Abstract
Protein phosphorylation is reversibly regulated by the interplay between kinases and phosphatases. Recent developments within the field of proteomics have revealed the extent of this modification in nature. To date there is still a lack of information about phosphatase specificity for different proteomes and their conditions to achieve maximum enzyme activity. This information is important per se, and in addition often requested in functional and biochemical in vitro studies, where a dephosphorylated sample is needed as a negative control to define baseline conditions. In this study, we have addressed the effectiveness of two phosphatases endogenously present in the heart (protein phosphatases 1 and 2A) and two generic phosphatases (alkaline phosphatase and lambda protein phosphatase) on three cardiac subproteomes known to be regulated by phosphorylation. We optimized the dephoshorylating conditions on a cardiac tissue fraction comprising cytosolic and myofilament proteins using 2-DE and MS. The two most efficient conditions were further investigated on a mitochondrial-enriched fraction. Dephosphorylation of specific proteins depends on the phosphatase, its concentration, as well as sample preparation including buffer composition. Finally, we analyzed the efficiency of alkaline phosphatase, the phosphatase with the broadest substrate specificity, using TiO2 peptide enrichment and 2DLC-MS/MS. Under these conditions, 95% of the detected cardiac cytoplasmic-enriched phospho-proteome was dephosphorylated. In summary, targeting dephosphorylation of the cardiac muscle subproteome or a specific protein will drive the selection of the specific phosphatase, and each requires different conditions for optimal performance.
Keywords: dephosphorylation, phosphatase, phosphorylation, cardiac muscle, 2-DE DIGE
Phosphorylation is strictly regulated by kinases and phosphatases [1]. Methods to identify protein phosphorylation have rapidly developed in recent years and it is now possible to identify hundreds to thousands of phospho-peptides using MS [2]. For intact proteins, phosphorylation can be assessed by MS based methods or other in vitro methods including radiolabeling, anti-phosphorylation antibodies, phospho-stains (e.g. ProQ Diamond), phosphate-binding agents (e.g. Phos-tag) and 2-DE [3–8]. To confirm phosphorylation, different types of in vitro dephosphorylation have been used [7;9], however the effectiveness of dephosphorylation is not always addressed. This has limited the use of negative control samples during proteomic and protein biochemistry experiments and reduced the ability to assign functional consequences based on comparison with dephosphorylated samples.
In the present study, we focus on the cardiac muscle proteome which is regulated by phosphorylation [10]. For example, highly abundant regulatory proteins like troponin I, C and T, myosin light chain 2 and ATP synthase are all modulated through phosphorylation [4;10;11]. These proteins are involved in regulation of myocardial functions including muscle contraction and energy metabolism [10]. Even so, the cardiac substrates for a number of phosphatases and the optimal conditions for dephosphorylation are not known. We therefore focused on in vitro dephosphorylation of the three main cardiac subproteomes (myofilament, soluble/cytoplasmic and mitochondrial-enriched fractions) assessing individual features of four phosphatases. Two of these phosphatases, protein phosphatases 1 and 2A (PP1 and PP2A), are endogenously present in the myocardium and the other two, alkaline phosphatase (AP) and lambda protein phosphatase (λ-PP), are generic phosphatases commonly used in in vitro studies. Our study has resulted in a set of optimized conditions applicable for downstream investigations. By optimizing conditions for intact proteins rather than peptides, we allow for proteomic, protein biochemistry and functional assays to be subsequently carried out. We show that dephosphorylation of individual cardiac proteins importantly depends on the specific phosphatase and its concentration, in addition to sample preparation and buffer composition.
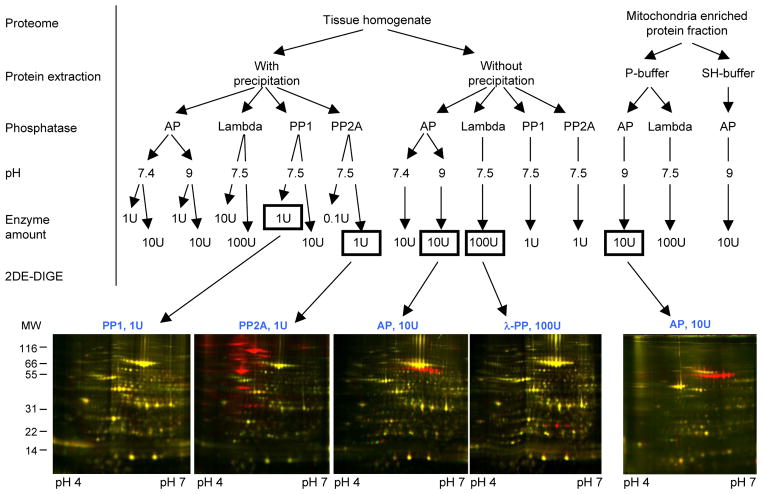
The experimental conditions are outlined in Figure 1 and the online supplement provides an extensive description of the methods. Briefly, mouse left ventricular (LV) myocardium (Pel-Freez Biologica, Rogers, AR) was homogenized in ice-cold 25mM HEPES buffer, pH 7.4, supplemented with protease (Complete, Roche) and phosphatase inhibitors (2.5mM EDTA, 50mM NaF, 0.25mM Na3VO4 and 0.25mM PMSF). The resulting suspension is dominated by the contractile myofilament subproteome and abundant cytosolic proteins [12]. Phosphatase inhibitors were removed from the sample by precipitation (using the 2D clean-up kit, GE Healthcare). The resulting pellets were resuspended in 0.2% SDS/25mM HEPES (termed “with precipitation”). Tissue was alternatively homogenized in the absence of phosphatase inhibitors in ice-cold 25mM HEPES buffer, pH 7.4, supplemented with protease inhibitors (termed “without precipitation”). A mitochondrial-enriched fraction was prepared using a well established centrifugation method [13] and either resuspended directly in phosphatase buffer (termed “P-buffer”) or in 0.2% SDS/25mM HEPES buffer (termed “SH-buffer”).
Figure 1.
Optimization of in vitro dephosphorylation of cardiac muscle by PP1, PP2A, AP and λ-PP monitored by 2-DE DIGE. The optimized condition for each phosphatase is boxed and the representative images are shown below (n≥2).
Dephosphorylation of the tissue homogenate and the mitochondria-enriched fraction (25μg protein) was carried out overnight (16 hrs) and samples without enzyme served as controls. Alkaline phosphatase (New England Biolabs [NEB]) and PP2A (Millipore) treatment were performed at 37°C, while PP1 (NEB) and λ-PP (NEB) treatment were carried out at 30°C. Enzyme quantities (units, U) are provided in Figure 1. Dephosphorylation with AP was performed both at pH 7.4 as recommended by manufacturer, and at pH 9 since the enzyme’s pH optimum range is 8.5–10 [14]. Each experimental condition was analyzed in biological (n=2) and technical replicates (n= 2–4).
The dephosphorylated samples and controls were labeled with different fluorescent dyes [13]. After quenching, samples were combined and diluted in IEF re-hydration buffer (8M urea, 2.5M thiourea, 4% CHAPS, 0.5% ampholytes, 50mM DTT, and 0.01% w/v bromophenol blue). An extra amount of non-labeled sample was also included for downstream silver staining and MS analysis and a total of 150μg proteins were separated using IPG strips (pH 4–7) by means of an IEF Protean apparatus (Bio-Rad). After reduction (1% DTT) and alkylation (2.5% iodoacetamide), proteins were separated by SDS-PAGE in the second dimension. Gels were then fixed and fluorescent images acquired using a laser scanner (9400 Typhoon, GE Healthcare). In merged images of a treated and non-treated sample, dephosphorylation was assessed as a clear shift in protein spot pI, towards a more basic pH independent of dye-labeling (dye swap) (Table 1, Supplementary Figure 2–7, n≥2). The gels were silver stained, and protein spots of interest excised (Supplementary Figure 1), destained and in-gel digested with trypsin (Promega) as previously described [13]. Digested samples were analyzed by either MALDI or ESI MS/MS (Supplementary Table 1 and 2).
Table 1.
Differences in substrate specificity of AP, λ-PP, PP1 and PP2A
| Spot no | Protein ID | DEPHOSPHORYLATIONa
|
Superior phosphataseb | Additional phosphataseb | AP pH preferencec | Non responsive | Phospho- proteind |
|---|---|---|---|---|---|---|---|
| 1 | ATP syntase δ (MW:15.5/pI: 4.3) |

|
AP (i) | ND | Indifferent | PP1, PP2A, λ-PP | Yes |
| 2 | 14 kDa phosphohistidine phosphatase Galectin-1 (MW:13.0/pI: 5.3) |
|
AP (s) | ND | Indifferent | PP1, PP2A, λ- PP | Yes (all) |
| 3 | Myosin regulatory light chain 2 ventricular/ cardiac muscle isoform (MW:16.5/pI: 5.0) |
|
AP (i) | ND | pH 9** | PP1, PP2A, λ- PP | Yes |
| 4 | Adenylate kinase isoenzyme 1 ATP synthase δ (MW:19.4/pI: 5.6) |
|
AP (p) | ND | Indifferent | PP1, PP2A, λ- PP | No |
| 5 | 39S ribosomal protein L12 (MW:18.5/pI: 5.7) |
|
AP (p) | ND | Indifferent | PP1, PP2A, λ- PP | No |
| 6 | Unknown 1 (MW:18.7/pI: 6.2) |

|
λ-PP (s) | PP1 (i), PP2A (i) | N/A | AP | N/A |
| 7 | Unknown 2 (MW:25.4/pI: 4.6) |
|
λ-PP (i) | ND | N/A | AP, PP1 PP2A | N/A |
| 8 | Protein DJ-1 (MW:23.5/pI: 6.5) |

|
AP (p) | ND | pH 9** | PP1, PP2A, λ- PP | Yes |
| 9 | Heat shock protein β-1 (MW:23.7/pI: 6.5) |
|
AP (p) | PP2A (p) | Indifferent | N/A | Yes |
| 10 | Tropomyosin α-1 (MW:33.4/pI: 4.7) |

|
PP2A (p) | PP1 (p), λ-PP (i) | N/A | AP | Yes |
| 11 | Electron transfer flavoprotein α (MW:34.7/pI: 6.2) |
|
AP (s) | ND | Indifferent | PP1, PP2A, λ- PP | Yes |
| 12 | Troponin T, cardiac muscle (MW:38.4.5/pI: 5.7) |
|
PP1 (p) | PP2A (p), λ-PP (s) | N/A | AP | Yes |
| 13 | Unknown 3 (MW:48.3/pI: 5.7) |
|
λ-PP (s) | AP (s) | Indifferent | PP1 | N/A |
| 14 | cAMP-dependent protein kinase type I-α (MW:49.0/pI: 5.6) |

|
λ-PP (i) | AP (p) | pH 9** | PP1 | Yes |
| 15 | Guanine deaminase α-2-macro-globulin α-2-HS-glycoprotein ATP synthase β Serum albumin (MW:53.3/pI: 5.6) |

|
λ-PP (i) | ND | N/A | AP, PP1 | Yes (all except Guanase) |
| 16 | Pyruvate dehydrogenase E1α (MW:45.4/pI: 6.7) |
|
AP (p) | PP1 (p), PP2A (p), λ-PP (p) | Indifferent | ND | Yes |
| 17 | Phosphoglucomutase-1 (MW:63.6/pI: 6.7) |
|
PP1 (p) PP2A (p) | AP (p), λ-PP (p) | Indifferent | ND | Yes |
| 18 | Heat shock protein HSP 90-β (MW:83.9/pI: 5.2) |
|
λ-PP (s) | AP (i) | Indifferent | PP1, | Yes |
| 19 | Stress-70 protein, mitochondrial Heat shock cognate 71 kDa protein (MW:75.3/pI: 5.8) |
|
AP (i) | ND | indifferent | PP1, PP2A, λ- PP | Yes |
| 20 | Propionyl-CoA carboxylase α (MW:78.9/pI: 6.6) |
|
AP (p) | ND | Indifferent | PP1, PP2A, λ- PP | Yes |
| 21 | Heat shock 70 kDa protein 4 (MW:98.5/pI: 5.4) |
|
AP (s) | ND | pH 9** | PP1, λ-PP | Yes |
| 22 | Vinculin (MW: 108.8/pI: 6.3) |
|
AP (p) | ND | pH 9** | PP1, PP2A, λ-PP | Yes |
| 23 | ATP synthase δ (MW:15.4/pI: 4.4) |

|
AP (i) | ND | N/A | λ -PP | Yes |
| 24 | Annexin A5 (MW:23.5/pI: 4.8) |

|
AP (s) | ND | N/A | λ -PP | Yes |
| 25 | 39S ribosomal protein L12 (MW:18.8/pI: 5.7) |
|
AP (i) | ND | N/A | λ -PP | No |
A representative DIGE image is given for every substrate. The dephosphorylated sample is labeled with cy5 (red color) *indicate the protein spot used for ID, if more than one spot, the experimental MW and pI is shown for the more basic. Arrow points at the dephosphorylated form of MRP-L12.
Specific phosphatases for a substrate and how SDS presence is affecting enzyme activity: (p) preferred, (s) sensitive or (i) indifferent (e.g. no notably difference with and without SDS).
Prefered pH for the AP buffer. **also dephosphorylated at pH 7.4, but more pronounced at pH 9.
As reported in the database UniprotKB.
ND: not detected; N/A: not applicable
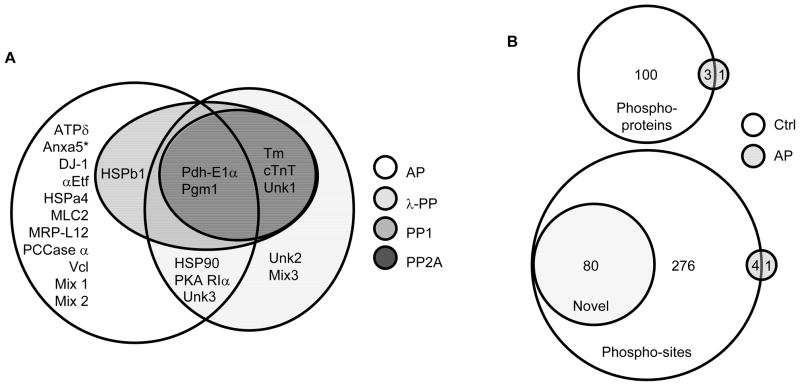
When studying PTMs there is always a balance between maintaining endogenous PTMs during the protein isolation procedure and successfully perform enzymatic assays downstream of the isolation. Initially, proteins were isolated in the presence of phosphatase inhibitors to capture their in vivo phosphorylation status. This required protein precipitation prior to phosphatase treatment (to remove the inhibitors) and resuspension of the proteins in 0.2% SDS. Under these conditions, the two cardiac serine/threonine-protein phosphatases (PP1 and PP2A) dephosphorylated 5 and 6 cardiac proteins respectively, including tropomyosin (Tm) and troponin T (cTnT) in addition to pyruvate dehydrogenase E1 (Pdh-E1α), phosphoglucomutase-1 (Pgm1) and an unknown protein (Unk1), which we were unable to identify by MS/MS (Table 1). PP2A also dephosphorylated heat shock protein beta-1 (HSPβ1). For PP2A, dephosphorylation efficiency was partly concentration-dependent and both Tm and cTnT showed a more distinct dephosphorylation pattern at the higher enzyme concentration (Supplementary Figure 2). Consequently, 1U of PP2A was chosen for the rest of the experiments (Figure 1). Surprisingly, the higher concentration of PP1 (10U) strongly interfered with the labeling efficiency of the fluorescent dye. This might be due to chemicals in the proprietary enzyme solution (e.g. primary amines and DTT) since the reduced labeling efficiency was not observed in the control sample. Since the lower concentration of PP1 (1U, boxed in Figure 1) was sufficient to dephosphorylate Tm, cTnT, Pdh-E1α, Pgm1 and Unk1, this concentration was used for the remainder of the experiments (Supplementary Figure 3).
AP and λ-PP are promiscuous phosphatases and displayed a reduced specificity (intended as an increase in number of substrates) compared to PP1 and PP2A. PP1 and PP2A are expected to have even higher substrate specificity in vivo, as they appear in multimeric complexes with multiple inhibitors. In the in vitro conditions used, only the catalytic (PP1) or catalytic and scaffolding subunits (PP2A) were included [15]. Combined, AP and λ-PP dephosphorylated all the identified substrates of PP1 and PP2A. However, there are important differences in the substrate selectivity between AP and λ-PP. While AP displayed the broadest substrate affinity, it was unable to effectively dephosphorylate Tm and cTnT, nor the unknown proteins Unk1, -2 and -3 (Table 1, Figure 2, Supplementary Figure 4 and 5). These were instead all substrates of λ-PP. In contrast, only AP was able to dephosphorylate other cardiac proteins like ATP synthase delta (ATPδ), myosin regulatory light chain 2 (MLC2) and some of the heat shock proteins (Stress 70 protein [GRP75] and heat shock cognate 71 kDa protein [Hsc70]). The only mutual substrates for AP and λ-PP not dephosphorylated by PP1 and PP2 were cAMP- dependent protein kinase type 1 (PKA RIα) and heat shock protein 90 β (HSP90). Also for AP and λ-PP a 10-fold increase in enzyme concentration (1 to 10U for AP and 10 to 100U for λ-PP) resulted in an increase in the number of dephosphorylated targets (30% and 75%, respectively) (Supplementary Figure 4 and 5). Consequently, the higher concentrations were used for the following experiments.
Figure 2.
A) Substrate selectivity of the four phosphatases AP, λ-PP, PP1 and PP2A analysed by 2-DE DIGE MS/MS. Mix: Spots assigned with more than one ID. Unk: Unknown proteins which we could not identify by MS/MS. *only tested by AP and λ-PP. B) Overlap of phospho-proteins (upper) and phospho-sites (lower) observed in control and AP samples after TiO2 peptide enrichment and 2DLC-MS/MS (Supplementary Tables 4 and 5). Husberg et al: Dephosphorylation of Cardiac Proteins in vitro - a Matter of Phosphatase Specificity
AP, in contrast to the other three phosphatases, is reported to have an optimal in vitro activity above physiological pH, between pH 8.5 and 10 [14]. However, it has been suggested that pH may be critical only at high substrate concentrations [14]. Since the enzyme was supplied with a buffer of pH 7.4, we also analyzed the substrate specificity at pH 9 (Supplementary Figure 6). By increasing the pH towards the reported optimum, the degree of dephosphorylation of some of the substrates increased (MLC-2, DJ-1, PKA RIα and vinculin [VCL]) (Table 1). No new substrates were identified with the increase of the buffer’s pH, supporting the idea that a pH shift cannot alter the phosphatase specificity, but rather its efficiency.
SDS is a detergent commonly used in preparations of protein samples for proteomic studies. Knowing that SDS can influence phosphatase activity [16], we wanted to analyze if the activities of the four phosphatases increased when SDS was not present (intended as an increased number of substrates). In this instance, the only action to preserve the in vivo phosphorylation status of the proteins was to perform the protein isolation on ice (samples termed “without precipitation”, Figure 1). PP1 and PP2A activities were negatively affected by SDS removal, and both phosphatases were unable to dephosphorylate Pdh-E1α and Pgm1. SDS removal had a more diverse effect on AP and λ-PP activities (Table 1). AP showed enhanced dephosphorylation for 8 out of 18 identified substrates comprising 39S ribosomal protein L12 (MRP-L12), DJ-1, HSPβ1, Pgm1, Pdh-E1α, PKA RIα propionyl-CoA carboxylase (PCCase α) and VCL in the presence of SDS. In contrast, SDS removal increased dephosphorylation of electron transfer flavoprotein (α-ETF) and heat shock 70 kDa protein 4 (HSPa4) (Table 1, Supplementary Figure 4). These effects of SDS were not influenced by the reaction buffer’s pH (Supplementary Figure 6). For the λ-PP substrates, SDS presence was preferred for the dephosphorylation of Pgm1 and Pdh-E1α while for other substrates like cTnT, SDS had a negative effect. Consequently, there is not a universal negative effect of SDS on phosphatase activity, but not unexpected, it is enzyme and substrate specific. The positive effect of SDS may be due to its ability to induce conformational changes in the protein substrates, thereby making the phospho-sites more accessible to the phosphatase.
Lately it has become evident that mitochondrial proteins in the heart can be extensively phosphorylated [17]. Having established efficient conditions for AP (10U, pH 9, without precipitation) and λ-PP (100U, without precipitation), we also analyzed the effects of these two phosphatases on the mitochondrial subproteome (Figure 1). The mitochondrial-enriched protein fraction was resuspended either in the specific phosphatase reaction buffer (P-buffer) or in 0.2% SDS/25mM HEPES buffer. Surprisingly, λ-PP did not notably dephosphorylate any of the mitochondrial proteins. The only AP substrates where ATPδ, MRP-L12 and annexin A5 (Anxa5), the latter being dependent on the resuspension buffer (Table 1, Supplementary Figure 7).
Finally, we used TiO2 chromatography followed by LC-MS/MS to analyze dephosphorylation using the optimized conditions for AP. Of the 101 phospho-proteins identified, comprising 277 phospho-sites, only 4 proteins and 5 sites were detected after AP treatment, respectively (Figure 2B, supplementary Table 3). There is a surprisingly little overlap between the phosphorylated proteins identified with 2-DE MS/MS and those identified using TiO2 peptide enrichment and 2DLC-MS/MS. Only 8 proteins where identified by both methods. Reasons for this can be differential sensitivity in the methods and enrichment of different types of proteins as reported previously [18].
Our results show that thorough optimization of the conditions for phosphatase treatment is important when aiming to produce a completely dephosphorylated sample. We used 2-DE DIGE to optimize the conditions for dephosphorylation, by monitoring phosphatase activity and specificity on a broad array of substrates constituted by three important cardiac subproteomes. In total, we identified differential dephosphorylation of 25 proteins. This number reflects the large dynamic range of the cardiac proteome (in particular the myofilament subproteome dominates and it consists of <20 proteins) which limits the number of proteins observed by 2-DE. The results showed that no single phosphatase was able to dephosphorylate all cardiac proteins, but AP had the broadest substrate affinity (summarized in Figure 2). One study limitation is the presence of the exogenous phosphatase in the 2-DE gels, which may mask some of the substrates (Supplementary Figure 8). Phosphatase activity and substrate selectivity changed with enzyme concentration, the presence of SDS and the subproteome. In general, we found a dose-dependent effect on protein dephosphorylation. The only exception was constituted by PP1 emphasizing the fact that some available phosphatases are not optimized for proteomic studies. For AP, the buffer’s pH was less important than originally thought, and we only observed an effect on dephosphorylation efficiency and no notable effect on the substrate selectivity. While SDS is known to inhibit enzyme activities, in our samples SDS presence was rather influencing phosphatase selectivity than activity. SDS presence importantly increased dephosphorylation efficiency of some substrates. Finally, the optimized conditions for the phosphatase with the broadest substrate specificity (AP) were analyzed after TiO2 peptide enrichment. Even though the results showed that 95% of the cardiac subproteome was dephosphorylated under these conditions, it is interesting to notice that a single phosphatase does not easily dephosphorylate the cardiac proteome completely. To achieve this, a combination of different phosphatases might be a fruitful strategy although not straightforward due to the specific requirements for individual enzymes (e.g. buffers and reaction temperatures). Another option to consider is chemical dephosphorylation [19]. However, the PTM-specificity [20] and functional implications of this method are unclear today. Most interestingly, our results show that conditions found to be favorable for one subproteome had little positive effect on another, emphasizing the importance of optimizing conditions based on the specific subproteome to be analyzed.
SUPPLEMENTARY METHODS
Reagents
Murine ventricular tissue was purchased from Pel-Freez Biologicals (Rogers, AR). Protease inhibitors Complete, mini, EDTA-free, from Roche (Mannheim, Germany). BCA protein assay kit from Thermo Scientific Pierce (Rockford, IL). Alkaline Phosphatase (AP; Calf Intestinal Phosphatase), Lambda Protein Phosphatase (λ-PP) and Protein Phosphatase 1 (PP1) from New England Biolabs (Beverly, MA). Protein phosphatase 2A (PP2A) from Millipore (Cork, Ireland). 2D Clean-up kit, CyDyes DIGE Fluors, IPG strips (Immobiline DryStrip gel, pH 4–7, 18cm, linear gradients), IPG buffers, agarose NA from GE Healthcare (Buckinghamshire, UK). Bis-acrylamide from BioRad Laboratories (Hercules, CA), LDS sample buffer from Invitrogen (Carlsbad, CA), Phos-tag acrylamide from Wako Chemicals (Neuss, Germany) and HSPB1 antibody from Enzo Life Science (New York, NY). Sequencing grade modified porcine trypsin from Promega (Madison, WI). Ultrapure SHCA Maldi Matrix (Protea Morgantown, WV). Titanium dioxide bulk media and graphite columns were from Glygen Corporation (Columbia, MD) and Sep-pak C18 columns were from Waters Corporation (Milford, MA). All other chemicals were purchased from Fischer Bioreagents and were of the highest analytical grade.
Experimental Outline
Tissue homogenate or mitochondrial-enriched protein fractions were isolated from murine myocardium. The tissue homogenate with precipitation was used to establish the optimal enzyme concentration of the four phosphatases tested (AP, λ-PP, PP1 and PP2A). These conditions were also tested with tissue homogenate without precipitation. In addition, the mitochondrial enriched fraction resuspended in phosphatase buffer (termed “P-buffer”) was treated with λ-PP and AP, the latter was also used on the mitochondrial enriched fraction resuspended in SDS/HEPES buffer (termed “SH-buffer”) before phosphatase treatment (summarized in Figure 1 in the manuscript). Samples treated without enzyme were used as controls. Enzyme-treated samples and respective controls, were individually labeled with Cy-Dyes (GE healthcare), then combined and analyzed by 2D-DIGE. Dye labeling was alternatively swapped for every sample so that two technical replicates were generated for each condition. A protein spot clearly and consistently shifting toward a more basic pI upon phosphatase treatment was regarded as phosphatase substrate. An extra amount of un-labeled sample (150 μg/sample) was mixed with labeled ones after quenching. This was used to allow for post-silver staining. The same gels were hence used for both analytical and preparative separation, improving identification and reducing uncertainty due to misalignment issues. After silver staining, protein spots of interest were excised, in-gel digested and protein identity was determined by MS/MS.
Preparation of Tissue Homogenates
All steps were performed on ice. Frozen pieces of heart tissues were transferred to a glass homogenizer filled with ice-cold homogenization buffer (25 mM HEPES, pH 7.4 supplemented with protease inhibitors) and briefly homogenized. The homogenization buffer was alternatively supplemented with phosphatase inhibitors (2.5 mM EDTA, 50 mM NaF, 0.25 mM Na3VO4, 0.25 mM PMSF, see main text). Tissue homogenates were centrifuged (18000 rcf, 15 min, 4°C) and rinsed with ice-cold homogenization buffer once. Supernatants were collected, and pellets discarded. If the homogenization was carried out without any phosphatase inhibitors the protein concentration was determined directly by the BCA protein assay (samples termed “without precipitation”). If the homogenization buffer was supplemented with phosphatase inhibitors, the supernatant was treated with the 2-DE Clean-up kit following the manufactures protocol for samples of more than 100 μg protein. After precipitation, the protein sample was resuspended in 25 mM HEPES, pH 7.4 with 0.2% SDS and protein concentration determined (samples termed “with precipitation”).
Preparation of Mitochondria enriched protein fractions
Isolation of mitochondria-enriched fraction was performed as previously described [1]. Briefly, pieces of frozen heart tissues were homogenized in a glass homogenizer filled with ice-cold homogenization-buffer (220 mM mannitol, 70 mM Sucrose, 20 mM HEPES, pH 7.4). Tissue homogenates were filtered through a 100 μm filter. The filtrate was centrifuged (1100 rcf, 5 min, 4°C) and rinsed with ice-cold homogenization-buffer four times. The supernatants were combined (pellets comprising cell debris, nuclei, and intact cells were discarded) and further centrifuged three times (1× 7000 rcf and 2× 18000 rcf, each 15 min, 4°C). After each centrifugation, the pellets were collected and resuspended in homogenization-buffer; gradually decreasing the buffer volume. Next, two centrifugation steps at low speed (3000 rcf, 3 min, 4°C) were applied to remove remnant myofilaments. Supernatants were collected and finally centrifuged at high speed (20000 rcf, 20 min, 4°C) to pellet the purified mitochondria. Pellets were aliquoted, snap frozen and stored at −80°C. A fraction of the mitochondria slurry was lysed in an equal volume of 4M Urea, 4% SDS, 300 mM NaCl and the protein concentration was determined using the BCA protein assay. For half of the samples, the mitochondria were resuspended directly in the phosphatase reaction buffer (termed “P-buffer”), for the other half, the mitochondria were resuspended in 25 mM HEPES buffer pH 7.4 supplemented with protease inhibitors and 0.2% SDS, sonicated and briefly centrifuged before protein concentration was determined (BCA assay) (termed “with SH-buffer”).
Dephosphorylation treatment
Protein (40 μg) was resuspended in phosphatase buffer supplemented with protease inhibitors and the phosphatase of interest. Samples treated with λ-PP and PP1 were also supplemented with MgCl2. All phosphatase treatments where carried out overnight (16 hrs) with two different concentrations (high and low, with a 10 fold difference). For the different phosphatases used activities were: AP 1 and 10 U, λ-PP 10 and 100 U, PP1 1 and 10 U and PP2A 0.1 and 1 U. λ-PP and PP1 treatment were performed at 30°C and AP and PP2A treatment at 37°C as recommended by the manufacturer. Control samples were treated identically and simultaneously, but without any enzyme. 1U of a given phosphatase is either the amount hydrolyzing 1 nmol of p-nitrophenyl phosphate in 1 minute at the optimal temperature (AP, λ-PP and PP1) or releasing 1 nmol of phosphate in 1 minute from phosphorylase A (PP2A) as described by the manufacturers.
Cy-dye labeling of protein samples
Dephosphorylated samples and controls (18 μg of each) were solubilized in CHAPS buffer (8M urea, 2M thiourea, 4% CHAPS), and labeled with Cy-Dyes (Cy3 or Cy5) for 30 minutes at room temperature. Each pair of samples (dephosphorylated and the respective control) were labeled in duplicates, switching the dye between the samples (dye-swap). The labeling was quenched with 20mM lysine (using 5× volume of the dye).
Protein separation by Two-dimensional Gel Electrophoresis
Fluorescent labeled samples were analyzed by 2-DE as previously described [1]. A pair of labeled samples (one dephosphorylated and the respective control) were combined, supplemented with un-labeled protein (up to a total of 150 μg) and diluted in IEF re-hydration buffer (8M urea, 2M thiourea, 4% CHAPS, 1% DTT, 0.5% IPG buffer and 0.002% bromophenol blue [BPB]). After 30 min incubation at room temperature and centrifugation (20000 rcf, 20 min), samples were loaded to IPG strips (first dimension) and separated by the means of a Protean® IEF cell (Bio-Rad). The strips were actively rehydrated at 50 V (10–12 hrs), followed by stepwise rapid voltage ramping up to 200 V, and 500 V, linear ramping up to 1000 V with a subsequent step at the same voltage (each sep was 1 hr) followed by linear ramping up to 10000 V (3–6 hrs) and the final step was 10000 V for 50 kVh. All steps were performed at 20°C.
After the first dimension, proteins in the IPG strips were reduced and alkylated (each step for 30 min) respectively in 1% DTT or 4% iodoacetamide in equilibration buffer (50mM Tris-HCl, pH 6.4, 6M urea, 30% glycerol, 4% SDS). The strips were loaded onto 10% bis-Tris SDS-PAGE gels (second dimension). Strips were sealed using agarose sealing solution (0.5% Agarose NA and traces of BPB in MES buffer (45mM MES, 50mM Tris base, 0.5% SDS, 0.8mM EDTA)). Proteins were separated for 12–16 hrs using a Protean® II XL system (Bio-Rad) at 90V and MES running buffer. When the dye front reached the end of the gel, the gels were fixed (50% methanol, 5% Acetic Acid), washed and fluorescent images were acquired. Cy3 and Cy5 images were scanned using a Typhoon 9400 (GE Healtcare). Representative gel images for all the conditions with dye-swap are given in Supplementary Figure 2–7. Supplementary Figure 8 shows a representative image of the separation of the endogenous phosphatases PP2A, AP and PP1. The color green was attributed to Cy3 whereas Cy5 signal is in red and Cy2 signal is in blue. Finally, gels were post-silver stained for MS analysis as previously described [2] (Supplementary Figure 1).
Protein separation by Phos-tag gels and Western blotting
Dephosphorylated samples were supplied with LDS sample buffer (Invitrogen, CA) and SDS (1%) and separated on a Phos-tag gels (10% bis-acrylamide, 0,5M Tris pH 8.8, 0,1% SDS, 5% glycerol, 100 μM Phos-tag acrylamide (Wako Chemicals, Germany) and 100μM MnCl2) were prepared and run according to a recently published protocol [3]. After separation proteins were transferred to a PVDF membrane by western blotting as previously described [4].
Protein digestion and peptide identification by Mass Spectrometry
Protein spots were excised from 2D gels and destained, reduced and alkylated as previously described [5] prior to in-gel digested. Briefly, after dehydration of the gel piece (100% ACN, 10 min), the gel was rehydrated in a trypsin solution (6.25μg/mL trypsin in 25mM NH4HCO3, 4°C) After 1 hr the excess of trypsin solution was replaced with 25mM NH4HCO3 and the gel plugs were incubated at 37°C for 12–16 hrs. After incubation, the NH4HCO3 solution contained most of the peptides. To maximize the recovery, additional peptide extraction from the gel plugs was performed, alternating between 50% ACN and 25mM NH4HCO3 (each for 20 min, both done twice). The combined extracts were dried under vacuum.
For MALDI-TOF-TOF analysis dried peptides were resuspended in 50% ACN, 0.1% TFA and plated (0.3μl) with matrix (0.3μl cyano-4-hydroxy-trans-cinnamic acid) on to a stainless steel mass spectrometry plate, and air-dried. Samples were analyzed using a 5800 MALDI-TOF TOF (ABI) and the MS/MS spectra obtained were analyzed using the Mascot search engine (www.matrixscience.com) using the IPI_Mouse_v3.52 database. The following criteria were used: Mass values: Monoisotopic; Fragment Mass Tolerance: 0,8 Da; Peptide Mass Tolerance: 0,3 Da; Fixed Modification: +57,021469) on C (Carbamidomethyl); Variable Modification: +15,9949 on M (Oxidation); Enzyme: Trypsin; Maximum Missed Cleavages: 2. Protein ID are listed in Supplementary Table 1 (ordered by spot number), accompanied by symbol, accession number, theoretical protein mass and pI, sequence coverage, total mascot ion score and number of unique peptides. The experimental protein mass and pI were calculated from the gel images.
Peptides analyzed by ESI MS/MS LTQ-Orbitrap mass spectrometer (Thermo, Waltham, MA, USA) were resuspended in 50% ACN and 0.1 % formic acid. MS/MS spectra obtained were analyzed using the SEQUEST Ver 27 (Rev 11) search engine using the ipi.MOUSE.v3.62 database. The following criteria was used: Fragment Tolerance: 1,00 Da (Monoisotopic); Parent Tolerance: 0.040–0.160 0,12 Da (Monoisotopic); Fixed Modification: +57 on C (Carbamidomethyl); Variable Modification: +16 on M (Oxidation); Digestion Enzyme: Trypsin; and Max Missed Cleavages: 2. Protein IDs, ordered by spot number, are listed in Supplementary Table 2, which also includes symbol, accession number, theoretical protein mass, sequence coverage, protein ID probability and number of peptides. The theoretical protein pI was calculated using the Expasy Compute pI/Mw tool. The experimental protein mass and pI were retrieved from the gel images.
All annotated spectra were manually evaluated and only good quality spectra with almost a complete list of b- and y-ions were accepted. A positive ID was based on a minimum of two peptides with a mascot score > 40 (MALDI-TOF-TOF) or an ID probability >95% (ESI-MS/MS). Additionally, all spectra of trypsin and keratin were excluded. For samples with multiple IDs, only one was considered correct if there was an 8-time or higher difference in the number of peptides.
Titanium Dioxide enrichment of phosphopeptides
Homogenization and AP treatment
Four hearts were individually homogenized using the method described in the presence of phosphatase inhibitors. Each homogenate was cleaned using the 2D Clean-up kit and the protein pellet was re-suspended in 25mM HEPES pH 7.4 containing 0.2% SDS. Protein concentration was determined using the BCA method. Dephosphorylation was preformed with 200 μg protein and AP as described above. A corresponding sample without AP was used as control.
Trypsin digestion and TiO2 enrichment
Each biological sample (n=4) was diluted with 200 mM NH4HCO3 pH 8, reduced by 200 mM DTT at 55° C for 30 minutes and alkylated with 200 mM iodoacetamide at room temperature for 1 hour in the dark. Each sample was then treated with 20 μg of trypsin and incubated for 3 hrs at 37°C with shaking, after which point an additional 20 μg of trypsin was added and samples were incubated overnight at 37°C with shaking. Digestion was stopped by the addition of TFA to 0.8% final concentration. Each sample was desalted using a Waters C18 Sep-pak 100mg cartridge and peptides were eluted in TiO2 starting buffer (80% ACN, 5% TFA, 1M glycolic acid) and treated with TiO2 enrichment solution (15 mg of TiO2 beads per 1 mL of TiO2 start buffer, incubated for 2 hrs at room temperature with shaking before use) and incubated at room temperature overnight with vigorous shaking. Samples were then quickly spun in a centrifuge to pellet the TiO2 beads and the supernatant was removed with a pipette tip. Beads were then washed three times with 80% ACN, 5% TFA and once with 80% ACN, 0.1% TFA. Phosphopeptides were eluted with 10% ACN, 3% NH4OH (pH ~10) with shaking for 10 minutes at room temperature. After centrifugation, the supernatant containing the phosphopeptides was treated with 0.5% TFA and desalted using graphite columns. Peptides were eluted in 70% ACN, 0.1% FA. Each sample was spotted twice for LC-MS/MS analysis.
LC-MS/MS analysis
Each sample was analyzed in duplicate on an Agilent 1200 nano-LC system (Agilent, Santa Clara, CA, USA) connected to an LTQ-Orbitrap mass spectrometer (Thermo, Waltham, MA, USA) equipped with a nanoelectrospray ion source. Peptides were separated on a C18 RP-HPLC column (75μm × 10cm self-packed with 5μm, 200 Å Magic C18; Michrom BioResources, Auburn, CA, USA) at a flow rate of 300 nl/min where mobile phase A was 0.1% formic acid in water and mobile phase B was 90% ACN, 0.1% formic acid in water. The linear gradient was 10–30% B over 100 minutes, 30–40% B over 10 minutes, 40–65% B over 16 minutes and 65–95% B over 10 minutes. Each MS1 scan was followed by collision induced dissociation (CID, acquired in the LTQ part) of the 10 most abundant precursor ions with dynamic exclusion for 30 seconds. Only MS1 signals exceeding 1000 counts triggered the MS2 scans and +1 and unassigned charge states were not selected for MS2 analysis. For MS1, 2×105 ions were accumulated in the Orbitrap over a maximum time of 500 ms and scanned at a resolution of 60,000 FWHM (from 375–2000 m/z). MS2 spectra (via collision induced dissociation (CID)) were acquired in normal scan mode in the LTQ with multistage activation turned on with a target setting of 104 ions and accumulation time of 30 ms. The normalized collision energy was set to 35% and one microscan was acquired for each spectra.
Data Searching and Processing
Raw MS/MS data was searched using the Sorcerer 2™-SEQUEST® algorithm (Sage-N Research, Milpitas, CA, USA) and searched against the Mouse IPI database version 3.62 using the following criteria: Fragment tolerance: 1.00 Da; Parent Tolerance: 0.040–0.160 Da; Fixed modification: +57 on C (carbamidomethyl); Variable modification: +16 on M (oxidation), +80 on S,T,Y (phosphorylation); Enzyme: Trypsin with 2 max missed cleavages. Post-search analysis was performed using Scaffold 3 version 1.4.1 (Proteome Software, Inc., Portland, OR, USA) with protein and peptide probability thresholds set to 95% and one peptide required for identification. Phosphosite analysis was performed using the Scaffold PTM software version 1.1.3 (Proteome Software) and a complete list of phosphoproteins and phosphosites observed is listed in Supplementary Table 4 and 5. For instances where a phosphosite was observed on multiple peptides or in multiple spectra only the peptide that generated the best Ascore was reported, but total spectral counts for each site and the number of replicates in which a site was observed are listed. The data is available in Pride (http://www.ebi.ac.uk/pride/) through accession numbers 19673–19674.
SUPPLEMENTARY RESULTS
Concentrations of phosphatases were selected based on the dephosphorylation of HSPβ1, a test cardiac protein, and analyzed by phospho-tag gels (see supplementary Figure 9). The conditions used in this study produced complete dephosphorylation of HSPβ1.
In general every phosphatase treatment and DIGE experiment was done in duplicate with swapping the Cy-dye assignment. The consistency of the method was confirmed by analyzing a subset of the conditions in triplicate (Supplementary Figure 10). The results show that the technical variability of the system is small, and independent of the number of dephosphorylated substrates (Supplementary Figure 10 A and B vs. C and D respectively).
Supplementary Material
Supplementary Figure 1: Representative images of silver stained 2-DE gels of tissue homogenate (A) and mitochondrial enriched protein fraction (B). The images were used to calculate experimental MW and pI. *Spots 6, 7 and 13 do not have any assigned ID, for the other spots see supplementary Table 1 and 2 for IDs.
Supplementary Figure 2: PP2A treated cardiac proteins together with untreated controls separated by 2-DE; samples with (left) or without (right) precipitation. Dephosphorylations induced by PP2A treatment are boxed. The amount of enzyme used are indicated (Cy3=green, Cy5=red).
Supplementary Figure 3: PP1 treated cardiac proteins together with untreated controls separated by 2-DE; samples with (left) or without (right) precipitation. Dephosphorylations induced by PP1 treatment are boxed. The amount of enzyme used are indicated (Cy3=green, Cy5=red).
Supplementary Figure 4: AP treated cardiac proteins at pH 7.4 together with untreated controls separated by 2-DE; samples with (left) or without (right) precipitation. Dephosphorylations induced by AP treatment are boxed. The amount of enzyme used are indicated (Cy3=green, Cy5=red).
Supplementary Figure 5: λ-PP treated cardiac proteins together with untreated controls separated by 2-DE; samples with (left) or without (right) precipitation. Dephosphorylations induced by λ-PP treatment are boxed. The amount of enzyme used are indicated (Cy3=green, Cy5=red).
Supplementary Figure 6: AP treated cardiac proteins at pH 9 together with untreated controls separated by 2-DE; samples with (left) or without (right) precipitation. Dephosphorylations induced by AP treatment are boxed. The amount of enzyme used are indicated (Cy3=green, Cy5=red).
Supplementary Figure 7: AP or λ-PP treated myocardial enriched fraction together with untreated controls separated by 2-DE; samples lysed directly in phosphatase buffer (left) or resuspended in HEPES buffer (right). Dephosphorylations induced by phosphatase treatment are boxed. The amount of enzyme used are indicated (Cy3=green, Cy5=red).
Supplementary Figure 8: Each of the three phosphatases AP, PP1 and PP2A labeled with Cy-dyes and separated by 2-DE, showing that especially PP2A solution comprises several protein forms.
Supplementary Figure 9: Dephosphorylation of HSPB1 using either 10 or 1U of AP or 1U of PP2A (lane 2, 4 and 6 respectively) with corresponding non-treated controls (lane 1, 3 and 5). Efficient dephosphorylation was evaluated as lack of visible phosphorylated forms on the western blot.
Supplementary Figure 10: Consistency of the method was confirmed by analyzing a subset of the experimental conditions in triplicate. Tissue homogenate without precipitation (A–C) dephosphorylated with 10U of AP (A), 100U of λ-PP and 1U PP1 or mitochondrial-enriched fraction resuspended in phosphatase buffer (P-buffer) treated with 100U of λ-PP.
Acknowledgments
The authors would like to thank Justyna Fert-Bober for the ESI LTQ Orbitrap MS/MS analysis. Financial support: Oslo University Hospital–Ullevaal and Anders Jahre’s Fund for the Promotion of Science (C.H.); NIH Program Project grant P01-HL077180 and RFO, University of Bologna (G.A.); NHLBI grants HV-10-05 (2) and P50 HL 084946-01, and the Clinical Translational Science Award at Johns Hopkins University (J.E.V.E).
Footnotes
Conflict of interest
None
Reference list
- 1.Brautigan DL. Flicking the switches: phosphorylation of serine/threonine protein phosphatases. Semin Cancer Biol. 1995;6:211–217. doi: 10.1006/scbi.1995.0028. [DOI] [PubMed] [Google Scholar]
- 2.Olsen JV, Vermeulen M, Santamaria A, Kumar C, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 3.Capriotti AL, Cavaliere C, Foglia P, Samperi R, et al. Intact protein separation by chromatographic and/or electrophoretic techniques for top-down proteomics. J Chromatogr A. 2011 doi: 10.1016/j.chroma.2011.05.094. [DOI] [PubMed] [Google Scholar]
- 4.Kane LA, Youngman MJ, Jensen RE, Van Eyk JE. Phosphorylation of the F(1)F(o) ATP synthase beta subunit: functional and structural consequences assessed in a model system. Circ Res. 2010;106:504–513. doi: 10.1161/CIRCRESAHA.109.214155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kinoshita E, Kinoshita-Kikuta E. Improved Phos-tag SDS-PAGE under neutral pH conditions for advanced protein phosphorylation profiling. Proteomics. 2011;11:319–323. doi: 10.1002/pmic.201000472. [DOI] [PubMed] [Google Scholar]
- 6.Raggiaschi R, Gotta S, Terstappen GC. Phosphoproteome analysis. Biosci Rep. 2005;25:33–44. doi: 10.1007/s10540-005-2846-0. [DOI] [PubMed] [Google Scholar]
- 7.Raggiaschi R, Lorenzetto C, Diodato E, Caricasole A, et al. Detection of phosphorylation patterns in rat cortical neurons by combining phosphatase treatment and DIGE technology. Proteomics. 2006;6:748–756. doi: 10.1002/pmic.200500064. [DOI] [PubMed] [Google Scholar]
- 8.Schulenberg B, Aggeler R, Beechem JM, Capaldi RA, et al. Analysis of steady-state protein phosphorylation in mitochondria using a novel fluorescent phosphosensor dye. J Biol Chem. 2003;278:27251–27255. doi: 10.1074/jbc.C300189200. [DOI] [PubMed] [Google Scholar]
- 9.Gevaert K, Staes A, Van DJ, De GS, et al. Global phosphoproteome analysis on human HepG2 hepatocytes using reversed-phase diagonal LC. Proteomics. 2005;5:3589–3599. doi: 10.1002/pmic.200401217. [DOI] [PubMed] [Google Scholar]
- 10.Agnetti G, Husberg C, Van Eyk JE. Divide and conquer: the application of organelle proteomics to heart failure. Circ Res. 2011;108:512–526. doi: 10.1161/CIRCRESAHA.110.226910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Solaro RJ, van der Velden J. Why does troponin I have so many phosphorylation sites? Fact and fancy. J Mol Cell Cardiol. 2010;48:810–816. doi: 10.1016/j.yjmcc.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kane LA, Neverova I, Van Eyk JE. Subfractionation of heart tissue: the “in sequence” myofilament protein extraction of myocardial tissue. Methods Mol Biol. 2007;357:87–90. doi: 10.1385/1-59745-214-9:87. [DOI] [PubMed] [Google Scholar]
- 13.Agnetti G, Kaludercic N, Kane LA, Elliott ST, et al. Modulation of mitochondrial proteome and improved mitochondrial function by biventricular pacing of dyssynchronous failing hearts. Circ Cardiovasc Genet. 2010;3:78–87. doi: 10.1161/CIRCGENETICS.109.871236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ross MH, Ely JO, Archer JG. Alkaline phosphatase activity and pH optima. J Biol Chem. 1951;192:561–568. [PubMed] [Google Scholar]
- 15.Virshup DM, Shenolikar S. From promiscuity to precision: protein phosphatases get a makeover. Mol Cell. 2009;33:537–545. doi: 10.1016/j.molcel.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 16.Rotenberg SA, Brautigan DL. Membrane protein phosphotyrosine phosphatase in rabbit kidney. Proteolysis activates the enzyme and generates soluble catalytic fragments. Biochem J. 1987;243:747–754. doi: 10.1042/bj2430747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng N, Zhang J, Zong C, Wang Y, et al. Phosphoproteome analysis reveals regulatory sites in major pathways of cardiac mitochondria. Mol Cell Proteomics. 2011;10:M110. doi: 10.1074/mcp.M110.000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDonald T, Sheng S, Stanley B, Chen D, et al. Expanding the subproteome of the inner mitochondria using protein separation technologies: one- and two-dimensional liquid chromatography and two-dimensional gel electrophoresis. Mol Cell Proteomics. 2006;5:2392–2411. doi: 10.1074/mcp.T500036-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Woo EM, Fenyo D, Kwok BH, Funabiki H, et al. Efficient identification of phosphorylation by mass spectrometric phosphopeptide fingerprinting. Anal Chem. 2008;80:2419–2425. doi: 10.1021/ac702059p. [DOI] [PubMed] [Google Scholar]
- 20.Du H, Simpson RJ, Moritz RL, Clarke AE, et al. Isolation of the protein backbone of an arabinogalactan-protein from the styles of Nicotiana alata and characterization of a corresponding cDNA. Plant Cell. 1994;6:1643–1653. doi: 10.1105/tpc.6.11.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCE LIST
- 1.Agnetti G, Kaludercic N, Kane LA, Elliott ST, et al. Modulation of mitochondrial proteome and improved mitochondrial function by biventricular pacing of dyssynchronous failing hearts. Circ Cardiovasc Genet. 2010;3:78–87. doi: 10.1161/CIRCGENETICS.109.871236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 3.Kinoshita E, Kinoshita-Kikuta E, Koike T. Separation and detection of large phosphoproteins using Phos-tag SDS-PAGE. Nat Protoc. 2009;4:1513–1521. doi: 10.1038/nprot.2009.154. [DOI] [PubMed] [Google Scholar]
- 4.Husberg C, Nygard S, Finsen AV, Damas JK, et al. Cytokine expression profiling of the myocardium reveals a role for CX3CL1 (fractalkine) in heart failure. J Mol Cell Cardiol. 2008;45:261–269. doi: 10.1016/j.yjmcc.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 5.Kane LA, Yung CK, Agnetti G, Neverova I, et al. Optimization of paper bridge loading for 2-DE analysis in the basic pH region: application to the mitochondrial subproteome. Proteomics. 2006;6:5683–5687. doi: 10.1002/pmic.200600267. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Representative images of silver stained 2-DE gels of tissue homogenate (A) and mitochondrial enriched protein fraction (B). The images were used to calculate experimental MW and pI. *Spots 6, 7 and 13 do not have any assigned ID, for the other spots see supplementary Table 1 and 2 for IDs.
Supplementary Figure 2: PP2A treated cardiac proteins together with untreated controls separated by 2-DE; samples with (left) or without (right) precipitation. Dephosphorylations induced by PP2A treatment are boxed. The amount of enzyme used are indicated (Cy3=green, Cy5=red).
Supplementary Figure 3: PP1 treated cardiac proteins together with untreated controls separated by 2-DE; samples with (left) or without (right) precipitation. Dephosphorylations induced by PP1 treatment are boxed. The amount of enzyme used are indicated (Cy3=green, Cy5=red).
Supplementary Figure 4: AP treated cardiac proteins at pH 7.4 together with untreated controls separated by 2-DE; samples with (left) or without (right) precipitation. Dephosphorylations induced by AP treatment are boxed. The amount of enzyme used are indicated (Cy3=green, Cy5=red).
Supplementary Figure 5: λ-PP treated cardiac proteins together with untreated controls separated by 2-DE; samples with (left) or without (right) precipitation. Dephosphorylations induced by λ-PP treatment are boxed. The amount of enzyme used are indicated (Cy3=green, Cy5=red).
Supplementary Figure 6: AP treated cardiac proteins at pH 9 together with untreated controls separated by 2-DE; samples with (left) or without (right) precipitation. Dephosphorylations induced by AP treatment are boxed. The amount of enzyme used are indicated (Cy3=green, Cy5=red).
Supplementary Figure 7: AP or λ-PP treated myocardial enriched fraction together with untreated controls separated by 2-DE; samples lysed directly in phosphatase buffer (left) or resuspended in HEPES buffer (right). Dephosphorylations induced by phosphatase treatment are boxed. The amount of enzyme used are indicated (Cy3=green, Cy5=red).
Supplementary Figure 8: Each of the three phosphatases AP, PP1 and PP2A labeled with Cy-dyes and separated by 2-DE, showing that especially PP2A solution comprises several protein forms.
Supplementary Figure 9: Dephosphorylation of HSPB1 using either 10 or 1U of AP or 1U of PP2A (lane 2, 4 and 6 respectively) with corresponding non-treated controls (lane 1, 3 and 5). Efficient dephosphorylation was evaluated as lack of visible phosphorylated forms on the western blot.
Supplementary Figure 10: Consistency of the method was confirmed by analyzing a subset of the experimental conditions in triplicate. Tissue homogenate without precipitation (A–C) dephosphorylated with 10U of AP (A), 100U of λ-PP and 1U PP1 or mitochondrial-enriched fraction resuspended in phosphatase buffer (P-buffer) treated with 100U of λ-PP.