Summary
Pristinamycin, produced by Streptomyces pristinaespiralis Pr11, is a streptogramin antibiotic consisting of two chemically unrelated compounds, pristinamycin I and pristinamycin II. The semi‐synthetic derivatives of these compounds are used in human medicine as therapeutic agents against methicillin‐resistant Staphylococcus aureus strains. Only the partial sequence of the pristinamycin biosynthetic gene cluster has been previously reported. To complete the sequence, overlapping cosmids were isolated from a S. pristinaespiralis Pr11 gene library and sequenced. The boundaries of the cluster were deduced, limiting the cluster size to approximately 210 kb. In the central region of the cluster, previously unknown pristinamycin biosynthetic genes were identified. Combining the current and previously identified sequence information, we propose that all essential pristinamycin biosynthetic genes are included in the 210 kb region. A pristinamycin biosynthetic pathway was established. Furthermore, the pristinamycin gene cluster was found to be interspersed by a cryptic secondary metabolite cluster, which probably codes for a glycosylated aromatic polyketide. Gene inactivation experiments revealed that this cluster has no influence on pristinamycin production. Overall, this work provides new insights into pristinamycin biosynthesis and the unique genetic organization of the pristinamycin gene region, which is the largest antibiotic ‘supercluster’ known so far.
Introduction
Streptomyces pristinaespiralis produces the streptogramin‐type antibiotic pristinamycin. Like other members of this family (also designated synergimycins), such as virginiamycin, mikamycin, vernamycin and others (Cocito, 1979), pristinamycin is a mixture of two different chemical classes of components, pristinamycin I (PI) and pristinamycin II (PII). PI is a branched cyclic hexadepsipeptide of the B group of streptogramins, while pristinamycin PII has the structure of a polyunsaturated cyclo‐peptidic macrolactone belonging to the A group of streptogramins (Figs 3A and 5). PI and PII are produced as different congeners. PIA is the major form of PI (usually 90–95%) containing a 4‐N,N‐dimethylamino‐l‐phenylalanine (DMAPA) residue instead of N‐methyl‐4‐(methylamino)‐l‐phenylalanine (MMAPA) in the PIB minor form (5%) (Blanc et al., 1997). The PII component is synthesized mainly in two forms, PIIA and PIIB, which are present in an 80:20 ratio. The difference between PIIA and PIIB is the presence of a dehydroproline instead of a d‐proline in the macrocycle respectively (Blanc et al., 1995).
Figure 3.
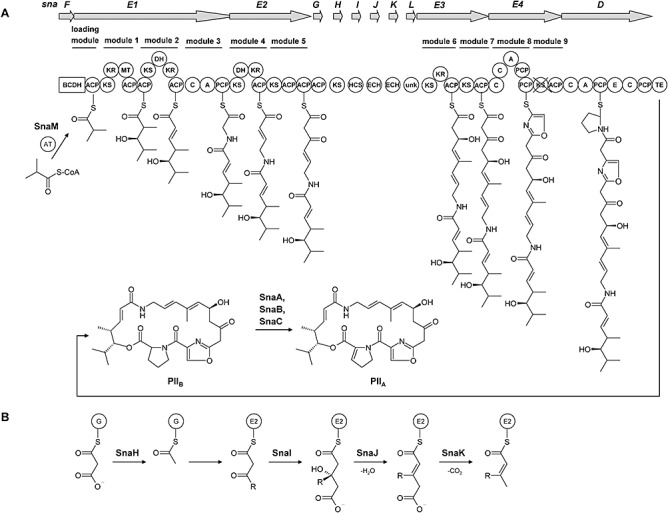
Schematic presentation of the PII biosynthetic pathway in S. pristinaespiralis. A. Domain arrangement of the PII biosynthetic enzymes. PKS domains: AT, acyl transferase; ACP, acyl carrier protein; KS, β‐ketoacyl‐ACP synthase; DH, dehydrogenase; KR, ketoreductase; MT, methyltransferase; TE, thioesterase. NRPS domains: A, adenylation; C, condensation; P, peptidyl carrier protein; BCDH, branched‐chain α‐keto acid dehydrogenase complex; HCS, hydroxymethylglutaryl‐CoA (HMG‐CoA) synthase; ECH, enoyl‐CoA hydratase; unk, unknown. B. HMG‐based model for the incorporation of the C12 methyl group in PII. G, SnaG; E2, ACP of SnaE2.
Figure 5.
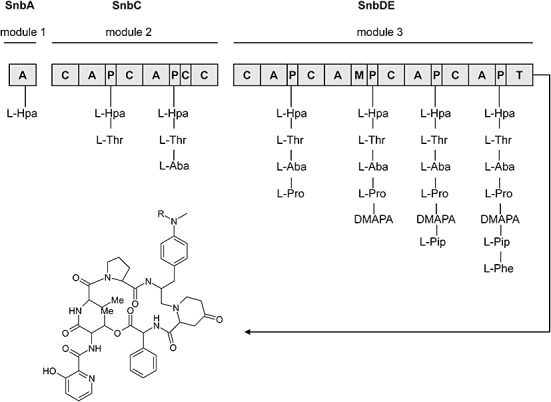
Schematic presentation of the PI biosynthetic pathway in S. pristinaespiralis. Domain arrangement of the PI NRPSs SnbA, SnbC and SnbDE. Domains: A, adenylation; C, condensation; P, peptidyl carrier protein; M, methyltransferase; T, thioesterase. Amino acids: L‐HPA, l‐hydroxypicolinic acid; L‐Thr, l‐threonine; L‐Aba, l‐aminobutyric acid; L‐Pro, l‐proline; DMAPA, 4‐N,N‐dimethylamino‐l‐phenylalanine; L‐Pip, 4‐oxo‐l‐pipecolic acid; L‐Phe, l‐phenylglycine. R = CH3 (PIA), R = H (PIB).
Pristinamycin I and pristinamycin II are coproduced by S. pristinaespiralis in a ratio of 30:70. Each compound binds to the bacterial 50 S ribosomal subunit and inhibits the elongation process of the protein synthesis, thereby exhibiting only a moderate bacteriostatic activity. However, the combination of both substances acts synergistically and leads to a potent bactericidal activity that can reach up to 100 times that of the separate components (Rehm et al., 2001). Pristinamycin is very active against a broad range of Gram‐positive bacteria, including methicillin‐resistant staphylococci, drug‐resistant Streptococcus pneumoniae and vancomycin‐resistant Enterococcus faecium as well as against some Gram‐negative bacteria, such as Haemophilus spp. Due to its antimicrobial capacities, pristinamycin is used as a therapeutic drug in human medicine, such as the semi‐synthetic streptogramin Synercid, which is a mixture of the PI derivative quinupristin and the PII derivative dalfopristin (Barrière et al., 1994).
Pristinamycin I is synthesized by non‐ribosomal peptide synthetases (NRPSs) that catalyse the stepwise condensation of seven amino acid precursors: 3‐hydroxypicolinic acid, l‐threonine, l‐aminobutyric acid, l‐proline, DMAPA, 4‐oxo‐l‐pipecolic acid and l‐phenylglycine (de Crécy‐Lagard et al., 1997). PII is suggested to be synthesized from isobutyryl‐CoA as a starter unit. Six malonyl‐CoA extender units and the amino acids glycine, serine and proline are subsequently added by the action of hybrid PKS/NRPS enzymes (Thibaut et al., 1995).
Only partial sequence information of the pristinamycin biosynthetic gene cluster has been previously reported, and only a few of the biosynthetic genes have been characterized (Blanc et al., 1994; Bamas‐Jacques et al., 1999). In previous studies, the cluster organization was elucidated by pulsed‐field gel electrophoresis and was mapped to four non‐overlapping chromosomal regions A, B, C and D (Bamas‐Jacques et al., 1999). Region B, C and D together harbour some of the pristinamycin structural genes and thus represent the antibiotic biosynthetic gene cluster (Fig. 1). Thereby, the PI and PII structural genes exhibit a unique organization because they are not arranged in discrete clusters but are more or less mixed among each other. Region A encompasses a pristinamycin resistance gene ptr that is located outside the biosynthetic cluster (Bamas‐Jacques et al., 1999). Due to its interspersed organization, we do not name this region a ‘cluster’ but rather the ‘pristinamycin biosynthetic gene region’.
Figure 1.
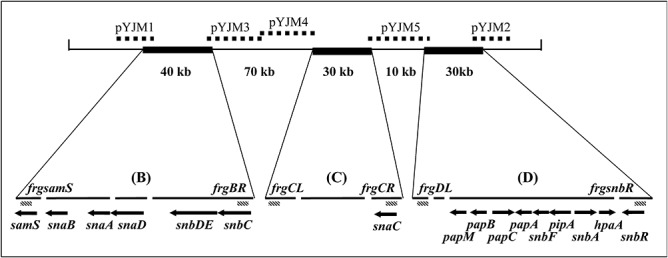
Schematic presentation of the primordial pristinamycin biosynthetic gene region. Gaps are shown as spaces. Cosmids pYJM1‐5 are indicated by broken lines. The fragments, amplified by PCR and used as probes in the screening approach, are demonstrated as diagonal boxes and denoted by the prefix frg.
So far, the overall organization of the pristinamycin biosynthetic gene region and its boundaries has not yet been described. Furthermore, the published sequence contains large gaps and several genes known to be essential for PI and PII biosynthesis have not been identified.
In this study, we report on the characterization of the complete pristinamycin biosynthetic gene region and the identification of a cryptic type II PKS gene cluster in S. pristinaespiralis Pr11. This provides information on the origin and the unique gene organization of the ‘pristinamycin supercluster’ and on pristinamycin biosynthesis.
Results and discussion
Isolation and characterization of DNA fragments covering the gaps and borders of the pristinamycin biosynthetic gene region
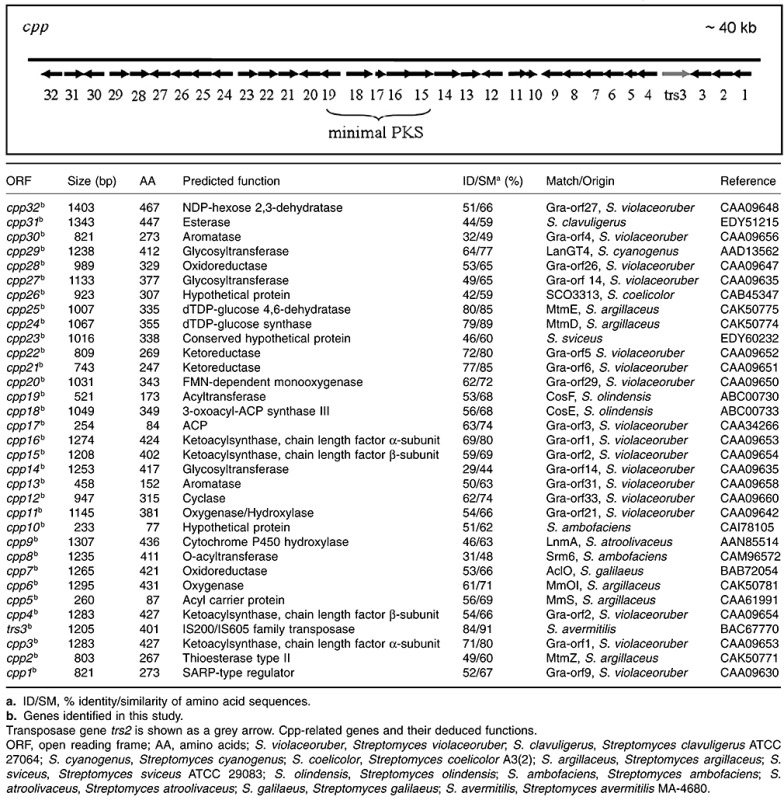
Thus far, only parts of the pristinamycin biosynthetic gene cluster have been isolated and characterized, and only 16 genes have been described to be involved in pristinamycin biosynthesis (Fig. 1) (Blanc et al., 1994; Bamas‐Jacques et al., 1999). To complete the pristinamycin biosynthetic gene region, a S. pristinaespiralis Pr11 cosmid library was constructed and used in hybridization experiments to search for cosmids that overlap with published sequences. The cosmids were analysed by restriction analyses, polymerase chain reaction (PCR) and Southern blotting experiments to select overlapping cosmids (see Experimental procedures). After sequencing the cosmids, the obtained sequence data were assembled to contigs and were analysed in silico.
Small gaps (< 10 kb) within regions B, C and D were closed by sequencing of PCR products spanning these gaps. To close the large gaps in between region B, C and D, DNA probes were derived from the inner cluster borders and used for screening approaches with the S. pristinaespiralis cosmid library (Fig. 1). Three cosmids were chosen to be sequenced covering the large gaps within the pristinamycin gene region: cosmid pYJM3 and pYJM4 span the gap between regions B and C, whereas pYJM5 covers the gap between regions C and D (Fig. 1). To find the boundaries of the pristinamycin biosynthetic gene region, probes from the 5′ (left border) and 3′ (right border) ends of the known sequence were designed, where the genes samS and snbR are localized respectively. Cosmid pYJM1 and pYJM2 were identified and sequenced, which overlap with the left and right boundaries of the gene region respectively (Fig. 1).
The sequence data were assembled to contigs. Still existing gaps were closed using the S. pristinaespiralis ATCC 25486 draft sequence data as a scaffold to align the S. pristinaespiralis Pr11 sequence. The genome of S. pristinaespiralis ATCC 25486 is sequenced in part (Accession No. ABJI00000000; M. Fischbach, P. Godfrey, D. Ward, S. Young, Q. Zeng, M. Koehrsen et al., unpublished) and preliminary sequence data are accessible in GenBank. In this way it was possible to obtain one scaffold containing three contigs, of which 210 kb constitute the pristinamycin biosynthetic gene region (Fig. 2).
Figure 2.
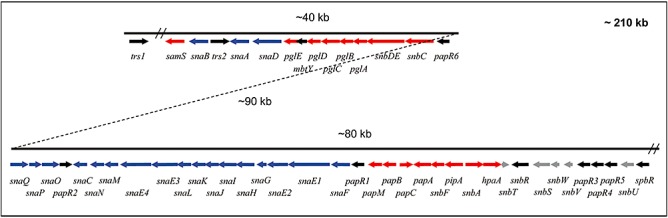
Schematic presentation of the pristinamycin biosynthetic gene region of S. pristinaespiralis Pr11. PI and PII biosynthetic genes are shown as red and blue arrows respectively. Genes for regulation, resistance and of general function are shown as black arrows. Genes of unknown function are shown as grey arrows. The ∼ 90 kb interjacent gene region is shown as a dashed line. Parallel dashes mark the gene region boundaries.
Taking the initial S. pristinaespiralis genome size of 7500 kb as predicted by Bamas‐Jacques and colleagues (1999), the cluster corresponds to 2.8% of the whole linear chromosome. Forty‐five genes covering a region of ∼ 120 kb were assigned as pristinamycin‐specific genes involved in the biosynthesis, regulation and resistance of pristinamycin, and 32 genes covering a region of ∼ 40 kb presumably code for the biosynthesis of a type II aromatic polyketide. The function of the remaining ∼ 50 kb, mainly located within the ∼ 90 kb interjacent sequence region, is unknown (see below; Figs 2 and 6, Tables 1 and 2). Altogether, this makes the pristinamycin biosynthetic gene region the largest antibiotic ‘supercluster’ that has been described so far.
Figure 6.
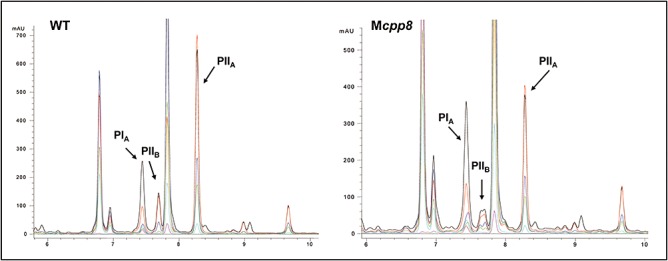
Production profiles of S. pristinaespiralis Pr11 wild‐type (left) and mutant Mcpp8 (right) respectively. Pristinamycin PIA (Rt = 7.5 min), PIIA (Rt = 8.3 min) and PIIB (Rt = 7.7 min) were detected by UV‐Vis spectrometry (data not shown). Corresponding UV‐VIS spectra of retention region 7.5–7.7 are listed below. Multiple wavelength monitoring was performed at 210 (black), 230 (red), 260 (blue), 280 (green), 310 (light blue), 360 (magenta) and 435 (yellow) nm.
Table 1.
Pristinamycin‐related genes and their deduced functions.
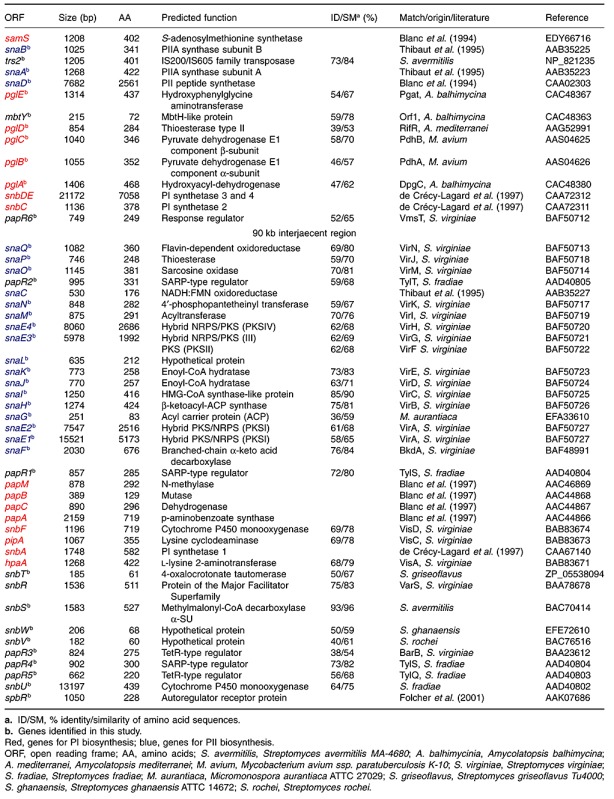
Table 2.
Schematic presentation of the S. pristinaespiralis Pr11 cpp cluster with a size of approximately 40 kb encoding the unknown substance.
Definition of the gene cluster boundaries
The left boundary of the pristinamycin gene cluster was previously defined by the gene samS, which is localized adjacent to the PII biosynthetic gene snaB (Fig. 1). samS encodes a putative S‐adenosylmethionine synthetase and was shown to be involved in PI but not in PII biosynthesis (Blanc et al., 1994; Huh et al., 2004).
The gene located directly downstream of samS encodes a deduced protein that shows similarity to an adenosine kinase. Because there is no nucleoside residue present in the pristinamycins, this adenosine kinase is most likely not involved in antibiotic biosynthesis but probably needed for purine synthesis during primary metabolism. Other genes identified on this cosmid code for primary metabolic enzymes such as methionine synthase, 5,10‐methylentetrahydrofolate reductase and S‐adenosyl‐l‐homocysteine hydrolase, which are enzymes of the folate pathway. Similarities were also found with a gene encoding a predicted ATP‐dependent DNA helicase (data not shown). Altogether, these similarities indicate that an involvement of the genes in the biosynthesis of pristinamycin is unlikely. Thus, we postulate that no further pristinamycin biosynthetic genes are located downstream of samS, which therefore assigns the left border of the cluster.
The right boundary was previously defined by the gene snbR that codes for a predicted ABC transporter protein probably involved in conferring resistance to pristinamycin (Blanc et al., 1994) (Fig. 1).
On cosmid pYJM2, five orfs of unknown function and four regulatory genes, designated as papR3, papR4, papR5 and spbR, were identified in the upstream region of snbR (Fig. 2). The orfs papR3 and papR5 both encode deduced proteins showing similarity to TetR repressor proteins, whereas the predicted gene product of papR4 revealed a high degree of similarity to Streptomyces antibiotic regulatory proteins (SARPs). The orfs papR3, papR4 and papR5 were already shown to be involved in the regulation of pristinamycin biosynthesis (Y. Mast, unpublished results). In addition, the gene spbR could be identified at the right border of the cluster. It encodes a γ‐butyrolactone receptor protein, whose regulatory function was demonstrated previously but whose genetic location was so far unknown (Folcher et al., 2001). SpbR acts as the global regulator of pristinamycin biosynthesis. The TetR repressor proteins PapR3 and PapR5 function as repressors of pristinamycin biosynthesis, whereas the SARP regulator PapR4, as well as PapR1 and PapR2 (Table 1), activates pristinamycin biosynthesis. All the regulators are part of a complex signalling cascade that is responsible for the fine‐tuned regulation of pristinamycin production (Y. Mast, unpublished results). After assigning functions to all identified genes (see below), we propose that all essential pristinamycin biosynthetic genes are identified. Thus, instead of snbR, spbR probably marks the right border of the pristinamycin gene region. However, we cannot exclude that upstream of spbR pristinamycin‐related genes are localized that may be involved in, but are not essential for pristinamycin biosynthesis.
PII biosynthetic genes
Between regions C and D, 16 orfs (snaF, snaE1, snaE2, snaG, snaH, snaI, snaJ, snaK, snaL, snaE3, snaE4, snaM, snaN, snaO, snaP and snaQ) were identified, whose predicted gene products show ∼ 60–90% amino acid identity to gene products that are putatively involved in the virginiamycin M (VM) biosynthesis of Streptomyces virginiae, published by Pulsawat and colleagues (2007a) (Table 1). To date 75 kb of the S. virginiae gene cluster have been described harbouring 34 genes (Pulsawat et al., 2007a). Due to the structural similarity of pristinamycin and virginiamycin, we expect that the respective gene products have a similar function in both strains. However, there are also several differences between both clusters. Below, we specify some of the predicted gene products of S. pristinaespiralis together with their corresponding as well as missing counterparts in S. virginiae and their predicted functions in streptogramin type A biosynthesis.
Genes for precursor supply. The newly identified PII structural gene snaF is located downstream of the regulatory gene papR1 and encodes a predicted protein with similarity to the branched α‐keto acid decarboxylase BkdA of S. virginiae. The gene bkdA was already shown to be essential for VM biosynthesis (Pulsawat et al., 2007b). It is part of the bkdAB operon, of which bkdB codes for a predicted dihydrolipoamide acyltransferase BkdB. BkdA and BkdB are suggested to convert valine into isobutyryl‐CoA, which serves as the primer for VM biosynthesis. In S. pristinaespiralis, the respective dihydrolipoamide acyltransferase is not encoded by a discrete gene but by the first ∼ 2 kb of the gene snaE1, which is located downstream of snaF. snaE1 codes for a hybrid PKS/NRPS, and through inactivation experiments, has been shown to be essential for PII biosynthesis (Y.‐F. Bizouerne, unpublished). SnaF and the dihydrolipoamide acyltransferase domain of SnaE1 together might constitute a branched‐chain α‐keto acid dehydrogenase complex, which is responsible for supplying the isobutyryl‐CoA precursor for PII biosynthesis (Fig. 3A).
Genes encoding PKS/NRPS and tailoring enzymes. Five genes (snaE1, snaE2, snaE3, snaE4 and snaD) encode multifunctional enzymes such as PKS, NRPS and hybrid PKS/NRPS. snaE1 codes for the deduced large hybrid PKS/NRPS complex SnaE1, which consists of the loading module, two PKS modules and one NRPS module. SnaE1 is probably responsible for attaching the isobutyryl‐CoA starter unit to the first acyl carrier protein (ACP) loading domain followed by the addition of two malonyl‐CoA molecules and a glycine residue. The snaE2 gene is located directly downstream of snaE1 and encodes the predicted PKS SnaE2 that contains two PKS modules that are responsible for the incorporation of two further malonyl‐CoA molecules (Fig. 3A). SnaE1 and SnaE2 together show similarity to the predicted large PKS/NRPS complex VirA of S. virginiae, whose coding gene has been shown to be essential for VM biosynthesis (Pulsawat et al., 2007a). SnaE2 contains two ACP domains, and a third one is encoded by a discrete gene named snaG. This ACP tridomain is part of a 3‐hydroxy‐3‐methylglutaryl (HMG) enzyme cassette as found in the curacin and the jamaicamide gene cluster of Lyngbya majuscula (Gu et al., 2009). Such HMG enzyme cassettes catalyse β‐branching reactions in the growing polyketide. They normally consist of a tandem ACP tridomain, a ketosynthase and a HMG‐CoA synthase, which is often followed by two enoyl‐CoA hydratase enzymes. In S. pristinaespiralis, snaH, snaI, snaJ and snaK code for predicted monofunctional enzymes that are similar to the deduced ketosynthase‐like carboxylase VirB, to HMG‐CoA synthase‐like protein VirC and the enoyl‐CoA hydratases VirD and VirE of S. virginiae respectively. Due to the respective amino acid identities, the two SnaE2 ACP domains together with SnaG‐I might constitute a HMG enzyme cassette, which is followed by the two enoyl‐CoA hydratases SnaJ and K. We predict SnaH‐K to insert a methyl group at the C12 position of PII (SnaI) and to catalyse the subsequent dehydration (SnaJ) and decarboxylation (SnaK) to yield a C11αβ‐unsaturated thioester (Fig. 3B). snaE3 encodes a putative PKS consisting of two PKS modules that probably are needed for the addition of another two malonyl‐CoA. The N‐terminal part of SnaE3 is similar to the deduced PKS VirF, whereas the C‐terminal part corresponds to the hybrid PKS/NRPS VirG. The snaE4 gene codes for a putative hybrid NRPS/PKS, which is similar to the deduced hybrid PKS/NRPS VirH of S. virginiae and which consists of an NRPS and a PKS domain. The SnaE4 NRPS is suggested to introduce a serine into the growing polyketide chain, whereas the PKS module is probably inactive because its ketosynthase domain is missing the conserved amino acid sequence pattern VDTACSSS, which is essential for activity (Long et al., 2002). Finally, snaD, which has already been shown to be essential for PII biosynthesis (Blanc et al., 1994), codes for the NRPS SnaD, which introduces the final residue proline into the PII precursor.
Accessory and tailoring genes. Twelve of the newly identified genes (snaF, snaG, snaH, snaI, snaJ, snaK, snaL, snaM, snaN, snaO, snaP and snaQ) within the pristinamycin gene region code for monofunctional polypeptides. The putative function of snaG‐snaK during PII biosynthesis is described above. Interestingly, in all PII PKSs (SnaE1, SnaE2, SnaE3 and SnaE4) the internal acyltransferase (AT) domains are missing. Generally, the AT domains of PKSs are responsible for the selection and loading of CoA‐activated extender units onto the ACP domains; subsequently, the ketosynthase (KS) domains finally catalyse the condensation of each extender unit, resulting in the elongation of the polyketide chain. Downstream of snaE4, we identified snaM whose deduced gene product is similar to the discrete AT VirI of S. virginiae. VirI is probably responsible for loading the several acyl units to the ‘AT‐less’ VM PKSs as first experiments indicate (Pulsawat et al., 2007a). Due to the striking similarity between SnaM and VirI (Table 1), we suggest that SnaM acts accordingly as a malonyl‐CoA specific iterative AT in loading the acyl precursors to the ‘AT‐less’S. pristinaespiralis PKSs during PII biosynthesis.
Phylogenetic analyses of acyltransferase domains of cis and trans‐AT PKS gene clusters demonstrate that SnaM is a member of a clade comprised exclusively of discrete ATs of trans‐AT‐PKS biosynthetic gene clusters such as LnmG from leinamycin biosynthesis (Cheng et al., 2003) or KirCI from kirromycin biosynthesis (Weber et al., 2008) (Fig. 4). It is noteworthy that these enzymes are phylogenetically more closely related to ATs from fatty acid synthases than to the internal AT domains of cis‐AT PKS.
Figure 4.
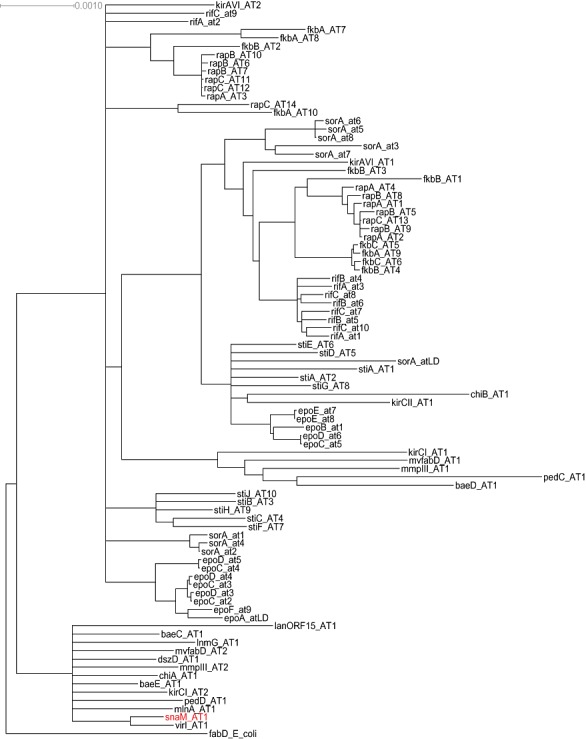
Consensus tree of alignment of acyltransferase core domains of cis‐ and trans‐AT‐type polyketide synthases. Outgroup is FabD from E. coli.
snaL is localized downstream of the enoyl‐CoA hydratase gene snaK that codes for a deduced protein with no similarity to any known protein. Furthermore, the gene snaN was identified encoding a predicted 4′‐phosphopantetheinyl transferase, which is similar to VirK.
Exclusive PII‐specific genes. One of the striking differences between the pristinamycin and virginamycin gene cluster is the presence of three PII‐specific genes –snaA, snaB and snaC– within the pristinamycin biosynthetic gene region whose homologous counterparts are missing in the virginiamycin cluster. It has previously been shown that the monooxygenase subunits SnaA and SnaB, together with the FMN reductase SnaC, are responsible for the conversion of the precursor PIIB to the final product PIIA (Blanc et al., 1995) (Fig. 3A). Together, these three enzymes catalyse the oxidation of the d‐proline residue in PIIB that leads to the formation of the unique dehydroproline residue in PIIA. Because no homologous snaA, snaB and snaC genes were yet identified in the virginiamycin producer, the genes virM and virN that code for a predicted sarcosine oxidase VirM and a flavin‐dependent oxidoreductase VirN, respectively, were suggested to catalyse the analogous reaction in S. virginiae, whereby VM2 is converted into VM1 (Pulsawat et al., 2007a). However, homologues (snaO and snaQ) are also present in S. pristinaespiralis, suggesting that the sarcosine oxidases and flavin‐dependent oxidoreductases have a different function during streptogramin biosynthesis that so far remains unknown.
A further discrepancy between both streptogramin producers is based on the gene snaD. In S. pristinaespiralis, snaD codes for the peptide synthetase SnaD, which introduces a proline residue into the PII framework (Fig. 3A). Proline is also a constituent of VM; however, no snaD homologous gene has been identified in S. virginiae so far. Sequence analysis revealed that SnaD contains a C‐terminal type I thioesterase (TE) domain (Y.‐F. Bizouerne, unpublished) that probably catalyses the release of the polyketide chain and the cyclization reaction resulting in the macrolactonic ring structure of PII. Since no snaD counterpart was found in S. virginiae, the final cyclization reaction was suggested to be catalysed by the predicted TE VirJ. However, also in S. pristinaespiralis a discrete gene, snaP, is present that encodes a predicted type II TE that is similar to VirJ. As type II TEs often have editing function during antibiotic biosyntheses (Heathcote et al., 2001; Schwarzer et al., 2002), we propose that SnaP has a corrective function during PII biosynthesis for instance by removing aberrant acyl residues from the PII PKSs and regenerating misprimed NRPS.
PI biosynthetic genes
The PI biosynthesis is catalysed by an NRPS system composed of three proteins, SnbA, SnbC and SnbDE. SnbA activates the starter molecule 3‐hydroxypicolinic acid, SnbC incorporates l‐threonine and l‐aminobutyric acid, and SnbDE adds l‐proline, DMAPA, 4‐oxo‐l‐pipecolic acid and l‐phenylglycine to the precursor molecule (Fig. 5) (Thibaut et al., 1997). The linear precursor is then cyclized to the final PI macrocycle under release from the synthetase.
Twelve genes are involved in the PI precursor supply: hpaA is required for 3‐hydroxypicolinic acid formation (Blanc et al., 1996), pipA and snbF are involved in the biosynthesis of 4‐oxo‐l‐pipecolic acid (Blanc et al., 1996), and the cluster of pap genes code for a DMAPA biosynthetic pathway starting from chorismic acid (Table 1) (Blanc et al., 1997). Furthermore, we identified a set of genes, designated as pglA, pglB, pglC, pglD and pglE, which show high amino acid identity to enzymes involved in the biosynthesis of aproteinogenic amino acids such as 4‐hydroxyphenylglycine and 3,5‐dihydroxyphenylglycine. The pgl genes have been shown to be responsible for the formation of l‐phenylglycine, which probably starts from phenylpyruvate, a compound from the primary metabolic shikimate pathway, which is converted to l‐phenylglycine via a phenylacetyl‐CoA intermediate (Y. Mast, unpublished). To date, no pap and pgl homologous genes have been found in the virginiamycin cluster. Together with the identification of the phenylglycine biosynthetic genes, all genes have likely been identified that are essential for pristinamycin biosynthesis.
Genes with similarity to actinorhodin‐like biosynthetic genes
Between regions B and C (Fig. 1), numerous orfs (cpp1‐cpp23) were identified whose deduced gene products exhibit similarities to proteins involved in aromatic polyketide biosyntheses, such as the biosynthesis of granaticin (gra) of S. violaceoruber, actinorhodin (act) of S. coelicolor A3(2) and mithramycin (mtm) of S. argillaceus that belong to the class of benzoisochromanequinones (BIQs) (Table 2). The orfs were designated as cpp for ‘cryptic pristinaespiralis polyketide’.
The genes cpp19 cpp18, cpp17, cpp16 and cpp15 presumably encode a minimal type II PKS (AT, KS, chain length‐determining factor and ACP) as their deduced gene products show similarity to minimal PKSs of the S. coelicolor A3(2) act and S. violaceoruber gra cluster. The genes cpp21, cpp22, cpp13 and cpp12 presumably code for early biosynthetic enzymes that are normally closely associated with the minimal PKS. Their deduced gene products are similar to early granaticin biosynthetic enzymes of S. violaceoruber. The predicted gene products of cpp21 and cpp22 show high amino acid identity to the ketoreductases (KR) Gra‐orf6 (80%) and Gra‐orf5 (85%) respectively, whereas cpp13 and cpp12 encode a deduced aromatase and a cyclase similar to Gra‐orf4 and Gra‐orf33 respectively. In the act mutant S. coelicolor CH999, the expression of an act gene set encoding for the minimal PKS, KR, aromatase (ARO) and cyclase (CYC) resulted in the formation of aloesaponarin II, which is a three‐ring aromatic aglycon (McDaniel et al., 1994). Thus, a similar compound might be the intermediate generated by the action of the homologous enzyme set in S. pristinaespiralis. This is also supported by the observation that the cyclases Cpp13 and Cpp12 belong to a family of cyclases involved in BIQ biosyntheses (Fritzsche et al., 2008). Furthermore, several orfs were identified, whose predicted proteins presumably are involved in tailoring reactions. For instance the deduced gene products of cpp25 and cpp24 show high amino acid identity to the dTDP‐glucose 4,6‐dehydratase MtmE (85%) and the dTDP‐glucose synthase MtmD (89%) of S. argillaceus, respectively, and thus might be involved in the formation of 6‐deoxyhexose (Lombóet al., 1999). The predicted gene products of cpp14, cpp27, cpp29 and cpp32 show similarity to several enzymes putatively involved in sugar synthesis and glycosyltransferase reactions during granaticin biosynthesis in S. violaceoruber. Therefore, we predict the unknown compound to have a granaticin/actinorhodin‐like skeleton and to be modified several times by glycosylation. Furthermore, cpp1 was identified, which codes for a deduced gene similar to regulators belonging to the SARP family and thus might be responsible for regulating the biosynthesis of the unknown compound. Altogether, the cryptic PKS II gene cluster of S. pristinaespiralis Pr11 covers a region of approximately 40 kb (Table 2). However, no actinorhodin‐like compound could be detected in S. pristinaespiralis cultures by high‐performance liquid chromatography (HPLC), suggesting that the interjacent gene cluster is not expressed under typical pristinamycin production conditions.
To prove that the orfs are not required for pristinamycin biosynthesis, some representative genes (cpp27, cpp7, cpp12, cpp8 and cpp1) were inactivated by gene insertion mutagenesis using plasmids pJcpp27apr, pJcpp7apr, pJcpp12apr, pJcpp8apr and pJcpp1apr respectively (see Experimental procedures). The genotypes of the generated mutants Mcpp27, Mcpp7, Mcpp12, Mcpp8 and Mcpp1, respectively, were proven by Southern hybridization (data not shown), and antibiotic production of the mutants was analysed by HPLC. Inactivation of any of these genes had no effect on pristinamycin biosynthesis (Fig. 6), suggesting that the genes lying in the interjacent PKS cluster are not involved in pristinamycin production. However, it would be interesting to find out if there is a co‐regulation between pristinamycin biosynthesis and the biosynthesis of the unknown substance maybe signalled via the SARP Cpp1, which could be a regulator interacting with the components of the pristinamycin signalling cascade.
Characteristics of the pristinamycin gene region
A characteristic feature of the pristinamycin biosynthetic gene region is that the PI and PII biosynthetic genes that encode the synthesis of two chemically distinct components are not clustered individually but are scattered across the complete 210 kb sequence region. This special genetic organization is suggested to be the result of evolutionary chromosomal rearrangement that could somehow be beneficial for the strain ensuring the production of both antibiotics in the synergistically active 70:30 ratio (Bamas‐Jacques et al., 1999). However, so far there has been no indication of such an evolutionary rearrangement process.
In the course of the sequence analyses, we identified a transposase gene trs2 between the PII monooxygenase subunit genes snaA and snaB as well as approximately 7 kb upstream of samS near the left border of the pristinamycin biosynthetic gene region (trs1; Fig. 2). A further transposase gene trs3 is localized to the right site of the PKS II type biosynthetic cluster between cpp3 and cpp4 (Table 2). The presence of transposase genes within and nearby the pristinamycin biosynthetic gene region strongly suggests that the original individual clusters have been shuffled during genomic rearrangement processes.
By alignment of protein sequences, we found that the putative Trs2 and Trs3 proteins show 67% amino acid identity to each other, whereas Trs1 shows no similarity to any of the transposases found in the pristinamycin biosynthetic gene region. Thus, trs3, which is localized to the cpp gene cluster, might be a duplicate of trs2, encoding the transposase that might have been responsible for the insertion of the type II PKS gene cluster.
In contrast, no such transposase genes have been reported for the virginiamycin cluster. Thus, there is no hint of a horizontal gene transfer event of the streptogramin antibiotic cluster in S. virginiae. However, so far, the virginiamycin biosynthetic gene cluster only comprises 75 kb and as some essential virginiamycin biosynthetic genes have not been identified, it seems that the cluster is still incomplete. Thus, it might be possible that there are transposase genes present in the entire cluster, which have not yet been identified (Fig. 7). In this context, it would be interesting to find out how the streptogramin biosynthetic gene clusters evolved. The high amino acid identity between numerous pristinamycin and virginiamycin specific proteins indicates that both clusters may originate from a common ancestor. Probably, a primordial actinomycete originally produced only one type of streptogramin antibiotic and the second cluster was acquired by horizontal gene transfer. To date, no strain is known that produces only one type of streptogramin. It would be interesting to identify such a strain to get more information about which cluster was acquired at first.
Figure 7.
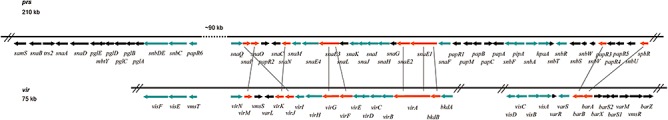
Comparison between the pristinamycin biosynthetic gene region and the virginiamycin cluster. Homologous genes are presented as coloured arrows: genes with similar orientations in both clusters are labelled turquoise; genes with different orientations are labelled orange and connected by lines due to their respective homologies. Genes with no homologous counterpart in the respective cluster are labelled black.
A unique feature of the pristinamycin biosynthetic gene region is that it is interrupted by a cryptic type II PKS gene cluster. Probably also this cluster has been acquired by horizontal gene transfer. In S. pristinaespiralis, the type II PKS gene cluster is localized between the response regulator gene papR6 and the gene snaQ encoding the putative flavin‐dependent oxidoreductase. In the virginiamycin cluster, the homologous counterparts of these genes are present (vmsT and virN respectively); however, there is no further cluster inserted at this position. Thus, we suggest the pristinamycin biosynthetic gene region with its integrated cpp cluster is of later evolutionary origin than the virginiamycin biosynthetic gene region. To our knowledge, no other streptogramin antibiotic gene cluster contains such an intercalary secondary metabolite biosynthetic gene cluster. Altogether, this makes the pristinamycin biosynthetic gene region the largest antibiotic ‘supercluster’ with a size of ∼ 210 kb harbouring genes for at least three different antibiotic biosynthesis pathways.
The advantage of such a supercluster organization could be to ensure the co‐regulation and thus co‐production of different small molecules, which could be beneficial for the producer strain as mentioned above. Evidence for the co‐regulation of the different pristinamycins includes the fact that the S. pristinaespiralis global regulator SpbR and several other regulators influence the biosynthesis of both compounds: PI and PII. For example, deletion of spbR leads to no pristinamycin production at all (Folcher et al., 2001), whereas the overexpression of any of the pristinamycin SARP regulators results in enhanced PI as well as PII biosynthesis (Y. Mast, unpublished). Further examples of co‐regulation were reported for S. virginiae, of which a VM biosynthetic gene and a virginiamycin S resistance gene are under the control of the same regulator (Namwat et al., 2001). Also the lankamycin and lankacidin productions of Streptomyces rochei are influenced by the same regulator (Mochizuki et al., 2003). A well‐understood system is the co‐regulation of the production of the β‐lactam antibiotic cephamycin and the β‐lactamase inhibitor clavulanic acid in Streptomyces clavuligerus, where the biosynthetic genes are organized in a concatenate supercluster and are regulated by the same transcriptional activator protein CcaR (Santamarta et al., 2002). However, there are some reports that claim that co‐regulation is not the driving force for clustering of secondary metabolite genes (Lawrence and Roth, 1996; Walton, 2000).
But then the question arises why those different antibiotic biosynthetic genes are all mixed up in one large cluster. In general, antibiotic genes tend to cluster in the genome of their microbial producers. These antibiotic gene clusters are ‘selfish genetic elements’ that evolve rapidly (Fischbach et al., 2007). The ‘selfish cluster’ hypothesis says that clustering, in the first place, does not have a selective advantage for the donor organism, but confers a new selective advantage to the unsophisticated recipient genome (Lawrence and Roth, 1996; Walton, 2000). In this context, the existence of clusters is beneficial because horizontal gene transfer requires the movement of contiguous DNA fragments, and clustering ensures that all essential genes needed for biosynthesis, regulation and resistance of the respective antibiotics can be transferred together to a recipient. The pristinamycin biosynthetic gene region would be one of the best examples for the ‘selfish cluster hypothesis’.
Experimental procedures
Bacterial strains, cosmids and plasmids
The bacterial strains, cosmids and plasmids used in this study are listed in Table S1.
Media and culture conditions
Escherichia coli XL1‐Blue (Bullock et al., 1987) was used as the host for subcloning. Escherichia coli strains were grown in Luria–Bertani (LB) medium at 37°C (Sambrook et al., 1989) supplemented with kanamycin, apramycin or ampicillin (50, 100 or 150 µg ml−1 respectively) when appropriate.
Streptomyces pristinaespiralis Pr11 (Aventis Pharma) was used throughout this study. This strain was isolated after spontaneous mutation of S. pristinaespiralis ATCC 25486. Streptomyces pristinaespiralis Pr11 was used for cosmid library construction and for the generation of gene insertion mutants. Streptomyces pristinaespiralis strains were grown on yeast malt agar or on MS agar for isolation of spores (Kieser et al., 2000). For isolation of genomic DNA S. pristinaespiralis strains were grown in 100 ml of S‐medium (Kieser et al., 2000) in 500 ml Erlenmeyer flasks (with steel springs) on an orbital shaker (180 r.p.m.) at 30°C. Liquid cultures were supplemented with kanamycin or apramycin (50 or 100 µg ml−1 respectively) when appropriate.
Cloning, restriction mapping and in vitro manipulation of DNA
The methods used for the isolation and manipulation of DNA from S. pristinaespiralis and E. coli were as described by Kieser and colleagues (2000) and Sambrook and colleagues (1989) respectively. Polymerase chain reactions were performed on a RoboCycler Gradient 40 thermocycler from Stratagene (La Jolla, CA, USA) or on a Primus 96plus (MWG Biotech) using Taq DNA polymerase (QIAGEN). Primers used for PCR are listed in Table S2. PCR fragments were isolated from 1% agarose gels and purified with GE Healthcare GFX columns. Isolation of Streptomyces genomic DNA was performed with the NucleoSpin Tissue Kit (Macherey‐Nagel). Restriction endonucleases were obtained from various suppliers and were used according to their specifications. Escherichia coli was transformed by the CaCl2 method (Sambrook et al., 1989). Plasmids were introduced into Streptomyces strains by polyethylene glycol‐mediated protoplast transformation (Kieser et al., 2000).
Construction and screening of a cosmid library
A cosmid library of S. pristinaespiralis Pr11 DNA was constructed by Combinature Biopharm AG using cosmid pOJ436 (Bierman et al., 1992). The cosmids were automatically spotted on a nylon membrane. This filter was used in hybridization experiments to identify cosmids that overlap with already published or patented sequences (Blanc et al., 1994; Bamas‐Jacques et al., 1999). Southern hybridizations with the digoxygenin DNA labelling and detection kit from Roche Biochemicals were carried out as described previously (Pelzer et al., 1997).
DNA fragments covering the gaps and borders of the pristinamycin biosynthetic gene region
The probes frgBR, frgCL, frgCR and frgDL were used to close the large gaps between region B, C and D. frgBR and frgCR hybridized with the right borders of regions C and B respectively, whereas probe frgCL and frgDL hybridized with the left border of regions C and D respectively (Fig. 1). The probes were used for screening the S. pristinaespiralis cosmid library. Altogether, 31 cosmids were isolated that overlapped with the ends of known cosmids, thus spanning the gaps from region B to C and C to D respectively. The cosmids were analysed in restriction, PCR and Southern blot experiments. Cosmid pYJM3, pYJM4 and pYJM5 showed minimal overlap with already known sequences and carried a large portion of new sequence data. Cosmid pYJM3 and pYJM4 span the gap between regions B and C, whereas pYJM5 covers the gap between regions C and D (Fig. 1).
The probes frgsamS and frgsnbR were used to find the left and right boundaries of the pristinamycin biosynthetic gene region respectively. The probes were used for screening the S. pristinaespiralis cosmid library. Twenty‐seven cosmids were isolated that hybridized with the frgsamS probe, thus overlapping the left border of the pristinamycin gene region, whereas 31 cosmids were isolated that hybridized with the frgsnbR probe, characterizing the right border of the cluster. Two cosmids were chosen to be sequenced: cosmid pYJM1 and pYJM2 that overlap with the left and right boundaries of the pristinamycin biosynthetic gene region respectively (Fig. 1).
DNA sequencing and analysis
DNA sequencing of cosmids pYJM1‐5 was performed by shotgun cloning followed by automated DNA sequencing carried out on double‐stranded DNA templates to obtain at least fourfold coverage. Sequencing was done by GATC, MWG, or Göttingen Genomics Laboratory (G2L) respectively, and the obtained raw sequence data were assembled using phred/phrap/consed and analysed with clusean (Weber et al., 2009), blast (Altschul et al., 1990) and Artemis (Rutherford et al., 2000). The sequences were deposited in the EMBL data library under accession numbers FR681999, FR682000 and FR682001.
Phylogenetic analysis
Acyltransferase domains were identified using the Pfam ‘Acyl_transf’ HMM Profile, extracted and aligned using muscle (Edgar, 2004). Prior to tree reconstruction, the alignment was manually edited. Trees were calculated using Splitstree 4.2 (Huson, 1998). Parameters were determined using Prottest (Abascal et al., 2005): WAG+I+G model; Pinv = 0.02; α = 1.32. Tree reconstruction was performed using Neighbor Joining with 1000 bootstrap replicates. The majority consensus method was used to calculate the consensus tree in Dendroscope (Huson et al., 2007).
Gene insertion mutagenesis of cpp genes
Internal fragments of the genes cpp27 (∼ 0.9 kb), cpp7 (∼ 0.8 kb), cpp12 (∼ 0.8 kb) and cpp8 (∼ 1.1 kb) were amplified by PCR using S. pristinaespiralis genomic DNA as a template and the primer pairs Pcpp27m1/m2, Pcpp7m1/m2, Pcpp12m1/m2 and Pcpp8m1/m2 respectively. The amplified fragments were designated cpp27*, cpp7*, cpp12* and cpp8* respectively. Fragments cpp27* and cpp8* were subcloned in EcoRV‐restricted E. coli vector pJOE890 resulting in the constructs pJOE890/cpp27* and pJOE890/cpp8* respectively, whereas fragments cpp7* and cpp12* were subcloned in EcoRV‐restricted E. coli vector pDrive, resulting in the constructs pDrive/cpp7* and pDrive/cpp12* respectively. Subsequently, a 1.5 kb aac(3)IV cassette was isolated as an EcoRV/SmaI fragment from pEH13 and cloned into the singular ScaI restriction site of pJOE890/cpp27* and pJOE890/cpp8*, respectively, resulting in the constructs pJOE890/cpp27*apr and pJOE890/cpp8*apr, as well as into the singular ScaI restriction site of pDrive/cpp7* and pDrive/cpp12*, resulting in the constructs pDrive/cpp7*apr and pDrive/cpp12*apr. For the construction of Mcpp1, a ∼ 2.5 kb fragment (cpp1*) was amplified by PCR, using S. pristinaespiralis genomic DNA as a template and the primer pairs Pcpp1m1/m2, which encompasses the entire cpp1 gene, 0.5 kb of the cpp1‐upstream and 0.8 kb of the cpp1‐downstream region. cpp1* was subcloned in EcoRV‐restricted E. coli vector pJOE890 resulting in construct pJOE890/cpp1*. Subsequently, a 1.5 kb aac(3)IV cassette was isolated as an EcoRV/SmaI fragment from pEH13 and cloned into the singular StuI restriction site within the cpp1 gene of pJOE890/cpp1* resulting in construct pJOE890/cpp1*apr.
The targeting plasmids pJOE890/cpp27*apr, pJOE890/cpp8*apr, pDrive/cpp7*apr, pDrive/cpp12*apr and pJOE890/cpp1*apr were transferred into S. pristinaespiralis Pr11 by protoplast transformation (Kieser et al., 2000). Apramycin‐resistant, kanamycin‐sensitive transformants were analysed with PCR and/or Southern hybridization experiments (data not shown) to identify those clones in which a double‐cross‐over event between the chromosomal copy of cpp27, cpp7, cpp12, cpp8 and cpp1 and the mutated fragment located on pJOE890/cpp27*apr, pJOE890/cpp8*apr, pDrive/cpp7*apr, pDrive/cpp12*apr and pJOE890/cpp1*apr, respectively, had occurred.
Pristinamycin production conditions and detection
For pristinamycin production, strains were cultivated in 100 ml inoculum medium consisting of corn steep powder (10 g l−1), saccharose (15 g l−1), (NH4)2SO4 (10 g l−1), K2HPO4 (1 g l−1), NaCl (3 g l−1), MgSO4 × 7H2O (0.2 g l−1), CaCO3 (1.25 g l−1) and tap water. The pH was adjusted to 6.9 prior to CaCO3 addition and sterilization. Cultures were incubated at 30°C in 100 ml of inoculum medium in 500 ml Erlenmeyer flasks (with steel springs) on an orbital shaker (180 r.p.m.). After 48–72 h, 17 ml of precultures was inoculated in 200 ml of production medium consisting of soybean flour (25 g l−1), starch (7.5 g l−1), glucose (22.5 g l−1), yeast extract (3.5 g l−1), ZnSO4 × 7H2O (0.5 g l−1), CaCO3 (6 g l−1) and tap water. The pH was adjusted to 6.0 prior to CaCO3 addition and sterilization. Production cultures were cultivated for 3–4 days at 30°C in 1 l Erlenmeyer flasks with steel springs on an orbital shaker (180 r.p.m.). Five millilitres of S. pristinaespiralis cultures was extracted with 5 ml ethyl acetate for 20 min and concentrated completely in vacuo. The extract was then redissolved in appropriate volumes of propan‐2‐ol (0.75 ml for 5 ml extraction volume), and samples were analysed by HPLC and diode‐array detection (Fiedler, 1993). HPLC analyses were performed on an HP1090M/HP3392A/HP7994B system (Hewlett Packard) with Nucleosil C18 columns (5 µm, 125 mm × 4.6 mm) (Grom) with a flow rate of 2 ml min−1. The following linear gradient for elution was applied using solvent PhoA (100% water with 0.1% phosphoric acid) and solvent AcCN (100% acetonitrile): at 0 min: 100% PhoA; at 15 min: 100% AcCN; at 16 min: PhoA, at 21 min: PhoA. Pristinamycin was detected at wavelength l = 230 nm and compared with a purified substance (provided by Sanofi‐Aventis) and to an HPLC‐UV/Vis spectra library (Fiedler, 1993).
Acknowledgments
We wish to thank H.‐P. Fiedler (University of Tübingen) for assistance in HPLC measurements. Y.M. is grateful for scholarships funded by the Landesgraduiertenförderungsgesetz des Landes Baden‐Württemberg and the DFG (Graduiertenkolleg ‘Infektionsbiologie’). This work was financed in part by Sanofi‐Aventis.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Bacterial strains, plasmids and cosmids.
Primer sequence and amplified fragments.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Abascal F., Zardoya R., Posada D. ProtTest: selection of best‐fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bamas‐Jacques N., Lorenzon S., Lacroix P., de Swetschin C., Crouzet J. Cluster organisation of the genes of Streptomyces pristinaespiralis involved in pristinamycin biosynthesis and resistance elucidated by pulsed‐field gel electrophoresis. J Appl Microbiol. 1999;87:939–948. doi: 10.1046/j.1365-2672.1999.00955.x. [DOI] [PubMed] [Google Scholar]
- Barrière J.C., Bouanchaud D.H., Desnottes J.F., Paris J.M. Streptogramin analogues. Expert Opin Investig Drugs. 1994;3:115–131. [Google Scholar]
- Bierman M., Logan R., O'Brien K., Seno E.T., Rao R.N., Schoner B.E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- Blanc V., Blanche F., Crouzet J., Jacques N., Lacroix P., Thibaut D. 1994. et al) Polypeptides involved in streptogramin biosynthesis, nucleotide sequences coding for said polypeptides and use thereof. Patent Cooperation Treaty International Publication no. WO 94/08014. Derwent, London, England.
- Blanc V., Lagneaux D., Didier P., Gil P., Lacroix P., Crouzet J. Cloning and analysis of structural genes from Streptomyces pristinaespiralis encoding enzymes involved in the conversion of pristinamycin IIB to pristinamycin IIA (PIIA): PIIA synthase and NADH:riboflavin 5′‐phosphate oxidoreductase. J Bacteriol. 1995;177:5206–5214. doi: 10.1128/jb.177.18.5206-5214.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc V., Thibaut D., Bamas‐Jacques N., Blanche F., Crouzet J., Barrière J.‐C. 1996. et al) New streptogramin derivatives useful as antibiotics produced by new mutants of Streptomyces having altered genes for streptogramin B biosynthesis. Patent Cooperation Treaty International Pubilcation no. WO 9601.901‐A1. Derwent, London, England.
- Blanc V., Gil P., Bamas‐Jacques N., Lorenzon S., Zagorec M., Schleuniger J. Identification and analysis of genes from S. pristinaespiralis encoding enzymes involved in the biosynthesis of the 4‐dimethylamino‐l‐phenylalanine precursor of pristinamycin I. Mol Microbiol. 1997;23:191–202. doi: 10.1046/j.1365-2958.1997.2031574.x. et al. [DOI] [PubMed] [Google Scholar]
- Bullock W.O., Fernandez J.M., Short J.M. Xl1‐Blue, a high efficiency plasmid transforming recA Escherichia coli strain with beta galactosidase selection. Focus. 1987;5:376–378. [Google Scholar]
- Cheng Y.Q., Tang G.L., Shen B. Type I polyketide synthase requiring a discrete acyltransferase for polyketide biosynthesis. Proc Natl Acad Sci USA. 2003;100:3149–3154. doi: 10.1073/pnas.0537286100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocito C.G. Antibiotics of the virginiamycin family, inhibitors which contain synergistic components. Microbiol Rev. 1979;43:145–198. doi: 10.1128/mr.43.2.145-192.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Crécy‐Lagard V., Saurin W., Thibaut D., Gil P., Naudin L., Crouzet J., Blanc V. Streptogramin B biosynthesis in Streptomyces pristinaespiralis and Streptomyces virginiae: molecular characterization of the last structural peptide synthetase gene. Antimicrob Agents Chemother. 1997;41:1904–1909. doi: 10.1128/aac.41.9.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler H.P. Screening for secondary metabolite products by HPLC and UV‐visible absorbance spectral libraries. Nat Prod Lett. 1993;2:99–128. [Google Scholar]
- Fischbach M.A., Walsh C.T., Clardy J. The evolution of gene collectives: how natural selection drives chemical innovation. Proc Natl Acad Sci USA. 2007;105:4601–4608. doi: 10.1073/pnas.0709132105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folcher M., Gaillard H., Nguyen L.T., Nguyen K.T., Lacroix P., Bamas‐Jacques N. Pleiotropic functions of a Streptomyces pristinaespiralis autoregulator receptor in development, antibiotic biosynthesis, and expression of a superoxide dismutase. J Biol Chem. 2001;276:44297–44306. doi: 10.1074/jbc.M101109200. et al. [DOI] [PubMed] [Google Scholar]
- Fritzsche K., Ishida K., Hertweck C. Orchestration of discoid polyketide cyclization in the resistomycin pathway. J Am Chem Soc. 2008;130:8307–8316. doi: 10.1021/ja800251m. [DOI] [PubMed] [Google Scholar]
- Gu L., Wang B., Kulkarni A., Geders T.W., Grindberg R.V., Gerwick L. Metamorphic enzyme assembly in polyketide diversification. Nature. 2009;459:731–735. doi: 10.1038/nature07870. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heathcote M.L., Staunton J., Leadlay P.F. Role of type II thioesterases: evidence for removal of short acyl chains produced by aberrant decarboxylation of chain extender units. Chem Biol. 2001;8:207–220. doi: 10.1016/s1074-5521(01)00002-3. [DOI] [PubMed] [Google Scholar]
- Huh J.H., Kim D.J., Zhao X.Q., Li M., Jo Y.Y., Yoon T.M. Widespread activation of antibiotic biosynthesis by S‐adenosylmethionine in streptomycetes. FEMS Microbiol Lett. 2004;238:439–447. doi: 10.1016/j.femsle.2004.08.009. et al. [DOI] [PubMed] [Google Scholar]
- Huson D.H. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- Huson D.H., Richter D.C., Rausch C., Dezulian T., Franz M., Rupp R. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser T., Bibb M.J., Buttner M.J., Chater K.F., Hopwood D.A. The John Innes Foundation; 2000. [Google Scholar]
- Lawrence J.G., Roth J.R. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics. 1996;143:1843–1860. doi: 10.1093/genetics/143.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombó F., Braña A.F., Méndez C., Salas J.A. The mithramycin gene cluster of Streptomyces argillaceus contains a positive regulatory gene and two repeated DNA sequences that are located at both ends of the cluster. J Bacteriol. 1999;181:642–647. doi: 10.1128/jb.181.2.642-647.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long P.F., Wilkinson C.J., Bisang C.P., Cortés J., Dunster N., Oliynyk M. Engineering specificity of starter unit selection by the erythromycin‐producing polyketide synthase. Mol Microbiol. 2002;43:1215–1225. doi: 10.1046/j.1365-2958.2002.02815.x. et al. [DOI] [PubMed] [Google Scholar]
- McDaniel R., Ebert‐Khosla S., Fu H., Hopwood D.A., Khosla C. Engineered biosynthesis of novel polyketides: influence of a downstream enzyme on the catalytic specificity of a minimal aromatic polyketide synthase. Proc Natl Acad Sci USA. 1994;91:11542–11546. doi: 10.1073/pnas.91.24.11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki S., Hiratsu K., Suwa M., Ishii T., Sugino F., Yamada K., Kinashi H. The large linear plasmid pSLA2‐L of Streptomyces rochei has an unusually condensed gene organization for secondary metabolism. Mol Microbiol. 2003;48:1501–1510. doi: 10.1046/j.1365-2958.2003.03523.x. [DOI] [PubMed] [Google Scholar]
- Namwat W., Lee C.K., Kinoshita H., Yamada Y., Nihira T. Identification of the varR gene as a transcriptional regulator of virginiamycin S resistance in Streptomyces virginiae. J Bacteriol. 2001;183:2025–2031. doi: 10.1128/JB.183.6.2025-2031.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelzer S., Reichert W., Huppert M., Heckmann D., Wohlleben W. Cloning and analysis of a peptide synthetase gene of the balhimycin producer Amycolatopsis mediterranei DSM5908 and development of a gene disruption/replacement system. J Biotechnol. 1997;56:115–128. doi: 10.1016/s0168-1656(97)00082-5. [DOI] [PubMed] [Google Scholar]
- Pulsawat N., Kitani S., Nihira T. Characterization of biosynthetic genecluster for the production of virginiamycin M, a streptogramin type A antibiotic, in Streptomyces virginiae. Gene. 2007a;393:31–42. doi: 10.1016/j.gene.2006.12.035. [DOI] [PubMed] [Google Scholar]
- Pulsawat N., Kitani S., Kinoshita H., Lee C.K., Nihira T. Identification of the bkdAB gene cluster, a plausible source of the starter‐unit for virginiamycin M production in Streptomyces virginiae. Arch Microbiol. 2007b;187:459–466. doi: 10.1007/s00203-007-0212-2. [DOI] [PubMed] [Google Scholar]
- Rehm S.J., Graham D.R., Srinath L., Prokocimer P., Richard M.P., Talbot G.H. Successful administration of quinupristin/dalfopristin in the outpatient setting. J Antimicrob Chemother. 2001;47:639–645. doi: 10.1093/jac/47.5.639. [DOI] [PubMed] [Google Scholar]
- Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M.A., Barrel B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch T., Maniatis T. 2nd. Cold Spring Harbour Laboratory Press; 1989. [Google Scholar]
- Santamarta I., Rodriguez‐Garcia A., Pérez‐Redondo R., Martin J.F., Liras P. CCaR is an autoregulatory protein that binds to the ccaR and cefD‐cmcI promoters of the cephamycin C‐clavulanic acid cluster in Streptomyces clavuligerus. J Bacteriol. 2002;184:3106–3113. doi: 10.1128/JB.184.11.3106-3113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer D., Mootz H.D., Linne U., Marahiel M.A. Regeneration of misprimed nonribosomal peptide synthetases by type II thioesterases. Proc Natl Acad Sci USA. 2002;99:4083–14088. doi: 10.1073/pnas.212382199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaut D., Ratet N., Bisch D., Faucher D., Debussche L., Blanche F. Purification of the two‐enzyme system catalyzing the oxidation of the d‐proline residue of pristinamycin IIB during the last step of pristinamycin IIA biosynthesis. J Bacteriol. 1995;177:5199–5205. doi: 10.1128/jb.177.18.5199-5205.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaut D., Bisch D., Ratet N., Maton L., Couder M., Debussche L., Blanche F. Purification of peptide synthetases involved in pristinamycin I biosynthesis. J Bacteriol. 1997;179:697–704. doi: 10.1128/jb.179.3.697-704.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton J.D. Horizontal gene transfer and the evolution of secondary metabolite gene clusters in fungi: an hypothesis. Fungal Genet Biol. 2000;30:167–171. doi: 10.1006/fgbi.2000.1224. [DOI] [PubMed] [Google Scholar]
- Weber T., Laiple K.J., Pross E.K., Textor A., Grond S., Welzel K. Molecular analysis of the kirromycin biosynthetic gene cluster revealed β‐alanine as precursor of the pyridone moiety. Chem Biol. 2008;15:175–188. doi: 10.1016/j.chembiol.2007.12.009. et al. [DOI] [PubMed] [Google Scholar]
- Weber T., Rausch C., Lopez P., Hoof I., Gaykova V., Huson D.H., Wohlleben W. CLUSEAN: a computer‐based framework for the automated analysis of bacterial secondary metabolite biosynthetic gene clusters. J Biotechnol. 2009;140:13–17. doi: 10.1016/j.jbiotec.2009.01.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bacterial strains, plasmids and cosmids.
Primer sequence and amplified fragments.


