Abstract
SUMMARY
Homology-dependent repair of double-strand breaks (DSBs) from non-sister templates has the potential to generate loss of heterozygosity or deleterious genome rearrangements. Here we show the Saccharomyces cerevisiae Mph1 helicase prevents crossovers between ectopic sequences by removing substrates for Mus81-Mms4 or Rad1-Rad10 cleavage. A role for Yen1 is only apparent in the absence of Mus81. Cells lacking Mph1 and the three nucleases are highly defective in the repair of a single DSB, suggesting the recombination intermediates that accumulate cannot be processed by the Sgs1-Top3-Rmi1 complex (STR). Consistent with this hypothesis, ectopic joint molecules accumulate transiently in the mph1Δ mutant and persistently when Mus81 is eliminated. Furthermore, the ectopic JMs formed in the absence of Mus81 are connected by a single HJ explaining why STR is unable to process them. We suggest that Mph1 and Mus81-Mms4 recognize a common early strand exchange intermediate and direct repair to non-crossover or crossover outcomes, respectively.
INTRODUCTION
Homologous recombination (HR) plays a critical role in maintaining genome integrity in mitotic and meiotic cells. The primary function of HR in mitotic cells is to repair double-strand breaks (DSBs) or single-stranded DNA (ssDNA) gaps that form as a result of replication fork stalling or fork collapse, from processing of spontaneous DNA damage and from exposure to DNA damaging agents. HR is activated during the S and G2 phases of the cell cycle when a sister chromatid is available and is the preferred repair template (Kadyk and Hartwell, 1992), but repair can occur using other homologous sequences. For some regions of the genome homology is available in the form of repeats that can be used for ectopic recombination, and a chromosome homolog is present in diploid cells (Baker et al., 1996; Jinks-Robertson and Petes, 1986; Kadyk and Hartwell, 1992; Lichten and Haber, 1989).
Although HR is considered to be an error-free mechanism, the fidelity of the process depends on the percent sequence identity between the recombining sequences. If the donor and recipient sequences are diverged, the template is rejected during homologous pairing/strand invasion in a process involving mismatch repair proteins and HR will be averted (Datta et al., 1996; Priebe et al., 1994; Selva et al., 1995). However, some sequence divergence is tolerated and can lead to gene conversion during HR (Krogh and Symington, 2004). When repair occurs between chromosome homologs or between ectopic repeats, in addition to the risk of limited loss of heterozygosity (LOH) by gene conversion, cells face another source of genome instability by crossing over (Krogh and Symington, 2004). Crossovers can lead to extensive LOH when a chromosome homolog is used as the repair template, or to translocation when HR occurs between dispersed repeats.
Homology-dependent repair of DSBs initiates by Rad51 binding to the 3′ ssDNA tails generated by end resection. The Rad51 nucleoprotein filament catalyzes homologous pairing and invasion of a donor duplex to form a displacement loop (D-loop, Figure S1) (San Filippo et al., 2008). After extension of the invading 3′ end by DNA synthesis, the invading strand can be displaced and pair with the resected strand on the other side of the DSB resulting in non-crossover products (synthesis-dependent strand annealing [SDSA]) (Ferguson and Holloman, 1996; Nassif et al., 1994; Paques et al., 1998). Several DNA helicases have been implicated in D-loop displacement to promote SDSA in different organisms. The Mph1/Fml1/FANCM 3′-5′ helicase, which dissociates D-loops and extended D-loops in vitro, reduces crossovers (CO) during mitotic DSB repair in budding and fission yeasts (Mitchel et al., 2013; Prakash et al., 2009; Sebesta et al., 2011; Sun et al., 2008; Tay et al., 2010), and during meiotic recombination in S. pombe and A. thaliana (Crismani et al., 2012; Knoll et al., 2012; Lorenz et al., 2012). The BLM helicase promotes SDSA during mitotic DSB repair in Drosophila, and the budding yeast ortholog, Sgs1, is important for non-crossovers (NCO) generated by meiotic recombination (Adams et al., 2003; De Muyt et al., 2012; Ira et al., 2003; Zakharyevich et al., 2012). Mutation of rtel-1 increases meiotic COs in C. elegans and the RTEL-1 helicase is able to dissociate Rad51-mediated D-loops in vitro, in agreement with a role in SDSA (Barber et al., 2008; Youds et al., 2010).
Capture of the second break end by the D-loop, followed by repair synthesis and ligation results in formation of a double Holliday junction (dHJ) intermediate (Figure S1); this structure was proposed as a key intermediate in the DSB repair model and has been detected experimentally during meiotic and mitotic recombination in budding yeast (Bzymek et al., 2010; Schwacha and Kleckner, 1995; Szostak et al., 1983). The dHJ intermediate can be “dissolved” by the Sgs1 helicase in collaboration with Top3 and Rmi1 (BLM-TopoIIIα-RMI1-RMI2 in humans) generating NCO products exclusively (Cejka et al., 2010; Plank et al., 2006; Wu et al., 2006; Wu and Hickson, 2003). Alternatively, the dHJ intermediate can be resolved by cleavage with structure-selective nucleases to yield NCO or CO products (De Muyt et al., 2012; Szostak et al., 1983; Zakharyevich et al., 2012). Generation of COs in S. pombe requires Mus81-Mms4/Eme1, an endonuclease that preferentially cleaves D-loops and nicked HJs, but has low activity towards intact HJs (Boddy et al., 2001; Ho et al., 2010; Osman et al., 2003; Schwartz and Heyer, 2011; Sun et al., 2008). These observations led to the proposal that Mus81-Mms4 cleaves the second-end capture intermediate, prior to gap filling and ligation, generating only CO products (Figure S1) (Osman et al., 2003; Schwartz and Heyer, 2011). In S. cerevisiae, the majority of meiotic COs form independently of Mus81-Mms4 (De Muyt et al., 2012; Zakharyevich et al., 2012). Yen1/GEN1, which serves as a back-up function for Mus81-Mms4 during mitotic recombination in budding yeast, is able to cleave intact HJs in vitro and might remove dHJ intermediates at mitosis that escape dissolution, or that have become converted to single HJs and cannot be dissolved by Sgs1-Top3-Rmi1 (Agmon et al., 2011; Blanco et al., 2010; Ho et al., 2010; Ip et al., 2008; Munoz-Galvan et al., 2012). S. pombe lacks Yen1 explaining the reliance on Mus81-Eme1 for COs in this organism (Boddy et al., 2001; Ip et al., 2008). In addition, the Rad1-Rad10 nuclease (XPF-ERCC1 in human) plays a role in processing recombination intermediates between ectopic repeats to form CO and NCO products, but has no apparent role in the formation of recombinants between chromosome homologs in yeast (Mazon et al., 2012). The role of Rad1-Rad10 in formation of NCOs is thought to be removal of heterologous flaps that would be formed by SDSA if DNA synthesis within the D-loop extended beyond the region of shared homology (Al-Minawi et al., 2008; Fishman-Lobell and Haber, 1992; Mazon et al., 2012). We previously suggested that Rad1-Rad10 facilitates CO formation by cleaving the leading edge of the captured D-loop at the heterology boundary creating a substrate for subsequent cleavage by Mus81-Mms4 or Yen1 (Mazon et al., 2012). The resolvase responsible for meiotic crossovers in budding yeast has not been identified biochemically, but genetic studies point to a role for the Exo1-Mlh1-Mlh3 complex (Zakharyevich et al., 2012).
Most of the products of DSB-induced mitotic recombination between ectopic repeats are NCOs suggesting that the mechanisms that enforce NCOs play an important role in this context (Inbar et al., 2000). Here we explored the functions of the Mph1 and Sgs1 helicases and the interplay between Mph1 and structure-selective nucleases involved in crossover formation using assays to identify intermediates and products of ectopic recombination.
RESULTS
The Mph1 helicase prevents crossovers between ectopic repeats
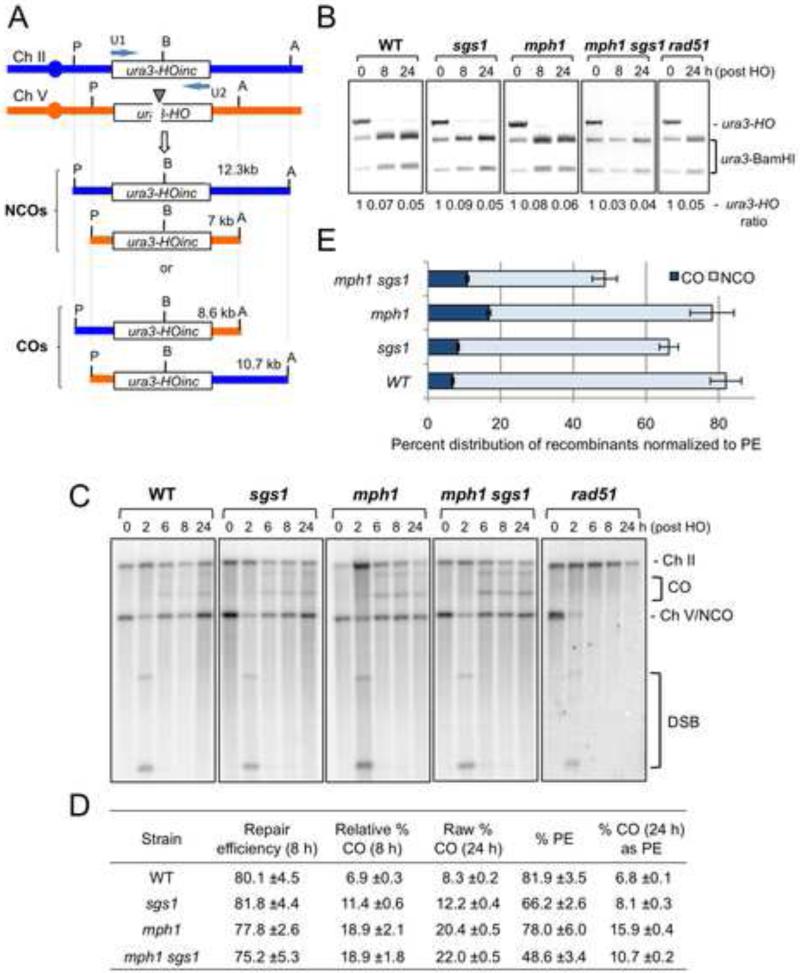
The Mph1 and Sgs1 helicases are proposed to prevent mitotic crossovers by D-loop dissociation and dHJ dissolution, respectively. To address their roles in DSB-induced recombination between dispersed chromosomal repeats we used a previously described assay that allows recovery of both NCO and CO (reciprocal translocation) products (Agmon et al., 2011; Aylon and Kupiec, 2003; Mazon et al., 2012). The haploid strains have a 39 bp HO endonuclease cut site (HOcs) inserted within the native URA3 locus on chromosome (Ch) V and a donor cassette containing 5.6 kb of the ura3 region, inserted in the LYS2 locus on Ch II (Figure 1A). The donor sequence has a 39 bp insertion of the non-cleavable HOcs-inc, including a unique BamHI site to monitor gene conversion repair, at the same position as the HOcs at the native ura3 locus. A PGAL1-HO cassette integrated at the ade3 locus enables induction of a DSB at the Ch V HOcs by addition of galactose to the growth media. Repair of the DSB by gene conversion transfers the HOcs-inc and BamHI sequence to the recipient locus. Cells are haploid and will only survive the induction of the DSB if they are able to repair the break without loss of the genetic information encoded in both arms of Ch V. Restriction endonuclease sites in heterologous sequences flanking the ura3 genes are used to distinguish between NCO and CO products (Figure 1A).
Figure 1. Mph1 prevents CO between ectopic repeats.
A. Schematic of the assay used to detect recombinants formed between ectopic ura3 repeats. CO products are detected by the formation of novel ApaLI (A)/PvuII (P) restriction fragments of 10.7 and 8.6 kb. B. The efficiency of HO cleavage and repair was determined by PCR amplification of the donor and recipient ura3 loci followed by BamHI digestion. C. Southern blot analysis of DNA extracted from cells of the indicated genotype following HO induction. D. The mean CO values at 8 and 24 h are shown with the 8 h value normalized to the repair efficiency and the 24 h value normalized to PE. PE was determined from 4-11 trials and COs were quantified from 3-8 trials. E. Recombinant distribution for the indicated strains normalized to the PE. Error bars show standard error of the mean (SEM) for both PE and CO. See also Figures S1 and S2.
The plating efficiency (PE) of mph1Δ cells on medium containing galactose (constitutive HO expression) relative to growth on glucose-containing medium was 78%, not significantly different to wild type (p=0.59). By contrast, the PE of the sgs1Δ mutant was significantly reduced (p=0.034), consistent with a previous study (Ira et al., 2003). To determine the fraction of repaired products with an associated CO we induced HO in liquid cultures and isolated DNA at different times after HO induction for restriction digestion and Southern blot hybridization (Mazon et al., 2012) (Figure 1A). By 6-8 h, >95% of the recipient loci in the population were cut by HO, as evidenced by loss of the Ch V band in the rad51Δ mutant, and repaired from the ectopic donor resulting in restoration of the Ch V band and sensitivity of the recipient ura3 locus to BamHI digestion (Figure 1B, C). The CO fraction was determined by the ratio of CO bands to NCO bands at 8 or 24 h post HO induction. COs accumulated to 8.3% of the DNA products 24 h after HO induction in wild-type cells, and were detected at higher levels in the mph1Δ (20.4%) and sgs1Δ (12.2%) mutants (Figure 1D). Mutants that exhibit an increased frequency of ectopic COs are expected to show decreased viability due to segregation of unbalanced chromosome translocations at mitosis. CO products increase to higher levels in the mph1Δ mutant as compared to sgs1Δ; however, the sgs1Δ mutant has lower cell viability in response to the DSB suggesting inviability is not due to unbalanced translocations. The decreased viability of the sgs1Δ mutant could be due to unresolved recombination intermediates or non-reciprocal COs that contribute to the CO bands detected by Southern blot, but are not viable in haploid survivors. To take into account the reduced plating efficiency of the sgs1Δ mutant, we normalized the CO level determined by Southern blot hybridization to cell survival on galactose medium (Figure 1E). By this measure COs increase by only 1.2 fold in the sgs1Δ mutant (8.1%) compared to wild type (6.8%) (p=0.02), whereas the mph1Δ mutant exhibits a 2.4-fold increase (15.9%, p=0.00005), similar to results obtained with other ectopic assays (Ira et al., 2003; Mitchel et al., 2013; Prakash et al., 2009; Sun et al., 2008; Tay et al., 2010). Thus, intermediates are channeled from a NCO to a CO outcome in the mph1Δ mutant, whereas elimination of Sgs1 results in loss of NCO products accompanied by a modest increase in COs.
Inactivation of both helicases resulted in a slight increase in the percent COs at 24 h (22.0%), but, when normalized for the PE, the relative CO level was lower than the mph1Δ single mutant (10.7% compared to 15.9%). The further reduction in PE of the mph1Δ sgs1Δ double mutant (48.6%) compared with the sgs1Δ single mutant (66.2%) is due to a preferential loss of NCO events (Figure 1E). This result points to a non-additive effect of Sgs1 and Mph1 for crossover avoidance, similar to studies of plasmid gap repair (Mitchel et al., 2013; Tay et al., 2010).
To verify use of PE to normalize the raw CO levels at 24 h, genomic DNA from individual recombinants recovered on galactose-containing medium was analyzed by Southern blot hybridization to determine the CO frequency among survivors (Figure S2A). The CO frequency was higher for the mph1Δ and mph1Δ sgs1Δ mutants than sgs1Δ, but after adjustment of the CO frequency to the reduced recovery of recombinants (PE) the CO value for the mph1Δ sgs1Δ double mutant was lower than the mph1Δ single mutant, similar to the frequencies obtained by normalizing raw CO values to the PE (Figure S2B and Figure 1E). Thus, the correction of raw CO values at 24 h to the PE agrees with the CO frequency of surviving colonies.
The ectopic recombination assay does not allow us to distinguish between reciprocal CO and non-reciprocal CO events, such as break-induced replication (BIR), that generate the same size restriction fragments in the cell population. BIR events are not viable after plating because genes downstream of the DSB site are indispensable, but could persist in the population until essential gene products are depleted. The contribution of BIR can be estimated by comparing the efficiency of repair at 8 h (ratio between total DNA detected in both CO and NCO bands at 8 h versus 0 h, normalized to an internal loading control using a fragment on Ch I) and the PE. Although wild type, sgs1Δ, mph1Δ and sgs1 mph1Δ cells have similar repair efficiencies, ranging from 75-80%, PEs drop for sgs1Δ and mph1 sgs1Δ cells, suggesting some repair products detected at 8 h are inviable. This explains the difference between the relative CO percent detected at 8 h for mph1 sgs1Δ (18.9%) and the amount after normalization by PE (10.7%), while this large difference is not detected in wild type and mph1Δ strains, which have PEs at similar levels to the repair efficiency at 8 h. The sgs1Δ mutant is reported to have a higher frequency of BIR than wild type and this could contribute to the reduced PE (Lydeard et al., 2010).
In addition, we monitored the early accumulation of CO products after HO induction (Figure S2C). The mph1Δ mutant accumulated COs rapidly after DSB formation with CO levels representing ~15% of the products at 3-4 h, while sgs1Δ and wild-type cells accumulated CO to ~5% at the same time points. Together, these results suggest that Mph1 plays an important anti-crossover role and acts on an early intermediate to avert nucleolytic processing.
Mus81 and Rad1 promote crossovers in the absence of Mph1
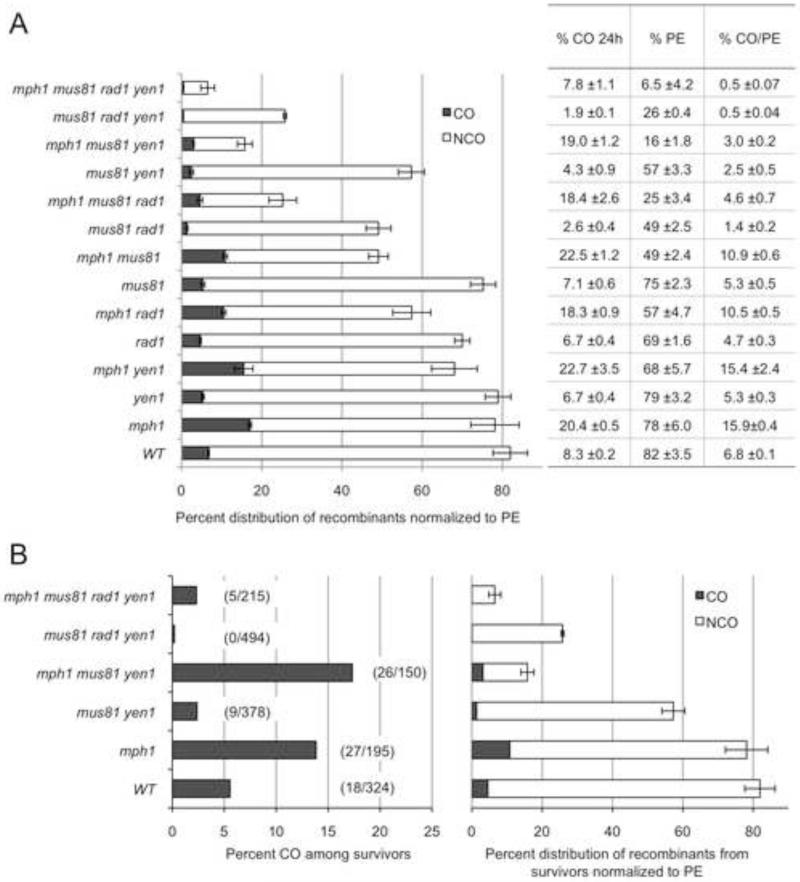
To identify the nucleases responsible for COs in the absence of Mph1 we introduced into the mph1Δ background null mutations of structure-selective nucleases previously implicated in CO formation between ectopic repeats (Mazon et al., 2012). For each strain, we determined PE in response to the HO-induced DSB and the CO fraction by Southern blot hybridization of DNA isolated 24 h after HO induction (Figure 2A). The mph1Δ mutation reduced the PE of all the nuclease mutants with the greatest decrease observed for the mph1Δ mus81Δ double mutant (49%). The PE of the mph1Δ mus81Δ mutant was further reduced by rad1Δ or yen1Δ mutations (25% and 16%, respectively), and only 6.5% of cells of the quadruple mutant were able to repair the DSB. The low recovery of recombinants in the mph1Δ mus81Δ rad1Δ yen1Δ mutant implies that Sgs1-Top3-Rmi1 is not able to process the DNA intermediates generated in these cells to form viable products.
Figure 2. Mph1, Mus81-Mms4, Rad1-Rad10 and Yen1 are required for DSB repair.
A. For each strain the fraction of CO products assessed 24 h after HO induction by Southern blot hybridization was normalized to the PE (see Figure 1). Mean CO values were determined from 2-8 trials and PE from 3-11 trials of each strain, and error bars show SEM. B. Physical analysis to determine the percent crossovers among survivors (left graph) and normalized to the PE (right graph). Mean PE was determined from 3-11 trials and error bars show SEM. See also Figure S3.
Surprisingly, the raw CO values of all of the nuclease mutants harboring the mph1Δ mutation were similar to the mph1Δ single mutant, and the quadruple mutant was the only one with a raw CO value of <8%. However, when the raw CO values were normalized to the PE the rad1Δ and mus81Δ mutations both reduced COs by ~40% in the mph1Δ mutant (p=0.002 and p=0.0005, respectively), whereas yen1Δ caused no loss of CO products in the mph1Δ background (p=0.87) except in the absence of Mus81 (p<0.0001). Eliminating Mus81 and Rad1 in the mph1Δ mutant reduced CO levels by an additional 50% as compared to mph1Δ mus81Δ and mph1Δ rad1Δ (p<0.0001 and p=0.03, respectively), and COs were further reduced in the quadruple mutant. These data are in agreement with the proposed role of Rad1-Rad10 to cleave the leading edge of the D-loop at the heterology boundary as a first step preceding Mus81-Mms4 or Yen1 cleavage to complete the CO event (Mazon et al., 2012).
To confirm that the PE-adjusted CO value from the Southern blot represents viable CO products, the frequency of COs was determined for individual survivors of the mph1Δ mus81Δ yen1Δ and mph1Δ mus81Δ rad1Δ yen1Δ mutants. Seventeen percent of the mph1Δ mus81Δ yen1Δ survivors contained COs, not significantly different to the mph1Δ single mutant, and when normalized by PE the value was similar to that obtained for the PE-adjusted raw CO value from Southern blots. Similarly, 2.3% of survivors of the quadruple mutant had CO products, a value similar to the Southern blot adjusted value when normalized to the PE. The inviable “CO” products detected by Southern blot are most likely due to BIR (see below), although we cannot exclude a contribution from unbalanced reciprocal translocations. The large decrease in the recovery of CO products by the rad1Δ mutation highlights the important role of Rad1-Rad10 to cleave the D-loop intermediate formed during ectopic recombination, particularly in the absence of Mph1 and Mus81.
The reduced ability of cells lacking Mph1 and structure-selective nucleases to repair DSBs is also seen in response to ionizing radiation (IR). The mph1Δ and yen1Δ mutants exhibited normal resistance to 400 Gy, and survival of the mus81Δ mutant was reduced by 3-fold in comparison to wild type (Figure S3). Survival of the mph1Δ mus81Δ double mutant was 5-fold lower than the mus81Δ single mutant and the triple mutant exhibited greatly reduced survival, even at the lower dose of 200 Gy. Sensitivity to methyl methanesulfonate (MMS) was also increased in the mus81Δ mph1Δ mutant, consistent with a previous report (Panico et al., 2010) (Figure S3).
Ectopic recombination intermediates accumulate in mph1Δ and mus81Δ mutants
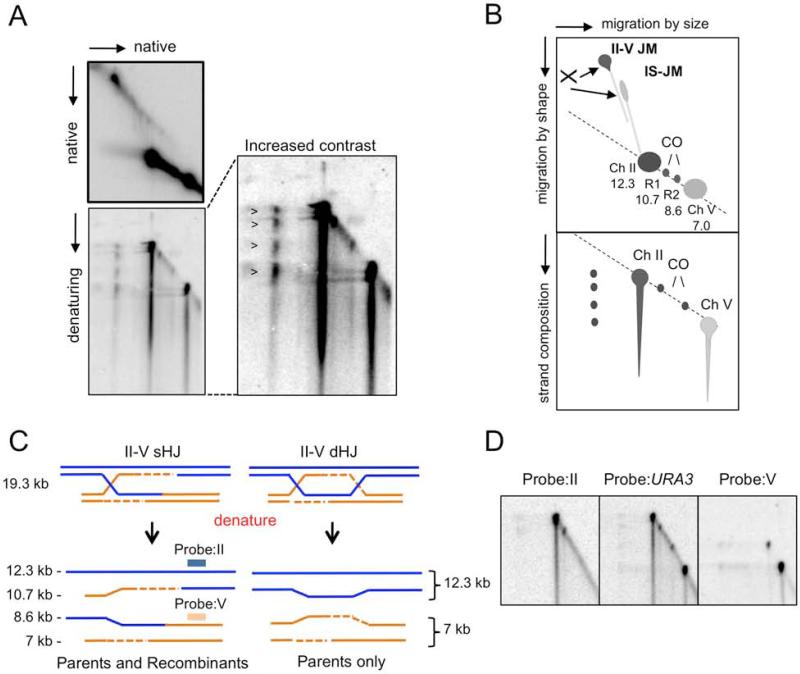
The altered distribution of recombination products points to a key role of Mph1 in preventing the nucleolytic processing of recombination intermediates that ultimately leads to CO formation. Therefore, we expected to observe an accumulation of the joint molecules (JMs) that are substrates for nucleolytic cleavage in the absence of Mph1. Two-dimensional (2D) neutral gel electrophoresis was used to separate JMs from bulk genomic DNA isolated from cells that had been arrested in G2/M with nocodazole prior to HO induction. Under these conditions, two distinct JMs were detected after HO cleavage: Ch V inter-sister JMs (due to asynchronous cleavage of the sister chromatids by HO) and Ch II-V ectopic JMs (Figure 3). JMs were barely detectable in the wild-type strain, presumably due to their rapid turnover (Mazon et al., 2012). Ectopic JMs transiently accumulated in the mph1Δ mutant with a peak at 2.5 h (Figure 3B), coinciding with the time CO products started to appear (Figure S2). Inter-sister and ectopic JMs were detected in the sgs1Δ mutant and persisted at later time points, in agreement with a study of DSB-induced JMs in diploid cells (Bzymek et al., 2010). Previously we showed the accumulation of ectopic and inter-sister JMs in the mus81Δ yen1Δ double mutant. Here, we analyzed the mus81Δ single mutant and found the accumulation of JMs to be similar to the mus81Δ yen1Δ double mutant indicating that Mus81-Mms4 is the primary activity responsible for JM resolution (Figure 3B). As the cells were arrested with nocodazole prior to HO induction Yen1 is expected to be inactive and Mus81-Mms4 hyper activated under these conditions (Gallo-Fernandez et al., 2012; Matos et al., 2011; Szakal and Branzei, 2013).
Figure 3. Joint molecules accumulate in mph1Δ, mus81Δ and sgs1Δ mutants.
A. Schematic showing the expected sizes of parental fragments and joint molecules after digestion with ApaLI and PvuII, and their expected migration by neutral 2D gel electrophoresis. B. 2D gel analysis of DNA isolated from the indicated strains 1.5, 2.5 or 4.5 h after HO induction. The orange and green triangles correspond to inter-sister and ectopic JM, respectively; * corresponds to HO collapsed inter-sister JMs. C. An IS-JM forms by invasion of an uncut sister chromatid by the HO cut sister. If the IS-JM is not fully gap filled and ligated then HO would cleave only the newly synthesized strand and its complement and not the D-loop. Branch migration of the junction towards the DSB would release the junction resulting in Y-shaped molecules of 11.5 or 9.5 kb. Ch-specific probes were used to verify formation of inter-sister JMs.
Loss of Mus81 in the mph1Δ background resulted in a ~40% greater accumulation of ectopic JMs (3.46%) at later time points than observed for the mus81Δ single mutant (2.5%) (p=0.034, paired student t-test) (Figure 3B). The profile of the mph1Δ mus81Δ yen1Δ triple mutant was similar to the mph1Δ mus81Δ double mutant. Accumulation of ectopic JMs was still dependent on Rad1-Rad10, confirming the role previously identified for Rad1-Rad10 in processing a D-loop intermediate at the heterology barrier to generate a substrate for the other nucleases (Figure 3B).
In contrast to mus81Δ and sgs1Δ, we observed no accumulation of inter-sister JMs in the mph1Δ mutant. Detection of inter-sister JMs could be due to low efficiency and/or asynchronous cleavage by HO or due to failure to process JMs. The percent total DNA cut by HO in the mph1Δ and mus81Δ mutants was similar suggesting Mph1 might be more important in D-loop dissociation when the substrates are flanked by heterology. We consistently observed more persistent inter-sister JMs when Mph1 and all nucleases were absent. This could be due to less synchronous cleavage by HO in the slower growing strains or possibly to a futile D-loop extension that is neither dissociated by Mph1 nor cleaved by Mus81-Mms4. In addition, we observed the accumulation of two branched DNA structures of 11.5 and 9.5 kb, which migrated on the Y-arc and hybridized specifically with a chromosome V probe, in all of the strains lacking Mus81. The Y-shaped JMs are most likely due to HO-collapsed inter-sister JMs (Figure 3C).
Ectopic JMs that accumulate in the mus81Δ mutant are connected by a single HJ
To determine the strand composition of ectopic JMs, genomic DNA from the mus81Δ mutant was first fractionated through neutral agarose and then denatured in the second dimension (Figure 4). Under these conditions we detected four spots of equal intensity corresponding to the two parental and two recombinant strands. The strand composition was verified by using probes specific to Ch II or Ch V that hybridize to only one of the two parental spots and one of the recombinants (Figure 4C). This observation provides strong support for the notion that the intermediate contains a single rather than double HJ intermediate. Furthermore, loss of Mph1 resulted in a greater accumulation of these intermediates than in the mus81Δ and mus81Δ yen1Δ strains (Figure 3B). A previous study demonstrated that DSB-induced JMs between chromosome homologs are connected by a dHJ (Bzymek et al., 2010). Our results are not necessarily contradictory to their findings. We propose Rad1-Rad10 plays a unique role during recombination between ectopic repeats by cleaving at the heterology barrier and as a consequence the potential to generate a sHJ.
Figure 4. Ectopic JMs recovered from the mus81Δ mutant are connected by a single HJ.
A. Native-native and native-denaturing 2D gel electrophoresis of genomic DNA isolated from the mus81Δ mutant. The ectopic JM resolves as four spots corresponding to parental and recombinant strands in the denaturing gel. B. Cartoon representation of the 2D gels. C. Equal representation of parental and recombinant strands is indicative of a sHJ, whereas dHJs are denatured to parental only strands by alkaline gel electrophoresis. D. The Southern blot membrane was sequentially probed with probes specific to Ch II or Ch V that hybridize to only one of the two parental spots and one of the two recombinant spots and to the URA3 probe that hybridizes with all parental and recombinant strands.
Mus81 is required for inter-homolog crossovers in the absence of Mph1
To learn whether Mph1 prevents Mus81-dependent crossovers between sequences with more extensive homology, we measured the association of crossing over with gene conversion between chromosome homologs in diploids using a previously described assay (Ho et al., 2010). The diploids used have heteroalleles of ade2, one of which was made by insertion of an I-SceI cut site, and heterozygous markers (his3::Hph/his3::Nat) 150 kb centromere distal to the ade2 locus to detect LOH associated with a G2 recombination event (Figure 5A and Figure S4). Heterozygous markers on the other chromosome arm were used to distinguish between LOH events caused by crossing over and chromosome loss.
Figure 5. Mph1 prevents inter-homolog recombination by removing substrates for Mus81-Mms4.
A. Schematic of chromosome (Ch.) XV showing the I-SceI cut site insertion creating the ade2-I allele, the asterisk notes the location of the ade2-n mutation present on the other homolog. Hph and Nat cassettes replace the HIS3 ORF 150 kb downstream of the ade2 locus. B. Distribution of NCO, CO and BIR products for red/white-sectored colonies normalized to the PE for each of the indicated strains. PE was determined from 4 trials and error bars show SEM. See also Figures S4-6.
The analysis of recombinants was conducted after inducing I-SceI for 1.5 h in the wild-type strain and mutants with an equivalent growth rate, and 3 h for mutants that exhibit slower growth (mus81Δ yen1Δ and mph1Δ mus81Δ yen1Δ). The PE of the different diploid strains growing on medium containing galactose (constitutive I-SceI expression) relative to growth on glucose-containing medium was used to normalize the fractions of recombination events. Recombination events were classified as NCO, CO or BIR based on LOH for the Nat and Hph markers (Figure S4). The red/white sectored colonies are generated by a G2 recombination event and can be scored with less ambiguity than solid white or solid red colonies (Ho et al., 2010).
Analysis of the red/white sectored recombinant colonies revealed a 1.3-fold increase of the relative CO fraction for the mph1Δ strain (30.1%) compared to wild type (22.4%, p=0.0001) (Figure 5B). In the mph1Δ background, loss of Mus81 resulted in a 3.5-fold reduction of the CO fraction (from 30.1% to 8.6%), compared to a 1.5-fold reduction in CO levels between wild type and mus81Δ (p=0.0001 and p=0.0053, respectively). This result is consistent with a much larger dependence on Mus81-Mms4 for CO formation with extensive homology substrates, where Rad1-Rad10 has no apparent role (Mazon et al., 2012). The decreased CO level of the mus81Δ mph1Δ mutant was accompanied by an increase in BIR events (non-reciprocal LOH for the Nat and Hph markers) to 8.6% of recombinants and reduced PE (76%). Notably, the frequency of COs and BIR are equivalent in the mph1Δ mus81Δ mutant, consistent with the proposal that half of the ectopic CO bands seen by Southern blot analysis are due to BIR and contribute to inviability.
The mph1Δ yen1Δ double mutant showed no reduction in PE and the same profile of recombinants as the mph1Δ single mutant suggesting Yen1 does not contribute appreciably to CO formation in the absence of Mph1. The severe growth defect of the mph1Δ mus81Δyen1Δ triple mutant suggests Yen1 might be required to alleviate the accumulation of some toxic intermediates in the absence of Mus81-Mms4 and Mph1, and is consistent with Yen1 overlapping to some extent with Mus81-Mms4 in the formation of COs (Agmon et al., 2011; Ho et al., 2010; Mazon et al., 2012; Munoz-Galvan et al., 2012). The triple mutant yielded a low number of red/white-sectored recombinants and a PE of 47% (Figures 5 and S5A), but in this limited pool the CO fraction was reduced to 2.8% and BIR increased to 13.8%. As noted previously (Ho et al., 2010), around 35% of the non-reciprocal LOH products recovered from the mus81Δ yen1Δ background exhibit loss of the Nat marker instead of Hyg suggesting they are due to breakage of unresolved intermediates and BIR repair in the next cell cycle. The low recovery of red/white-sectored colonies could be due to lethal sectoring as a consequence of the DSB repair defect. Of note, 11% of the colonies recovered from the triple mutant after I-SceI induction exhibited chromosome loss consistent with the reduced DSB repair capacity.
In order to increase the number of events analyzed from the mph1Δ mus81Δ yen1Δ triple mutant we pooled data from the three types of recombinant colonies (red, white and red/white sectored) (Figures S5B and S6). The triple mutant showed a high number of unassigned LOH (22%) and BIR (6.3%) events, and a small number of COs (3%), consistent with the important roles of Mph1 and resolvases in preventing accumulation of toxic recombination intermediates that result in BIR and extensive LOH.
DISCUSSION
Mph1-catalyzed D-loop dissociation is proposed to be a major crossover avoidance mechanism during mitotic and meiotic DSB repair in several organisms (Crismani et al., 2012; Knoll et al., 2012; Lorenz et al., 2012; Prakash et al., 2009; Sun et al., 2008; Tay et al., 2010). The crossovers formed in the mph1Δ mutant could arise from direct processing of the D-loop intermediate or by resolution of a dHJ intermediate by structure-selective nucleases. To identify the critical substrate, we analyzed the interplay between the Mph1 helicase and three structure-selective nucleases involved in crossover formation during ectopic and inter-homolog mitotic recombination in budding yeast. Several important findings were made. First, DSB repair efficiency is unaffected by mph1Δ but COs are significantly increased, indicating that intermediates are channeled from a NCO to a CO outcome in the absence of Mph1. The NCO products that are still formed in the mph1Δ mutant could be due to dHJ dissolution by STR, alternative D-loop displacement activities yet to be identified, or nucleolytic resolution. Second, the elevated frequency of COs between ectopic repeats in the mph1Δ mutant is primarily due to the activities of Mus81-Mms4 and Rad1-Rad10; a role for Yen1 is only apparent in the absence of the Mus81. Third, the increase in inter-homolog COs seen in the mph1Δ mutant is largely due to Mus81-Mms4 activity. Fourth, ectopic JMs accumulate to high levels in the mus81Δ mutant (2.5%) suggesting a prominent role for Mus81-Mms4 in processing DSB-induced recombination intermediates formed between dispersed repeats. Furthermore, we show the ectopic JMs that accumulate in the absence of Mus81 are connected by a single HJ. As discussed below, these findings are most consistent with the generation of ectopic COs by Mus81-Mms4 cleavage of an early strand exchange intermediate instead of by dHJ resolution.
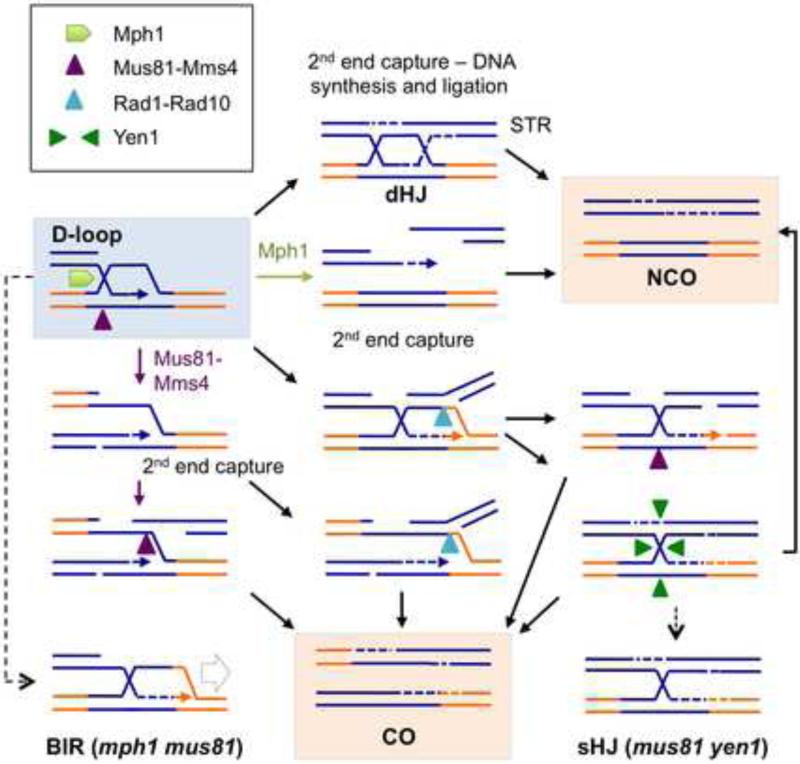
We previously proposed a two-step cleavage mechanism to generate COs during ectopic recombination (Mazon et al., 2012). First, the leading edge of the captured D-loop is cut by Rad1-Rad10 at the heterology boundary. Second, cleavage of the nicked or intact HJ by Mus81-Mms4 or Yen1, respectively, completes the reaction (Figure 6). We suggest that Mus81-Mms4 replaces Rad1-Rad10 when the captured D-loop is within homologous sequence to explain the redundancy between Mus81 and Rad1 for ectopic CO formation, and requirement for Mus81 for inter-homolog COs. As predicted by the model, sHJ intermediates accumulate in the absence of Mus81. JMs with a sHJ cannot be dissolved by Sgs1-Top3-Rmi1 and the accumulation of these structures is likely to contribute to poor viability of the mph1Δ mus81Δ yen1Δ mutant after HO induction. We suggest the COs formed in the absence of Mus81 and Yen1 are due to segregation of Ch II-V JMs to the same daughter cell and replication through the sHJ in the next cell cycle. In agreement with this proposal, rad1Δ reduces JM accumulation and CO products among survivors. The reduced IR resistance and sensitivity to I-SceI of the mph1Δ mus81Δ yen1Δ mutant in the inter-homolog assay indicate that the requirement for Mph1, Mus81 and Yen1 is not restricted to ectopic repeats. JMs with a sHJ were identified by electron microscopy as intermediates of meiotic recombination between chromosome homologs in S. pombe (Cromie et al., 2006; Oh et al., 2008). The fission yeast nuclease responsible for cleaving the D-loop to create a sHJ between chromosome homologs has not been identified. Although we propose Rad1-Rad10 acts at the heterology boundary because the captured D loop would not be fully paired with the other DSB end resulting in a ssDNA gap, it is possible that Rad1-Rad10 could cut within homologous sequences if a ssDNA gap formed transiently adjacent to the branch point.
Figure 6. Model showing the predicted substrates for Mph1, Mus81-Mms4, Rad1-Rad10 and Yen1 during ectopic recombination.
NCOs result from D-loop disassembly by Mph1 or dHJ dissolution by STR. Mus81-Mms4 cuts the D-loop intermediate and captured cleaved D-loop to yield COs. Rad1-Rad10 cuts at the heterology boundary of the captured D-loop before or after cleavage by Mus81-Mms4 to generate COs. If Mus81-Mms4 fails to cut, the intact HJ can be cut by Yen1 to generate NCO or CO products. In the absence of Mph1 and Mus81, the stable D-loop intermediate can undergo extensive DNA synthesis (BIR); without Mus81 and Yen1, sHJ intermediates persist.
The abundance of JMs is too low in the wild-type strain to determine the strand composition, therefore, we cannot rule out the possibility that sHJ intermediates arise from aberrant processing of D-loop intermediates in the absence of Mus81. One possible scenario is that Mus81-Mms4 normally acts in a coordinated fashion to cleave second-end capture intermediates (at the leading edge of the D-loop and the gapped HJ at the trailing end of the D-loop), prior to gap filling and ligation, resulting in CO products (Figure S1). In the absence of Mus81, the captured D-loop could mature to a dHJ by repair synthesis and ligation and then be dissolved by STR to generate NCOs. By having Mus81-Mms4 activity precede STR, COs could be generated during inter-homolog recombination without the requirement for dHJ cleavage. In the case of ectopic repeats the coordination of the two cleavages might be lost if Rad1-Rad10 acts independently to incise the leading edge of the D-loop at the heterology boundary (Figure 6). The nicked HJ resulting from Rad1-Rad10 cleavage could then be cut by Mus81-Mms4 preventing the accumulation of sHJs in wild-type cells. Alternatively, Mus81-Mms4 cleavage at the trailing end of the D-loop, prior to second-end capture and cleavage by Rad1-Rad10, would also ensure sHJs do not form. In the mus81Δ mutant, repair synthesis and ligation of the captured D-loop that had been cut by Rad1-Rad10 would result in a sHJ that could only be resolved by Yen1 to produce either COs or NCOs. Mus81-Mms4 would then be cutting an earlier intermediate than Yen1, and with a biased CO preference. This would explain why Yen1 has no obvious role in processing mitotic or meiotic recombination intermediates, except in the absence of Mus81 (Agmon et al., 2011; De Muyt et al., 2012; Ho et al., 2010; Zakharyevich et al., 2012).
If Rad1-Rad10 is largely responsible for sHJ formation between ectopic repeats, and sHJs are a source of lethality of the mph1Δ mus81Δ yen1Δ mutant, it might have been predicted that the rad1Δ mutation would partially suppress the DSB repair defect of the mph1Δ mus81Δ yen1Δ mutant. Instead, the rad1Δ mutation enhances the DSB repair defect. We propose the early function of Mph1 in disassembling extended D-loop intermediates serves two roles: first, to prevent Mus81-Mms4 cleavage of the D-loop or captured D-loop, and second, to limit the extent of DNA synthesis. There would be a greater necessity for Rad1-Rad10 flap-clipping activity to complete repair If DNA synthesis more frequently extended beyond the region of homology shared by ectopic repeats in the mph1Δ mutant. Interestingly, we found a significant increase in BIR products from inter-homolog recombination in the mph1Δ mus81Δ mutant, suggesting the stable extended D-loop can transition more readily to a BIR intermediate in the absence of Mus81. Consistent with a role for Mph1 to prevent BIR, over-expression of Mph1 was shown to eliminate BIR but not gene conversion repair (Luke-Glaser and Luke, 2012). BIR is not a viable outcome in the ectopic assay and is likely to contribute to the reduced DSB repair efficiency of mph1Δ mus81Δ derivatives. BIR could be more of a problem in the ectopic system because the opportunity for second end capture is lost once the D-loop migrates beyond the region of shared homology.
The plating efficiency of the sgs1Δ mutant was significantly reduced compared to wild type indicating that Sgs1 plays an important role in the generation of ectopic recombinants. By contrast to mph1Δ, the decrease in NCO products observed for the sgs1Δ mutant is accompanied by a minor increase in COs suggesting the intermediates generated are not efficiently resolved. We suggest these intermediates are dHJs that are poor substrates for Mus81-Mms4 cleavage. Analysis of heteroduplex DNA in recombination intermediates formed during plasmid gap repair (analogous to the ectopic system used here) revealed no evidence for cleavage of a dHJ intermediate (Mitchel et al., 2010) . Instead, the CO products were most compatible with cleavage of a nicked or sHJ intermediate. Ten percent of the NCO products were consistent with dHJ dissolution and this class was reduced in the sgs1Δ mutant (Mitchel et al., 2013), suggesting some strand invasion intermediates mature to a dHJ. The reduced yield of NCO products in the sgs1Δ mutant, coupled with the lack of CO products expected by dHJ resolution, is most consistent with Mus81-Mms4 cleavage to form COs prior to dHJ formation.
The dHJ intermediates that accumulate in Ndt80-deficient meiotic cells are rapidly dissolved by Sgs1 to form NCOs when the cells are transferred to vegetative growth conditions (Dayani et al., 2011). These intermediates persist for several hours in the absence of Sgs1 and late resolution to form COs is largely the result of Mus81-Mms4 activity (Dayani et al., 2011). Mms4 is phosphorylated in late G2 and the phosphorylated form of the heterodimer has higher activity on all branched DNA structures, including intact HJs (Gallo-Fernandez et al., 2012; Matos et al., 2011; Szakal and Branzei, 2013). Thus, the delayed resolution could be due to the time taken to reach late G2 and to activate Mms4.
The asynchronous cutting of both sister chromatids by HO allowed us to detect inter-sister intermediates at early time points. Inter-sister JMs accumulated to similar levels in mus81Δ and sgs1Δ mutants suggesting Mus81-Mms4 and Sgs1 have equally important roles in processing sister-chromatid strand-invasion intermediates. The accumulation of inter-sister JMs has also been reported for the S. pombe mus81Δ mutant (Roseaulin et al., 2008). In addition, we detected the accumulation of two chromosome V-derived branched DNA structures of 11.5 and 9.5 kb in all of the mus81Δ derivatives, but not in the sgs1Δ mutant. We speculate these structures correspond to inter-sister D-loop intermediates that after DNA synthesis over the region encoding the sequence of the HOcs and second end capture are cleaved by HO-endonuclease. Branch migration of one or the other junction would collapse the JM to a Y-shaped intermediate of 11.5 or 9.5 kb (Mazon et al., 2012). Importantly, these structures are only predicted to occur if there is a delay in the initiation of repair synthesis from the captured 3′ end because a mature dHJ intermediate would be converted to two X-forms of 9 and 5 kb. The 11.5 and 9.5 kb Y-shaped molecules are expected to derive from the same substrate normally cut by Mus81-Mms4, explaining why they are only detected in the absence of Mus81. Thus, the 2D gel analysis provides additional evidence for Mus81 and Sgs1 acting on different structures and for Mus81-Mms4 cleavage of a structure prior to maturation to a dHJ intermediate.
In summary, the results presented here confirm the important role of Mph1 in disassembly of strand invasion intermediates to prevent crossovers by removing substrates for Mus81-Mms4 cleavage and to limit DNA synthesis that can lead to BIR and extensive LOH. These functions of Mph1 could contribute to the genome instability and cancer predisposition associated with loss of FANCM (Whitby, 2010). In addition, we show the ectopic JMs that accumulate in the absence of Mus81 (and Yen1) are connected by single HJ, explaining the inability of STR to dissolve them to form viable products. We suggest Mph1 and the mitotic resolvases collaborate to remove toxic structures that would otherwise lead to increased LOH, translocations, chromosome mis-segregation and BIR.
EXPERIMENTAL PROCEDURES
Yeast strains
S. cerevisiae strains used in this study are listed in Table S1. All strains are in W303 background and were generated by crossing strains with different recombination reporters to strains with mph1::KanMX6, mus81::KanMX6, rad1::LEU2, sgs1::HphMX or yen1::HIS3 alleles.
Media, growth conditions and genetic methods
YPR or YPL medium (2% raffinose or 3% lactic acid substituted for glucose) was used for galactose induction of I-SceI or HO in the DSB-induced recombination assays. The ectopic and diploid recombination assays were performed as described previously (Aylon et al., 2003; Ho et al., 2010). Recombination outcomes are presented relative to the galactose vs glucose PE of the strains and significance was determined by t test or Fisher exact test (diploid assay). Methyl methane sulfonate (MMS) was added to YPD medium at the designated dose for MMS sensitivity assays. For IR sensitivity, YPD plates were irradiated in a Gammacell-220 irradiator containing 60Co for the designated dose. For quantitation, the mean percent survival from at least three independent experiments is presented.
Physical analysis of ectopic recombination
The distribution of CO and NCO products was determined by Southern blot hybridization using a URA3 probe of ApaLI-PvuII digested genomic DNA from cell populations growing in YPL after galactose induction. Repair efficiency at 8 h was determined by normalization to a Ch I probe. Survivor analysis was performed by pooling genomic DNA from 5 independent colonies obtained from plating cells on YPGal after an 8 h induction of HO. Genomic DNA was digested with ApaLI and PvuII and probed with URA3 sequence. Samples for neutral-neutral and neutral-denaturing 2D-gel electrophoresis were obtained from 30ml aliquots of cultures arrested with nocodazole (15 ug/ml) and induced with 2% galactose. Cells were embedded in agarose, lysed and DNA digested as described previously (Hyppa and Smith, 2009; Mazon et al., 2012). Conditions for 2D gels were as described (Oh et al., 2009). For neutral-denaturing 2D gels, lanes sliced from the first dimension were soaked in 5mM EDTA, rinsed with water and soaked twice in 250mM NaOH, 5mM EDTA for 30min, then soaked twice in 50mM NaOH 1mM EDTA for 30min before pouring around the agarose gel at 0.8% (50mM NaOH, 1 mM EDTA). The gel was run at 4°C at 1V/cm for 48hrs and the DNA transferred to a nylon membrane prior to hybridization with URA3, Ch II or Ch V probes.
Supplementary Material
-
■
mph1 prevents formation of Mus81-dependent crossovers
-
■
Mus81-Mms4 acts on an earlier intermediate than Sgs1
-
■
Single HJ intermediates accumulate in the mus81Δ mutant
-
■
Break-induced replication increases in the absence of Mph1 and Mus81
ACKNOWLEDGEMENTS
We thank M. Kupiec and R. Rothstein for gifts of yeast strains, and W.K. Holloman and members of the Symington lab for comments on the manuscript. This study was supported by grants from the National Institutes of Health (GM041784 and GM094386) to L.S.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adams MD, McVey M, Sekelsky JJ. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science. 2003;299:265–267. doi: 10.1126/science.1077198. [DOI] [PubMed] [Google Scholar]
- Agmon N, Yovel M, Harari Y, Liefshitz B, Kupiec M. The role of Holliday junction resolvases in the repair of spontaneous and induced DNA damage. Nucleic Acids Res. 2011;39:7009–7019. doi: 10.1093/nar/gkr277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Minawi AZ, Saleh-Gohari N, Helleday T. The ERCC1/XPF endonuclease is required for efficient single-strand annealing and gene conversion in mammalian cells. Nucleic Acids Res. 2008;36:1–9. doi: 10.1093/nar/gkm888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y, Kupiec M. The checkpoint protein Rad24 of Saccharomyces cerevisiae is involved in processing double-strand break ends and in recombination partner choice. Mol Cell Biol. 2003;23:6585–6596. doi: 10.1128/MCB.23.18.6585-6596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y, Liefshitz B, Bitan-Banin G, Kupiec M. Molecular dissection of mitotic recombination in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 2003;23:1403–1417. doi: 10.1128/MCB.23.4.1403-1417.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MD, Read LR, Beatty BG, Ng P. Requirements for ectopic homologous recombination in mammalian somatic cells. Mol Cell Biol. 1996;16:7122–7132. doi: 10.1128/mcb.16.12.7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber LJ, Youds JL, Ward JD, McIlwraith MJ, O’Neil NJ, Petalcorin MI, Martin JS, Collis SJ, Cantor SB, Auclair M, et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco MG, Matos J, Rass U, Ip SC, West SC. Functional overlap between the structure-specific nucleases Yen1 and Mus81-Mms4 for DNA-damage repair in S. cerevisiae. DNA Repair. 2010;9:394–402. doi: 10.1016/j.dnarep.2009.12.017. [DOI] [PubMed] [Google Scholar]
- Boddy MN, Gaillard PH, McDonald WH, Shanahan P, Yates JR, 3rd, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- Bzymek M, Thayer NH, Oh SD, Kleckner N, Hunter N. Double Holliday junctions are intermediates of DNA break repair. Nature. 2010;464:937–941. doi: 10.1038/nature08868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Plank JL, Bachrati CZ, Hickson ID, Kowalczykowski SC. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nat Struct Mol Biol. 2010;17:1377–1382. doi: 10.1038/nsmb.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crismani W, Girard C, Froger N, Pradillo M, Santos JL, Chelysheva L, Copenhaver GP, Horlow C, Mercier R. FANCM limits meiotic crossovers. Science. 2012;336:1588–1590. doi: 10.1126/science.1220381. [DOI] [PubMed] [Google Scholar]
- Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, Smith GR. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Adjiri A, New L, Crouse GF, Jinks Robertson S. Mitotic crossovers between diverged sequences are regulated by mismatch repair proteins in Saccaromyces cerevisiae. Mol Cell Biol. 1996;16:1085–1093. doi: 10.1128/mcb.16.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayani Y, Simchen G, Lichten M. Meiotic recombination intermediates are resolved with minimal crossover formation during return-to-growth, an analogue of the mitotic cell cycle. PLoS genetics. 2011;7:e1002083. doi: 10.1371/journal.pgen.1002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A, Jessop L, Kolar E, Sourirajan A, Chen J, Dayani Y, Lichten M. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol Cell. 2012;46:43–53. doi: 10.1016/j.molcel.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson DO, Holloman WK. Recombinational repair of gaps in DNA is asymmetric in Ustilago maydis and can be explained by a migrating D-loop model. Proc Natl Acad Sci USA. 1996;93:5419–5424. doi: 10.1073/pnas.93.11.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman-Lobell J, Haber JE. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science. 1992;258:480–484. doi: 10.1126/science.1411547. [DOI] [PubMed] [Google Scholar]
- Gallo-Fernandez M, Saugar I, Ortiz-Bazan MA, Vazquez MV, Tercero JA. Cell cycle-dependent regulation of the nuclease activity of Mus81-Eme1/Mms4. Nucleic Acids Res. 2012;40:8325–8335. doi: 10.1093/nar/gks599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CK, Mazon G, Lam AF, Symington LS. Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genome integrity in budding yeast. Mol Cell. 2010;40:988–1000. doi: 10.1016/j.molcel.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyppa RW, Smith GR. Using Schizosaccharomyces pombe meiosis to analyze DNA recombination intermediates. Methods Mol Biol. 2009;557:235–252. doi: 10.1007/978-1-59745-527-5_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inbar O, Liefshitz B, Bitan G, Kupiec M. The relationship between homology length and crossing over during the repair of a broken chromosome. J Biol Chem. 2000;275:30833–30838. doi: 10.1074/jbc.C000133200. [DOI] [PubMed] [Google Scholar]
- Ip SC, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456:357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinks-Robertson S, Petes TD. Chromosomal translocations generated by high-frequency meiotic recombination between repeated yeast genes. Genetics. 1986;114:731–752. doi: 10.1093/genetics/114.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk LC, Hartwell LH. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics. 1992;132:387–402. doi: 10.1093/genetics/132.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll A, Higgins JD, Seeliger K, Reha SJ, Dangel NJ, Bauknecht M, Schropfer S, Franklin FC, Puchta H. The Fanconi anemia ortholog FANCM ensures ordered homologous recombination in both somatic and meiotic cells in Arabidopsis. Plant Cell. 2012;24:1448–1464. doi: 10.1105/tpc.112.096644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh BO, Symington LS. Recombination proteins in yeast. Ann Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Lichten M, Haber JE. Position effects in ectopic and allelic mitotic recombination in Saccharomyces cerevisiae. Genetics. 1989;123:261–268. doi: 10.1093/genetics/123.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz A, Osman F, Sun W, Nandi S, Steinacher R, Whitby MC. The fission yeast FANCM ortholog directs non-crossover recombination during meiosis. Science. 2012;336:1585–1588. doi: 10.1126/science.1220111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke-Glaser S, Luke B. The Mph1 helicase can promote telomere uncapping and premature senescence in budding yeast. PloS one. 2012;7:e42028. doi: 10.1371/journal.pone.0042028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydeard JR, Lipkin-Moore Z, Jain S, Eapen VV, Haber JE. Sgs1 and exo1 redundantly inhibit break-induced replication and de novo telomere addition at broken chromosome ends. PLoS genetics. 2010;6:e1000973. doi: 10.1371/journal.pgen.1000973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J, Blanco MG, Maslen S, Skehel JM, West SC. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell. 2011;147:158–172. doi: 10.1016/j.cell.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazon G, Lam AF, Ho CK, Kupiec M, Symington LS. The Rad1-Rad10 nuclease promotes chromosome translocations between dispersed repeats. Nat Struct Mol Biol. 2012;19:964–971. doi: 10.1038/nsmb.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchel K, Lehner K, Jinks-Robertson S. Heteroduplex DNA position defines the roles of the Sgs1, Srs2, and Mph1 helicases in promoting distinct recombination outcomes. PLoS genetics. 2013;9:e1003340. doi: 10.1371/journal.pgen.1003340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchel K, Zhang H, Welz-Voegele C, Jinks-Robertson S. Molecular structures of crossover and noncrossover intermediates during gap repair in yeast: implications for recombination. Mol Cell. 2010;38:211–222. doi: 10.1016/j.molcel.2010.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvan S, Tous C, Blanco MG, Schwartz EK, Ehmsen KT, West SC, Heyer WD, Aguilera A. Distinct roles of Mus81, Yen1, Slx1-Slx4, and Rad1 nucleases in the repair of replication-born double-strand breaks by sister chromatid exchange. Mol Cell Biol. 2012;32:1592–1603. doi: 10.1128/MCB.00111-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassif N, Penney J, Pal S, Engels WR, Gloor GB. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol Cell Biol. 1994;14:1613–1625. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SD, Jessop L, Lao JP, Allers T, Lichten M, Hunter N. Stabilization and electrophoretic analysis of meiotic recombination intermediates in Saccharomyces cerevisiae. Methods Mol Biol. 2009;557:209–234. doi: 10.1007/978-1-59745-527-5_14. [DOI] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Taylor AF, Smith GR, Hunter N. RecQ helicase, Sgs1, and XPF family endonuclease, Mus81-Mms4, resolve aberrant joint molecules during meiotic recombination. Mol Cell. 2008;31:324–336. doi: 10.1016/j.molcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman F, Dixon J, Doe CL, Whitby MC. Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol Cell. 2003;12:761–774. doi: 10.1016/s1097-2765(03)00343-5. [DOI] [PubMed] [Google Scholar]
- Panico ER, Ede C, Schildmann M, Schurer KA, Kramer W. Genetic evidence for a role of Saccharomyces cerevisiae Mph1 in recombinational DNA repair under replicative stress. Yeast. 2010;27:11–27. doi: 10.1002/yea.1727. [DOI] [PubMed] [Google Scholar]
- Paques F, Leung WY, Haber JE. Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol Cell Biol. 1998;18:2045–2054. doi: 10.1128/mcb.18.4.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plank JL, Wu J, Hsieh TS. Topoisomerase IIIalpha and Bloom’s helicase can resolve a mobile double Holliday junction substrate through convergent branch migration. Proc Natl Acad Sci U S A. 2006;103:11118–11123. doi: 10.1073/pnas.0604873103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R, Satory D, Dray E, Papusha A, Scheller J, Kramer W, Krejci L, Klein H, Haber JE, Sung P, et al. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 2009;23:67–79. doi: 10.1101/gad.1737809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priebe SD, Westmoreland J, Nilsson-Tillgren T, Resnick MA. Induction of recombination between homologous and diverged DNAs by double-strand gaps and breaks and role of mismatch repair. Mol Cell Biol. 1994;14:4802–4814. doi: 10.1128/mcb.14.7.4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseaulin L, Yamada Y, Tsutsui Y, Russell P, Iwasaki H, Arcangioli B. Mus81 is essential for sister chromatid recombination at broken replication forks. EMBO J. 2008;27:1378–1387. doi: 10.1038/emboj.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Ann Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- Schwartz EK, Heyer WD. Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma. 2011;120:109–127. doi: 10.1007/s00412-010-0304-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebesta M, Burkovics P, Haracska L, Krejci L. Reconstitution of DNA repair synthesis in vitro and the role of polymerase and helicase activities. DNA Repair (Amst) 2011;10:567–576. doi: 10.1016/j.dnarep.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selva EM, New L, Crouse GF, Lahue RS. Mismatch correction acts as a barrier to homeologous recombination in Saccharomyces cerevisiae. Genetics. 1995;139:1175–1188. doi: 10.1093/genetics/139.3.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Nandi S, Osman F, Ahn JS, Jakovleska J, Lorenz A, Whitby MC. The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol Cell. 2008;32:118–128. doi: 10.1016/j.molcel.2008.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakal B, Branzei D. Premature Cdk1/Cdc5/Mus81 pathway activation induces aberrant replication and deleterious crossover. EMBO J. 2013;32:1155–1167. doi: 10.1038/emboj.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Tay YD, Sidebotham JM, Wu L. Mph1 requires mismatch repair-independent and -dependent functions of MutSalpha to regulate crossover formation during homologous recombination repair. Nucleic Acids Res. 2010;38:1889–1901. doi: 10.1093/nar/gkp1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby MC. The FANCM family of DNA helicases/translocases. DNA Repair. 2010;9:224–236. doi: 10.1016/j.dnarep.2009.12.012. [DOI] [PubMed] [Google Scholar]
- Wu L, Bachrati CZ, Ou J, Xu C, Yin J, Chang M, Wang W, Li L, Brown GW, Hickson ID. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci USA. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Youds JL, Mets DG, McIlwraith MJ, Martin JS, Ward JD, NJ ON, Rose AM, West SC, Meyer BJ, Boulton SJ. RTEL-1 enforces meiotic crossover interference and homeostasis. Science. 2010;327:1254–1258. doi: 10.1126/science.1183112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K, Tang S, Ma Y, Hunter N. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell. 2012;149:334–347. doi: 10.1016/j.cell.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.