Summary
Natural products are traditionally the main source of drug leads. In particular, many antitumour compounds are either natural products or derived from them. However, the search for novel antitumour drugs active against untreatable tumours, with fewer side‐effects or with enhanced therapeutic efficiency, is a priority goal in cancer chemotherapy. Microorganisms, particularly actinomycetes, are prolific producers of bioactive compounds, including antitumour drugs, produced as secondary metabolites. Structural genes involved in the biosynthesis of such compounds are normally clustered together with resistance and regulatory genes, which facilitates the isolation of the gene cluster. The characterization of these clusters has represented, during the last 25 years, a great source of genes for the generation of novel derivatives by using combinatorial biosynthesis approaches: gene inactivation, gene expression, heterologous expression of the clusters or mutasynthesis. In addition, these techniques have been also applied to improve the production yields of natural and novel antitumour compounds. In this review we focus on some representative antitumour compounds produced by actinomycetes covering the genetic approaches used to isolate and validate their biosynthesis gene clusters, which finally led to generating novel derivatives and to improving the production yields.
Introduction
The production of bioactive compounds (antibiotic, antifungal, antitumour, immunosuppressant, antiviral or antiparasitic agents) from living organisms is mainly supported by microorganisms. From them the most prolific producers are fungi, cyanobacteria, myxobacteria and actinobacteria, mainly actinomycetes. About half of all bioactive microbial metabolites discovered to date are produced by actinomycetes, with the genus Streptomyces as the primary bioactive metabolite‐producing organism exploited by the pharmaceutical industry (Bérdy, 2005; Newman and Cragg, 2007; Butler, 2008; Olano et al., 2009a). This has stimulated for half a century the study of actinomycetes at all levels: taxonomy, genetics and physiology.
Actinomycetes have large genomes (five to nine megabases) and use 5–10% of their DNA for the production of secondary metabolites (Baltz, 2008); a good number of them are antitumour drugs (Figs 1 and 2). These belong to different structural groups: polyketides such as anthracyclines daunomycin and doxorubicin, both promoting DNA cleavage mediated by DNA topoisomerase II, or aureolic acids mithramycin and chromomycin A3 that cause DNA‐dependent inhibition of RNA synthesis and inhibit topoisomerase I causing DNA cleavage; non‐ribosomal peptides such as glycopeptide actinomycin D, which inhibits DNA‐directed RNA synthesis by binding to double stranded DNA; mixed polyketide/non‐ribosomal peptides as glycopeptide bleomycin, which inhibits DNA synthesis through a metal‐dependent oxidative cleavage of DNA; heterocyclic quinones as mitomycin C that cause intra‐ and inter‐strand DNA cross‐linking leading to selective inhibition of DNA synthesis; indolocarbazoles such as protein kinases inhibitor staurosporine and topoisomerase I‐dependent DNA‐damaging agent rebeccamycin, etc. However, even considering the enormous number of natural products discovered to date, there is still a need for generating novel drugs to be used in cancer chemotherapy due to the rapid development of resistance to multiple chemotherapeutic drugs and the high toxicity usually associated with this type of drugs and their undesirable side‐effects. In addition, there is a demand for novel antitumour drugs active against untreatable tumours, with fewer side‐effects or with greater therapeutic efficiency. To this respect, enormous efforts are under way to discover new drugs from actinomycetes by searching uncommon actinomycetes in special ecological niches, mining genomes for cryptic pathways, and applying combinatorial biosynthesis to generate novel secondary metabolites related to existing pharmacophores (Baltz, 2008).
Figure 1.
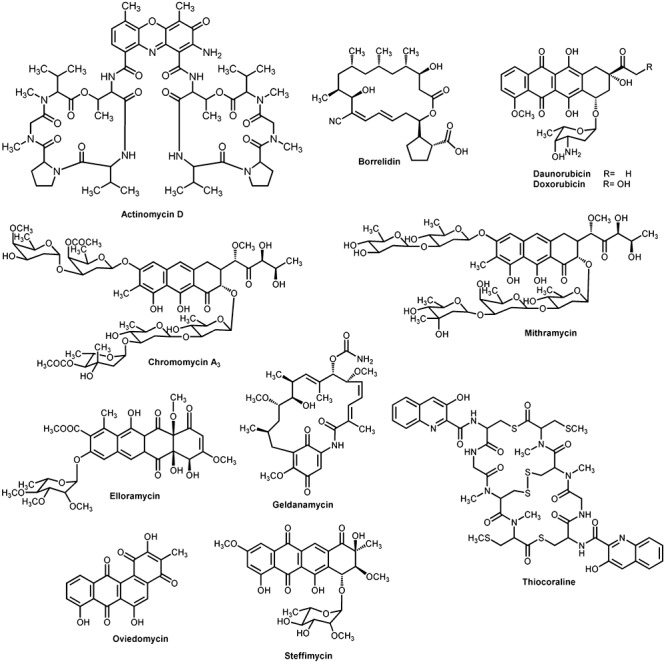
Structures of some polyketide and non‐ribosomal peptide antitumour compounds produced by actinomycetes.
Figure 2.
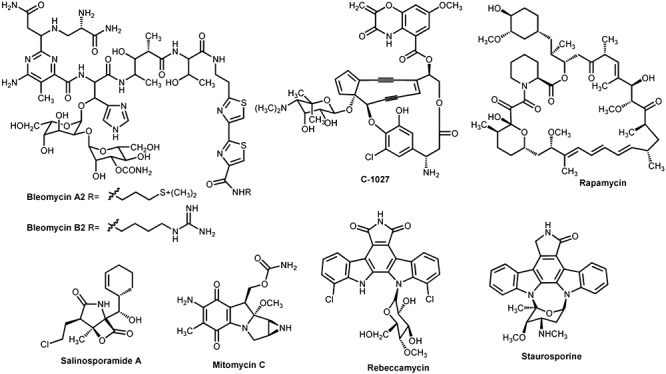
Structures of some mixed polyketide/non‐ribosomal peptide, indolocarbazole and heterocyclic quinone antitumour compounds produced by actinomycetes.
The utilization of combinatorial biosynthesis has become an important alternative approach for developing novel derivatives and for improving production yields of known antitumour compounds (Olano et al., 2008a; 2010). The strategies used in combinatorial biosynthesis (Fig. 3) involve gene inactivation to generate mutant strains that might accumulate novel products, expression of genes or combination of genes in different heterologous hosts including mutant strains or producers of related compounds, and mutasynthesis (Méndez and Salas, 2003; Floss, 2006; Menzella and Reeves, 2007; Kennedy, 2008; Salas and Méndez, 2009). Obviously, the application of this technology requires the isolation and characterization of the gene cluster involved in the biosynthesis of a particular antitumour compound to unravel its biosynthetic pathway.
Figure 3.
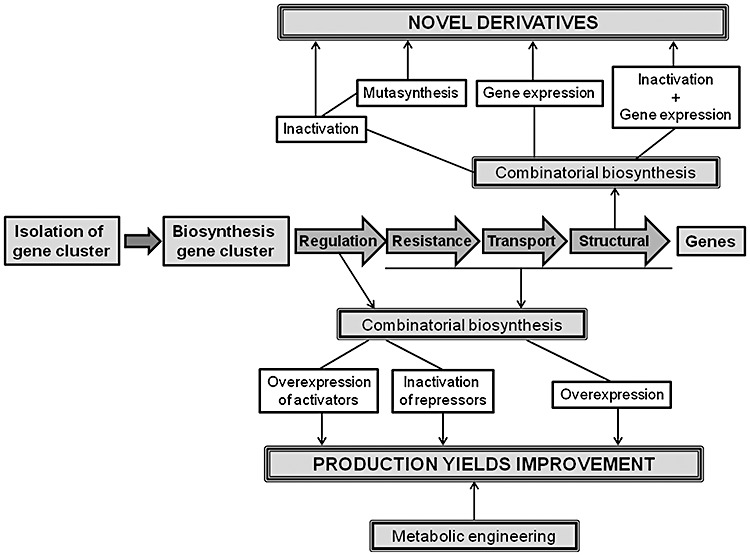
Strategies used for generating novel antitumour derivatives and improving production yields by combinatorial biosynthesis.
A large number of antitumour biosynthesis gene clusters have been characterized from producing actinomycetes during the last 25 years (Olano et al., 2009b). In this review, we focus on the approaches used for the isolation and validation of secondary metabolite biosynthesis gene clusters from antitumour‐producing actinomycetes (Table 1). We will summarize the genetic information that can be gathered from those clusters. We will also address the subsequent use of this information for the development of novel derivatives through combinatorial biosynthesis as well as for the improvement of production titres.
Table 1.
Some characteristics of gene clusters involved in the biosynthesis of different antitumour compounds by actinomycetes.
| Antitumour | Organism | Type | Strategy for cloning | Expressed in | Size (kb)a | Genes in the cluster | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genes | Reg. | R/T | Struc. | Other | ||||||
| Actinomycin D | S. chrysomallus | NPR | Homology | NE | 50 | 28 | 4 | 8 | 10 | 6 |
| Bleomycin | S. verticillus | PK/NPR | Resistance | NE | 62 | 28 | 1 | 3 | 16 | 8 |
| Borrelidin | S. parvulus | PK (I) | Homology | NE | 52 | 20 | – | 1 | 19 | – |
| C‐1027 | S. globisporus | PK/NRP | Homology | NE | 73 | 56 | 3 | 4 | 34 | 15 |
| Chromomycin A3 | S. griseus | PK (II) | Homology | NE | 43 | 36 | 2 | 3 | 29 | 2 |
| Daunorubicin | S. peucetius | PK (II) | Homology | NE | 39 | 37 | 3 | 4 | 28 | 2 |
| Doxorubicin | ||||||||||
| Elloramycin | S. olivaceus | PK (II) | Homology | S. lividans | 17 | 17 | – | 1 | 15 | 1 |
| Geldanamycin | S. hygroscopicus | PK (I) | Homology | NE | 70 | 23 | 4 | 1 | 15 | 3 |
| Mithramycin | S. argillaceus | PK (II) | Homology | NE | 40 | 34 | 2 | 3 | 25 | 4 |
| Mitomycin C | S. lavendulae | HQ | Resistance | NE | 55 | 47 | 2 | 4 | 19 | 22 |
| Oviedomycin | S. antibioticus | PK (II) | Homology | S. albus S. lividans | 20 | 17 | 2 | 1 | 13 | 1 |
| Rapamycin | S. hygroscopicus | PK/NRP | Homology | NE | 107 | 26 | 5 | 3 | 12 | 6 |
| Rebeccamycin | L. aerocolonigenes | IC | Homology | S. albus E. coli | 18 | 11 | 1 | 2 | 8 | – |
| Salinosporamide A | Sa. tropica | PK/NPR | Genome mining | NE | 41 | 30 | 3 | – | 17 | 10 |
| Staurosporine | S. longisporoflavus | IC | Homology | S. lividans S. albus | 23 | 15 | 1 | – | 14 | – |
| Steffimycin | S. steffisburgensis | PK (II) | Inactivation | S. albus | 27 | 24 | 3 | 2 | 16 | 3 |
| Thiocoraline | Micromonospora sp. ML1 | NPR | Homology | S. albus S. lividans | 53 | 26 | 5 | 3 | 18 | – |
Approximate estimation of the size of the gene cluster.
L., Lechevaleria; S., Streptomyces; Sa., Salinispora; PK, polyketide; PK (I), polyketide type I; PK (II), polyketide type II; PK/NPR, mixed polyketide and non‐ribosomal peptide; NRP, non‐ribosomal peptide; HQ, heterocyclic quinone; IC, indolocarbazole; NE, no heterologous expression of the cluster has been reported; E., Escherichia; Reg., Regulatory genes; R/T, Resistance and transport genes; Struc., Structural genes.
Isolation and characterization of gene clusters
Strategies used for the isolation of the gene clusters covered by this review do not differ from those used for the identification of clusters involved in the biosynthesis of non‐antitumour secondary metabolites. Furthermore, in most cases more than one strategy has been applied for the isolation of a single cluster. One of these approaches is based in the fact that structural biosynthetic genes are generally clustered all together with resistance and regulatory genes. This has facilitated the identification of several biosynthetic clusters by cloning resistance genes in an appropriate sensitive host as in the case of these involved in the biosynthesis of bleomycin (Sugiyama et al., 1994a) or mitomycin C (August et al., 1994). In addition, it has been observed that genes from biosynthetically related pathways show cross‐hybridization each other. This has prompted the homology‐based approach as the most widely method applied for the identification of biosynthesis gene clusters (Table 1). Heterologous DNA probes were used for the isolation of several clusters as for those involved in the biosynthesis of daunorubicin/doxorubicin (Otten et al., 1990), anthracycline‐like elloramycin (Decker et al., 1995), serine‐threonine kinase mTOR inhibitor rapamycin (Schwecke et al., 1995), mithramycin (Lombóet al., 1996), angucycline oviedomycin (Méndez et al., 2002), chromomycin A3 (Menéndez et al., 2004a) and angiogenesis inhibitor borrelidin (Olano et al., 2004a). Homologous probes, previously identified or generated by using degenerated oligonucleotides, have also been used for the isolation of actinomycin D (Schauwecker et al., 1998), the DNA‐cleaving agent enediyne C‐1027 (Liu and Shen, 2000), rebeccamycin (Sánchez et al., 2002), staurosporine (Onaka et al., 2002), the chaperone Hsp90 inhibitor geldanamycin (Rascher et al., 2003) and the DNA polymerase α inhibitor depsipeptide thiocoraline (Lombóet al., 2006) biosynthesis gene clusters. On the other hand, the method for isolating anthracycline steffimycin gene cluster was the direct inactivation of a gene involved in its biosynthesis using a PCR fragment that was amplified from steffimycin producer chromosomal DNA and used to generate a non‐producing mutant (Gullón et al., 2006). Finally, genome sequencing and mining is now a consolidated tool to identify novel biosynthesis gene clusters. Sequencing of Salinispora tropica CNB‐440 genome was used to locate the 20S proteasome inhibitor salinosporamide A gene cluster (Udwary et al., 2007).
Once a gene cluster has been isolated, it is necessary to confirm its involvement in the biosynthesis of the bioactive compound. The validation process is especially important for clusters identified by homology‐based approaches and genome mining and can be achieved by several methods that include gene inactivation or the heterologous expression of the cluster. The main objective of gene inactivation is the generation of non‐producing mutants as a proof of the involvement of the cluster in the biosynthesis of the compound under study. The expression of gene clusters involves the utilization of genetic tools to mobilize large DNA fragments into a heterologous host to produce the pursued compound. It takes advantage of the availability of a number of Streptomyces strains [mainly S. coelicolor A3(2), S. lividans 66, S. venezuelae YJ003 or S. albus J1074] to be used as hosts due to the deep knowledge accumulated about their physiology and growth conditions. In addition, some of these strains have been engineered to enhance the expression of heterologous gene clusters. Streptomyces lividans was the host used for the expression of elloramycin (Decker et al., 1995; Ramos et al., 2008), staurosporine (Onaka et al., 2002), oviedomycin (Méndez et al., 2002) and thiocoraline (Lombóet al., 2006) gene clusters. Streptomyces albus was used for the production of oviedomycin (Méndez et al., 2002), rebeccamycin (Sánchez et al., 2002), staurosporine (Salas et al., 2005), thiocoraline (Lombóet al., 2006) and steffimycin (Gullón et al., 2006). Furthermore, low levels of rebeccamycin production were also achieved in Escherichia coli (Hyun et al., 2003). In addition, gene inactivation and heterologous expression of the cluster are also used to establish the cluster boundaries.
Structural genes
Most of the genes in a cluster code for enzymes involved in the biosynthetic process, including the biosynthesis of precursor units, assembling of precursors, generation of other structural elements and tailoring modifications by attachment of the different structural elements or introduction of chemical functionalities (Fig. 4).
Figure 4.
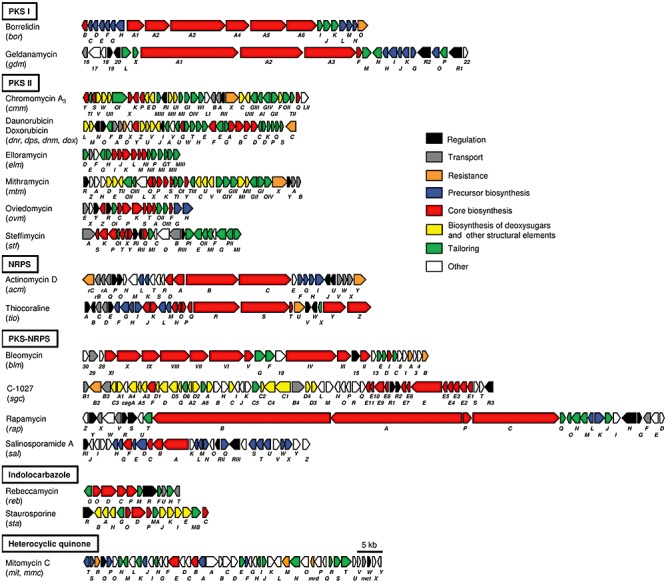
Genetic organization of antitumour biosynthesis gene clusters summarized in Table 1.
Biosynthesis of precursors. Some secondary metabolites are built using, as scaffold elements, precursors not supplied by primary metabolism or, when supplied, an extra amount is required at due time for the production of the secondary metabolite. These elements serve as starter units for the initiation of the pathway or as extender units. Genes belonging to this group are found mainly in polyketide and non‐ribosomal peptide biosynthetic pathways (Fig. 4).
Nine genes are present in the borrelidin cluster for the biosynthesis of cyclopentane dicarboxylic acid which is used as starter unit by a type I PKS. This precursor is proposed to derive from tyrosine catabolism through 5‐oxo‐pent‐3‐ene‐1,2,5‐tricarboxyilic acid (Olano et al., 2004a). In other cases, such as in the geldanamycin pathway, only one gene (gdmO) of those required for the biosynthesis of the type I PKS starter unit 3‐amino‐5‐hydroxybenzoic acid (AHBA) is present in the cluster (Rascher et al., 2003). The remaining genes are located elsewhere in the chromosome as it happens in the ansamitocin biosynthesis gene cluster (Yu et al., 2002). However, in the case of mitomycin C, all genes required for the biosynthesis of AHBA are located in the cluster (Mao et al., 1999a). The geldanamycin gene cluster contains, in addition, five genes proposed to be involved in the biosynthesis of PKS extender unit methoxy‐malonyl‐CoA (Rascher et al., 2003). In the case of type II PKS systems, there is no need for a special starter unit since the biosynthesis of these polyketides usually starts with an acetyl‐CoA unit. However, an extra supply of the extender malonyl‐CoA units is occasionally required. This might be the case of the oviedomycin pathway since genes encoding the three subunits of an acetyl‐CoA carboxylase involved in the production of malonyl‐CoA are present in the cluster (Lombóet al., 2004a; D. Zabala, pers. comm.).
NRPS systems for the biosynthesis of peptide or mixed polyketide‐peptide compounds occasionally require special starter units synthesized by dedicated enzymes whose genes are present in the cluster. Thiocoraline and actinomycin D starter units, 3‐hydroxy‐quinaldic acid and 4‐methyl‐3‐hydroxyanthranilic acid, respectively, derive from tryptophan. Both are produced through six enzymatic steps, the corresponding genes being located in their respective gene clusters (Lombóet al., 2006; Keller et al., 2010). Rapamycin PKS uses a shikimate‐derived 4,5‐dihydroxycyclohex‐1‐enecarboxylic acid as starter unit, while its NRPS incorporates l‐lysine‐derived l‐pipecolic acid. Two genes of this cluster are involved in the biosynthesis of these precursor units: pteridine‐dependent dioxygenase rapK and l‐lysine cyclodeaminase rapL (Molnár et al., 1996; Gregory et al., 2005). In the case of salinosporamide A, there are 13 genes involved in the biosynthesis of two precursors, substrates of the NRPS and PKS systems. On one hand, eight genes are involved in the production of 3‐(cyclohex‐2‐enyl)alanine derived from prephenate, probably originated in the shikimic acid pathway. On the other hand, five genes are required for the production of chloroethylmalonyl‐CoA, derived from S‐adenosyl‐methionine (SAM) (Beer and Moore, 2007; Gulder and Moore, 2009).
Inactivation of genes involved in biosynthesis of starter units generally leads to non‐producing mutants and has been used for abrogating the production of mitomycin C (Mao et al., 1999b), borrelidin (Olano et al., 2004a), geldanamycin (Gregory et al., 2005) or salinosporamide A (Eustáquio and Moore, 2008). In addition, some of these mutant strains have been used afterwards for mutasynthesis approaches in the generation of novel derivatives.
Biosynthesis of the structural core. A good number (in some cases up to 50%) of genes in each cluster are involved in the assembling of precursor units leading to the formation of the structural core of compounds. PKS and NRPS encoding genes are some examples. The carbon skeleton of polyketides are synthesized by step‐wise decarboxylative Claisen‐type condensation of acyl‐CoA precursors that are further reduced and modified. Type I PKSs are multifunctional enzymes that are organized into modules, each harbouring a set of distinct activities (ketosynthase, acyltransferase and acyl‐carrier protein domains) responsible for the catalysis of one cycle of polyketide chain elongation. These modules can contain, in addition, further domains to reduce keto groups (ketoreductase, dehydratase or enoylreductase) generated during the condensation process, and a thioesterase domain in the last module for polyketide release and cyclization (Fischbach and Walsh, 2006). Most type I PKSs act non‐iteratively, thus maintaining the co‐linearity rule of ‘one extension cycle–one PKS module’. This is the case of PKSs involved in geldanamycin (Rascher et al., 2003) or rapamycin (Schwecke et al., 1995) biosynthesis. Other type I PKSs are iterative, as those involved in enediyne C‐1027 biosynthesis, where a single module performs all cycles of polyketide chain elongation (Liu et al., 2002). There are also type I PKSs that contain both type of modules: non‐iterative and iterative, such as borrelidin PKS (Olano et al., 2003). On the other hand, type II PKSs are multienzyme complexes composed by ketosynthase, chain length factor and acyl‐carrier proteins, which carry out a single set of iteratively acting activities followed by enzymes dedicated to cyclization and aromatization processes (Shen, 2003). This type of PKS is involved in the biosynthesis of aromatic polyketides such as daunorubicin/doxorubicin, tetracycline, mithramycin, chromomycin A3, oviedomycin, steffimycin or elloramycin.
NRPSs synthesize non‐ribosomal peptides using as monomeric building blocks proteinogenic and non‐proteinogenic amino acids, including d‐amino acids, and other carboxylic acids. The NRPS assembly lines follow the same chemical logic as PKSs for chain elongation with several domains (in this case adenylation, condensation and peptidyl‐carrier protein) organized into modules. Additional domains with epimerization, methyltransferase, reductase or oxidase activity might be also present. These multimodular enzymes are required for the biosynthesis of several antitumour compounds such as thiocoraline (Lombóet al., 2006), actinomycin D (Keller et al., 2010) and mixed polyketide–peptide compounds such as rapamycin (Schwecke et al., 1995), bleomycin (Shen et al., 2002) or salinosporamide A (Udwary et al., 2007). The presence of type I PKSs and/or NRPSs coding genes determine the large genetic extension of the biosynthesis clusters that harbour these systems. Considering that a regular PKS or NRPS module is encoded by approximately 5 kb of DNA, in general the length of the cluster gets bigger as the number of modules grows (Table 1 and Fig. 4).
Heterocyclic quinone mitomycin C and indolocarbazoles staurosporine and rebeccamycin use other types of enzymes to assemble their structural cores. Mitomycin C pathway requires a glycosyltransferase, MitB, and an acyl‐CoA ligase, MitE, to condense AHBA and d‐glucosamine to generate the mitosane core (Mao et al., 1999a). On the other hand, indolocarbazole cores arcyriaflavin (the non‐chlorinated aglycone for rebeccamycin) and K252c (for staurosporine) are synthesized through the condensation of l‐tryptophan moieties after an oxidative deamination and several oxidation and decarboxylation steps (Sánchez et al., 2002; Salas et al., 2005).
Inactivation of genes involved in the assembling of precursor units also leads to non‐producing mutants. This has been shown by inactivating C‐1027 (Liu et al., 2002), mithramycin (Lombóet al., 1996), oviedomycin (Lombóet al., 2004a) and borrelidin (Olano et al., 2004a) PKS‐encoding genes, thiocoraline (Lombóet al., 2006) NRPS‐encoding genes, and mitomycin C mitB and mitE (Mao et al., 1999a).
Biosynthesis of deoxysugars and other structural elements. Attached to the structural core there are usually other structural elements that, in most cases, correspond to 6‐deoxyhexoses derived from d‐glucose‐1‐phosphate. The sugar moieties contribute to the structural biodiversity of compounds and participate in the interaction between the drug and the cellular target (Salas and Méndez, 2007; Méndez et al., 2008). These structures can be composed by monosaccharides (C‐10127, daunorubicin/doxorubicin, elloramycin, steffimycin and staurosporine), disaccharides (bleomycin, mithramycin and chromomycin A3), trisaccharides (mithramycin and chromomycin A3) and oligosaccharides (landomycin A) (Figs 1 and 2).
The absence of some or all genes involved in the biosynthesis of the sugar from the cluster is the exception rather than the rule. l‐rhamnose‐containing clusters constitute such an exception. The gene clusters for steffimycin (Gullón et al., 2006) and elloramycin (Ramos et al., 2008) do not contain any l‐rhamnose biosynthesis genes. This is a common feature to other clusters containing l‐rhamnose in the structures of the bioactive compounds and involved in the biosynthesis of polyketides as spinosad (Waldron et al., 2001) and aranciamycin (Luzhetskyy et al., 2007) or the diterpenoid brasilicardin A (Hayashi et al., 2008). In two cases, spinosyn (Madduri et al., 2001) and elloramycin (Ramos et al., 2008), a rhamnose biosynthesis gene cluster has been identified in other chromosomal location and most probably they use l‐rhamnose synthesized by the cell for other cellular functions or structures. The involvement of this rhamnose gene cluster in elloramycin biosynthesis was demonstrated by inactivation of the rhaB gene, encoding a 4,6‐dehydratase, thus generating a non‐producer mutant that accumulated the elloramycin aglycon 8‐demethyl‐tetracenomycin C (Ramos et al., 2008). In addition, a Streptomyces peucetius rhamnose biosynthesis gene cluster has been recently found to be involved in daunorubicin/doxorubicin biosynthesis providing TDP‐4‐keto‐6‐deoxy‐d‐glucose as precursor for the biosynthesis of daunosamine (Singh et al., 2010). Inactivation of genes involved in the production of deoxysugars from daunorubicin and mithramycin clusters has also led to identify non‐glycosylated compounds (Lombóet al., 1997; Otten et al., 1997) and derivatives with different patterns of glycosylation (Remsing et al., 2002).
In the case of enediyne C‐1025, in addition to a deoxyamino sugar, genes for the biosynthesis of two further structural elements are present in the cluster. These elements are the β‐amino acid (S)‐3‐chloro‐4,5‐dihydroxy‐β‐phenylalanine (derived from l‐tyrosine) and benzoxazolinate (derived from chorismate). The biosynthesis of the β‐amino acid moiety involves, in addition to other enzymes, different NRPS free standing domains such as an adenylation (SgcC1) and a peptidyl carrier protein (SgcC2) (Liu et al., 2002; Van Lanen et al., 2005; 2008). The inactivation of sgcA or sgcC1 involved in early steps in the biosynthesis of the deoxyamino or β‐amino acid moieties led to abolish production of the final compound and no intermediates or derivatives were accumulated (Liu and Shen, 2000; Liu et al., 2002). On the other hand, inactivation of sgcC, encoding a phenol hydroxylase catalysing a late step for the biosynthesis of the β‐amino acid moiety, led to a new C‐1027 derivative (Liu et al., 2002; Van Lanen et al., 2005). These results suggest that C‐1027 structural elements are required to stabilize the enediyne polyketide core. The lack of any of these structural elements, including the benzoxazolinate moiety (as will be described in the next section), determine the complete absence of any enediyne derivative.
Tailoring modifications. The introduction of additional chemical modifications into the structural core is performed by tailoring enzymes whose genes are found within the biosynthesis gene cluster. Group transferases such as glycosyl‐, methyl‐, acyl‐, prenyl‐, carbamoyl‐ or amino‐transferases belong to this class. Attachment of sugars is carried out by glycosyltransferases, which catalyses the transfer of the sugar from a NDP‐activated sugar donor to an acceptor molecule. In most of the cases, attachment of the sugars to the aglycone occurs through O‐glycosidic linkages, but C‐ and N‐glycosidic bonds also takes place. Methyltransferases generally use SAM as methyl donor to methylate functional groups either at the compound core or at other structural moieties. Acyltransferases are involved in the attachment of either short (C2–C6) or medium (C8–C12) acyl chains to both amine and alcohol moieties. The acyl transfer process is often one of the latest biosynthetic events representing a key step in generating the final biologically active molecule and, in some cases, it is an important event for self‐protection of the producing organism. Prenyltransferases catalyse the transfer of prenyl moieties derived from allylic isoprenyl diphosphates (PP) such as dimethylallyl‐PP (C5), geranyl‐PP (C10) or farnesyl‐PP (C15). Carbamoyltransferases catalyse the attachment of carbamoyl moieties, and aminotransferases incorporate amino groups. On the other hand, enzymes that normally participate in the biosynthesis of structural cores such NRPS can occasionally act as group transferases. This is the case of SgcC5, a NRPS free standing condensation domain, which incorporates β‐amino acid (S)‐3‐chloro‐4,5‐dihydroxy‐β‐phenylalanine to the C‐1027 polyketide core (Liu et al., 2002).
Other tailoring modifications introduce oxygen‐containing functionalities, such as hydroxy, epoxide, aldehyde and keto groups, or modify these moieties by addition or removal of hydrogen atoms. These reactions, catalysed by oxygenases, oxidases, peroxidases, reductases and dehydrogenases, can provide a base for additional modifications such as methyl‐ amino‐ or glycosyl‐transfer. In addition, the introduction of chloride and bromide in secondary metabolites is frequently performed by halogenating enzymes known as flavin‐dependent halogenases.
Tailoring modification genes have also been subjected to inactivation for validating gene clusters. This approach usually generates mutants that accumulate biosynthetic intermediates or novel derivatives. This is the case of inactivation of several group transferases such as glycosyltransferases, methyltransferases, acyltransferases, carbamoyltransferases, aminotransferases or halogenases involved in the biosynthesis of daunorubicin (Otten et al., 1995a), mithramycin (Fernández et al., 1998; Blanco et al., 2000; Nur‐e‐Alam et al., 2005), chromomycin A3 (Menéndez et al., 2004b; 2006), borrelidin (Olano et al., 2004b), mitomycin C (Grüschow et al., 2007; Sitachitta et al., 2007), C‐1027 (Liu et al., 2002), geldanamycin (Shin et al., 2008), salinosporamide A (Eustáquio and Moore, 2008) and bleomycin (Galm et al., 2008). However, the inactivation of acyltransferase sgcD6 involved in the attachment of benzoxazolinate moiety to C‐1027 polyketide core abolished the production of either the final compound or any other related compound (Liu et al., 2002). This, as was mentioned before, points to C‐1027 structural elements, deoxyamino sugar, β‐amino acid and benzoxazolinate, to have a role as stabilizing units for the polyketide core integrity. Furthermore, several oxidoreductases have been subjected to inactivation for validating biosynthesis clusters, leading to novel compounds derived from mithramycin (Remsing et al., 2003), chromomycin A3 (Menéndez et al., 2004a), borrelidin (Olano et al., 2004b), salinosporamide (McGlinchey et al., 2008) or geldanamycin (Shin et al., 2008).
Regulatory genes
The expression of genes from biosynthesis clusters is normally controlled by pathway‐specific regulatory genes. They can have either positive (activators) or negative (repressors) effects on gene expression. The number of putative pathway‐specific regulatory genes in different clusters range from one in bleomycin, rebeccamycin or staurosporine pathways to five in the rapamycin or thiocoraline pathways (Table 1). However, in some pathways no regulatory genes have been identified, as it is the case of the borrelidin or elloramycin gene clusters.
Approximately half of the pathway‐specific activators belong to three groups of transcriptional factors containing DNA binding helix–turn–helix (HTH) signatures: SARP‐ (seven cases), LuxR‐ (six cases) and TetR‐family (six cases) regulators. The S treptomycesAntibiotic Regulatory Protein family (SARP) share sequence similarity with several members of the OmpR family of DNA‐binding domains. In particular, regulatory elements of this family are mainly present in aromatic polyketide gene clusters such as those involved in the biosynthesis of daunorubicin (Stutzman‐Engwall et al., 1992), mithramycin (Lombóet al., 1999), chromomycin A3 (Menéndez et al., 2004a) and steffimycin (Gullón et al., 2006). OmpR‐ and LuxR‐like regulators generally act as activators and TetR‐family transcriptional regulators generally act as repressors (Ramos et al., 2005), but there are examples of TetR‐like positive regulators such as AtrA from Streptomyces griseus (Hirano et al., 2008). Only two clusters (bleomycin and C‐1027) do not contain any regulatory element of these families (Liu et al., 2002; Shen et al., 2002), while daunorubicin cluster contains one of each (Stutzman‐Engwall et al., 1992; Otten et al., 1995b). The regulation of the daunorubicin/doxorubicin production has been studied in detail. Transcriptional regulator DnrO (BirA/TetR‐like) binds specifically to the overlapping dnrN/dnrO promoter region activating the expression of response regulator DnrN that contains a LuxR‐like HTH motif. Then, DnrN activates the transcription of dnrI whose product DnrI, a SARP regulator, activates the production of daunorubicin biosynthesis and resistance genes (Madduri and Hutchinson, 1995; Otten et al., 1995b; 2000; Furuya and Hutchinson, 1996; Sheldon et al., 2002). In addition, DnrO has been shown to repress its own expression, effect that could be relieved by the presence of rhodomycin D, one of the first glycosylated intermediates in the daunorubicin/doxorubicin pathway, and enhanced by the final products (Jiang and Hutchinson, 2006; Ajithkumar and Prasad, 2010; Srinivasan et al., 2010). In most cases, inactivation of positive regulatory genes led to completely abolish the biosynthesis of the drug, the exception being mtrY of the mithramycin pathway: its inactivation considerably decreased (but did not abolish) mithramycin biosynthesis (Garcia‐Bernardo et al., 2000). On the other hand, inactivation of negative regulatory genes led to an increase in the production of the respective secondary metabolite. This has been shown for mitomycin C mmcW (Mao et al., 1999a) and chromomycin A3cmmRII (Menéndez et al., 2007). Some of these regulatory elements, both activators and repressors, might be specifically involved in the control of secondary metabolite resistance/transport systems since they are located next to these elements in several clusters. This is probably the case of regulatory elements mmcW, mtrY and cmmRII from mitomycin C (Mao et al., 1999a), mithramycin (Garcia‐Bernardo et al., 2000) and chromomycin A3 (Menéndez et al., 2007) gene clusters respectively.
Resistance and transport genes
Most antitumour compounds produced by actinomycetes interact with DNA. Therefore, producer microorganisms have developed mechanisms to survive during the biosynthesis of the drug. These resistance mechanisms could be divided into two groups: resistance systems and transport systems dedicated to self‐defence and/or extrusion of the final compound or intermediates. Most clusters (except those of salinosporamide A and staurosporine) contain elements of both groups, and only in the case of borrelidin cluster there is not transport system present.
The most extended resistance mechanism is a UvrA‐like protein found in daunorubicin/doxorubicin (Lomovskaya et al., 1996), mithramycin (Fernández et al., 1996; Garcia‐Bernardo et al., 2000), C‐1027 (Liu et al., 2002), chromomycin A3 (Menéndez et al., 2004a; 2007) and thiocoraline (Lombóet al., 2006) gene clusters. The UvrA protein is an E. coli UvrABC complex homologue involved in excision repair of UV‐induced DNA lesions (Truglio et al., 2006). This system has been shown to repair DNA damaged by the free oxygen radicals produced from the interaction of anthracyclines to DNA. The UvrA‐like encoding genes have been inactivated in different gene clusters showing various effects. In the daunorubicin gene cluster the inactivation of drrC could only be obtained in a non‐producing mutant (Lomovskaya et al., 1996). Furthermore, the expression of drrC is induced by daunorubicin, and DrrC has been shown to bind DNA in the presence of ATP and daunorubicin (Furuya and Hutchinson, 1998). The inactivation of mtrX has no apparent effect on mithramycin resistance but the expression of this gene into heterologous host S. albus conferred certain level of resistance to mithramycin (Garcia‐Bernardo et al., 2000). The inactivation of cmrX led to a chromomycin A3 sensitive strain that, in addition, showed a decreased production of the compound (Menéndez et al., 2007). Moreover, this gene is required to confer a high level of chormomycin A3 resistance in heterologous hosts. Oxidases have also been reported as resistance mechanisms to mitomycin C and anthracyclines. McrA was shown to be able of re‐oxidizing activated mitomycin C before it can initiate the event leading to DNA cross‐linking (August et al., 1994; Johnson et al., 1997). Two McrA homologues have been identified in the mitomycin gene cluster: MitR and MmcM. The inactivation of mitR and mmcM resulted in reduced mitomycin C production in the mitR mutant strain while mmcM mutant displayed a normal production level (Mao et al., 1999a). By analogy to McrA, DrrD has been proposed to exert a re‐oxidizing activity to counteract the effects of the reduced antitumour drugs daunorubicin and doxorubicin in S. peucetius (Hutchinson, 1997). Furthermore, the bleomycin resistance mechanism involves the inactivation of the drug by an N‐acetyltransferase (BlmB) (Sugiyama et al., 1994a,b). On the other hand, the expression of borO, encoding a borrelidin‐resistant threonyl‐tRNA synthetase, into S. albus was shown to confer resistance to the drug (Olano et al., 2004a).
Regarding the transport/resistance systems, these can be grouped in two different classes. The first corresponds to membrane proteins that mediate the extrusion of secondary metabolites in a coupled exchange with protons or sodium ions. The second are ABC (ATP Binging Cassette) transporter systems that couple the transport of the antibiotic through the cell membrane to ATP hydrolysis. The most widely distributed transport systems among the clusters covered in this review are ABC transporters, which have been found as the unique transporters in daunorubicin/doxorubicin (Guilfoile and Hutchinson, 1991), rapamycin (Schwecke et al., 1995), mithramycin (Fernández et al., 1996), chromomycin A3 (Menéndez et al., 2004a), thiocoraline (Lombóet al., 2006) and actinomycin D (Keller et al., 2010) biosynthesis gene clusters. In the C‐1027 cluster there are genes encoding a Na+/H+ efflux pump and a membrane protein in addition to the ATP‐binding protein of an ABC transporter system (Liu et al., 2002). In some cases the effect of these systems on the resistance/transport of several compounds has been investigated. The daunorubicin ABC transporter composed by the DrrA ATP‐binding protein and the DrrB membrane protein has been shown to confer resistance to daunorubicin/doxorubicin when expressed in S. lividans (Guilfoile and Hutchinson, 1991). On the other hand, disruption of the drrAB genes in S. peucetius resulted in a mutant strain highly sensitive to daunorubicin and with 10‐fold decrease in drug production (Srinivasan et al., 2010). On the contrary, inactivation of mtrA, encoding the ATP‐binding component of mithramycin ABC transporter, is lethal to Streptomyces argillaceus and could be only achieved in a non‐producing mutant. The expression of mtrAB into S. albus conferred high level of resistance to mithramycin (Fernández et al., 1996). In the case of chromomycin A3 ABC transporter, expression of cmrAB in S. albus conferred only low level of chromomycin A3 resistance, but it confers a high level of resistance to 4A,4E‐O‐dideacetyl‐chromomycin A3, a biosynthetic intermediate of chromomycin A3. To achieve a chromomycin A3 high level of resistance in S. albus it was necessary to coexpress the ABC transporter cmrAB together with the UvrA‐like encoding gene cmrX. These results pointed to the participation of the ABC transporter CmrAB in the chromomycin A3 biosynthesis process by extrusion of an intermediate that might be then acetylated (final tailoring modification) by the membrane‐bound CmmA acetyltransferase, and thus converted into the fully biologically active chromomycin A3 (Menéndez et al., 2007).
In some cases, there are drug‐binding proteins that confer resistance to the compound by drug sequestering or by participating in the transport process as ligand‐binding proteins. Genes encoding these drug‐binding proteins have been found in actinomycin D (Keller et al., 2010), bleomycin (Sugiyama et al., 1994a; Shen et al., 2002) and mitomycin C (Sheldon et al., 1997; 1999; Mao et al., 1999a) gene clusters. BlmA is a bleomycin‐binding protein that confers resistance in E. coli by drug sequestering. In the case of mitomycin C Mrd is a resistance protein that binds the drug conferring 30‐fold increase resistance when expressed into E. coli (Sheldon et al., 1997; 1999). On the other hand, the mitomycin C translocase Mct confers fivefold increase in resistance to mitomycin C in E. coli, while disruption of mct in Streptomyces lavendulae resulted in a strain more sensitive to the drug. Coexpression of mct and mrd in E. coli resulted in a 150‐fold increase in resistance (Sheldon et al., 1999).
Other genes
To this group belong genes to which no specifc function has been ascribed. This is the case of several genes from the mitomycin C biosynthesis cluster (mitI, mmcA, mmcB and mmcP). Its inactivation led to non‐producer mutants thus confirming their involvement in the biosynthesis of mitomycin C (Mao et al., 1999a). Another example is cagA found in C‐1027 biosynthesis cluster (Liu et al., 2002). C‐1027 is a chromoprotein antibiotic composed of a chromophore (Fig. 2) and an apoprotein encoded by cagA (Liu and Shen, 2000). CagA is required for the antibiotic activity of C‐1027 due to the extreme lability of the chromophore. However, inactivation of cagA has no effect on the biosynthesis of the chromophore itself (Cui et al., 2009).
Biotechnology processes using combinatorial biosynthesis and metabolic engineering
Once a biosynthesis gene cluster has been isolated and characterized, the available genetic information can be used for the generation of novel derivatives or to improve the production of both the native natural product and the novel compounds generated (Fig. 3).
Generation of novel derivatives
The non‐producer mutant strains generated by inactivating genes involved in the generation of starter units can be used for precursor‐directed biosynthesis of novel compounds. This approach, known as mutational biosynthesis or mutasynthesis, couples the utilization of chemical synthesis with molecular biology (Kennedy, 2008). Several non‐natural compounds derived from different metabolites such as rapamycin, borrelidin or salinosporamide have been generated following this method (Fig. 5). BC210 is a mutasynthetic derivative of rapamycin generated by feeding cyclohexane carboxylic acid to Streptomyces hygrocopicus mutant MG2‐10. This compound was found to be a better inhibitor of mTOR kinase activity and to show anticancer activity both in vivo and in vitro (Gregory et al., 2005). In addition, BC210 is not a substrate of the multidrug resistance efflux pump P‐glycoprotein, has good oral bioavailability and has a highly effective penetration of the blood–brain barrier, which makes this compound a potential therapeutic drug for the treatment of brain tumours (Zhang and Wilkinson, 2007). Six borrelidin derivatives were generated by mutasynthesis using different dicarboxylic acids as starter units (Moss et al., 2006). One of these derivatives, containing a carboxyl‐cyclobutane instead of a carboxyl‐cyclopentane moiety, was found to be 15‐fold less cytotoxic than borrelidin but sixfold more antiangiogenic (Wilkinson et al., 2006). Mutasynthesis has been used, in addition, to generate fluorosalinosporamide, a salinosporamide A analogue produced by a Sa. tropica chlorinase salL mutant that was supplemented with synthetic 5′‐fluoro‐5′‐deoxyadenosine. This compound is the most potent salinoporamide analogue showing high proteasome binding properties in which the fluoro group significantly extends its residence time on the proteasome (Eustáquio and Moore, 2008).
Figure 5.
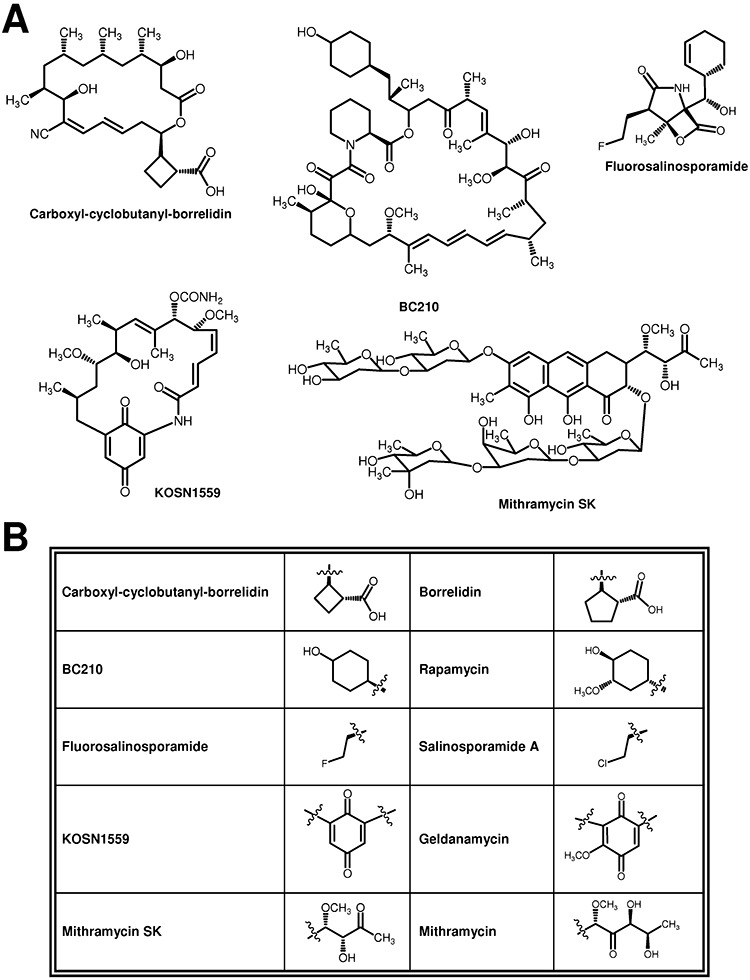
A. Structures of novel antitumour derivatives generated by mutasynthesis or modification of the structural core. B. Altered features and their comparison with the unmodified compound.
Genetic manipulation of genes encoding condensing enzymes has been a successful approach to generate novel compounds with modifications in the structural core from different natural products. As an example, acyltransferase substitution in geldanamycin PKS module 7 (GdmA3) by the rapamycin equivalent from module 2 led to the production of several novel geldanamycin derivatives. In particular, compound KOSN1559 (Fig. 5) showed fourfold higher binding affinity towards Hsp90, a chaperone overproduced in several types of human cancers and new target for cancer therapy (Patel et al., 2004). Occasionally, modifications in the structural core can be obtained by targeting genes involved in tailoring modification. This is the case of insertional inactivation of mithramycin mtmW, encoding a ketoreductase involved in side‐chain modification, which led to the production of several new mithramycin derivatives (Remsing et al., 2003). Among them, mithramycin SK (Fig. 5) showed an antitumour activity ninefold higher than that of mithramycin (Remsing et al., 2003), being especially effective against colon carcinoma cells (Bataller et al., 2008; 2010).
Targeting genes involved in the generation of structural elements attached to the core structure, including those tailoring modification, is the most productive approach to generate novel derivatives. Examples are the production of 22‐dehydroxy‐C‐1027 and 3‐morpholino‐propylamine‐decarbamoyl‐bleomycin (Fig. 6). The first one was generated by inactivating Streptomyces globisporus FAD‐dependent monooxygenase sgcC (Liu et al., 2002). The second compound was produced by inactivating Streptomyces verticillus carbamoyltransferase blmD (Galm et al., 2008). In addition, the expression of different gene combinations in a heterologous host can bypass the eventuality of working with actinomycetes refractory to genetic manipulation. This was the method used to obtain several oviedomycin derivatives in S. albus by combining type II PKS and cyclase genes together with those of three FAD‐dependent oxygenase genes from its own cluster. Three of these derivatives: prejadomycin‐2‐carboxylate, prejadomycin and 4a‐12b‐dehydro‐UWM6 (Fig. 6), showed significant improved antitumour activity (Lombóet al., 2009).
Figure 6.
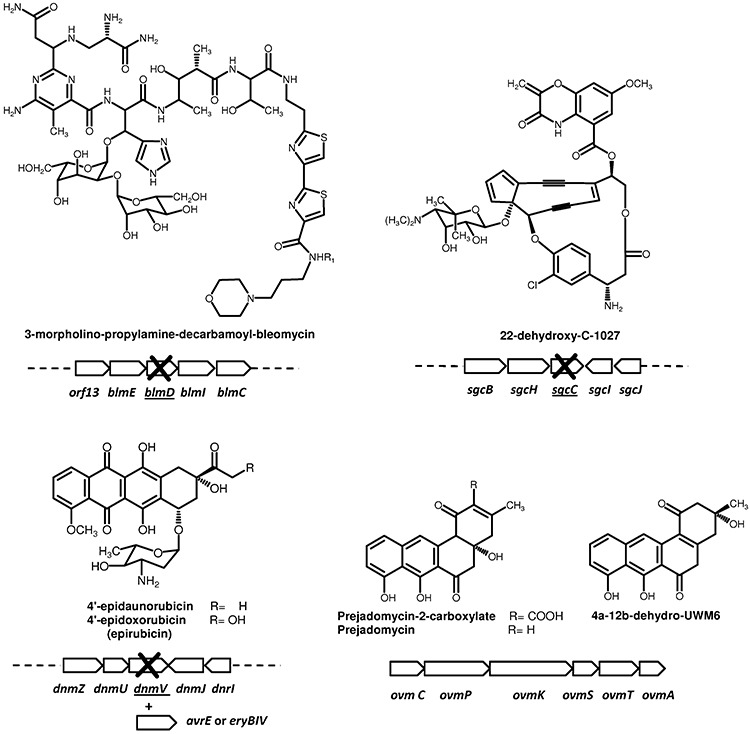
Structures of novel antitumour derivatives generated by tailoring modification showing the genes inactivated (crossed) or expressed for the generation of the novel compound.
The combination of targeting gene disruption and expression of genes or combinations of genes has been widely used to modify the glycosylation pattern of different antitumour compounds (Figs 6 and 7). The important antitumour drug 4′‐epidoxorubicin (epirubicin), traditionally obtained semisynthetically, along with its precursor 4′‐epidaunorubicin, was generated using a genetically engineered S. peucetius strain defective in the daunosamine biosynthesis gene dnmV. This strain was used to express the avermectin biosynthetic gene avrE from Streptomyces avermitilis or the erythromycin biosynthetic gene eryBIV from Saccharopolyspora erythraea, which led to 4′‐epianthracyclines (Madduri et al., 1998) (Fig. 6). A number of anthracyclines with modification in the glycosylation pattern have been obtained by expressing the steffimycin gene cluster in S. albus together with plasmids directing the biosynthesis of different deoxysugars. Three of these compounds showed higher antitumour activity than the parental natural product. In particular, d‐digitoxosyl‐8‐demethoxy‐10‐deoxysteffimycin (Fig. 7) was found to be 24‐fold more active than steffimycin when tested against different human tumour cell lines (Olano et al., 2008b). Analogues with improved activity of other aromatic compounds such as mithramycin and elloramycin have been also obtained by modification of the sugar moieties. The expression of genes involved in the biosynthesis of different deoxysugars in S. argillaceus led to 11 new mithramycin derivatives with altered glycosylation patterns (Baig et al., 2008; Pérez et al., 2008). Two of these, demycarosyl‐3D‐β‐d‐digitoxosyl‐mithramycin and deoliosyl‐3C‐β‐d‐mycarosyl‐mithramycin, showed improved activity against an oestrogen receptor‐positive human breast cancer cell line. In addition, demycarosyl‐3D‐β‐d‐digitoxosyl‐mithramycin was highly active against an oestrogen receptor‐negative human breast cancer cell line that is not strongly inhibited by mithramycin (Baig et al., 2008). On the other hand, coexpression of elloramycin gene cluster in S. albus with plasmids directing the biosynthesis of different deoxysugars has led to the isolation of new elloramycin (R1=CH3) and 8‐demethyl‐tetracenomycin C (R1=H) derivatives (Fig. 7) with different sugar moieties (Fischer et al., 2002; Rodríguez et al., 2002; Lombóet al., 2004b; Pérez et al., 2005; 2006). From these compounds, l‐mycarosyl‐elloramycin showed improved activity against three cancer cell lines (Lombóet al., 2004b). The modification of glycosylation patterns has not only been applied to polyketide natural products. Several indolocarbazole derivatives were generated by using this approach (Salas et al., 2005). Rebeccamycin, a DNA topoisomerase I inhibitor, and staurosporine, a promiscuous kinase inhibitor, contain a sugar moiety (Fig. 2) attached through a single N‐glycosidic bond (rebeccamycin) or by two N‐glycosidic bonds (staurosporine). The coexpression of rebeccamycin and staurosporine biosynthesis genes together with plasmids directing the biosynthesis of deoxysugars in S. albus has allowed the generation of novel derivatives from both compounds (Fig. 7). Some of these analogues were found highly active against specific kinases. l‐olivosyl‐arcyriaflavin A is a potent inhibitor of Ikkb, kinase that prevents activation of the NF‐κB pathway and thus has potential application on inflammation and cancer. On the other hand, l‐digitoxosyl‐k252c is a subnanomolar JAK2 inhibitor, kinase with potential application in myeloproliferative disorders (Sánchez et al., 2009).
Figure 7.
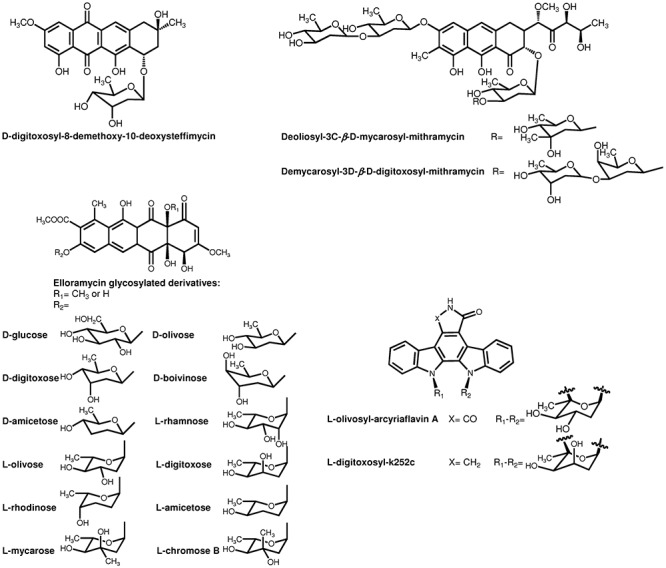
Structures of novel antitumour derivatives with an altered glycosylation pattern by using plasmids directing the biosynthesis of different deoxysugars.
Improving production yields
Current methods to increase the productivity of industrial microorganisms are classical random mutagenesis and optimization of large‐scale industrial fermentations. Metabolic engineering can help, in addition, to improve the production of bioactive metabolites (Fig. 3). Overproduction of a particular compound can be achieved by engineering different aspects of cellular metabolism at the level of primary metabolism such as precursor supply or ribosome engineering. The production of bioactive metabolites can also be improved by engineering genes from their specific biosynthesis clusters: pathway‐specific regulators, structural and self‐resistance genes. In addition, the expression of heterologous genes or the expression of the gene cluster in a heterologous host can be used to enhance production yields.
At the level of primary metabolism, it has been observed that overexpression of SAM synthetase results in higher secondary metabolite yields. The characterization of two S. peucetius SAM synthetase genes, metK1‐sp and metK2‐sp, and their independent overexpression in the same organism resulted in 2.1‐ and 1.4‐fold increase of doxorubicin production (Oh et al., 2010). Ribosome engineering has been used as a rational approach to enhance antibiotic production in different Streptomyces spp. by selecting mutants resistant to several antibiotics. In particular, ribosomes containing a mutation in ribosomal protein S12 gene, rpsL, which confers resistance to streptomycin, led to high increases in secondary metabolite production due to a high level of protein synthesis and ppGpp accumulation (Wang et al., 2008). Actinomycin D production in Streptomyces antibioticus has been enhanced 5.2‐fold by this method (Hosoya et al., 1998). On the other hand, transport/resistance systems encoded in the biosynthesis gene cluster have been observed to be involved not only in self‐protection but also in the export of biologically active intermediates (Menéndez et al., 2007). The production of doxorubicin by S. peucetius has been enhanced by overexpressing the resistance genes drrA, drrB and drrC in different combinations. The expression of drrAB led to a 2.2‐fold increase in doxorubicin yields while the drrC led to a 5.1‐fold increase. Surprisingly, only a 2.4‐fold increase was obtained by expressing the three genes together (Malla et al., 2010a). Consequently, increasing self‐resistance levels in producing organisms can be also used as an approach for increasing production yields.
Engineering pathway‐specific regulators is a useful method to bypass the imposed constrains to the expression of biosynthetic and transport/regulatory genes thus deregulating the production of secondary metabolites. Overexpression of pathway‐specific activators under the control of constitutive promoters and using multicopy plasmids has been used to improve the production of mithramycin (Lombóet al., 1999; Garcia‐Bernardo et al., 2000), daunorubicin (Stutzman‐Engwall et al., 1992; Otten et al., 1995b), doxorubicin (Parajuli et al., 2005; Malla et al., 2010b), rapamycin (Kuscer et al., 2007) and C‐1025 (Wang et al., 2009). Furthermore, inactivation of pathway‐specific repressors can also lead to overproduction of secondary metabolites. This is the case of mitomycin C (Mao et al., 1999a) and chromomycin A3 (Menéndez et al., 2007) that were overproduced when pathway‐specific transcriptional repressors mmcW and cmmRII were inactivated in their respective producer strains.
Biosynthesis structural genes can also be used to improve secondary metabolite production. Overexpression of sgcA1, encoding a NDP‐glucose synthase, resulted in a twofold improvement for enediyne C‐1027 production, and up to fourfold if cagA coding for C‐1027 apoprotein was coexpressed (Murrell et al., 2004). On the other hand, the expression of cagA alone, either as an extra copy under a constitutive promoter or as multiple copies of the gene under its native promoter, also resulted in a significant increase in enediyne production (Cui et al., 2009). The production of doxorubicin has also been improved by expressing genes involved in the biosynthesis and attachment of deoxyamino sugar daunosamine. By expressing the daunorubicin glycosyltransferase genes dnrS/dnrQ together with glucose‐1‐phosphate thymidylyltransferase desIII and TDP‐d‐glucose‐4,6‐dehydratase desIV from S. venezuelae involved in the formation of the intermediate TDP‐4‐keto‐6‐deoxy‐d‐glucose a 5.6‐fold increase in doxorubicin yield was obtained (Malla et al., 2009). The utilization of heterologous genes has also allowed improving 4′‐epidaunorubicin yields in Streptomyces coeruleorubidus. In this case, by increasing threefold the aveBIV (formerly avrE) copy number, 4′‐epidaunorubicin yields increased in the same proportion (Shao et al., 2010). The expression of heterologous genes in mutant strains can also be used to bypass the utilization of mutasynthesis approaches for the generation of certain derivatives. The production of fluorosalinosporamide, generated by mutasynthesis, has been induced in Sa. tropica by replacing the chlorinase gene salL with the fluorinase gene flA from Streptomyces cattleya (Eustáquio et al., 2010).
Another approach for improving metabolite production is to remove genes involved in the generation of undesired compounds thus enhancing the yields of the desired one. Following this philosophy, inactivation of S. peucetius dnrX and dnrH improved 3‐ and 8.5‐fold the biosynthesis of doxorubicin and daunorubicin respectively (Scotti and Hutchinson, 1996; Lomovskaya et al., 1998). These genes are involved in the transformation of daunorubicin and doxorubicin into their polyglycosylated forms known as baumycins. In addition, disruption of dnrU, involved in the transformation of daunorubicin into 13‐dihydrodaunorubicin, has a similar effect on doxorubicin production since the production of an undesired compound is abolished (Lomovskaya et al., 1999). The combination of these beneficial gene disruptions further improved doxorubicin yield, up to sevenfold in the dnrU/dnrX double mutant and to 26‐fold in the dnrU/dnrX/dnrH triple mutant. Furthermore, an additional 1.3‐ to 2.8‐fold increase was obtained by overexpressing in the double or triple mutants mentioned above dnrV and doxA genes involved in late steps during doxorubicin biosynthesis (Lomovskaya et al., 1999). Using the same approach, overexpression of dnmT, involved in the biosynthesis of daunorubicin deoxysugar l‐daunosamine, in the dnrH mutant led to improve daunorubicin production (Scotti and Hutchinson, 1996). The production of 4′‐epidaunorubicin in S. peucetius dnmV::avrE was enhanced also by inactivating dnrH and overexpressing dnmT (Madduri et al., 1998).
Occasionally, it is important to narrow the substrate flexibility of an enzyme to make it more efficient for the generation of certain derivatives. This objective can be accomplished by site‐directed mutagenesis targeting specific residues in the substrate binding region. Using this approach, elloramycin glycosyltransferase ElmGT that possesses remarkable donor substrate flexibility has been engineered to enhance the production of different 8‐demethyl‐tetracenomycin C derivatives. As mentioned above ElmGT has been shown to transfer at least 11 different sugars, including both l‐ and d‐isomeric forms, to its aglycon. Site‐directed mutagenesis in certain residues located in the α/β/α motif within the ElmGT nucleoside diphosphate‐sugar binding region has led to obtain several versions of the enzyme with improved ability to transfer particular deoxysugars: d‐boivinose 1.5‐fold, l‐rhamnose twofold and l‐olivose up to sixfold (Ramos et al., 2009).
Finally, in some cases production titres can be enhanced by expressing the biosynthesis gene cluster in a heterologous host. This was the case of the indolocarbazole rebeccamycin produced by Lechevaleria aerocolonigenes. The expression of the cluster in S. albus led to production levels several folds higher than those of the original strain (Sánchez et al., 2002).
Future perspectives
A great effort has been made in the last decades for the discovery of new natural products, antitumour drugs included. However, many of the novel compounds identified usually present some undesired biological and chemical properties, i.e. unspecific cytotoxicity, side‐effects, no effect on resistant strains or lack of solubility for oral or parenteral administration, which represent a step back in the expectations for their commercial use. The development of recombinant DNA technology during the second part of the last century has set up the foundations for unravelling the information encoded in microbial genomes and to access the information regarding the biosynthesis of natural products. All the efforts executed for the isolation, characterization and manipulation of biosynthesis gene clusters have crystallized in the generation of a large number of analogues of bioactive compounds. Some of these compounds show improved biological properties, which make them lead compounds for their application in clinic. In addition, these gene clusters contain elements that can be used to increase the production of both natural and engineered products. Keeping on with combinatorial biosynthesis approaches will provide increasing numbers of safer and more specific compounds that might enhance the arsenal of antitumour drugs available for chemotherapy.
Acknowledgments
This research was supported by the Spanish Ministry of Science and Innovation (BFU2006‐00404 and BIO2009‐07643 to J.A.S. and BIO2005‐04115 and BIO2008‐00269 to C.M.), Red Temática de Investigación Cooperativa de Centros de Cáncer (Ministry of Health, ISCIII‐RETIC RD06/0020/0026) and the EU FP6 (ActinoGen; Integrated project No. 005224). We thank Obra Social Cajastur for financial support to Carlos Olano.
References
- Ajithkumar V., Prasad R. The activator/repressor protein DnrO of Streptomyces peucetius binds to DNA without changing its topology. Int J Biol Macromol. 2010;46:380–384. doi: 10.1016/j.ijbiomac.2010.01.019. [DOI] [PubMed] [Google Scholar]
- August P.R., Flickinger M.C., Sherman D.H. Cloning and analysis of a locus (mcr) involved in mitomycin C resistance in Streptomyces lavendulae. J Bacteriol. 1994;176:4448–4454. doi: 10.1128/jb.176.14.4448-4454.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baig I., Perez M., Braña A.F., Gomathinayagam R., Damodaran C., Salas J.A. Mithramycin analogues generated by combinatorial biosynthesis show improved bioactivity. J Nat Prod. 2008;71:199–207. doi: 10.1021/np0705763. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz R.H. Renaissance in antibacterial discovery from actinomycetes. Curr Opin Pharmacol. 2008;8:557–563. doi: 10.1016/j.coph.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Bataller M., Méndez C., Salas J.A., Portugal J. Mithramycin SK modulates polyploidy and cell death in colon carcinoma cells. Mol Cancer Ther. 2008;7:2988–2997. doi: 10.1158/1535-7163.MCT-08-0420. [DOI] [PubMed] [Google Scholar]
- Bataller M., Méndez C., Salas J.A., Portugal J. Cellular response and activation of apoptosis by mithramycin SK in p21(WAF1)‐deficient HCT116 human colon carcinoma cells. Cancer Lett. 2010;292:80–90. doi: 10.1016/j.canlet.2009.11.008. [DOI] [PubMed] [Google Scholar]
- Beer L.L., Moore B.S. Biosynthetic convergence of salinosporamides A and B in the marine actinomycete Salinispora tropica. Org Lett. 2007;9:845–848. doi: 10.1021/ol063102o. [DOI] [PubMed] [Google Scholar]
- Bérdy J. Bioactive microbial metabolites. J Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- Blanco G., Fernández E., Fernández M.J., Braña A.F., Weissbach U., Künzel E. Characterization of two glycosyltransferases involved in early glycosylation steps during biosynthesis of the antitumor polyketide mithramycin by Streptomyces argillaceus. Mol Gen Genet. 2000;262:991–1000. doi: 10.1007/pl00008667. et al. [DOI] [PubMed] [Google Scholar]
- Butler M.S. Natural products to drugs: natural product‐derived compounds in clinical trials. Nat Prod Rep. 2008;25:475–516. doi: 10.1039/b514294f. [DOI] [PubMed] [Google Scholar]
- Cui Z., Wang L., Wang S., Li G., Hong B. Disruption of cagA, the apoprotein gene of chromoprotein antibiotic C‐1027, eliminates holo‐antibiotic production, but not the cytotoxic chromophore. FEMS Microbiol Lett. 2009;301:57–68. doi: 10.1111/j.1574-6968.2009.01800.x. [DOI] [PubMed] [Google Scholar]
- Decker H., Rohr J., Motamedi H., Zähner H., Hutchinson C.R. Identification of Streptomyces olivaceus Tü2353 genes involved in the production of the polyketide elloramycin. Gene. 1995;166:121–126. doi: 10.1016/0378-1119(95)00573-7. [DOI] [PubMed] [Google Scholar]
- Eustáquio A.S., Moore B.S. Mutasynthesis of fluorosalinosporamide, a potent and reversible inhibitor of the proteasome. Angew Chem Int Ed Engl. 2008;47:3936–3938. doi: 10.1002/anie.200800177. [DOI] [PubMed] [Google Scholar]
- Eustáquio A.S., O'Hagan D., Moore B.S. Engineering fluorometabolite production: fluorinase expression in Salinispora tropica yields fluorosalinosporamide. J Nat Prod. 2010;73:378–382. doi: 10.1021/np900719u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández E., Lombó F., Méndez C., Salas J.A. An ABC transporter is essential for resistance to the antitumor agent mithramycin in the producer Streptomyces argillaceus. Mol Gen Genet. 1996;251:692–698. doi: 10.1007/BF02174118. [DOI] [PubMed] [Google Scholar]
- Fernández E., Weissbach U., Sánchez Reillo C., Braña A.F., Méndez C., Rohr J., Salas J.A. Identification of two genes from Streptomyces argillaceus encoding glycosyltransferases involved in transfer of a disaccharide during biosynthesis of the antitumor drug mithramycin. J Bacteriol. 1998;180:4929–4937. doi: 10.1128/jb.180.18.4929-4937.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach M.A., Walsh C.T. Assembly‐line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- Fischer C., Rodríguez L., Patallo E.P., Lipata F., Braña A.F., Méndez C. Digitoxosyltetracenomycin C and glucosyltetracenomycin C, two novel elloramycin analogues obtained by exploring the sugar donor substrate specificity of glycosyltransferase ElmGT. J Nat Prod. 2002;65:1685–1689. doi: 10.1021/np020112z. et al. [DOI] [PubMed] [Google Scholar]
- Floss H.G. Combinatorial biosynthesis‐potential and problems. J Biotechnol. 2006;124:242–257. doi: 10.1016/j.jbiotec.2005.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya K., Hutchinson C.R. The DnrN protein of Streptomyces peucetius, a pseudo‐response regulator, is a DNA binding protein involved in the regulation of daunorubicin biosynthesis. J Bacteriol. 1996;178:6310–6318. doi: 10.1128/jb.178.21.6310-6318.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya K., Hutchinson C.R. The DrrC protein of Streptomyces peucetius, a UvrA‐like protein, is a DNA‐binding protein whose gene is induced by daunorubicin. FEMS Microbiol Lett. 1998;168:243–249. doi: 10.1111/j.1574-6968.1998.tb13280.x. [DOI] [PubMed] [Google Scholar]
- Galm U., Wang L., Wendt‐Pienkowski E., Yang R., Liu W., Tao M. In vivo manipulation of the bleomycin biosynthetic gene cluster in Streptomyces verticillus ATCC 15003 revealing new insights into its biosynthetic pathway. J Biol Chem. 2008;283:28236–28245. doi: 10.1074/jbc.M804971200. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Bernardo J., Braña A.F., Méndez C., Salas J.A. Insertional inactivation of mtrX and mtrY genes from the mithramycin gene cluster affects production and growth of the producer organism Streptomyces argillaceus. FEMS Microbiol Lett. 2000;186:61–65. doi: 10.1111/j.1574-6968.2000.tb09082.x. [DOI] [PubMed] [Google Scholar]
- Gregory M.A., Petkovic H., Lill R.E., Moss S.J., Wilkinson B., Gaisser S. Mutasynthesis of rapamycin analogues through the manipulation of a gene governing starter unit biosynthesis. Angew Chem Int Ed Engl. 2005;44:4757–4760. doi: 10.1002/anie.200462784. et al. [DOI] [PubMed] [Google Scholar]
- Grüschow S., Chang L.C., Mao Y., Sherman D.H. Hydroxyquinone O‐methylation in mitomycin biosynthesis. J Am Chem Soc. 2007;129:6470–6476. doi: 10.1021/ja0700193. [DOI] [PubMed] [Google Scholar]
- Guilfoile P.G., Hutchinson C.R. A bacterial analog of the mdr gene of mammalian tumor cells is present in Streptomyces peucetius, the producer of daunorubicin and doxorubicin. Proc Natl Acad Sci USA. 1991;88:8553–8557. doi: 10.1073/pnas.88.19.8553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulder T.A., Moore B.S. Chasing the treasures of the sea – bacterial marine natural products. Curr Opin Microbiol. 2009;12:252–260. doi: 10.1016/j.mib.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullón S., Olano C., Abdelfattah M.S., Braña A.F., Rohr J., Méndez C., Salas J.A. Isolation, characterization, and heterologous expression of the biosynthesis gene cluster for the antitumor anthracycline steffimycin. Appl Environ Microbiol. 2006;72:4172–4183. doi: 10.1128/AEM.00734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y., Matsuura N., Toshima H., Itoh N., Ishikawa J., Mikami Y., Dairi T. Cloning of the gene cluster responsible for the biosynthesis of brasilicardin A, a unique diterpenoid. J Antibiot. 2008;61:164–174. doi: 10.1038/ja.2008.126. [DOI] [PubMed] [Google Scholar]
- Hirano S., Tanaka K., Ohnishi Y., Horinouchi S. Conditionally positive effect of the TetR‐family transcriptional regulator AtrA on streptomycin production by Streptomyces griseus. Microbiology. 2008;154:905–914. doi: 10.1099/mic.0.2007/014381-0. [DOI] [PubMed] [Google Scholar]
- Hosoya Y., Okamoto S., Muramatsu H., Ochi K. Acquisition of certain streptomycin‐resistant (str) mutations enhances antibiotic production in bacteria. Antimicrob Agents Chemother. 1998;42:2041–2047. doi: 10.1128/aac.42.8.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson C.R. Biosynthetic studies of daunorubicin and tetracenomycin C. Chem Rev. 1997;97:2525–2536. doi: 10.1021/cr960022x. [DOI] [PubMed] [Google Scholar]
- Hyun C.G., Bililign T., Liao J., Thorson J.S. The biosynthesis of indolocarbazoles in a heterologous E. coli host. Chembiochem. 2003;4:114–117. doi: 10.1002/cbic.200390004. [DOI] [PubMed] [Google Scholar]
- Jiang H., Hutchinson C.R. Feedback regulation of doxorubicin biosynthesis in Streptomyces peucetius. Res Microbiol. 2006;157:666–674. doi: 10.1016/j.resmic.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Johnson D.A., August P.R., Shackleton C., Liu H.W., Sherman D.H. Microbial resistance to mitomycins involves a redox relay mechanism. J Am Chem Soc. 1997;119:2576–2577. [Google Scholar]
- Keller U., Lang M., Crnovcic I., Pfennig F., Schauwecker F. The actinomycin biosynthetic gene cluster of Streptomyces chrysomallus: a genetic hall of mirrors for synthesis of a molecule with mirror symmetry. J Bacteriol. 2010;192:2583–2595. doi: 10.1128/JB.01526-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy J. Mutasynthesis, chemobiosynthesis, and back to semi‐synthesis: combining synthetic chemistry and biosynthetic engineering for diversifying natural products. Nat Prod Rep. 2008;25:25–34. doi: 10.1039/b707678a. [DOI] [PubMed] [Google Scholar]
- Kuscer E., Coates N., Challis I., Gregory M., Wilkinson B., Sheridan R., Petković H. Roles of rapH and rapG in positive regulation of rapamycin biosynthesis in Streptomyces hygroscopicus. J Bacteriol. 2007;189:4756–4763. doi: 10.1128/JB.00129-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Shen B. Genes for production of the enediyne antitumor antibiotic C‐1027 in Streptomyces globisporus are clustered with the cagA gene that encodes the C‐1027 apoprotein. Antimicrob Agents Chemother. 2000;44:382–392. doi: 10.1128/aac.44.2.382-392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Christenson S.D., Standage S., Shen B. Biosynthesis of the enediyne antitumor antibiotic C‐1027. Science. 2002;297:1170–1173. doi: 10.1126/science.1072110. [DOI] [PubMed] [Google Scholar]
- Lombó F., Blanco G., Fernández E., Méndez C., Salas J.A. Characterization of Streptomyces argillaceus genes encoding a polyketide synthase involved in the biosynthesis of the antitumor mithramycin. Gene. 1996;172:87–91. doi: 10.1016/0378-1119(96)00029-7. [DOI] [PubMed] [Google Scholar]
- Lombó F., Siems K., Braña A.F., Méndez C., Bindseil K., Salas J.A. Cloning and insertional inactivation of Streptomyces argillaceus genes involved in the earliest steps of biosynthesis of the sugar moieties of the antitumor polyketide mithramycin. J Bacteriol. 1997;179:3354–3357. doi: 10.1128/jb.179.10.3354-3357.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombó F., Braña A.F., Méndez C., Salas J.A. The mithramycin gene cluster of Streptomyces argillaceus contains a positive regulatory gene and two repeated DNA sequences that are located at both ends of the cluster. J Bacteriol. 1999;181:642–647. doi: 10.1128/jb.181.2.642-647.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombó F., Braña A.F., Salas J.A., Méndez C. Genetic organization of the biosynthetic gene cluster for the antitumor angucycline oviedomycin in Streptomyces antibioticus ATCC 11891. Chembiochem. 2004a;5:1181–1187. doi: 10.1002/cbic.200400073. [DOI] [PubMed] [Google Scholar]
- Lombó F., Gibson M., Greenwell L., Braña A.F., Rohr J., Salas J.A., Méndez C. Engineering biosynthetic pathways for deoxysugars: branched‐chain sugar pathways and derivatives from the antitumor tetracenomycin. Chem Biol. 2004b;11:1709–1718. doi: 10.1016/j.chembiol.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Lombó F., Velasco A., Castro A., de la Calle F., Braña A.F., Sánchez‐Puelles J.M. Deciphering the biosynthesis pathway of the antitumor thiocoraline from a marine actinomycete and its expression in two Streptomyces species. Chembiochem. 2006;7:366–376. doi: 10.1002/cbic.200500325. et al. [DOI] [PubMed] [Google Scholar]
- Lombó F., Abdelfattah M.S., Braña A.F., Salas J.A., Rohr J., Méndez C. Elucidation of oxygenation steps during oviedomycin biosynthesis and generation of derivatives with increased antitumor activity. Chembiochem. 2009;10:296–303. doi: 10.1002/cbic.200800425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomovskaya N., Hong S.K., Kim S.U., Fonstein L., Furuya K., Hutchinson R.C. The Streptomyces peucetius drrC gene encodes a UvrA‐like protein involved in daunorubicin resistance and production. J Bacteriol. 1996;178:3238–3245. doi: 10.1128/jb.178.11.3238-3245.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomovskaya N., Doi‐Katayama Y., Filippini S., Nastro C., Fonstein L., Gallo M. The Streptomyces peucetius dpsY and dnrX genes govern early and late steps of daunorubicin and doxorubicin biosynthesis. J Bacteriol. 1998;180:2379–2386. doi: 10.1128/jb.180.9.2379-2386.1998. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomovskaya N., Otten S.L., Doi‐Katayama Y., Fonstein L., Liu X.C., Takatsu T. Doxorubicin overproduction in Streptomyces peucetius: cloning and characterization of the dnrU ketoreductase and dnrV genes and the doxA cytochrome P‐450 hydroxylase gene. J Bacteriol. 1999;181:305–318. doi: 10.1128/jb.181.1.305-318.1999. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzhetskyy A., Mayer A., Hoffmann J., Pelzer S., Holzenkämper M., Schmitt B. Cloning and heterologous expression of the aranciamycin biosynthetic gene cluster revealed a new flexible glycosyltransferase. Chembiochem. 2007;8:599–602. doi: 10.1002/cbic.200600529. et al. [DOI] [PubMed] [Google Scholar]
- McGlinchey R.P., Nett M., Eustáquio A.S., Asolkar R.N., Fenical W., Moore B.S. Engineered biosynthesis of antiprotealide and other unnatural salinosporamide proteasome inhibitors. J Am Chem Soc. 2008;130:7822–7823. doi: 10.1021/ja8029398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madduri K., Hutchinson C.R. Functional characterization and transcriptional analysis of the dnrR1 locus, which controls daunorubicin biosynthesis in Streptomyces peucetius. J Bacteriol. 1995;177:1208–1215. doi: 10.1128/jb.177.5.1208-1215.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madduri K., Kennedy J., Rivola G., Inventi‐Solari A., Filippini S., Zanuso G. Production of the antitumor drug epirubicin (4′‐epidoxorubicin) and its precursor by a genetically engineered strain of Streptomyces peucetius. Nat Biotechnol. 1998;16:69–74. doi: 10.1038/nbt0198-69. et al. [DOI] [PubMed] [Google Scholar]
- Madduri K., Waldron C., Merlo D.J. Rhamnose biosynthesis pathway supplies precursors for primary and secondary metabolism in Saccharopolyspora spinosa. J Bacteriol. 2001;183:5632–5638. doi: 10.1128/JB.183.19.5632-5638.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malla S., Niraula N.P., Liou K., Sohng J.K. Enhancement of doxorubicin production by expression of structural sugar biosynthesis and glycosyltransferase genes in Streptomyces peucetius. J Biosci Bioeng. 2009;108:92–98. doi: 10.1016/j.jbiosc.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Malla S., Niraula N.P., Liou K., Sohng J.K. Self‐resistance mechanism in Streptomyces peucetius: overexpression of drrA, drrB and drrC for doxorubicin enhancement. Microbiol Res. 2010a;165:259–267. doi: 10.1016/j.micres.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Malla S., Niraula N.P., Liou K., Sohng J.K. Improvement in doxorubicin productivity by overexpression of regulatory genes in Streptomyces peucetius. Res Microbiol. 2010b;161:109–117. doi: 10.1016/j.resmic.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Mao Y., Varoglu M., Sherman D.H. Molecular characterization and analysis of the biosynthetic gene cluster for the antitumor antibiotic mitomycin C from Streptomyces lavendulae NRRL 2564. Chem Biol. 1999a;6:251–263. doi: 10.1016/S1074-5521(99)80040-4. [DOI] [PubMed] [Google Scholar]
- Mao Y., Varoglu M., Sherman D.H. Genetic localization and molecular characterization of two key genes (mitAB) required for biosynthesis of the antitumor antibiotic mitomycin C. J Bacteriol. 1999b;181:2199–2208. doi: 10.1128/jb.181.7.2199-2208.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez C., Salas J.A. On the generation of novel anticancer drugs by recombinant DNA technology: the use of combinatorial biosynthesis to produce novel drugs. Comb Chem High Throughput Screen. 2003;6:513–526. doi: 10.2174/138620703106298699. [DOI] [PubMed] [Google Scholar]
- Méndez C., Künzel E., Lipata F., Lombó F., Cotham W., Walla M. Oviedomycin, an unusual angucyclinone encoded by genes of the oleandomycin‐producer Streptomyces antibioticus ATCC 11891. J Nat Prod. 2002;65:779–782. doi: 10.1021/np010555n. et al. [DOI] [PubMed] [Google Scholar]
- Méndez C., Luzhetskyy A., Bechthold A., Salas J.A. Deoxysugars in bioactive natural products: development of novel derivatives by altering the sugar pattern. Curr Top Med Chem. 2008;8:710–724. doi: 10.2174/156802608784221532. [DOI] [PubMed] [Google Scholar]
- Menéndez N., Nur‐e‐Alam M., Braña A.F., Rohr J., Salas J.A., Méndez C. Biosynthesis of the antitumor chromomycin A3 in Streptomyces griseus: analysis of the gene cluster and rational design of novel chromomycin analogs. Chem Biol. 2004a;11:21–32. doi: 10.1016/j.chembiol.2003.12.011. [DOI] [PubMed] [Google Scholar]
- Menéndez N., Nur‐e‐Alam M., Braña A.F., Rohr J., Salas J.A., Méndez C. Tailoring modification of deoxysugars during biosynthesis of the antitumour drug chromomycin A3 by Streptomyces griseus ssp. griseus. Mol Microbiol. 2004b;53:903–915. doi: 10.1111/j.1365-2958.2004.04166.x. [DOI] [PubMed] [Google Scholar]
- Menéndez N., Nur‐e‐Alam M., Fischer C., Braña A.F., Salas J.A., Rohr J., Méndez C. Deoxysugar transfer during chromomycin A3 biosynthesis in Streptomyces griseus subsp. griseus: new derivatives with antitumor activity. Appl Environ Microbiol. 2006;72:167–177. doi: 10.1128/AEM.72.1.167-177.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menéndez N., Braña A.F., Salas J.A., Méndez C. Involvement of a chromomycin ABC transporter system in secretion of a deacetylated precursor during chromomycin biosynthesis. Microbiology. 2007;153:3061–3070. doi: 10.1099/mic.0.2007/007922-0. [DOI] [PubMed] [Google Scholar]
- Menzella H.G., Reeves C.D. Combinatorial biosynthesis for drug development. Curr Opin Microbiol. 2007;10:238–245. doi: 10.1016/j.mib.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Molnár I., Aparicio J.F., Haydock S.F., Khaw L.E., Schwecke T., König A. Organisation of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of genes flanking the polyketide synthase. Gene. 1996;169:1–7. doi: 10.1016/0378-1119(95)00799-7. et al. [DOI] [PubMed] [Google Scholar]
- Moss S.J., Carletti I., Olano C., Sheridan R.M., Ward M., Math V. Biosynthesis of the angiogenesis inhibitor borrelidin: directed biosynthesis of novel analogues. Chem Commun. 2006;22:2341–2343. doi: 10.1039/b602931k. et al. [DOI] [PubMed] [Google Scholar]
- Murrell J.M., Liu W., Shen B. Biochemical characterization of the SgcA1 alpha‐d‐glucopyranosyl‐1‐phosphate thymidylyltransferase from the enediyne antitumor antibiotic C‐1027 biosynthetic pathway and overexpression of sgcA1 in Streptomyces globisporus to improve C‐1027 production. J Nat Prod. 2004;67:206–213. doi: 10.1021/np0340403. [DOI] [PubMed] [Google Scholar]
- Newman D.J., Cragg G.M. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- Nur‐e‐Alam M., Méndez C., Salas J.A., Rohr J. Elucidation of the glycosylation sequence of mithramycin biosynthesis: isolation of 3A‐deolivosylpremithramycin B and its conversion to premithramycin B by glycosyltransferase MtmGII. Chembiochem. 2005;6:632–636. doi: 10.1002/cbic.200400309. [DOI] [PubMed] [Google Scholar]
- Oh T.J., Niraula N.P., Liou K., Sohng J.K. Identification of the duplicated genes for S‐adenosyl‐l‐methionine synthetase (metK1‐sp and metK2‐sp) in Streptomyces peucetius var. caesius ATCC 27952. J Appl Microbiol. 2010;109:398–407. doi: 10.1111/j.1365-2672.2010.04688.x. [DOI] [PubMed] [Google Scholar]
- Olano C., Wilkinson B., Moss S.J., Braña A.F., Méndez C., Leadlay P.F., Salas J.A. Evidence from engineered gene fusions for the repeated use of a module in a modular polyketide synthase. Chem Commun. 2003;22:2780–2782. doi: 10.1039/b310648a. [DOI] [PubMed] [Google Scholar]
- Olano C., Wilkinson B., Sánchez C., Moss S.J., Sheridan R., Math V. Biosynthesis of the angiogenesis inhibitor borrelidin by Streptomyces parvulus Tü4055: cluster analysis and assignment of functions. Chem Biol. 2004a;11:87–97. doi: 10.1016/j.chembiol.2003.12.018. et al. [DOI] [PubMed] [Google Scholar]
- Olano C., Moss S.J., Braña A.F., Sheridan R.M., Math V., Weston A.J. Biosynthesis of the angiogenesis inhibitor borrelidin by Streptomyces parvulus Tü4055: insights into nitrile formation. Mol Microbiol. 2004b;52:1745–1756. doi: 10.1111/j.1365-2958.2004.04090.x. et al. [DOI] [PubMed] [Google Scholar]
- Olano C., Lombó F., Méndez C., Salas J.A. Improving production of bioactive secondary metabolites in actinomycetes by metabolic engineering. Metab Eng. 2008a;10:281–292. doi: 10.1016/j.ymben.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Olano C., Abdelfattah M.S., Gullón S., Braña A.F., Rohr J., Méndez C., Salas J.A. Glycosylated derivatives of steffimycin: insights into the role of the sugar moieties for the biological activity. Chembiochem. 2008b;9:624–633. doi: 10.1002/cbic.200700610. [DOI] [PubMed] [Google Scholar]
- Olano C., Méndez C., Salas J.A. Antitumor compounds from marine actinomycetes. Mar Drugs. 2009a;7:210–248. doi: 10.3390/md7020210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olano C., Méndez C., Salas J.A. Antitumor compounds from actinomycetes: from gene clusters to new derivatives by combinatorial biosynthesis. Nat Prod Rep. 2009b;26:628–660. doi: 10.1039/b822528a. [DOI] [PubMed] [Google Scholar]
- Olano C., Méndez C., Salas J.A. Post‐PKS tailoring steps in natural product‐producing actinomycetes from the perspective of combinatorial biosynthesis. Nat Prod Rep. 2010;27:571–616. doi: 10.1039/b911956f. [DOI] [PubMed] [Google Scholar]
- Onaka H., Taniguchi S., Igarashi Y., Furumai T. Cloning of the staurosporine biosynthetic gene cluster from Streptomyces sp. TP‐A0274 and its heterologous expression in Streptomyces lividans. J Antibiot. 2002;55:1063–1071. doi: 10.7164/antibiotics.55.1063. [DOI] [PubMed] [Google Scholar]
- Otten S.L., Stutzman‐Engwall K.J., Hutchinson C.R. Cloning and expression of daunorubicin biosynthesis genes from Streptomyces peucetius and S. peucetius subsp. caesius. J Bacteriol. 1990;172:3427–3434. doi: 10.1128/jb.172.6.3427-3434.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten S.L., Liu X., Ferguson J., Hutchinson C.R. Cloning and characterization of the Streptomyces peucetius dnrQS genes encoding a daunosamine biosynthesis enzyme and a glycosyl transferase involved in daunorubicin biosynthesis. J Bacteriol. 1995a;177:6688–6692. doi: 10.1128/jb.177.22.6688-6692.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten S.L., Ferguson J., Hutchinson C.R. Regulation of daunorubicin production in Streptomyces peucetius by the dnrR2 locus. J Bacteriol. 1995b;177:1216–1224. doi: 10.1128/jb.177.5.1216-1224.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten S.L., Gallo M.A., Madduri K., Liu X., Hutchinson C.R. Cloning and characterization of the Streptomyces peucetius dnmZUV genes encoding three enzymes required for biosynthesis of the daunorubicin precursor thymidine diphospho‐l‐daunosamine. J Bacteriol. 1997;179:4446–4450. doi: 10.1128/jb.179.13.4446-4450.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten S.L., Olano C., Hutchinson C.R. The dnrO gene encodes a DNA‐binding protein that regulates daunorubicin production in Streptomyces peucetius by controlling expression of the dnrN pseudo response regulator gene. Microbiology. 2000;146:1457–1468. doi: 10.1099/00221287-146-6-1457. [DOI] [PubMed] [Google Scholar]
- Parajuli N., Viet H.T., Ishida K., Tong H.T., Lee H.C., Liou K., Sohng J.K. Identification and characterization of the afsR homologue regulatory gene from Streptomyces peucetius ATCC 27952. Res Microbiol. 2005;156:707–712. doi: 10.1016/j.resmic.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Patel K., Piagentini M., Rascher A., Tian Z.Q., Buchanan G.O., Regentin R. Engineered biosynthesis of geldanamycin analogs for Hsp90 inhibition. Chem Biol. 2004;11:1625–1633. doi: 10.1016/j.chembiol.2004.09.012. et al. [DOI] [PubMed] [Google Scholar]
- Pérez M., Lombó F., Zhu L., Gibson M., Braña A.F., Rohr J. Combining sugar biosynthesis genes for the generation of l‐ and d‐amicetose and formation of two novel antitumor tetracenomycins. Chem Commun. 2005;12:1604–1606. doi: 10.1039/b417815g. et al. [DOI] [PubMed] [Google Scholar]
- Pérez M., Lombó F., Baig I., Braña A.F., Rohr J., Salas J.A., Méndez C. Combinatorial biosynthesis of antitumor deoxysugar pathways in Streptomyces griseus: reconstitution of ‘unnatural natural gene clusters’ for the biosynthesis of four 2,6‐d‐dideoxyhexoses. Appl Environ Microbiol. 2006;72:6644–6652. doi: 10.1128/AEM.01266-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez M., Baig I., Braña A.F., Salas J.A., Rohr J., Méndez C. Generation of new derivatives of the antitumor antibiotic mithramycin by altering the glycosylation pattern through combinatorial biosynthesis. Chembiochem. 2008;9:2295–2304. doi: 10.1002/cbic.200800299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A., Lombó F., Braña A.F., Rohr J., Méndez C., Salas J.A. Biosynthesis of elloramycin in Streptomyces olivaceus requires glycosylation by enzymes encoded outside the aglycon cluster. Microbiology. 2008;154:781–788. doi: 10.1099/mic.0.2007/014035-0. [DOI] [PubMed] [Google Scholar]
- Ramos A., Olano C., Braña A.F., Méndez C., Salas J.A. Modulation of deoxysugar transfer by the elloramycin glycosyltransferase ElmGT through site‐directed mutagenesis. J Bacteriol. 2009;191:2871–2875. doi: 10.1128/JB.01747-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos J.L., Martínez‐Bueno M., Molina‐Henares A.J., Terán W., Watanabe K., Zhang X. The TetR family of transcriptional repressors. Microbiol Mol Biol Rev. 2005;69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rascher A., Hu Z., Viswanathan N., Schirmer A., Reid R., Nierman W.C. Cloning and characterization of a gene cluster for geldanamycin production in Streptomyces hygroscopicus NRRL 3602. FEMS Microbiol Lett. 2003;218:223–230. doi: 10.1016/S0378-1097(02)01148-5. et al. [DOI] [PubMed] [Google Scholar]
- Remsing L.L., Garcia‐Bernardo J., Gonzalez A., Künzel E., Rix U., Braña A.F. Ketopremithramycins and ketomithramycins, four new aureolic acid‐type compounds obtained upon inactivation of two genes involved in the biosynthesis of the deoxysugar moieties of the antitumor drug mithramycin by Streptomyces argillaceus, reveal novel insights into post‐PKS tailoring steps of the mithramycin biosynthetic pathway. J Am Chem Soc. 2002;124:1606–1614. doi: 10.1021/ja0105156. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remsing L.L., González A.M., Nur‐e‐Alam M., Fernández‐Lozano M.J., Braña A.F., Rix U. Mithramycin SK, a novel antitumor drug with improved therapeutic index, mithramycin SA, and demycarosyl‐mithramycin SK: three new products generated in the mithramycin producer Streptomyces argillaceus through combinatorial biosynthesis. J Am Chem Soc. 2003;125:5745–5753. doi: 10.1021/ja034162h. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez L., Aguirrezabalaga I., Allende N., Braña A.F., Méndez C., Salas J.A. Engineering deoxysugar biosynthetic pathways from antibiotic‐producing microorganisms. A tool to produce novel glycosylated bioactive compounds. Chem Biol. 2002;9:721–729. doi: 10.1016/s1074-5521(02)00154-0. [DOI] [PubMed] [Google Scholar]
- Salas A.P., Zhu L., Sánchez C., Braña A.F., Rohr J., Méndez C., Salas J.A. Deciphering the late steps in the biosynthesis of the anti‐tumour indolocarbazole staurosporine: sugar donor substrate flexibility of the StaG glycosyltransferase. Mol Microbiol. 2005;58:17–27. doi: 10.1111/j.1365-2958.2005.04777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas J.A., Méndez C. Engineering the glycosylation of natural products in actinomycetes. Trends Microbiol. 2007;15:219–232. doi: 10.1016/j.tim.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Salas J.A., Méndez C. Indolocarbazole antitumour compounds by combinatorial biosynthesis. Curr Opin Chem Biol. 2009;13:152–160. doi: 10.1016/j.cbpa.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Sánchez C., Butovich I.A., Braña A.F., Rohr J., Méndez C., Salas J.A. The biosynthetic gene cluster for the antitumor rebeccamycin: characterization and generation of indolocarbazole derivatives. Chem Biol. 2002;9:519–531. doi: 10.1016/s1074-5521(02)00126-6. [DOI] [PubMed] [Google Scholar]
- Sánchez C., Salas A.P., Braña A.F., Palomino M., Pineda‐Lucena A., Carbajo R.J. Generation of potent and selective kinase inhibitors by combinatorial biosynthesis of glycosylated indolocarbazoles. Chem Commun. 2009;27:4118–4120. doi: 10.1039/b905068j. et al. [DOI] [PubMed] [Google Scholar]
- Schauwecker F., Pfennig F., Schröder W., Keller U. Molecular cloning of the actinomycin synthetase gene cluster from Streptomyces chrysomallus and functional heterologous expression of the gene encoding actinomycin synthetase II. J Bacteriol. 1998;180:2468–2474. doi: 10.1128/jb.180.9.2468-2474.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwecke T., Aparicio J.F., Molnar I., Konig A., Khaw L.E., Haydock S.F. The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin. Proc Natl Acad Sci USA. 1995;92:7839–7843. doi: 10.1073/pnas.92.17.7839. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti C., Hutchinson C.R. Enhanced antibiotic production by manipulation of the Streptomyces peucetius dnrH and dnmT genes involved in doxorubicin (adriamycin) biosynthesis. J Bacteriol. 1996;178:7316–7321. doi: 10.1128/jb.178.24.7316-7321.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L., Huang J., Jing L., Chen J.Y., Kan S.D., Wang M. Overexpression of aveBIV leading to the improvement of 4′‐epidaunorubicin production in Streptomyces coeruleorubidus strain SIPI‐A0707. Appl Microbiol Biotechnol. 2010;87:1057–1064. doi: 10.1007/s00253-010-2541-3. et al. [DOI] [PubMed] [Google Scholar]
- Sheldon P.J., Johnson D.A., August P.R., Liu H.W., Sherman D.H. Characterization of a mitomycin‐binding drug resistance mechanism from the producing organism, Streptomyces lavendulae. J Bacteriol. 1997;179:1796–1804. doi: 10.1128/jb.179.5.1796-1804.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon P.J., Mao Y., He M., Sherman D.H. Mitomycin resistance in Streptomyces lavendulae includes a novel drug‐binding‐protein‐dependent export system. J Bacteriol. 1999;181:2507–2512. doi: 10.1128/jb.181.8.2507-2512.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon P.J., Busarow S.B., Hutchinson C.R. Mapping the DNA‐binding domain and target sequences of the Streptomyces peucetius daunorubicin biosynthesis regulatory protein, DnrI. Mol Microbiol. 2002;44:449–460. doi: 10.1046/j.1365-2958.2002.02886.x. [DOI] [PubMed] [Google Scholar]
- Shen B. Biosynthesis of aromatic polyketides. Curr Opin Chem Biol. 2003;7:285–295. doi: 10.1016/s1367-5931(03)00020-6. [DOI] [PubMed] [Google Scholar]
- Shen B., Du L., Sanchez C., Edwards D.J., Chen M., Murrell J.M. Cloning and characterization of the bleomycin biosynthetic gene cluster from Streptomyces verticillus ATCC 15003. J Nat Prod. 2002;65:422–431. doi: 10.1021/np010550q. [DOI] [PubMed] [Google Scholar]
- Shin J.C., Na Z., Lee D.H., Kim W.C., Lee K., Shen Y.M. Characterization of tailoring genes involved in the modification of geldanamycin polyketide in Streptomyces hygroscopicus JCM4427. J Microbiol Biotechnol. 2008;18:1101–1108. et al. [PubMed] [Google Scholar]
- Singh B., Lee C.B., Sohng J.K. Precursor for biosynthesis of sugar moiety of doxorubicin depends on rhamnose biosynthetic pathway in Streptomyces peucetius ATCC 27952. Appl Microbiol Biotechnol. 2010;85:1565–1574. doi: 10.1007/s00253-009-2225-z. [DOI] [PubMed] [Google Scholar]
- Sitachitta N., Lopanik N.B., Mao Y., Sherman D.H. Analysis of a parallel branch in the mitomycin biosynthetic pathway involving the mitN‐encoded aziridine N‐methyltransferase. J Biol Chem. 2007;282:20941–20947. doi: 10.1074/jbc.M702456200. [DOI] [PubMed] [Google Scholar]
- Srinivasan P., Palani S.N., Prasad R. Daunorubicin efflux in Streptomyces peucetius modulates biosynthesis by feedback regulation. FEMS Microbiol Lett. 2010;305:18–27. doi: 10.1111/j.1574-6968.2010.01905.x. [DOI] [PubMed] [Google Scholar]
- Stutzman‐Engwall K.J., Otten S.L., Hutchinson C.R. Regulation of secondary metabolism in Streptomyces spp. and overproduction of daunorubicin in Streptomyces peucetius. J Bacteriol. 1992;174:144–154. doi: 10.1128/jb.174.1.144-154.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama M., Thompson C.J., Kumagai T., Suzuki K., Deblaere R., Villarroel R., Davies J. Characterisation by molecular cloning of two genes from Streptomyces verticillus encoding resistance to bleomycin. Gene. 1994a;151:11–16. doi: 10.1016/0378-1119(94)90626-2. [DOI] [PubMed] [Google Scholar]
- Sugiyama M., Kumagai T., Shionoya M., Kimura E., Davies J.E. Inactivation of bleomycin by an N‐acetyltransferase in the bleomycin‐producing strain Streptomyces verticillus. FEMS Microbiol Lett. 1994b;121:81–85. doi: 10.1111/j.1574-6968.1994.tb07079.x. [DOI] [PubMed] [Google Scholar]
- Truglio J.J., Croteau D.L., Van Houten B., Kisker C. Prokaryotic nucleotide excision repair: the UvrABC system. Chem Rev. 2006;106:233–252. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- Udwary D.W., Zeigler L., Asolkar R.N., Singan V., Lapidus A., Fenical W. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc Natl Acad Sci USA. 2007;104:10376–10381. doi: 10.1073/pnas.0700962104. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lanen S.G., Dorrestein P.C., Christenson S.D., Liu W., Ju J., Kelleher N.L., Shen B. Biosynthesis of the β‐amino acid moiety of the enediyne antitumor antibiotic C‐1027 featuring β‐amino acyl‐S‐carrier protein intermediates. J Am Chem Soc. 2005;127:11594–11595. doi: 10.1021/ja052871k. [DOI] [PubMed] [Google Scholar]
- Van Lanen S.G., Linm S., Shen B. Biosynthesis of the enediyne antitumor antibiotic C‐1027 involves a new branching point in chorismate metabolism. Proc Natl Acad Sci USA. 2008;105:494–499. doi: 10.1073/pnas.0708750105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron C., Matsushima P., Rosteck P.R., Jr, Broughton M.C., Turner J., Madduri K. Cloning and analysis of the spinosad biosynthetic gene cluster of Saccharopolyspora spinosa. Chem Biol. 2001;8:487–499. doi: 10.1016/s1074-5521(01)00029-1. et al. [DOI] [PubMed] [Google Scholar]
- Wang G., Hosaka T., Ochi K. Dramatic activation of antibiotic production in Streptomyces coelicolor by cumulative drug resistance mutations. Appl Environ Microbiol. 2008;74:2834–2840. doi: 10.1128/AEM.02800-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Hu Y., Zhang Y., Wang S., Cui Z., Bao Y. Role of sgcR3 in positive regulation of enediyne antibiotic C‐1027 production of Streptomyces globisporus C‐1027. BMC Microbiol. 2009;9:1–12. doi: 10.1186/1471-2180-9-14. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson B., Gregory M.A., Moss S.J., Carletti I., Sheridan R.M., Kaja A. Separation of anti‐angiogenic and cytotoxic activities of borrelidin by modification at the C17 side chain. Bioorg Med Chem Lett. 2006;16:5814–5817. doi: 10.1016/j.bmcl.2006.08.073. et al. [DOI] [PubMed] [Google Scholar]
- Yu T.W., Bai L., Clade D., Hoffmann D., Toelzer S., Trinh K.Q. The biosynthetic gene cluster of the maytansinoid antitumor agent ansamitocin from Actinosynnema pretiosum. Proc Natl Acad Sci USA. 2002;99:7968–7973. doi: 10.1073/pnas.092697199. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.Q., Wilkinson B. Drug discovery beyond the ‘rule‐of‐five’. Curr Opin Biotechnol. 2007;18:478–488. doi: 10.1016/j.copbio.2007.10.005. [DOI] [PubMed] [Google Scholar]


