Abstract
Single-chain variable fragment (scFv) is a class of engineered antibodies generated by the fusion of the heavy (VH) and light chains (VL) of immunoglobulins through a short polypeptide linker. ScFv play a critical role in therapy and diagnosis of human diseases, and may in fact also be developed into a potential diagnostic and/or therapeutic agent. However, the fact that current scFv antibodies have poor stability, low solubility, and affinity, seriously limits their diagnostic and clinical implication. Here we have developed four different expression vectors, and evaluated their abilities to express a soluble scFv protein. The solubility and binding activity of the purified proteins were determined using both SDS-PAGE and ELISA. Amongst the four purified proteins, the Skp co-expressed scFv showed the highest solubility, and the binding activity to antigen TLH was 3-4 fold higher than the other three purified scFv. In fact, this scFv is specific for TLH and does not cross-react with other TLH-associated proteins and could be used to detect TLH directly in real samples. These results suggest that the pACYC-Duet-skp co-expression vector might be a useful tool for the production of soluble and functional scFv antibody.
Keywords: Vibrio parahaemolyticus, scFv, co-expression, solubility, production
Introduction
Single-chain variable fragment (scFv) is a fusion protein formed by engineering the association of the VH and VL domains of the antibodies with a short polypeptide linker. scFv specific to any particular antigen may be easily generated by Phage display (Wang et al., 2006), and this scFv have been developed to select molecular targets in cancer research such as in lymphatic invasion vessels, colon cancer and hepatocarcinoma (Rinderknecht et al., 2010; Sakai et al., 2010; Sommaruga et al., 2011). Furthermore, scFv has been extensively used to generate ligands for detecting pathogenic germs in vitro or in vivo (Wang et al., 2007, 2008a, b; Zhang et al., 2010; Cattepoel et al., 2011). In comparison to polyclonal antibodies or the hybridoma technology, scFv antibody may be easily manipulated for improving specificity and affinity, thereby reducing the production cost (Coia et al., 2001; Krag et al., 2006). Combing scFv with selection panning strategies, we were able to character the binding properties of scFv and investigate the potential use of these scFv as diagnostic tools or therapeutic agents (Eisenhardt et al., 2007; Rothe et al., 2007). However, these above mentioned applications of scFv were limited by drawbacks such the formation of inclusion bodies, which often lead to low binding activity, unstable structure and are cytotoxic to host cells.
Currently, the soluble expression of scFv antibody remains an awkward plight, so the majority of the work in this field focuses on developing a strategy based on molecular manipulation to improve the stability and solubility of scFv antibody. Till today, a number of methods have been used to express the scFv antibody, including expression of affinity tag fusion (Esposito and Chatterjee, 2006), co-expression of molecular chaperones, and folding modulators (De Marco and De Marco, 2004; Sonoda et al., 2011), extracellular accumulation in a defined medium (Fu, 2010), refolding scFv using detergent and additive (Kudou et al., 2011) and expression in different host systems (Goulding and Perry, 2003). Amongst of these methods, expression of affinity tags fusion protein is the common method to improve the solubility of target proteins. Previously, some affinity tags such as thioredoxin (TRX) (Nygren et al., 1994), maltose binding protein (MBP) (Nallamsetty and Waugh, 2006), N-utilization substance A (NusA) (Fox and Waugh, 2003), bacteriophage T7 protein kinase gene (T7PK) (Jurado et al., 2006), small peptide tags (SET) (Davis et al., 1999), monomeric mutant of the Ocr protein of bacteriophage T7 (Mocr) and glutathione S-transferase (GST) were used to enhance the solubility of some of the partner proteins to which they were attached (DelProposto et al., 2009). Unfortunately, the tags needed to be cleaved as the large tags usually interfered with the folding of their partner protein and made them more difficult to assay for activity and for functional research (Esposito and Chatterjee, 2006). Besides, the partner proteins often remained insoluble when the fusion tags were removed, and the entire process of tags removal is costly and laborious (Esposito and Chatterjee, 2006). Though the use of detergents and additives to refold the target protein can assist in making protein soluble, there is still no guarantee that these methods will be suitable for every protein of interest. When it comes to expression system, though a number of them, such as E. coli, Yeast, mammalian, insect cell, wheat germ cell-free expression system, and plant-based expression system (Goulding and Perry, 2003; Greene, 2004; Daly and Hearn, 2005; Guild et al., 2011), are available for protein production, the E. coli host system is widely regarded as the most suitable host for the expression of recombinant antibody fragments (Wang et al., 2008a, b). Compared to other host systems, the E. coli system is an economical, shows faster growth and is easier to manipulate genetically (Sushma et al., 2011).
It was also reported that the solubility and affinity of scFv was improved by co-expression of molecular chaperones such as Skp, Dsbc, and FkpA (Ow et al., 2010; Sonoda et al., 2011). In some cases, co-expression of molecular chaperone not only improves the soluble expression but also increases the cell viability (Ow et al., 2010). Skp is a key periplasmic chaperone (18 kDa) that plays an important role in folding and assembling of outer membrane proteins in E. coli, and reports have shown that the co-expression of Skp facilitates correct folding of the expressed protein and enhances the solubility and affinity of proteins (Hayhurst and Harris, 1999; Hayhurst et al., 2003; Sonoda et al., 2011).
In this study, we investigate the effects of the molecular chaperone Skp on the level of expression, solubility and antigen-binding activity of scFv. The co-expression vector pACYC-Duet containing two multiple cloning sites (MCS), was used for the construction and expression of two target genes in an appropriate host strain. By co-expression of a periplasmic chaperone (Skp) with scFv, a relatively high amount of soluble scFv antibody was successfully obtained.
Materials and methods
Materials
V. parahaemolyticusCGMCC1.2164, V. parahaemolyticusCGMCC1.1614, V. parahaemolyticusCGMCC1.1615, V. parahaemolyticusCGMCC1.1616 were purchased from the Institute of Microbiology, Chinese Academy of Sciences (Beijing, China). All other strains were from Fujian Agriculture and Forestry University (Fujian, China). The plasmids pET28a(+) and pET32a(+) were purchased from Novagen (Madison, WI, USA), and the vector pACYC-Duet-1 was generously provided by Professor Lijun Bi (Institute of Biophysics, Chinese Academy of Sciences, Beijing China). The vector pGEPi was generously provided by Dr Mengfei Ho (University of Illinois at Urbana-Champaign, USA). DNA restriction enzymes were purchased from Promega (USA). Taq DNA polymerase and T4 DNA ligase were purchased from Takara (Dalian, China). Anti 6×His tag monoclonal antibody was purchased from Abgent (USA), and horseradish peroxidase (HRP)-labeled goat anti-mouse IgG was from Boster Biological Technology Co. (Wuhan, China). All oligonucleotides were listed in Table 1, and all other reagents used were of analytical reagent grade.
Table 1.
Primers used in the present study.
| Primers | DNA sequence |
|---|---|
| Skp-F | GGCGAGATCTGACGAAAAAGTGGTTATTAGCTG |
| Skp-R | CGGCTCGAGTTATTTAACCTGTTTCAGTACGTC |
| scFv-1-F | AAGAATTCATGGCCCAGGTCAAACTGCAGGAG |
| scFv-1-R | CCCGCAAGCTTCCGTTTTATTTCCAGCT |
| scFv-2-F | AAGAATTCAGCCCAGGTCAAACTGCAGGAG |
| scFv-2-R | CCCGCAAGCTTCCGTTTTATTTCCAGCT |
| TLH-F | AAACTGGTACCGGTAGAAATGATGAAAAAAACAATCAC ACTATTAACTGCATTAC |
| TLH-R | GAACCTGCGGCCGCACCAGAACCGAAACGGTACTCGG CTAAGTTGTTGCTAC |
Expression and purification of TLH antigen
The V. parahaemolyticus(CGMCC1.2164) genome was used as a template to amplify the tlh gene[GeneBank:GU971665.1] (Wang et al., 2011). Primers with Kpn I and Not I restriction enzymatic sites were designed for cloning the tlh gene into pGEPi vector. The constructed pGEPi-tlh vector was transformed into E. coli BL21 by electroporation, and a single colony from the selection plate was inoculated into 5 mL LB liquid media containing 100 μg/mL ampicillin for the expression of TLH antigen. The expressed protein contains HA and Myc tags at the C-terminal (without 6×His tag). Expression of the target protein was induced by adding 1 mM IPTG when the culture reached an OD600 of 0.8, and the induced cells were grown at 37°C for overnight, and then harvested by centrifugation. After SDS-PAGE, the expressed proteins were stained using 0.3 M CuCl2 by shaking for 5 min at 37°C, and visualized against a black background. The target TLH protein bands were cut and mashed, and mixed with 1 mL PBS. The mixture solution was dialyzed with Gly/Tris buffer and the supernatant was harvested by centrifugation, and the extracted supernatant containing the TLH protein was visualized by SDS-PAGE. Protein concentration was determined by the BCA protein assay kit.
Construction of expression vectors for scFv
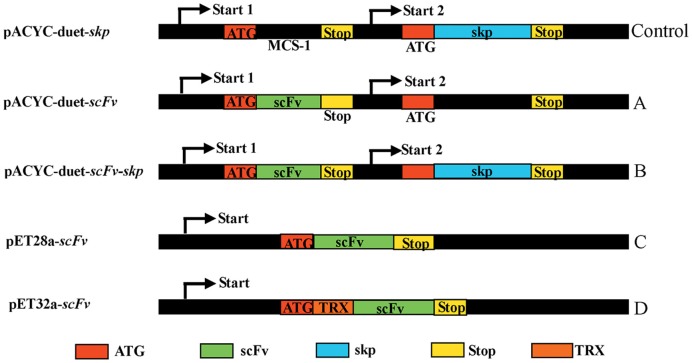
pET28a(+), pET32a(+), and pACYC-Duet vectors were used for the construction of expression vectors with the scFv proteins fused to 6×His tag, 6×His-TRX, and 6×His tag, respectively. For co-expression, a recombinant vector pACYC-Duet-skp was first constructed. The DNA skp gene[GeneBank:U00096.2] encoding the molecular chaperone was amplified using the primers Skp-F and Skp-R, and JM109 genomic DNA was used as template. The amplified DNA fragment was digested by Bgl II and Xho I, and ligated the pACYC-Duet vector digested with the same restriction enzymes. To construct four different formats of expression vectors, the segment encoding anti-TLH scFv was amplified using the template vector pCANTAB-5E-scFv (Wang et al., 2012) and the following primers (scFv-1-F and scFv-1-R for scFv-1; scFv-2-F and scFv-2-R for scFv-2). The amplified scFv-1 fragment was digested with EcoR I and Hind III, and ligated with pET28a(+) and pET32a(+) vectors digested with EcoR I and Hind III, and the resultant vectors were designated as pET28a-scFv and pET32a-scFv, respectively. The amplified scFv-2 fragment was digested with EcoR I and Hind III, and ligated with pACYC-Duet and pACYC-Duet-skp vectors digested with the EcoR I and Hind III, and the resultant vectors were designated as pACYC-Duet-scFv and pACYC-Duet-scFv-skp, respectively.
Expression and soluble analysis of scFv antibodies
For protein expression, the recombinant plasmids were transformed into E. coli BL21 by electroporation, and a single colony from the selection plate was inoculated into 5 mL LB liquid media containing either 100 μ g/mL ampicillin or 50 μ g/mL kanamycin or 34 μ g/mL chloramphenicol. The culture was incubated overnight with shaking at 37°C, and then transferred to a larger-scale LB media (1 mL culture transferred into 500 mL fresh LB). Expression of the target protein was induced by adding 1 mM IPTG when the culture reached an OD600 of 0.8. Cells were grown for an additional 12 or 32 h (only for pACYC-Duet-scFv-skp/BL21) at 16°C, and then harvested by centrifugation. To ensure the accuracy of the experiment, four different derived cultures were adjusted to the same concentration for samples treatment, and the treated proteins were analyzed by SDS-PAGE using 12% (v/v) polyacrylamide gels.
Purification and identification of anti-TLH scFv antibodies
The purification of the expressed anti-TLH scFv was performed using Ni2+ affinity chromatography. The expressed product was first harvested by centrifugation (10000 r/min) at 4°C for 10 min, followed by addition of 5 mL of the binding buffer (50 mM NaH2PO4, 300 mM NaCl, 1 mM imidazole, 0.05% Tween 20) after the removal of the supernatant. The complex was then mixed gently, sonicated and then centrifuged at 4°C for 10 min. The supernatant was collected and loaded onto a 3 mL Ni2+-NTA column, which was equilibrated with the binding buffer before the purification. After allowing the sample to flow-through, the column was washed twice with the wash buffer (100 mL/time), and the wash fraction was collected for SDS-PAGE analysis. After the wash, the retained protein of interest was eluted with 0.5 mL elution buffer 4 times, and the eluate was collected and analyzed by SDS-PAGE and verified by Western blotting. Protein concentration was determined by the BCA protein assay kit.
Elisa analyses
The activity of the purified scFv products was determined by ELISA. The dialyzed TLH antigen (without 6×His tag) was used to coat the 96 well plates at 4°C for overnight. After blocking and washing the ELISA plate, the purified scFv products derived from four different formats of plasmids were added to the reaction wells and incubated at 37°C for 2 h. Then, the anti-His tag antibody was added to the reaction wells and incubated at 37°C for 2 h. The binding activity of the purified scFv was detected by using a HRP conjugated anti-mouse IgG antibody. The enzyme reaction was then performed with TMB as a substrate and color development was terminated with 2 M H2SO4. Absorbance at 450 nm was measured using a microplate reader.
Similarly, ELISA was also used for the detection of bacterial TLH. Totally, eleven different types of V. parahaemolyticus strains, seven Vibrio strains from other species, and four non-Vibrio bacterial strains were tested using ELISA. The bacterial cultures were grown in 5 mL APW medium without shaking at 37°C overnight, and then their supernatants were collected by centrifugation (8000 r/min, 15 min). The supernatants were added to a 96 well plate (100 μL/well) to perform ELISA as described before.
Western blotting
To further confirm the binding activity of scFv (co-expression), western blotting was performed as described by Singh et al. (2010) with minor modifications. Briefly, the extracted TLH was transferred from a SDS-PAGE gel onto a polyvinylidene difluoride (PVDF) membrane, and the membrane was treated with purified scFv antibody and anti-His tag antibody. After washing and blocking, the membrane was subsequently incubated with HRP-conjugated anti-mouse IgG antibody. Signals were visualized by enhanced chemiluminescence (ECL).
Analyses of anti-TLH scFv specificity
ELISA was performed to determine the specificity of the expressed anti-TLH scFv. BSA, KLH, and associated V. parahaemolyticus antigens (Vp1668 and YscF) were coated on 96-well plates (5 μg/mL, 100 μL/well) at 4°C for overnight, respectively. After the blocking and washing, the purified scFv (pACYC-Duet-scFv-skp) was added to the reaction wells and incubated at 37°C for 2 h. Then the anti-His tag antibody was added to the reaction wells and incubated at 37°C for 2 h. The specificity of the scFv was detected with the HRP conjugated anti-mouse IgG antibody.
Affinity determination of skp co-expression scFv by microcalorimetry
The bio-molecular interaction and affinity of the scFv to TLH was measured using a TA-instrument Nano ITC-LV. The measurement was automatically monitored by data acquisition software supplied along with the instrument. All the measurements were carried out at 25°C. The affinity constant was calculated using Origin software supplied with the Nano ITC-LV.
Results
Construction of fusion expression vectors carrying scFv
In this study, four different expression vectors pACYC-Duet-scFv, pACYC-Duet-scFv-skp, pET28a-scFv, and pET32a-scFv were constructed (Figure 1), and transformed into E. coli BL21 to investigate their effects on solubility and activity of the expressed scFv. Among of four expression vectors, pET28a is the basic vector for protein expression, pET32a contains a TRX tags that can enhance the solubility of protein. While pACYC-Duet-skp contains two MCS sites for different DNA insertions, and all the expression vectors contains a 6×His tag for protein purification and ELISA detection. To analyze the function of Skp on soluble expression, the vector pACYC-Duet-skp was first constructed by introducing the skp gene into the multiple cloning sites 2 with restriction enzyme sites Bgl II and Xho I. Then the scFv gene was cloned into the multiple cloning sites 1 via EcoR I and Hind III, and the resulting pACYC-Duet-scFv-skp was used for the expression of both proteins (Skp and scFv) in E. coli BL21, with the expressed scFv contains a 6×His tag at C-terminal. The expression vector pET28a-scFv encodes scFv with two 6×His tags, one each at the N- and C-terminal, respectively, while the expression vector pET32a-scFv encodes a TRX-scFv fusion protein with two 6×His tags, one each at the N-terminal and C-terminal, respectively.
Figure 1.
Constructs of scFv with different fusion formats used in this study. MCS, Multiple cloning sites; Stop, stop codon; scFv, single chain variable fragment; Skp, molecular chaperone Skp; TRX, thioredoxin.
Expression and purification of TLH antigen and scFV
After the TLH protein was expressed, the total protein was analyzed by SDS-PAGE, and the gel was stained using 0.3 M CuCl2, and visualized against a black background. The target TLH protein bands were cut and mashed with PBS buffer. After harvested by centrifugation, the extracted supernatant containing the TLH protein was visualized by SDS-PAGE. As shown in Figure 2A, the TLH antigen protein was expressed successfully (Figure 2A, lane 2), and the target TLH protein was extracted with a high purity (Figure 2A, lane 3, 4). The expressed TLH protein contains HA and Myc tags (without 6×His tag), and was used as the immobilized antigen during ELISA.
Figure 2.
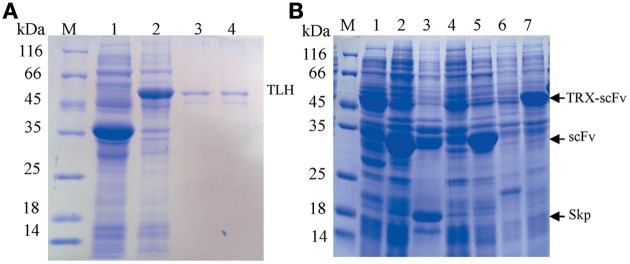
SDS-PAGE analysis. (A) SDS-PAGE analysis of expressed TLH antigen protein. Lane 1: Control (empty vector pGEPi, IPTG induced); Lane 2: pGEPi-tlh expressed product (IPTG induced); Lane 3, 4: Extracted TLH protein (without 6×His tag); (B) Expression of different scFv constructs in E. coli BL21. M: Molecular weight of marker proteins; Lanes 1, 4, and 6: negative control (pACYC-Duet, pET28a, and pET32a, respectively); Lane 2, 3, 5, and 7: the expressed products of pACYC-Duet-scFv, pACYC-Duet-scFv-skp, pET28a-scFv, and pET32a-scFv, respectively.
Four constructed vectors were used to transform E. coli BL21 (DE3) by electroporation for the expression of their respective target proteins. The transformants were grown in test-tube cultures and induced with 0.5 mM IPTG at 16°C for 12 or 36 h, and the expressed products were analyzed by SDS-PAGE. As shown in Figure 2B, the target proteins were expressed successfully in every kind of vector, and that the apparent molecular weight of each target scFv was consistent with its corresponding theoretical molecular weight.
Expression and analysis of solubility of scFv
To test the solubility of expressed proteins, we lysed the cells by ultrasonication after protein expression, and the results were analyzed by SDS-PAGE and OD600 value. As shown in Figure 3A, Skp co-expression (Figure 3A, lane 3) and TRX fusion expression (Figure 3A, lane 7) were the best choices with respect to protein solubility, in comparison to the other two fusions (Figure 3A, lane 1, 5) where a large portion proteins were expressed in an insoluble forms (Figure 3A, lane 2, 6). This was further confirmed by the measure of OD600 (Figure 3B). These results demonstrated that the solubility of expressed protein was improved by either TRX fusion or Skp co-expression.
Figure 3.
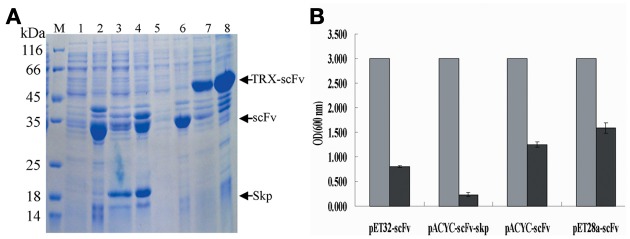
Analysis of solubility of expressed products. (A) Analysis of solubility of the expressed protein by SDS-PAGE. Lane 1, 3, 5, and 7: the soluble cell lysates for pACYC-Duet-scFv, pACYC-Duet-scFv-skp, pET28a-scFv, and pET32a-scFv, respectively; Lane 2, 4, 6, and 8: the insoluble cell lysates for pACYC-Duet-scFv, pACYC-Duet-scFv-skp, pET28a-scFv, and pET32a-scFv, respectively. (B) Cell lysates analysis by OD600 assay (gray bar indicates the concentration of cells after protein expression; black bar indicates the concentration of cell lysates after ultrasonication).
Purification and identification of anti-TLH scFv
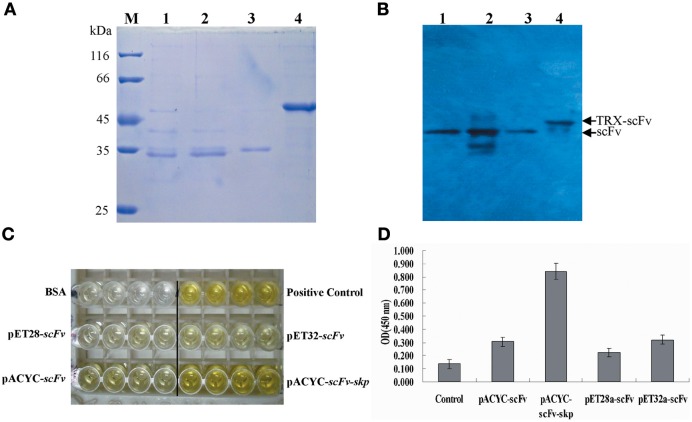
To purify scFv protein, cells were grown for an additional 12 or 32 h with IPTG induction at 16°C and then harvested by centrifugation. Protein purification was performed using Ni2+ affinity chromatography, and the purified proteins were visualized by SDS-PAGE using 12% (v/v) polyacrylamide gels. The concentrations of purified proteins from four different constructs are 0.13, 0.29, 0.115, and 0.38 mg/mL, respectively. After SDS-PAGE analysis, the target protein was further transferred onto a PVDF membrane, and anti-6×His tag antibody was used to confirm the presence of the expressed scFv protein. Protein purification and western blotting results showed that all four different fusion proteins were purified successfully (Figure 4A), and that all four could be recognized by the anti-6×His tag antibody during western blotting (Figure 4B).
Figure 4.
Protein purification and identification. (A) Protein purification. The purified protein was determined by SDS-PAGE. M: Molecular weight of marker proteins; Lane 1–4: the purified proteins for pACYC-Duet-scFv, pACYC-Duet-scFv-skp, pET28a-scFv, and pET32a-scFv, respectively. (B) Western blotting results. Lane 1–4: the detected bands for pACYC-Duet-scFv, pACYC-Duet-scFv-skp, pET28a-scFv, and pET32a-scFv, respectively. (C) ELISA. TLH antigen were coated on 96-well plates in triplicate (5 μ g/mL, 100 μ L/well), and the purified scFv proteins were added to the reaction wells after blocking and washing. The binding activities of the four purified proteins were determined using an anti-6×His tag antibody. (D) Quantitative result of ELISA*, P < 0.05, compared with control antibody treatment.
To compare the effects of different fusion tags on the production of functional protein, the four purified proteins were assessed for their binding activity to the TLH antigen by ELISA. As seen in Figures 4C,D, amongst of four different proteins, the purified protein derived from the pACYC-Duet-scFv-skp vector showed the highest binding activity to TLH antigen, with the binding activity being 3–4 folds higher than the others. Although TRX fusion expressed scFv had higher solubility, the binding activity of TRX fusion protein was barely satisfactory. It was lower than that of scFv obtained by Skp co-expression, and only showed the similar binding activity similar to the scFv expressed from the plasmids pACYC-Duet-scFv. In fact, the scFv expressed from the pET28a-scFv showed a very low binding activity to antigen TLH.
Specificity analysis and affinity determination of scFv obtained by Skp co-expression
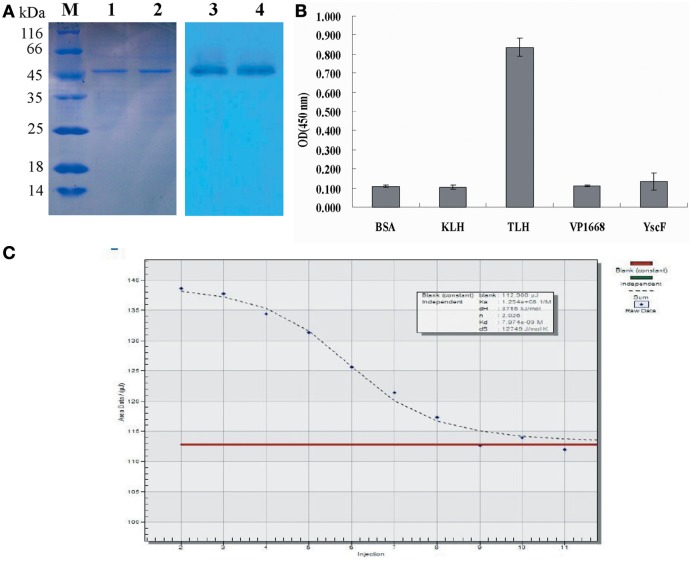
As shown in Figure 5A, a clear band was revealed via western blotting (Figure 5A, lane 3, 4), and the result demonstrated that the TLH band at 47 kDa could be recognized by scFv obtained by Skp co-expression. To further determine the specificity of Skp co-expressed scFv protein to TLH antigen, ELISA was carried out. As shown in Figure 5B, Skp co-expressed scFv was found to be specific to TLH, while there was no cross binding to any other antigen-associated proteins such as BSA, KLH, Vp1668, and YscF. These results indicated that the co-expressed scFv could recognize TLH specifically and be used as an antibody reagent to detect TLH.
Figure 5.
Western blotting and affinity determination. (A) Western blotting analysis of the binding activity of scFv obtained by Skp co-expression to the TLH antigen. Left panel: SDS-PAGE results for the extracted TLH; Lane M: Protein molecular weight markers; Lanes 1, 2: extracted TLH. Right panel: Western blotting results; Lanes 3, 4: TLH band at 47 kDa bound by scFv obtained by Skp co-expression. (B) Specificity analysis of scFv obtained by Skp co-expression. BSA, KLH, and associated antigen proteins (VP1668 and YscF) of V. parahaemolyticus were coated on 96-well plates in triplicate (5 μ g/mL, 100 μ L/well). The purified scFv were added to the reaction wells and incubated for 2 h at 37°C. Specificity of Skp co-expressed scFv was determined using an anti-6×His tag antibody. (C) Affinity Determination of Skp co-expressed scFv. The affinity of anti-TLH Skp co-expressed scFv for the TLH antigen was studied by Microcalorimetry using the Nano ITC-LV. The measured data was used for the quantitative determination of the affinity constant of the anti-TLH Skp co-expressed scFv antibody.
To further probe the interaction of Skp co-expressed scFv and TLH antigen, the affinity of anti-TLH scFv for the TLH antigen was studied by Microcalorimetry using the Nano ITC-LV. The measured data was used for the quantitative determination of the affinity constant of the anti-TLH scFv antibody. It was observed that the TLH antigen could be recognized by Skp co-expressed scFv, and the binding activity was dose dependent. The calculated affinity constant of anti-TLH scFv was 1.254 × 108 L/mol as determined by Origin software (Figure 5C). The above result was consistent with our earlier results obtained by ELISA, and the magnitude of the affinity constants consistently remain at the level of 108 (Wang et al., 2012).
Elisa detection of TLH in bacteria
We were interested in determining whether the co-expressed scFv could serve as a good antibody reagent to directly detect TLH from bacterial strains. In the first detection, three TLH positive V. parahaemolyticus strains, three TLH negative Vibrio strains and two other E. coli strains were used to identify the practicality and feasibility of co-expressed scFv. As shown in Figure 6, three V. parahaemolyticus strains gave the positive signal, and other five non V. parahaemolyticus strains all gave the negative results. To further detect the accuracy of co-expressed scFv, total 22 different bacterial strains (eleven different types V. parahaemolyticus strains, seven Vibrio strains from other species containing no TLH, and four other bacterial strains) were tested. Table 2 showed that all the eleven V. parahaemolyticus strains were successfully detected by Skp co-expressed scFv. In contrast, seven Vibrio strains from other species and four other bacterial strains of non-Vibrio all gave negative detection results. These results showed that this Skp co-expressed scFv could specifically recognize TLH positive V. parahaemolyticus strains, and might be used as a potential reagent for V. parahaemolyticus diagnosis.
Figure 6.
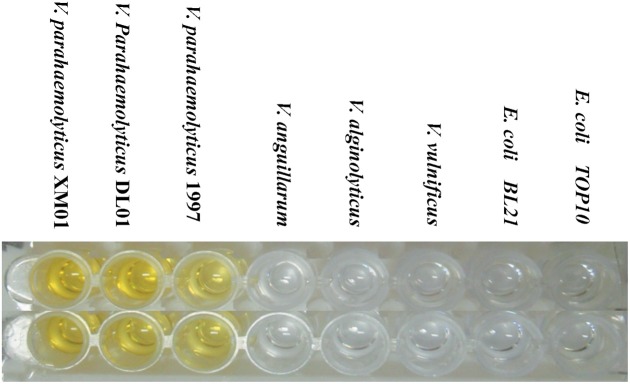
ELISA Detection of TLH in Bacteria. Eight different bacterial strains (three TLH positive V. parahaemolyticus strains, three TLH negative Vibrio strains and two other E. coli strains) were cultured in APW medium without shaking for 16 h, and the supernatant of culture was used to coat the 96 wells plate at 4°C for overnight. The ELISA assay was determined using an anti 6×His tag antibody. (PC, positive control; NC, negative control).
Table 2.
Detection of bacterial samples*.
| Strains | TLH | No. of | Detection |
|---|---|---|---|
| Content | tested | result | |
| TLH | |||
| Vibrio parahaemolyticus CGMCC1.2164 | + | 1 | + |
| Vibrio parahaemolyticus F2002 | + | 1 | + |
| Vibrio parahaemolyticus DL01 | + | 1 | + |
| Vibrio parahaemolyticus CGMCC1.1997 | + | 1 | + |
| Vibrio parahaemolyticus CGMCC1.1997A | + | 1 | + |
| Vibrio parahaemolyticus 121 | + | 1 | + |
| Vibrio parahaemolyticus 124 | + | 1 | + |
| Vibrio parahaemolyticus CGMCC1.1614 | + | 1 | + |
| Vibrio parahaemolyticusATCC17802 | + | 1 | + |
| Vibrio parahaemolyticus CGMCC1.1615 | + | 1 | + |
| Vibrio parahaemolyticus CGMCC1.1616 | + | 1 | + |
| Vibrio alginolyticus CGMCC1.1833 | − | 2 | − |
| Vibrio vulnificus CGMCC1.1597 | − | 2 | − |
| Vibrio anguillarium CCTCCM204066 | − | 2 | − |
| Vibrio harveyi CGMCC1.1601 | − | 1 | − |
| Escherichia coli BL21 | − | 1 | − |
| Escherichia coli DH5a | − | 1 | − |
| Escherichia coli TOP10 | − | 1 | − |
| Escherichia coli JM109 | − | 1 | − |
Each strain sample had two replicates.
Discussion
Although scFv have been used as therapeutic agents and plays a useful role in diagnosis of diseases (Saerens et al., 2008; Tong et al., 2010), but the extensive application of scFv has been limited by the misfolding of scFv, leading to a loss of binding activity and the formation of inclusion bodies. To effectively solve the problem, a variety of strategies have been employed to improve the soluble expression in different host systems. However, to date, the bottleneck of scFv soluble expression has not been resolved, and it is necessary to develop an effective method for scFv soluble expression (Esposito and Chatterjee, 2006; Kudou et al., 2011).
Co-expression of scFv with a molecular chaperon effectively improved the correct folding, and enhanced the solubility of scFv (Hayhurst and Harris, 1999; Sonoda et al., 2011). It is well known that Skp is a periplasmic chaperone that can effective improve the folding and assembly of outer membrane proteins in E. coli (Hayhurst et al., 2003; Sonoda et al., 2011). In this study, a double expression vector pACYC-Duet-skp was chosen for construction of co-expression vector pACYC-Duet-scFv-skp. Two target proteins were co-expressed simultaneously in a host strain, but the expression of the two target proteins were independent, and had different fusion protein tags. Compared to other expression formats, this method neither requires removal of the fusion protein tags, nor the need for co-transformation of plasmids, and makes the entire process of protein purification simple, convenient, and cost-effective. Besides, the expressed target scFv folded correctly and retained the full binding activity with the help of molecular chaperone, and the purified protein could be used directly for different purposes without refolding of protein in vitro after purification.
In this study, four different format expression vectors were constructed for soluble expression of scFv. Amongst the four purified protein, the Skp co-expressed scFv showed a higher solubility, and a 3–4 fold higher binding to antigen TLH than the other three purified proteins. Besides, the Skp co-expressed scFv was specific for TLH and did not cross-react with other TLH-associated proteins. Although TRX fusion expressed scFv had higher solubility, but the binding activity of TRX fusion protein was lower than Skp co-expressed scFv (Marblestone et al., 2006; Subedi et al., 2012). Together, the results demonstrate that the pACYC-Duet-Skp vector system is an effective expression system for soluble expression of scFv.
Low temperature is an important factor in soluble protein expression (Qing et al., 2004; Sørensen and Mortensen, 2011). To effectively express the soluble scFv, different temperatures (such as 37, 30, 25, and 16°C) were used. The result demonstrated that the soluble expression of scFv was improved greatly at low temperature (16°C), the expressed scFv had the highest solubility and binding activity (data not shown). This improvement of solubility can be attributed to the following three advantages of lower expression temperature: (1) The expression rate of protein is greatly reduced, leading to enhanced the folding efficiency of protein; (2) The endogenous proteases are usually inactive or have low-activity, leading to an enhancement of target protein stability (Subedi et al., 2012); (3) The metabolic pressure of host cells is decreased owing to the reduced the cytotoxicity of the expressed protein. However, the true mechanism of efficient expression at low temperature is still unclear, and additional works would be needed to understand this.
Conclusions
We constructed four different expression vectors, and tested their ability to express the soluble scFv protein. The solubility and binding activity of the purified protein were analyzed by SDS-PAGE and ELISA, respectively. Amongst the four purified protein, the Skp co-expressed scFv showed the highest solubility, and the binding activity to antigen TLH was 3–4 fold higher than other three purified scFv. Also, the scFv obtained by Skp co-expression was specific for TLH and did not cross-react with other TLH-associated proteins. Notably, this scFv could be used to detect TLH directly in real samples, suggesting that the pACYC-Duet-skp vector system is an effective system for soluble expression of scFv.
Author contributions
Rongzhi Wang, Youjun Feng, and Shihua Wang designed the experiments and wrote the manuscript. Rongzhi Wang, Shuangshuang Xiang, Yonghui Zhang performed all the experiments. DL Wu, Youjun Feng, Swaminath Srinivas, and Mingshen Lin performed a few experiments and data analysis. All authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Program for New Century Excellent Talents in University (grant NCET-10-0010), the Fujian Fund for Distinguished Young Scientists (grant 2009J06008), the National Agricultural Achievements Transformation Fund (grant 2011GB2C400012), Agricultural Five-new Engineering Projects of Fujian Development and Reform Commission. We would like to thank Dr. Xuanxian Peng (Xiamen University, China) for the gift of V. parahaemolyticus strains and Dr. Mengfei Ho (University of Illinois at Urbana-Champaign) for the donation of pGEPi vector. We are grateful to Prof. Lijun Bi (Institute of Biophysics, Chinese Academy of Sciences) for technical discussions and the donation of pACYC-Duet-1 vector, as well as Dr. Bing Wang (Dalian Fisheries University, China) for technical discussions and the gift of Vibrio spp.
Glossary
Abbreviations
- V.p
Vibrio parahaemolyticus
- TLH
Thermolabile Hemolysin
- IPTG
Isopropyl β-D-1-thiogalactopyranoside
- scFv
single chain variable fragment.
References
- Cattepoel S., Hanenberg M., Kulic L., Nitsch R. M. (2011). Chronic intranasal treatment with an anti-A¦Â30-42 scFv antibody ameliorates amyloid pathology in a transgenic mouse model of alzheimer's disease. PLoS ONE 6:e18296 10.1371/journal.pone.0018296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coia G., Hudson P. J., Irving R. A. (2001). Protein affinity maturation in vivo using E. coli mutator cells. J. Immun. Methods 251, 187–193 10.1016/S0022-1759(01)00300-3 [DOI] [PubMed] [Google Scholar]
- Daly R., Hearn M. T. (2005). Expression of heterologous proteins in Pichia pastoris: a useful experimental tool in protein engineering and production. J. Mol. Recognit. 18, 119–138 10.1002/jmr.687 [DOI] [PubMed] [Google Scholar]
- Davis G. D., Elisee C., Newham D. M., Harrison R. G. (1999). New fusion protein systems designed to give soluble expression in Escherichia coli. Biotechnol. Bioeng. 65, 382–388 [DOI] [PubMed] [Google Scholar]
- DelProposto J., Majmudar C. Y., Smith J. L., Brown W. C. (2009). Mocr: a novel fusion tag for enhancing solubility that is compatible with structural biology applications. Protein Expr. Purif. 63, 40–49 10.1016/j.pep.2008.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marco A., De Marco V. (2004). Bacteria co-transformed with recombinant proteins and chaperones cloned in independent plasmids are suitable for expression tuning. J. Biotechnol. 109, 45–52 10.1016/j.jbiotec.2003.10.025 [DOI] [PubMed] [Google Scholar]
- Eisenhardt S. U., Schwarz M., Schallner N., Soosairajah J., Bassler N., Huang D. X., et al. (2007). Generation of activation-specific human anti-{alpha} M {beta} 2 single-chain antibodies as potential diagnostic tools and therapeutic agents. Blood 109, 3521–3528 10.1182/blood-2006-03-007179 [DOI] [PubMed] [Google Scholar]
- Esposito D., Chatterjee D. K. (2006). Enhancement of soluble protein expression through the use of fusion tags. Curr. Opin. Biotechnol. 17, 353–358 10.1016/j.copbio.2006.06.003 [DOI] [PubMed] [Google Scholar]
- Fox J. D., Waugh D. S. (2003). Maltose-binding protein as a solubility enhancer. Methods Mol. Biol. 205, 99–117 10.1385/1-59259-301-1:99 [DOI] [PubMed] [Google Scholar]
- Fu X. Y. (2010). Extracellular accumulation of recombinant protein by Escherichia coli in a defined medium. Appl Microbiol. Biotechnol. 88, 75–86 10.1007/s00253-010-2718-9 [DOI] [PubMed] [Google Scholar]
- Goulding C. W., Perry L. J. (2003). Protein production in Escherichia coli for structural studies by X-ray crystallography. J Struct Biol 142, 133–143 10.1016/S1047-8477(03)00044-3 [DOI] [PubMed] [Google Scholar]
- Greene J. J. (2004). Host cell compatibility in protein expression. Methods Mol. Biol. 267, 3–14 10.1385/1-59259-774-2:003 [DOI] [PubMed] [Google Scholar]
- Guild K., Zhang Y., Stacy R., Mundt E., Benbow S., Green A., et al. (2011). Wheat germ cell-free expression system as a pathway to improve protein yield and solubility for the SSGCID pipeline. Acta Cryst. F67, 1027–1031 10.1107/S1744309111032143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayhurst A., Happe S., Mabry R., Koch Z., Iverson B. L., Georgiou G. (2003). Isolation and expression of recombinant antibody fragments to the biological warfare pathogen Brucella melitensis. J. Immunol. Methods 276, 185–196 10.1016/S0022-1759(03)00100-5 [DOI] [PubMed] [Google Scholar]
- Hayhurst A., Harris W. J. (1999). Escherichia coli skp chaperone co-expression improves solubility and phage display of single-chain antibody fragments. Protein Expr. Purif. 15, 336–343 10.1006/prep.1999.1035 [DOI] [PubMed] [Google Scholar]
- Jurado P., de Lorenzo V., Fernández L. A. (2006). Thioredoxin fusions increase folding of single chain Fv antibodies in the cytoplasm of E. coli: evidence that chaperone activity is the prime effect of thioredoxin. J. Mol. Biol. 357, 49–61 10.1016/j.jmb.2005.12.058 [DOI] [PubMed] [Google Scholar]
- Krag D. N., Shukla G. S., Shen G. P., Pero S., Ashikaga T., Fuller S., et al. (2006). Selection of tumor-binding ligands in cancer patients with phage display libraries. Cancer Res. 66, 7724–7733 10.1158/0008-5472.CAN-05-4441 [DOI] [PubMed] [Google Scholar]
- Kudou M., Ejima D., Sato H., Yumioka R., Arakawa T., Tsumoto K. (2011). Refoldin g single-chain antibody (scFv) using lauroyl-L-glutamate as a solubilization detergent and arginine as a refolding additive. Protein Expr. Purif. 77, 68–74 10.1016/j.pep.2010.12.007 [DOI] [PubMed] [Google Scholar]
- Marblestone J. G., Edavettal S. C., Lim Y., Lim P., Zuo X., Butt T. R. (2006). Comparison of SUMO fusion technology with traditional gene fusion systems: enhanced expression and solubility with SUMO. Protein Sci. 15, 182–189 10.1110/ps.051812706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallamsetty S., Waugh D. S. (2006). Solubility-enhancing proteins MBP and NusA play a passive role in the folding of their fusion partners. Protein Expr. Purif. 45, 175–182 10.1016/j.pep.2005.06.012 [DOI] [PubMed] [Google Scholar]
- Nygren P. A., Stahl S., Uhlen M. (1994). Engineering proteins to facilitate bioprocessing. Trends Biotechnol. 12, 184–188 10.1016/0167-7799(94)90080-9 [DOI] [PubMed] [Google Scholar]
- Ow D. S. W., Lim D. Y. X., Nissom P. M., Camattari A., Wong V. V. T. (2010). Co-Expression of Skp and FkpA chaperones improves cell viability and alters the global expression of stress response genes during scFvD1.3 production. Microb. Cell Fact. 9, 22–36 10.1186/1475-2859-9-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing G., Ma L. C., Khorchid A., Swapna G. Y. T., Mal T. K., Takayama M. M., et al. (2004). Cold-shock induced high yield protein production in Escherichia coli. Nat. Biotechnol. 22, 877–882 10.1038/nbt984 [DOI] [PubMed] [Google Scholar]
- Rinderknecht M., Villa A., Ballmer-Hofer K., Neri D., Detmar M. (2010). Phage-derived fully human monoclonal antibody fragments to human vascular endothelial growth factor-C block its interaction with VEGF receptor-2 and 3. PLoS ONE 5:e11941 10.1371/journal.pone.0011941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe A., Nathanielsz A., Hosse R. J., Oberh F., Strandmann E. P., Engert A., et al. (2007). Selection of human anti-CD28 scFvs from a T-NHL related scFv library using ribosome display. J. Biotechnol. 130, 448–454 10.1016/j.jbiotec.2007.05.012 [DOI] [PubMed] [Google Scholar]
- Saerens D., Conrath K., Govaert J., Muyldermans S. (2008). Disulfide bond introduction for general stabilization of immunoglobulin heavy-chain variable domains. J. Mol. Biol. 377, 478–488 10.1016/j.jmb.2008.01.022 [DOI] [PubMed] [Google Scholar]
- Sakai K., Yuasa N., Tsukamoto K., Takasaki-Matsumoto A., Yajima Y., Sato R., et al. (2010). Isolation and characterization of antibodies against three consecutive Tn-antigen clusters from a phage library displaying human single-chain variable fragments. J. Biochem. 147, 809–817 10.1093/jb/mvq014 [DOI] [PubMed] [Google Scholar]
- Singh P. K., Agrawal R., Kamboj D. V., Gupta G., Boopathi M., Goel A. K., et al. (2010). Construction of a single-chain variable-fragment antibody against the superantigen staphylococcal enterotoxin B. Appl. Environ. Microbiol. 76, 8184–8191 10.1128/AEM.01441-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommaruga S., Lombardi A., Salvadè A., Mazzucchelli S., Corsi F., Galeffi P., et al. (2011). Highly efficient production of anti-HER2 scFv antibody variant for targeting breast cancer cells. Appl. Microbiol. Biotechnol. 91, 613–621 10.1007/s00253-011-3306-3 [DOI] [PubMed] [Google Scholar]
- Sonoda H., Kumada Y., Katsuda T., Yamaji H. (2011). Effects of cytoplasmic and periplasmic chaperones on secretory production of single-chain Fv antibody in Escherichia coli. J. Biosci. Bioeng. 111, 465–470 10.1016/j.jbiosc.2010.12.015 [DOI] [PubMed] [Google Scholar]
- Sørensen H. P., Mortensen K. K. (2011). Advanced genetic strategies for recombinant protein expression in Escherichia coli. J. Biotechnol. 115, 113–128 10.1016/j.jbiotec.2004.08.004 [DOI] [PubMed] [Google Scholar]
- Subedi G. P., Satoh T., Hanashima S., Ikefda A., Nakada H., Sato R., et al. (2012). Overproduction of anti-Tn antibody MLS128 single-chain Fv fragment in Escherichia coli cytoplasm using a novel pCold-PDI vector. Protein Expr. Purif. 82, 197–204 10.1016/j.pep.2011.12.010 [DOI] [PubMed] [Google Scholar]
- Sushma K., Vijayalakshmia M. A., Krishnana V., Satheeshkumara P. K. (2011). Cloning, expression, purification and characterization of a single chain variable fragment specific to tumor necrosis factor alpha in Escherichia coli. J. Biotechnol. 156, 238–244 10.1016/j.jbiotec.2011.06.039 [DOI] [PubMed] [Google Scholar]
- Tong Q., Liu K., Lu X. M., Shu X. G., Wang G. B. (2010). Construction and characterization of a novel fusion protein MG7-scFv/SEB against gastric cancer. J. Biomed. Biotechnol. 1, 121094–121102 10.1155/2010/121094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. J., Dai J. X., Li B. H., Fan K. X., Peng L., Zhang D. P., et al. (2008a). Expression, purification, and characterization of an immunotoxin containing a humanized anti-CD25 single chain fragment variable antibody fused to a modified truncated Pseudomonas exotoxin A. Protein Expr. Purif. 58, 140–147 10.1016/j.pep.2007.09.009 [DOI] [PubMed] [Google Scholar]
- Wang S. H., Du X. Y., Lin L., Huang Y. M., Wang Z. H. (2008b). Zearalenone (ZEN) detection by a single chain fragment variable (scFv) antibody. World J. Microbiol. Biotechnol. 24, 1681–1685 10.1007/s11274-008-9657-y 18575809 [DOI] [Google Scholar]
- Wang R. Z., Fang S., Wu D. L., Lian J. W., Fan J., Zhang Y. F., et al. (2012). Screening of a single-chain variable- fragment antibody that can neutralize effectively the cytotoxicity of Vibrio parahaemolyticus TLH. Appl. Environ. Microbiol. 78, 4967–4975 10.1128/AEM.00435-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. Z., Huang J. D., Zhang W., Lin G. M., Lian J. W., Jiang L. B., et al. (2011). Detection and identification of Vibrio parahaemolyticus by multiplex PCR and DNA-DNA hybridization on a microarray. J. Genet. Genomics 38, 129–135 10.1016/j.jgg.2011.02.002 [DOI] [PubMed] [Google Scholar]
- Wang S. H., Du X. Y., Huang Y. M., Lin D. S., Hart P. L., Wang Z. H. (2007). Detection of deoxynivalenol based on a single chain fragment variable(scFv) of the antideoxynivalenol antibody. FEMS Microbiol. Lett. 272, 214–219 10.1111/j.1574-6968.2007.00765.x [DOI] [PubMed] [Google Scholar]
- Wang S. H., Zhang J. B., Zhang Z. P., Zhou Y. F., Yang R. F., Chen J., et al. (2006). Construction of Single chain variable fragment (ScFv) and biscFv-alkaline phosphatase fusion protein for detection of Bacillus anthracis. Anal. Chem. 78, 997–1004 10.1021/ac0512352 [DOI] [PubMed] [Google Scholar]
- Zhang G., Liu Y., Hu H. (2010). Preparation and cytotoxicity effect of anti- hepatocellular carcinoma scFv immunoliposome on hepatocarcinoma cell in vitro. Eur. J. Inflamm. 8, 75–82 [Google Scholar]