-09-0836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Borugian MJ, Spinelli JJ, Sun Z, Kolonel LN, Oakley-Girvan I, Pollak MD, et al. Prostate cancer risk in relation to insulin-like growth factor (IGF)-I and IGF-binding protein-3: a prospective multiethnic study. Cancer Epidemiol Biomarkers Prev. 2008;17:252–4. doi: 10.1158/1055-9965.EPI-07-2694. [DOI] [PubMed] [Google Scholar]
- 44.Chan JM, Stampfer MJ, Giovannucci E, Gann PH, Ma J, Wilkinson P, et al. Plasma insulin-like growth factor-I and prostate cancer risk: a prospective study. Science. 1998;279:563–6. doi: 10.1126/science.279.5350.563. [DOI] [PubMed] [Google Scholar]
- 45.Chan JM, Stampfer MJ, Ma J, Gann P, Gaziano JM, Pollak M, et al. Insulin-like growth factor-I (IGF-I) and IGF binding protein-3 as predictors of advanced-stage prostate cancer. J Natl Cancer Inst. 2002;94:1099–106. doi: 10.1093/jnci/94.14.1099. [DOI] [PubMed] [Google Scholar]
- 46.Roddam AW, Allen NE, Appleby P, Key TJ, Ferrucci L, Carter HB, et al. Insulin-like growth factors, their binding proteins, and prostate cancer risk: analysis of individual patient data from 12 prospective studies. Ann Intern Med. 2008;149:461–71. W83–8. doi: 10.7326/0003-4819-149-7-200810070-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stark JR, Perner S, Stampfer MJ, Sinnott JA, Finn S, Eisenstein AS, et al. Gleason score and lethal prostate cancer: does 3 + 4 = 4 + 3? J Clin Oncol. 2009;27:3459–64. doi: 10.1200/JCO.2008.20.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]